Abstract
Objectives/Hypothesis
Single nucleotide polymorphisms (SNPs) in the 8q24 chromosomal region identified from genome-wide scans have been associated with risk of several cancers including breast (rs1562430), prostate and colon (rs1447295 and rs6983267). A genome-wide scan in 26 families with papillary thyroid cancer (PTC) also found susceptibility loci in 8q24, supporting a closer evaluation of this chromosomal region in relation to risk of sporadic PTC.
Study Design
We evaluated 157 tag SNPs in the 8q24 chromosomal region between 120.91 Mb and 128.78 Mb (including rs1562430, rs1447295, and rs6983267) in a case-control study of 344 PTC cases and 452 age and gender frequency-matched controls.
Methods
We used logistic regression to estimate odds ratios and compute P-values of linear trend for PTC with genotypes of interest. To account for multiple comparisons, we applied the false discovery rate (FDR) method.
Results
We did not find a significant association between rs1562430, rs1447295, or rs6983267, and PTC risk. We found that one SNP (rs4733616) was associated with PTC risk at P = 0.003, and twelve other SNPs were associated with PTC risk at P < 0.05. However, no SNPs remained significant after FDR correction.
Conclusions
Our findings do not support a strong association between SNPs in the 8q24 chromosomal region and risk of sporadic PTC but several SNPs with small effects might exist.
Keywords: 8q24, thyroid cancer
Introduction
Single nucleotide polymorphisms (SNPs) in the 8q24 chromosomal gene-poor region, known as ‘gene desert’, have been consistently implicated in genome-wide association studies as susceptibility loci for several cancer sites including breast, prostate and colorectal cancers 1-6. Some of these SNPs (rs6983267) appear to confer susceptibility to multiple cancers 7,8, and others (rs1562430 and rs1447295) have been repeatedly identified in several genome-wide association studies 2,7-9. One case-control study of 485 sporadic thyroid cancer cases found a weak association between rs6983267 and thyroid cancer risk, but this result has not been reproduced and only the single marker in the 8q24 region was examined, leaving the possibility for other susceptibility loci in 8q24 8. Interestingly, a genome-wide association study examining the risk of heritable papillary thyroid cancer (PTC) among 26 families identified additional susceptibility loci in the 8q24 region 10. To evaluate whether the 8q24 region might contain genetic variants associated with risk of sporadic thyroid cancer, we genotyped 157 tag SNPs between 120.91 Mb and 128.78 Mb in a case-control study of 344 PTC cases and 452 controls, using a platform that was created to analyze genetic susceptibility to a variety of rare cancers.
Materials and Methods
Our subjects included 202 incident PTC cases diagnosed within the U.S. Radiologic Technologists (USRT) cohort 11, 142 incident PTC cases diagnosed and treated at the University of Texas M. D. Anderson Cancer Center (UTMDACC) 12, and 452 age and gender frequency-matched controls from the USRT study, matched to USRT cases (90.6% female). The proportion of females among the UTMDACC cases was lower (64.1%) than in the USRT cohort. All study subjects were of European ancestry. All PTC cases included in this study were histologically confirmed. To ensure that the cases from the two studies were comparable, we compared average age at diagnosis, smoking status and tumor size, and found that the two case series were similar (Supplemental Table 1). Both studies were reviewed and approved by their respective Institutional Review Boards and all subjects provided informed consent.
Genotyping of 157 tag SNPs in the 8q24 region located between 120.91 and 128.79 Mb was performed at the National Cancer Institute's Core Genotyping Facility (Advanced Technology Center, Gaithersburg, MD; http://cgf.nci.nih.gov/) using the custom-designed iSelect Infinium assay (Illumina, www.illumna.com). Tag SNPs were selected from the common SNPs (minor allele frequency, MAF > 5%) genotyped by the HapMap Project (Data Release 20/Phase II, NCBI Build 36.1, assembly dbSNPb126) in the Caucasian population (CEU) using TagZilla, which is a part of the GLU software package (http://code.google.com/p/glu-genetics/) with a binning threshold of r2 > 0.8. Previously identified hits from genome-wide association studies of other common cancers, including breast (rs1562430), and prostate and colon (rs1447295 and rs6983267) were also included. In the analysis, SNPs were excluded if they failed quality control measures: <95% concordance, <90% completion, or had evidence of a departure from Hardy-Weinberg equilibrium in controls (P < 0.00001). Allele frequencies for PTC cases were largely similar between the USRT and the UTMDACC cases and between males and females, so these groups were combined for genetic analyses.
