Abstract
Clostridium difficile infection (CDI) has become one of the most prevalent and costly nosocomial infections. In spite of the importance of CDI, our knowledge of the pathogenesis of this infection is still rudimentary. Although previous use of antibiotics is generally considered to be the sine qua non of CDI, the mechanisms by which antibiotics render the host susceptible to C. difficile are not well defined. In this review we will explore what is known about how the indigenous microbiota acts in concert with the host to prevent colonization and virulence of C. difficile and how antibiotic administration disturbs host–microbiota homeostasis, leading to CDI.
Keywords: Colonization resistance, C. difficile, microbiome, microbial ecology, antibiotics
Antibiotic-mediated changes in the gut microbiota in Clostridium difficile infection
Clostridium difficile infection (CDI) following the administration of antibiotics has been estimated to be the most costly healthcare-associated infection, responsible for an estimated 3 billion dollars in increased healthcare cost annually [1]. The recent rise in the incidence and apparent severity of CDI has prompted a renewed interest in the pathogenesis of this infection. The combination of the fact that CDI is most frequently associated with the administration of antibiotics and the recent interest in the role of the microbiome in human health and disease [2] has lead to a renewed interest in understanding the relationship between C. difficile and the indigenous gut microbiota. We will review what is known about how colonization resistance against pathogens is mediated by the gut microbiome and how antibiotic administration alters colonization resistance through changes in the structure and function of the gut microbiota. Although the host immune response is also important in the pathogenesis of CDI, we will focus our discussion of this topic to recent findings on the role of innate immunity in C. difficile infection. Readers are referred to a recent review for a more in depth discussion of the roll of the host immune response and C. difficile disease [3].
Description of ‘antibiotic-associated colitis’
Shortly after the development of broad-spectrum antimicrobial agents, it was noted that patients treated with these agents would develop gastrointestinal illness. Diarrhea was a well-known side effect of the administration of almost any antibiotic, but a subset of patients treated with broad-spectrum antibiotics were noted to develop a more clinically severe colitis. Clindamycin was one of the first antimicrobial agents associated with this severe form of antibiotic-associated colitis [4]. In 1977, Bartlett and colleagues fulfilled Koch’s postulates and implicated a toxin producing Clostridium, later identified as C. difficile, as the etiologic agent of clindamycin-associated colitis [5]. Simultaneous with this discovery came the clinical recognition that CDI could be treated by the administration of vancomycin or metronidazole. Furthermore, the dogma was established that antibiotics lead to CDI through the disruption of so-called ‘colonization resistance’ that is normally provided by the indigenous microbiota [6, 7]. Although a few investigators have pursued investigation of the mechanisms by which colonization resistance is mediated by the indigenous microbiota and how antibiotics disrupt this, the availability of a relatively efficacious treatment for CDI likely discouraged others from following this path. More recently, however, an increase in the prevalence and severity of C. difficile infection has renewed interest in understanding the basic mechanisms underlying the pathogenesis of CDI. In particular, an increase in recurrent C. difficile infection, occurring after an initially successful treatment with metronidazole or vancomycin, has rekindled efforts to understand how this pathogen is normally suppressed by the host and indigenous microbiota [8].
Colonization resistance to C. difficile
Early work in streptomycin-treated animal models
The concept that the indigenous gut microbiota is able to prevent colonization by potential pathogens predated the recognition of C. difficile as the cause of antibiotic-associated colitis. Shortly after streptomycin became available in the early 1940s as an antibiotic, researchers noted that oral administration of the drug would significantly alter the cultivatable bacteria that could be isolated from the feces of mice [9]. In the 1950s Rolf Freter exploited the ability of streptomycin to alter the gut microbiota to develop a model of infection with Vibrio cholerae in guinea pigs. Pretreatment with streptomycin would render the animals susceptible to a fatal infection with cholera whereas untreated animals would not even be colonized. In the manuscript where he described this model, Freter proposed concepts with regards to colonization resistance that remain prescient to this day [10]. He suggested that:
“…It might be worthwhile to consider the possibility of inhibitory action on the part of the normal human enteric flora as a factor in the resistance of humans to enteric diseases.”
He noted that this theory had been discussed previously, perhaps most notably by Nissle in 1916. The fact that antibiotics could overcome colonization resistance to V. cholerae provided experimental evidence for this theory. Furthermore, he noted guinea pigs that were derived from two different breeding lines exhibited markedly different susceptibility to V. cholerae challenge after streptomycin treatment. There was a 4 log difference in the LD50 of the two guinea pig strains and Freter hypothesized [10]:
“This difference in susceptibility might be due to a different composition of the normal flora in the two strains.”