We calculated the linear Ptrend for each SNP genotype, coded as 0, 1, 2, with 0 denoting the homozygous common allele genotype as the referent category, in logistic regression models adjusted for sex, attained age at diagnosis in four categories (<35, 35-44, 45-54, 55+ years), and year of birth (<1940, 1940-1949, 1950+) as an ordinal variable. We also evaluated the effect of additional adjustment for cigarette smoking. The results from the models with additional adjustments were similar and we chose to present the results from basic models adjusted for sex, attained age and year of birth only. We corrected for multiple comparisons using the false discovery rate control (FDR) method 13. Statistical analyses were conducted in SAS version 9.1 (SAS Institute, Cary, NC) and in R.
Results
Demographic characteristics of the 344 PTC cases and 452 controls have been reported previously 14 and are presented in supplemental material here (Supplemental Table 2). The age distribution was similar between cases and controls. Cases were more likely to report increased BMI and a family history of any cancer or of thyroid cancer specifically; but less likely to report history of smoking.
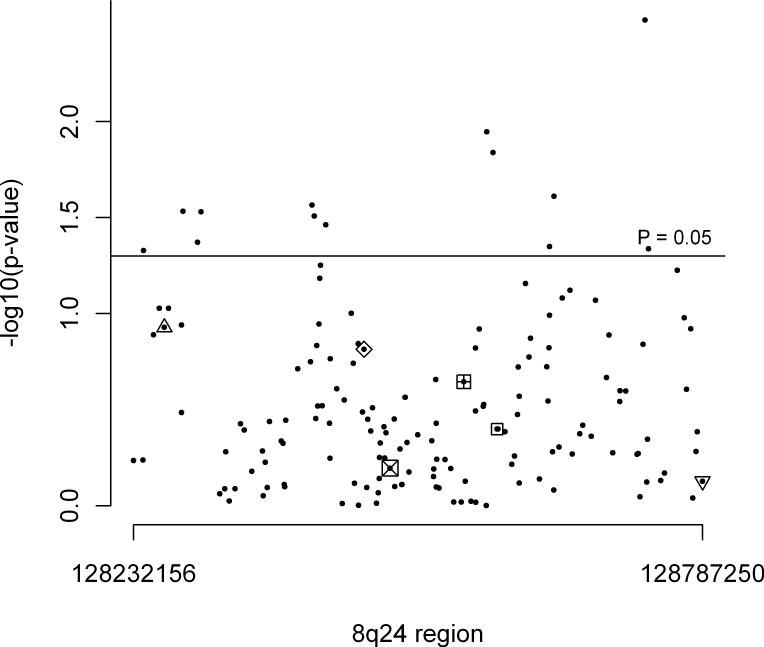
In logistic regression analyses, polymorphisms at rs1562430, rs6983267, and rs1447295 were not associated with PTC risk (P = 0.15, P = 0.64, and P = 0.23, respectively) (Figure 1). One SNP (rs4733616) was associated with PTC risk at P = 0.003, and 12 additional SNPs at P <0.05 (Table 1). None of these SNPs were in linkage disequilibrium with the previously identified loci in genome-wide association studies (Figure 1). After FDR correction, no SNPs were significantly associated with PTC risk. Raw and FDR corrected P-values for all 157 tag SNPs are presented in supplemental material (Table S3).
Figure 1.
Significance levels (P values) from logistic regression analyses of tag SNPs in the intergenic region of 8q24 and risk of papillary thyroid cancer.
For markers above the P = 0.05 line, see Table 1. For markers below the P = 0.05 line:
 rs6983267 previously associated with prostate and colorectal, and thyroid (not GWAS).
rs6983267 previously associated with prostate and colorectal, and thyroid (not GWAS).
 rs1447295 previously associated with prostate cancer.
rs1447295 previously associated with prostate cancer.
 rs9642880 previously associated with bladder and urinary cancer.
rs9642880 previously associated with bladder and urinary cancer.
 rs4242382 and rs4242384 previously associated with prostate cancer.
rs4242382 and rs4242384 previously associated with prostate cancer.
 rs1562430 previously associated with breast cancer 2.
rs1562430 previously associated with breast cancer 2.
 rs2456449 previously associated with chronic lymphocytic leukemia 19.
rs2456449 previously associated with chronic lymphocytic leukemia 19.
For more information on these SNPs in genome-wide association studies, see http://hugenavigator.net/HuGENavigator/gWAHitStartPage.do.
Table 1.