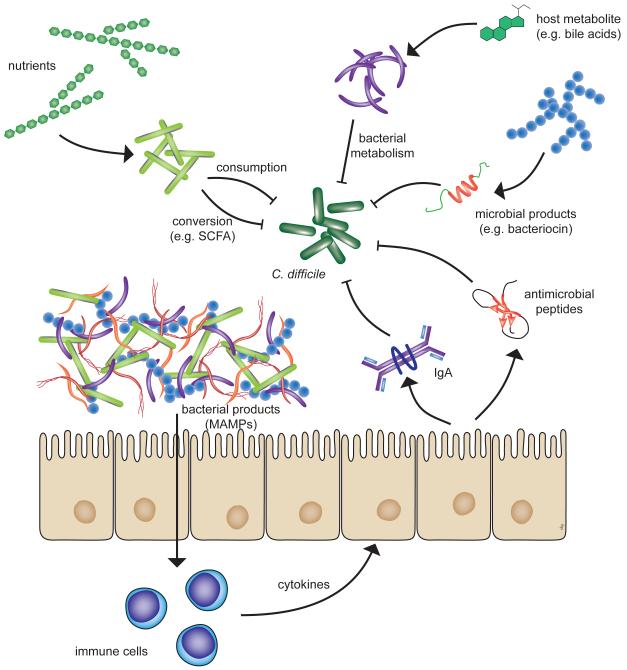
Thus, these observations by Freter laid the groundwork for the study of interactions between the intestinal microbiota, host and potential pathogens building the foundation of the more recent work that we will discuss in this review. It should be noted that later in his career, Freter and his colleague Kenneth Wilson investigated the role of the indigenous microbiota in colonization resistance against C. difficile. They were able to show that the indigenous microbiota of hamsters was able to suppress and/or outcompete C. difficile in continuous flow cultures and in germ-free mice [7, 11]. For the remainder of this review, we will discuss possible mechanisms by which the indigenous gut microbiota may mediate colonization resistance against C. difficile (Figure 1) and describe some of the studies that have provided evidence for these proposed mechanisms.
Figure 1.
Potential mechanisms by which the indigenous microbiota can mediate colonization resistance against C. difficile. Specific members of the microbiome can either directly, or indirectly interfere with the proliferation of C. difficile within the gut. Direct inhibition can occur via competition for nutrients, the conversion of nutrients or host metabolites to compounds that are inhibitory to C. difficile or by the production of primary microbial products that inhibit C. difficile. Indirect control of C. difficile can occur via interactions between the indigenous microbiota and the host that results in the expression of host products that control C. difficile colonization and proliferation. Detection of microbial-associated molecular patterns (MAMPs) can trigger the host immune signaling cascades leading to the production of innate (e.g. antimicrobial peptides) or adaptive (e.g. IgA) immune effectors. Abbreviations: SCFA, short-chain fatty acids.
Antibiotic use and the development of C. difficile infection
As noted above, development of diarrhea following antibiotic administration is a common outcome. In the majority of cases of antibiotic-associated diarrhea (AAD), no pathogen is isolated and the disease is mild, with diarrhea that resolves shortly after the completion of the antibiotic course and a lack of associated systemic signs and symptoms of disease. C. difficile infection occurs in about 25% of all cases of AAD and is the most common cause of severe forms of the disease that is accompanied by systemic illness and the development of pseudomembranes [12].
Of the various risk factors for developing nosocomial infection, antibiotic administration has been shown to be a key risk factor for contracting CDI. However, not all classes of these drugs carry equal risk of predisposing to CDI. Previously, the antibiotics associated with the highest risk of leading to CDI were noted to be clindamycin, cephalosporins, penicillins (especially aminopenicillins) and more recently, fluoroquinolones [13]. A recent report noted that although particular classes of antibiotics can predispose to CDI, increasing cumulative antibiotic exposure over time increases the risk of subsequently developing disease [14]. This finding is interesting in that it implies that the total ‘collateral damage’ [15] due to antibiotic-induced alteration of the indigenous gut microbiota is one of the main determinants of decreasing colonization resistance to C. difficile.
Antibiotic-induced invasion of C. difficile, presumably due to loss of colonization resistance, has been demonstrated in hamsters [5, 16], mice [17, 18], and in human fecal bioreactors [19]. Reduced diversity of the intestinal community associated with C. difficile colonization in these models has been demonstrated, although the community members responsible for suppressing C. difficile have yet to be identified.
Antibiotic administration and changes in microbiome structure and function
The study of complex microbial communities has been revolutionized by the application of culture-independent techniques, many of which utilize retrieval of the phylogenetic information contained within the 16S rRNA-encoding gene as a surrogate for individual bacterial species [20]. The application of these methods allows for in depth investigation of the impact of antibiotic administration on the microbiome. While not all of these studies have specifically examined antibiotic-induced changes in the context of C. difficile infection, the below studies have clearly demonstrated the profound effects that antibiotics can have on the structure and function of the gut microbiome.
As noted previously, CDI is responsible for about 25% of all cases of AAD. Cases of non-C. difficile-associated AAD are also postulated to arise from alterations in the indigenous microbiota [12]. This proposed model was examined by studying the dynamics of the fecal microbiota in a patient who developed non-C. difficile-associated AAD during a course of treatment with amoxicillin-clavulanate [21]. The development of AAD coincided with a decrease in carbohydrate-fermenting, butyrate-producing members of the phylum Firmicutes. These changes in the community structure largely resolved following discontinuation of antibiotic treatment. In follow-up animal studies, it was found that antibiotic-induced changes in the microbiota were not necessarily reversible. Both animal and human studies indicate that short-term antibiotic administration can result in long-term impacts on the gut microbiome [22-25].
Recent studies have used culture-independent analysis of the microbiome to follow how various antimicrobials alter the gut microbiota and how certain changes are associated with lowered colonization resistance and risk of subsequent CDI. One in vitro study used a fermentation system to study the effect of broad- and narrow-spectrum antimicrobials directed against C. difficile and the gut microbiota. These fermenters were set up with human feces in the presence and absence of C. difficile [19]. The various antibiotics varied in their activity against C. difficile and in their ability to alter the gut microbiota. By using a narrow-spectrum bacteriocin directed against C. difficile (for more details see below), the authors demonstrated that they could suppress C. difficile without significant alteration of the background microbiota. This finding may have implications for preventing relapse of CDI.
Two recent translational studies have used molecular microbiota analysis to begin to define the specific alterations in the intestinal microbiome that increase the risk of the development of AAD and CDI. In both of these studies, 16S rRNA-targeted methods were used to define ‘microbial risk factors’ that were associated with development of disease. One study employed 16S rRNA temporal temperature gradient gel electrophoresis to study the fecal microbiota in 156 patients undergoing antibiotic treatment for upper-respiratory tract infections, 44 of whom developed AAD (6 due to C. difficile) [26]. These authors identified 8 specific 16S rRNA-encoding genes in the fecal specimens of patients obtained prior to the initiation of antibiotic therapy that were associated with predisposition to AAD and could be used to predict the risk of development of this disease with an estimated error of 2%. Another study looked specifically at alteration in the gut microbiome associated with the risk of developing nosocomial CDI [27]. This study used a 16S rRNA-encoding gene microarray (combined with pyrosequencing) to detect differences in the pre-disease composition of the gut microbiome that predicted risk of CDI. The statistical model described in this study suggested that a combination of clinical variables [non-steroidal anti-inflammatory drug (NSAID) use and fluoroquinolone use] were associated with changes in the microbiome that led to subsequent CDI. It will be interesting to see if follow-up prospective studies confirm the use of microbiome profiling as described in these reports as clinically useful in determining the risk of developing CDI.
Recurrent CDI, variably defined as diarrhea associated with a positive test for C. difficile that occurs within days to weeks (generally less than 2 months) after completing clinically successful treatment of an episode of CDI, is becoming an increasingly alarming and important problem [8, 28]. It has been proposed that patients with recurrent CDI have sufficiently altered indigenous microbiota such that colonization resistance is not restored after treatment directed against C. difficile. Support for reduced microbiome diversity in recurrent CDI was found in a small sample of patients recovering from CDI infection [29]. Patients that suffered only a single episode of CDI displayed fecal microbial communities that were as diverse as control subjects, while patients that had multiple episodes of C. difficile infection had markedly reduced microbial diversity. It is likely that the inability to restore a normal intestinal microbiota in these individuals is the main driver of recurrent CDI, which is supported by the recent findings that stool transplants are currently the most effective treatment for recurrent CDI (92% effective in curing recurrent CDI from a meta-analysis [30]). Additionally, fecal transplantation was demonstrated to have a profound effect on the fecal microbiota of recipients [31]. It should be noted that despite the clinical efficacy of fecal transplantation, this is not an FDA-approved therapy, and does carry the small but finite risk of inadvertent transmission of pathogens. However, these observations demonstrate that disruption of the intestinal microbial community plays an important role in CDI and suggests that a more complete understanding of the mechanisms behind resistance to C. difficile infection will spearhead future therapeutic interventions. The next sections will review potential mechanisms by which the intestinal microbiota impact C. difficile physiology and virulence.
Proposed mechanisms for the mediation of C. difficile colonization resistance by the normal microbiota
Microbiota modulation of a host metabolite–bile acid transformation
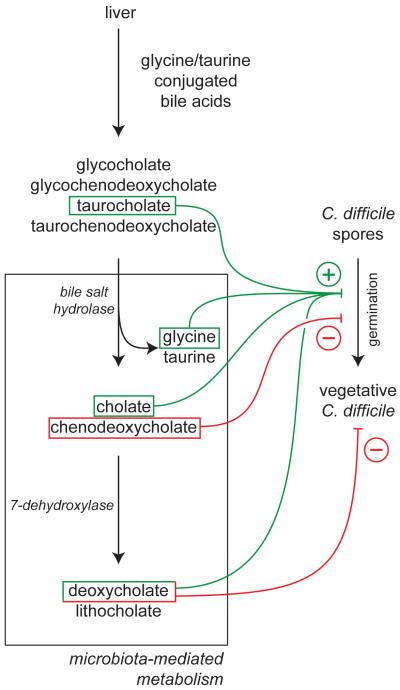
An example of a mechanism by which the microbiota may impact C. difficile invasion of the intestinal tract is via the transformation of bile acids, which have profound effects on spore germination and vegetative growth of C. difficile in vitro (Figure 2). Humans secrete bile acids into the small intestine in response to consumption of a meal; these acids aid in the absorption of fats and fat-soluble vitamins and nutrients. Humans synthesize two main primary bile acids, cholate and chenodeoxycholate, which are conjugated to an amino acid (glycine or taurine) [32]. The microbiota plays two important roles in bile acid transformation. First, bile salt hydrolase enzymes that are produced by bacteria in the intestinal tract insure that all of the bile acids are deconjugated from their amino acid in the intestinal lumen [32]. Why bacteria produce bile salt hydrolases is still not entirely clear but two theories are they detoxify the antimicrobial activity of bile to support colonization or they allow the bacteria to utilize the amino acid as a nutrient source (summarized in [33]). Second, bacteria mediate the transformation of primary bile acids to secondary bile acids via the enzyme 7-dehydroxylase, converting cholate and chenodeoxycholate into deoxycholate and lithocholate, respectively [32, 34]. Thus the intestinal microbiota plays a profound role in the bile composition of the intestine and may impact the antimicrobial properties of bile.