SNPs in intergenic region of 8q24 with an uncorrected P <0.05 for an association with papillary thyroid cancer.
| SNP | Location | Gene | P valuea | PFDR valueb |
|---|---|---|---|---|
| rs4733616 | 128731277 | 0.003 | 0.464 | |
| rs1562432 | 128576784 | 0.011 | 0.566 | |
| rs4871808 | 128582727 | 0.015 | 0.566 | |
| rs12543106 | 128642480 | 0.024 | 0.566 | |
| rs377649 | 128406423 | 0.027 | 0.566 | |
| rs2466035 | 128280411 | 0.029 | 0.566 | |
| rs10087719 | 128298045 | 0.030 | 0.566 | |
| rs424281 | 128408608 | 0.031 | 0.566 | |
| rs13249993 | 128419697 | 0.034 | 0.566 | |
| rs7816475 | 128294622 | 0.043 | 0.566 | |
| rs4733655 | 128638038 | 0.045 | 0.566 | |
| rs4385433 | 128734662 | 0.046 | 0.566 | |
| rs2124600 | 128241868 | SRRM1L | 0.047 | 0.566 |
SNP-based linear Ptrend calculated based on the three-level genotype (0, 1, and 2) in logistic regression models adjusted for sex, attained age, and year of birth.
False discovery rate (FDR) corrected linear Ptrend.
Discussion
Our study is the first to closely examine sporadic PTC risk in relation to densely selected SNPs in the 8q24 chromosomal region previously associated with risk of multiple cancers including breast cancer. The latter association is particularly intriguing given increased risk of breast cancer in women with a history of prior thyroid cancer reported by the Surveillance Epidemiology and End Results database studies 15. Several observational studies have also reported an increased risk of thyroid cancer with a history of benign breast disease 16,17. While the association between thyroid and breast diseases in epidemiologic studies could in part be explained by greater medical surveillance of cancer survivors, it may also be due to common underlying genetic mechanisms.
We found that none of the SNPs in the 8q24 region previously associated with other cancers were significantly associated with risk of sporadic PTC in our study, but several other SNPs across the region were. While none of the significant associations persisted after correction for multiple comparisons, our FDR adjustment may be overly conservative because SNPs in the 8q24 region appear to have a higher prior probability of being associated with PTC risk. Nonetheless, our findings do not provide evidence for a strong association between SNPs in the 8q24 region and risk of thyroid cancer.
Moreover, we found that PTC risk was not associated with the SNP rs6983267 consistently linked with prostate cancer, for which results from a previous thyroid cancer study were suggestive 8. The results of our study are consistent with a recent case-control study of 398 cases of differentiated thyroid cancer (339 PTC) that examined two SNPs in the 8q24 region (rs6983267 and rs1447295) and found no association 18.
Given that PTC cases were less likely to be cigarette smokers, we explored whether cigarette smoking may have influenced the associations of interest. When added to the models, cigarette smoking did not meaningfully change the risk estimates, and therefore, is unlikely to confound our main findings.
A limitation of our study is that while we had extensive coverage for a particular region of 8q24, we did not scan the entire 8q24 region, leaving the possibility that other susceptibility loci beyond the genotyped area might have been missed. Moreover, we had limited statistical power to detect weak associations. However, by narrowing our scope to areas of 8q24 previously implicated in a variety of other common cancers in genome-wide scans, we tried to increase the prior probability and to minimize the multiple testing problems.
Conclusion
In summary, our study is the first study to examine a large number of SNPs in the 8q24 chromosomal region in relation to risk of sporadic PTC. We found little evidence for a strong association of 157 tag SNPs in the 8q24 chromosomal region with risk of PTC, but small effects might exist for several SNPs.
Supplementary Material
Acknowledgments
This research was supported in part by the Intramural Research Program of the Division of Cancer Epidemiology and Genetics, National Cancer Institute, National Institutes of Health and by a grant from the American Thyroid Association (PI: E. M. Sturgis). We are grateful to the radiologic technologists who participated in the U. S. Radiologic Technologists Study; Jerry Reid of the American Registry of Radiologic Technologists for continued support of this study; Diane Kampa and Allison Iwan of the University of Minnesota for data collection and study coordination; Liliana Mugartegui for patient recruitment, data collection, and study coordination at the University of Texas M. D. Anderson Cancer Center; Laura Bowen of Information Management Systems for biomedical computing statistical support. This project has been funded in whole or in part with federal funds from the National Cancer Institute, National Institutes of Health, under contract number HHSN261200800001E. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government.