Figure 2.
Bile acid metabolism and C. difficile. Conjugated bile salts produced by the liver are metabolized in the intestine, primarily by activities carried out by members of the gut microbiota. Primary bile salts as well as their metabolites can have direct effects on C. difficile, primarily affecting germination of spores as well as the activity of vegetative cells. The balance of the positive and negative effects on C. difficile, which in turn reflect the overall community structure of the gut microbiota, may determine the ultimate clinical outcome of C. difficile exposure.
Nearly 30 years ago it was shown that bile was able to stimulate the germination of C. difficile spores in vitro [35]. Sorg and Sonenshein recently revisited the role of individual bile acids and found that taurocholate and the amino acid glycine served as co-germinants for spores, enhancing germination by 1000-fold [36]. Deoxycholate, a secondary bile acid generated by the action of 7-dehydroxylase on cholate, was also shown to be a potent germinant but was highly toxic to vegetative cells. This led to the hypothesis that antibiotic treatment reduces the members of the microbiota that generate deoxycholate, causing an increase in the levels of cholate derivatives and a reduction in deoxycholate. Increased levels of cholate stimulate spore germination and vegetative C. difficile can now flourish in the colon without the inhibitory effects of deoxycholate on cell growth [36]. In support of this model, small intestine and cecal contents from mice that had been treated with antibiotics were able to support C. difficile spore germination at higher levels than control mice [37]. Cholestyramine, a bile acid binding resin, suppressed this stimulation. These results support the hypothesis that bile acids may act as an environmental cue to allow spores to sense that they are in the intestinal tract.
In contrast to cholate, chenodeoxycholate (CDCA) has a strong inhibitory effect on spore germination [38]. This suggests that the ratio of cholate to chenodeoxycholate may have an important role in whether or not spores attempt to germinate. CDCA acts as a competitive inhibitor of germination with an IC50 of ~350 μM, and this has led to the idea that non-metabolizable derivatives of CDCA could serve as therapeutics that inhibit the germination of spores in the intestinal tract [39]. Chenodeoxycholate is absorbed by the small intestine 10 times faster than cholate, which may explain why antibiotic treated animals and people have higher levels of cholate vs CDCA [38].
Although the model of how bile plays a role in the ability of C. difficile to invade the intestinal microbiota is intriguing, it still remains to be tested in vivo. One intriguing prediction of this model is that providing bacteria that contain 7-dehydroxylase activity to animals or humans susceptible to CDI should increase the levels of deoxycholate and thereby prevent C. difficile invasion. Conversion of cholate to deoxycholate appears to be, in clostridia species, a multistep process that requires the action of several enzymes [34]. If bile metabolism turns out to be a key player in CDI, then bacteria harboring 7-dehydroxylase activity may serve as next-generation commensal derived probiotics tailored to ameliorating CDI.
Niche exclusion: exclusion of toxigenic C. difficile by non-toxigenic C. difficile
Another mechanism being pursued for the prevention, and possibly treatment, of C. difficile infection is the use of non-toxigenic C. difficile (NTCD) to prevent toxigenic C. difficile (TCD) from becoming established in the intestinal tract. Experiments in hamsters have previously established that pre-treatment of hamsters with NTCD protected hamsters from subsequent lethal challenge with TCD [40, 41]. The success of administration of NTCD to prevent CDI in animal models has lead to human clinical trials that are in early stages.