Footnotes
Conflict of Interest: None
References
- 1.Ghoussaini M, Song H, Koessler T, et al. Multiple loci with different cancer specificities within the 8q24 gene desert. J Natl Cancer Inst. 2008;100:962–966. doi: 10.1093/jnci/djn190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Turnbull C, Ahmed S, Morrison J, et al. Genome-wide association study identifies five new breast cancer susceptibility loci. Nat Genet. 2010;42:504–507. doi: 10.1038/ng.586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Thomas G, Jacobs KB, Yeager M, et al. Multiple loci identified in a genome-wide association study of prostate cancer. Nat Genet. 2008;40:310–315. doi: 10.1038/ng.91. [DOI] [PubMed] [Google Scholar]
- 4.Tomlinson I, Webb E, Carvajal-Carmona L, et al. A genome-wide association scan of tag SNPs identifies a susceptibility variant for colorectal cancer at 8q24.21. Nat Genet. 2007;39:984–988. doi: 10.1038/ng2085. [DOI] [PubMed] [Google Scholar]
- 5.Tomlinson IP, Webb E, Carvajal-Carmona L, et al. A genome-wide association study identifies colorectal cancer susceptibility loci on chromosomes 10p14 and 8q23.3. Nat Genet. 2008;40:623–630. doi: 10.1038/ng.111. [DOI] [PubMed] [Google Scholar]
- 6.Yeager M, Orr N, Hayes RB, et al. Genome-wide association study of prostate cancer identifies a second risk locus at 8q24. Nat Genet. 2007;39:645–649. doi: 10.1038/ng2022. [DOI] [PubMed] [Google Scholar]
- 7.Haiman CA, Le Marchand L, Yamamato J, et al. A common genetic risk factor for colorectal and prostate cancer. Nat Genet. 2007;39:954–956. doi: 10.1038/ng2098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wokolorczyk D, Gliniewicz B, Sikorski A, et al. A range of cancers is associated with the rs6983267 marker on chromosome 8. Cancer Res. 2008;68:9982–9986. doi: 10.1158/0008-5472.CAN-08-1838. [DOI] [PubMed] [Google Scholar]
- 9.Fletcher O, Johnson N, Orr N, et al. Novel breast cancer susceptibility locus at 9q31.2: results of a genome-wide association study. J Natl Cancer Inst. 2011;103:425–435. doi: 10.1093/jnci/djq563. [DOI] [PubMed] [Google Scholar]
- 10.He H, Nagy R, Liyanarachchi S, et al. A susceptibility locus for papillary thyroid carcinoma on chromosome 8q24. Cancer Res. 2009;69:625–631. doi: 10.1158/0008-5472.CAN-08-1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sigurdson AJ, Doody MM, Rao RS, et al. Cancer incidence in the US radiologic technologists health study, 1983-1998. Cancer. 2003;97:3080–3089. doi: 10.1002/cncr.11444. [DOI] [PubMed] [Google Scholar]
- 12.Ho T, Li G, Lu J, Zhao C, Wei Q, Sturgis EM. Association of XRCC1 polymorphisms and risk of differentiated thyroid carcinoma: a case-control analysis. Thyroid. 2009;19:129–135. doi: 10.1089/thy.2008.0153. [DOI] [PubMed] [Google Scholar]
- 13.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. Journal of the Royal Statistical Society Series B (Methodological) 1995;57:289–300. [Google Scholar]
- 14.Neta G, Brenner AV, Sturgis EM, et al. Common genetic variants related to genomic integrity and risk of papillary thyroid cancer. Carcinogenesis. 2011 doi: 10.1093/carcin/bgr100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Curtis RE, Freedman DM, Ron E, et al. New Malignancies Among Cancer Survivors: SEER Cancer Registries, 1973–2000. National Cancer Institute; Bethsda: 2006. [Google Scholar]
- 16.Meinhold CL, Ron E, Schonfeld SJ, et al. Nonradiation risk factors for thyroid cancer in the US Radiologic Technologists Study. Am J Epidemiol. 2010;171:242–252. doi: 10.1093/aje/kwp354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ron E, Kleinerman RA, Boice JD, Jr., LiVolsi VA, Flannery JT, Fraumeni JF., Jr A population-based case-control study of thyroid cancer. J Natl Cancer Inst. 1987;79:1–12. [PubMed] [Google Scholar]
- 18.Akdi A, Pérez G, Pastor S, et al. Common Variants of the Thyroglobulin Gene Are Associated with Differentiated Thyroid Cancer Risk. Thyroid. 2011 doi: 10.1089/thy.2010.0384. [DOI] [PubMed] [Google Scholar]
- 19.Crowther-Swanepoel D, Broderick P, Di Bernardo MC, et al. Common variants at 2q37.3, 8q24.21, 15q21.3 and 16q24.1 influence chronic lymphocytic leukemia risk. Nat Genet. 2010;42:132–136. doi: 10.1038/ng.510. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.