How might precolonization with NTCD prevent colonization and disease formation by TCD? Studies investigating the mechanisms of colonization resistance of commensal Escherichia coli strains suggest that competition for nutrients may be critical for colonization resistance [42, 43]. Pre-colonization of the intestinal tract with NTCD may exclude TCD by the ability the NTCD strain to outcompete TCD for a limiting nutrient. In the intestinal tract, akin to what occurs in a chemostat, the strain that more successfully competes for a single limiting resource will outcompete other strains dependent on that resource. Previous work investigating the intestinal microbiota and interactions with C. difficile suggested that colonization resistance is mediated, in part, by microbiota degrading carbon sources that favor growth of C. difficile. Thus it is possible that NTCD simply outcompetes TCD by utilizing a limiting carbon source more efficiently. If this is the case, this brings up many questions about the physiology of the NTCD strain being used in clinical trials as well as the ability of different clades of TCD strains to utilize carbon sources. Should certain clades of TCD strains now utilize alternative nutrients that NTCD does not, then there will be no colonization resistance conferred by the NTCD strain. Other possibilities exist, including that NTCD may prevent TCD colonization by inhibition of TCD germination or by direct antagonism of TCD growth. However, little is known about the effects of the interactions between closely related C. difficile species, in vitro or in vivo.
Direct antagonism by the intestinal microbiota–bacteriocins
Another activity of the microbiota that could be impacted by antibiotics is the ability of certain gut bacteria to produce antimicrobials that directly inhibit the growth of C. difficile. Several groups have ‘mined’ the human intestinal microbiota for antibacterial activities produced by the microbiota that are specifically targeted to C. difficile with the idea that such therapeutics would make the host less susceptible to recurrent disease (including phage, bacteriocins, and small molecules). One elegant example is the isolation of a bacteriocin with narrow spectrum activity against C. difficile. An in vitro screen for bacteria that antagonize C. difficile identified an intestinal Bacillus thuringensis strain that secreted a bacteriocin later named Thuricin CD [44]. This bacteriocin, which is composed of two peptides that act synergistically, was shown to have narrow spectrum activity against spore forming Gram-positive bacteria including C. difficile. Thuricin CD was shown to be as effective as metronidazole at inhibiting C. difficile in an in vitro model of the human colon and, given its narrow spectrum, it may be an alternative therapy for CDI [45]. Regardless of its potential as a therapeutic, the identification of the B. thuringensis strain that produces it may indicate that the intestinal microbiota possesses intrinsic resistance mechanisms that target pathogens such as C. difficile.
Toll-like receptor (TLR) signaling and CDI
The development of mouse models of C. difficile infection will offer a number of genetic tools for understanding how the host immune response impacts C. difficile colonization and virulence. Mice deficient in MyD88, a key node in the TLR signaling cascade, were shown to have increased susceptibility to experimental CDI [46]. Wild-type mice that were treated with a single dose of clindamycin were colonized by C. difficile and shed high concentrations of spores, yet did not show significant signs of disease. Deletion of MyD88 caused the mice to develop disease upon C. difficile infection, suggesting that TLR signaling may be important to protect the host from disease.
Subsequently, other investigators have looked for the specific TLRs that were important in recognition of C. difficile. Jarchum et al. reported that TLR5 activation by flagellin (from Salmonella) was sufficient to prevent C. difficile infection after antibiotic treatment [47]. They noted a 5 log reduction in C. difficile cell numbers, indicating that TLR5 activation either prevents spore germination or elicits a bactericidal effect against vegetative C. difficile. TLR5 mutants were unable to prevent C. difficile colonization in response to flagellin activation. However, the role of TLR5 in CDI remains unclear as TLR5 mutants show no increased susceptibility to C. difficile infection in the absence of flagellin treatment. Overexpression of the cationic peptide defensin RegIIIγ was proposed as a possible mechanism by which TLR5 activation suppresses C. difficile colonization, similar to previous work with vancomycin resistant Enterococcus infection [48]. However, no evidence for RegIIIγ upregulation was presented and a subsequent study of CDI in mice showed no alteration of the expression of this antimicrobial peptide upon antibiotic treatment [18]. While this does not rule out a role for this cationic peptide in TLR5 mediated suppression of C. difficile it suggests that it is not responsible for limiting C. difficile in the presence of a normal or disrupted microbiota.
In contrast to TLR5, TLR4 appears to have a direct role in the recognition and response to C. difficile infection. Using purified surface layer (S-layer) proteins from C. difficile it was shown that the S-layer could induce the maturation of dendritic cells (DCs), with the induction of cytokines similar to what is observed when DCs were stimulated with the TLR4 agonist lipopolysaccharide [49]. DCs lacking TLR4 did not respond to S-layer proteins. When wild-type, TLR4-deficient and TLR2-deficient mice were treated with antibiotics and then subsequently challenged with C. difficile, TLR4-deficient mice displayed more severe disease than wild-type and TLR2-deficient mice. The data support the hypothesis that the host immune system responds to C. difficile by responding to S-layer proteins via a TLR4-dependent mechanism, which activates innate and adaptive immune responses that are critical for elimination of the pathogen. However, TLR4 activation is not sufficient to prevent C. difficile infection in antibiotic treated mice, as they do become colonized and show signs of disease. The important role of the microbiota in colonization resistance was also highlighted in this study by the demonstration that TLR4-deficient mice were not susceptible to disease in the absence of antibiotic treatment.
As research with mouse models of C. difficile infection proceeds, it is incumbent on groups performing this work to be cognizant of the potential pitfalls of the starting microbiota of the various mice utilized in these studies. The microbiotas of animals will be influenced by supplier (and barriers), cleanliness of individual mouse facilities, diet, strength and types of antibiotics being used to induce CDI, and the genetic background of the mice being used. Therefore monitoring the intestinal microbial ecology of these mice is of critical importance to the interpretation of results obtained in these studies to aid in the resolution of conflicting results that may arise between laboratories. For example, C57BL/6 mice treated with a single dose of clindamycin have different responses following infection with C. difficile. Our group was unable to stably infect mice following disruption with just clindamycin [18], while other groups could demonstrate chronic colonization [46] and colitis [50] following the same treatment. It is possible that differences in the challenge C. difficile strain could contribute to the differential responses seen, but both groups used C. difficile strain VPI 10463. Thus, while our understanding of the intestinal microbiota may not be sufficient yet to understand key differences in the function of different communities, we expect that this will be overcome in the near future. Further experimentation with murine models of CDI should help with our understanding of how the indigenous microbiota interacts with C. difficile within the gastrointestinal tract.
Concluding remarks
It is clear that antibiotic administration is the main risk factor for the development of CDI. We are beginning to understand how the indigenous gut microbiota mediates colonization resistance against potential pathogens and how antibiotic disruption can alter colonization resistance. Going forward, there are still key, basic questions that need to be addressed regarding the interaction between the gut microbiota and C. difficile (Box 1). We still need to understand which of the possible mechanisms by which the intestinal microbiota inhibits C. difficile are the most important in vivo. Such information could lead to novel treatments and preventative strategies for CDI. For example, knowledge of specific organisms or groups of organisms that possess inhibitory action against C. difficile could lead to novel probiotics that could be administered to patients undergoing antibiotic treatment who are at high risk of developing CDI. Additionally, antibiotic regimens that spare organisms important for colonization resistance could be preferentially employed to decrease the risk of CDI. The recent development of murine models as described above will be important in this discovery process and in the initial testing of novel therapeutic strategies for CDI. Almost a century after it was proposed that the indigenous gut microbiota plays a beneficial role for the host by interfering with the invasion of pathogens, we are poised not only to understand the mechanisms by which this happens but to also employ this as a novel treatment for a variety of infectious diseases.
Box 1. Outstanding questions.
Which of the various possible mechanisms by which the intestinal microbiota inhibits C. difficile are the most important in vivo?
Can we identify specific members of the indigenous gut microbiota that can interfere with C. difficile when administered therapeutically or prophylactically?
How important is the role of the host immune system in mediating colonization resistance against C. difficile? What are the relative roles of innate and adaptive immunity in the pathogenesis of CDI?
Can the relative risk of CDI for different antibiotic regimens be predicted from the stereotypic effects that they have on the indigenous gut microbiota?
Acknowledgments
The authors thank David Aronoff and Jennifer Auchtung for critically reading the manuscript. This work was funded through NIH awards AI090872 (RAB), DK083993, AI090871 and DK070875 (VBY).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
VBY has the following declaration to make about conflicts of interest: he is on the advisory board of ViroPharma regarding the development of NTCD and the management of Clostridium difficile infection.
References
- 1.Dubberke ER, Wertheimer AI. Review of current literature on the economic burden of Clostridium difficile infection. Infect Control Hosp Epidemiol. 2009;30:57–66. doi: 10.1086/592981. [DOI] [PubMed] [Google Scholar]
- 2.Proctor LM. The Human Microbiome Project in 2011 and beyond. Cell Host Microbe. 2011;10:287–291. doi: 10.1016/j.chom.2011.10.001. [DOI] [PubMed] [Google Scholar]
- 3.Kelly CP, Kyne L. The host immune response to Clostridium difficile. J Med Microbiol. 2011;60:1070–1079. doi: 10.1099/jmm.0.030015-0. [DOI] [PubMed] [Google Scholar]
- 4.Tedesco FJ, et al. Clindamycin-associated colitis. A prospective study. Ann Intern Med. 1974;81:429–433. doi: 10.7326/0003-4819-81-4-429. [DOI] [PubMed] [Google Scholar]
- 5.Bartlett JG, et al. Clindamycin-associated colitis due to a toxin-producing species of Clostridium in hamsters. J Infect Dis. 1977;136:701–705. doi: 10.1093/infdis/136.5.701. [DOI] [PubMed] [Google Scholar]
- 6.Fekety R, et al. Antibiotic-associated colitis: effects of antibiotics on Clostridium difficile and the disease in hamsters. Rev Infect Dis. 1979;1:386–397. doi: 10.1093/clinids/1.2.386. [DOI] [PubMed] [Google Scholar]
- 7.Wilson KH, et al. Suppression of Clostridium difficile by normal hamster cecal flora and prevention of antibiotic-associated cecitis. Infect Immun. 1981;34:626–628. doi: 10.1128/iai.34.2.626-628.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Johnson S. Recurrent Clostridium difficile infection: causality and therapeutic approaches. Int J Antimicrob Agents. 2009;33(Suppl 1):S33–36. doi: 10.1016/S0924-8579(09)70014-7. [DOI] [PubMed] [Google Scholar]
- 9.Smith DG, Robinson HJ. The influence of streptomycin and streptothricin on the intestinal flora of mice. J Bacteriol. 1945;50:613–621. doi: 10.1128/JB.50.6.613-621.1945. [DOI] [PubMed] [Google Scholar]
- 10.Freter R. The fatal enteric cholera infection in the guinea pig, achieved by inhibition of normal enteric flora. J Infect Dis. 1955;97:57–65. doi: 10.1093/infdis/97.1.57. [DOI] [PubMed] [Google Scholar]
- 11.Wilson KH, Freter R. Interaction of Clostridium difficile and Escherichia coli with microfloras in continuous-flow cultures and gnotobiotic mice. Infect Immun. 1986;54:354–358. doi: 10.1128/iai.54.2.354-358.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bartlett JG. Clinical practice. Antibiotic-associated diarrhea. N Engl J Med. 2002;346:334–339. doi: 10.1056/NEJMcp011603. [DOI] [PubMed] [Google Scholar]
- 13.Bartlett JG. Clostridium difficile: progress and challenges. Ann N Y Acad Sci. 2010;1213:62–69. doi: 10.1111/j.1749-6632.2010.05863.x. [DOI] [PubMed] [Google Scholar]
- 14.Stevens V, et al. Cumulative antibiotic exposures over time and the risk of Clostridium difficile infection. Clin Infect Dis. 2011;53:42–48. doi: 10.1093/cid/cir301. [DOI] [PubMed] [Google Scholar]
- 15.Paterson DL. Collateral damage” from cephalosporin or quinolone antibiotic therapy. Clin Infect Dis. 2004;38(Suppl 4):S341–345. doi: 10.1086/382690. [DOI] [PubMed] [Google Scholar]
- 16.Razaq N, et al. Infection of hamsters with historical and epidemic BI types of Clostridium difficile. J Infect Dis. 2007;196:1813–1819. doi: 10.1086/523106. [DOI] [PubMed] [Google Scholar]
- 17.Chen X, et al. A mouse model of Clostridium difficile-associated disease. Gastroenterology. 2008;135:1984–1992. doi: 10.1053/j.gastro.2008.09.002. [DOI] [PubMed] [Google Scholar]
- 18.Reeves AE, et al. The interplay between microbiome dynamics and pathogen dynamics in a murine model of Clostridium difficile Infection. Gut Microbes. 2011;2:145–158. doi: 10.4161/gmic.2.3.16333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rea MC, et al. Microbes and Health Sackler Colloquium: Effect of broad- and narrow-spectrum antimicrobials on Clostridium difficile and microbial diversity in a model of the distal colon. Proc Natl Acad Sci U S A. 2010 doi: 10.1073/pnas.1001224107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Robinson CJ, et al. From structure to function: the ecology of host-associated microbial communities. Microbiol Mol Biol Rev. 2010;74:453–476. doi: 10.1128/MMBR.00014-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Young VB, Schmidt TM. Antibiotic-associated diarrhea accompanied by large-scale alterations in the composition of the fecal microbiota. J Clin Microbiol. 2004;42:1203–1206. doi: 10.1128/JCM.42.3.1203-1206.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jakobsson HE, et al. Short-term antibiotic treatment has differing long-term impacts on the human throat and gut microbiome. PLoS One. 2010;5:e9836. doi: 10.1371/journal.pone.0009836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dethlefsen L, et al. The pervasive effects of an antibiotic on the human gut microbiota, as revealed by deep 16S rRNA sequencing. PLoS Biol. 2008;6:e280. doi: 10.1371/journal.pbio.0060280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dethlefsen L, Relman DA. Incomplete recovery and individualized responses of the human distal gut microbiota to repeated antibiotic perturbation. Proc Natl Acad Sci U S A. 2011;108(Suppl 1):4554–4561. doi: 10.1073/pnas.1000087107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Antonopoulos DA, et al. Reproducible community dynamics of the gastrointestinal microbiota following antibiotic perturbation. Infect Immun. 2009;77:2367–2375. doi: 10.1128/IAI.01520-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.de La Cochetiere MF, et al. Human intestinal microbiota gene risk factors for antibiotic-associated diarrhea: perspectives for prevention. Risk factors for antibiotic-associated diarrhea. Microb Ecol. 2010;59:830–837. doi: 10.1007/s00248-010-9637-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Manges AR, et al. Comparative metagenomic study of alterations to the intestinal microbiota and risk of nosocomial Clostridum difficile-associated disease. J Infect Dis. 2010;202:1877–1884. doi: 10.1086/657319. [DOI] [PubMed] [Google Scholar]
- 28.Maroo S, Lamont JT. Recurrent Clostridium difficile. Gastroenterology. 2006;130:1311–1316. doi: 10.1053/j.gastro.2006.02.044. [DOI] [PubMed] [Google Scholar]
- 29.Chang JY, et al. Decreased diversity of the fecal microbiome in recurrent Clostridium difficile-associated diarrhea. J Infect Dis. 2008;197:435–438. doi: 10.1086/525047. [DOI] [PubMed] [Google Scholar]
- 30.Gough E, et al. Systematic review of intestinal microbiota transplantation (fecal bacteriotherapy) for recurrent Clostridium difficile infection. Clin Infect Dis. 2011;53:994–1002. doi: 10.1093/cid/cir632. [DOI] [PubMed] [Google Scholar]
- 31.Khoruts A, et al. Changes in the composition of the human fecal microbiome after bacteriotherapy for recurrent Clostridium difficile-associated diarrhea. J Clin Gastroenterol. 2010;44:354–360. doi: 10.1097/MCG.0b013e3181c87e02. [DOI] [PubMed] [Google Scholar]
- 32.Ridlon JM, et al. Bile salt biotransformations by human intestinal bacteria. J Lipid Res. 2006;47:241–259. doi: 10.1194/jlr.R500013-JLR200. [DOI] [PubMed] [Google Scholar]
- 33.Begley M, et al. Bile salt hydrolase activity in probiotics. Appl Environ Microbiol. 2006;72:1729–1738. doi: 10.1128/AEM.72.3.1729-1738.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ridlon JM, et al. Isolation and characterization of a bile acid inducible 7 alpha-dehydroxylating operon in Clostridium hylemonae TN271. Anaerobe. 2010;16:137–146. doi: 10.1016/j.anaerobe.2009.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wilson KH. Efficiency of various bile salt preparations for stimulation of Clostridium difficile spore germination. J Clin Microbiol. 1983;18:1017–1019. doi: 10.1128/jcm.18.4.1017-1019.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sorg JA, Sonenshein AL. Bile salts and glycine as cogerminants for Clostridium difficile spores. J Bacteriol. 2008;190:2505–2512. doi: 10.1128/JB.01765-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Giel JL, et al. Metabolism of bile salts in mice influences spore germination in Clostridium difficile. PLoS One. 2010;5:e8740. doi: 10.1371/journal.pone.0008740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sorg JA, Sonenshein AL. Chenodeoxycholate is an inhibitor of Clostridium difficile spore germination. J Bacteriol. 2009;191:1115–1117. doi: 10.1128/JB.01260-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sorg JA, Sonenshein AL. Inhibiting the initiation of Clostridium difficile spore germination using analogs of chenodeoxycholic acid, a bile acid. J Bacteriol. 2010;192:4983–4990. doi: 10.1128/JB.00610-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Merrigan MM, et al. Prevention of fatal Clostridium difficile-associated disease during continuous administration of clindamycin in hamsters. J Infect Dis. 2003;188:1922–1927. doi: 10.1086/379836. [DOI] [PubMed] [Google Scholar]
- 41.Sambol SP, et al. Colonization for the prevention of Clostridium difficile disease in hamsters. J Infect Dis. 2002;186:1781–1789. doi: 10.1086/345676. [DOI] [PubMed] [Google Scholar]
- 42.Chang DE, et al. Carbon nutrition of Escherichia coli in the mouse intestine. Proc Natl Acad Sci U S A. 2004;101:7427–7432. doi: 10.1073/pnas.0307888101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Leatham MP, et al. Precolonized human commensal Escherichia coli strains serve as a barrier to E. coli O157:H7 growth in the streptomycin-treated mouse intestine. Infect Immun. 2009;77:2876–2886. doi: 10.1128/IAI.00059-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rea MC, et al. Thuricin CD, a posttranslationally modified bacteriocin with a narrow spectrum of activity against Clostridium difficile. Proc Natl Acad Sci U S A. 2010;107:9352–9357. doi: 10.1073/pnas.0913554107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rea MC, et al. Effect of broad- and narrow-spectrum antimicrobials on Clostridium difficile and microbial diversity in a model of the distal colon. Proc Natl Acad Sci U S A. 2011;108(Suppl 1):4639–4644. doi: 10.1073/pnas.1001224107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lawley TD, et al. Antibiotic treatment of Clostridium difficile carrier mice triggers a supershedder state, spore-mediated transmission, and severe disease in immunocompromised hosts. Infect Immun. 2009;77:3661–3669. doi: 10.1128/IAI.00558-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jarchum I, et al. Toll-like receptor-5 stimulation protects mice from acute Clostridium difficile colitis. Infect Immun. 2011;79:1498–1503. doi: 10.1128/IAI.01196-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Brandl K, et al. Vancomycin-resistant enterococci exploit antibiotic-induced innate immune deficits. Nature. 2008;455:804–807. doi: 10.1038/nature07250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ryan A, et al. A role for TLR4 in Clostridium difficile infection and the recognition of surface layer proteins. PLoS Pathog. 2011;7:e1002076. doi: 10.1371/journal.ppat.1002076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Buffie CG, et al. Profound alterations of intestinal microbiota following a single dose of clindamycin results in sustained susceptibility to Clostridium difficile-induced colitis. Infect Immun. 2012;80:62–73. doi: 10.1128/IAI.05496-11. [DOI] [PMC free article] [PubMed] [Google Scholar]