Abstract
NR4A2 (Nurr 1) is an orphan nuclear receptor with no known endogenous ligands and is highly expressed in many cancer cell lines including Panc1 and Panc28 pancreatic cancer cells. Structure-dependent activation of NR4A2 by a series of 1,1-bis(3′-indolyl)-1-(aromatic)methane (C-DIM) analogs was determined in pancreatic cancer cells transfected with yeast GAL4-Nurr1 chimeras and a UASx5-luc reporter gene or constructs containing response elements that bind NR4A2. Among 23 different structural analogs, phenyl groups containing p-substituted trifluoromethyl, t-butyl, cyano, bromo, iodo and trifluoromethoxy groups were the most active compounds in transactivation assay. The p-bromophenyl analog (DIM-C-pPhBr) was used as a model for structure-activity studies among a series of ortho-, meta- and para-bromophenyl isomers and the corresponding indole 2- and N-methyl analogs. Results show that NR4A2 activation was maximal with the p-bromophenyl analog and methylation of the indole NH group abrogated activity. Moreover, using GAL4-Nurr1 (full length) or GAL-Nurr1-A/B and GAL4-Nurr1-(C-F) chimeras expressing N- and C-terminal domains of Nurr1, respectively, DIM-C-pPhBr activated all three constructs and these responses were differentially affected by kinase inhibitors. DIM-C-pPhBr also modulated expression of several Nurr1-regulated genes in pancreatic cancer cells including vasoactive intestinal peptide (VIP), and the immunohistochemical and western blot analyses indicated that DIM-C-pPhBr activates nuclear NR4A2.
Keywords: DIM analogs, NR4A2/Nurr1, structure-activity
1. Introduction
The nuclear receptor (NR) superfamily of transcription factors are characterized by their structural homology which includes N- and C-terminal domains (A/B and E/F, respectively), a DNA binding domain (C), and an adjacent hinge region (D) [1-3]. Both N- and C-terminal regions may contain activation functions (AFs), and the ligand binding domain (LBD) resides in the C-terminus of NRs [1-4]. The 48 members of the NR superfamily can be subdivided into three broad categories, namely, the endocrine nuclear receptors, adopted orphan receptors, and orphan receptors such as NR4A for which cognate ligands have not yet been identified [1-4]. The NR4A orphan receptor subfamily includes NR4A1 (Nur77, NGFI-B, TR3), NR4A2 (Nurr1, NOT), and NR4A3 (Nor-1, MINOR) [5-8], and it has been suggested that the failure to identify an endogenous ligand may be due to the lack of a typical NR ligand binding pocket in the LBD domain of NR4A receptors [9].
NR4A receptors are immediate-early genes that are induced by diverse stimuli in multiple tissues, and there is increasing evidence that these receptors play important roles in maintaining tissue homeostasis and in pathophysiological processes including cancer [5-8]. NR4A receptors have specific functions in the brain, T-cells/thymocytes, adipose tissue, steroidgenesis, muscle, blood vessels, macrophages, and cardiovascular system; however, the role of NR4A receptors in carcinogenesis is less well understood [7, 8]. Knockdown of NR4A1 by RNA interference in cancer cell lines induced apoptosis or inhibited growth in multiple cell lines [reviewed in 8] and NR4A2 knockdown also induced apoptosis and decreased metastasis in cancer cells lines [10-12].
The effects and mechanisms of action of drug-induced activation or deactivation of NR4A receptors is complex and is dependent on both cell context and structure. Studies with phorbol esters, retinoids and other apoptosis-inducing agents have unraveled a novel NR4A1-dependent proapoptotic pathway that involves nuclear export of the receptor which in some cell lines forms a mitochondrial proapoptotic bcl-2-NR4A1 complex [13, 14]. Studies in this laboratory have demonstrated that among 1,1-bis(3′-indolyl)-1-(p-substituted phenyl) methanes (C-DIMs), the p-methoxy (DIM-C-pPhOCH3) and p-hydroxy (DIM-C-pPhOH) derivatives induce nuclear NR4A1-dependent apoptosis and growth inhibition of colon, pancreatic and bladder cancer cells through activation or deactivation of the receptor [15-18]. Cytosporone B and related compounds bind directly the receptor and appear to activate nuclear NR4A1-mediated transcription; these receptor agonists also induce nuclear NR4A1 export [19, 20].
6-Mercaptopurine (6-MP) activates NR4A2 in CV1 and HEK293 cells through the N-terminal A/B domain and this pathway involves metabolic activation of 6-MP [21]. Several benzimidazoles also induce NR4A2-dependent transactivation, and a p-chloro-substituted C-DIM analog (DIM-C-pPhCI) also activates Nurr1 in bladder cancer cells [22, 23]. In this study, we have investigated the structure-dependent activation of NR4A2 in pancreatic cancer cells by twenty-three C-DIM analogs containing various p-substituted phenyl and heteroaromatic substituents and their activity was compared to 6-MP. Among the most active compounds were the p-trifluoromethyl (DIM-C-pPhCF3), p-bromo (DIM-C-pPhBr), p-t-butyl (DIM-C-pPhtBu), p-cyano (DIM-C-pPhCN), p-iodo (DIM-C-pPhI), and p-trifluoromethoxy (DIM-C-phOCF3) analogs. Using one or more NR4A2-active C-DIMs as models, these compounds also induced transactivation in cells transfected with constructs containing three copies of an NGFI-B response element (NBRE3-luc) and three copies of a Nur response element (NuREx3-luc). Nurr1-active C-DIMs also activated a wild-type GAL4-Nurr1 variant and GAL4-Nurr1-(A/B) and GAL4-Nurr1-(C-F) chimeras containing N- and C-terminal regions of NR4A2, respectively, and the prototypical model NR4A2 activator DIM-C-pPhBr induced expression of several NR4A2-dependent genes in Panc1 and Panc28 that were confirmed by RNA interference. These studies demonstrate that the C-DIM structure is an excellent scaffold for developing NR4A2-active compounds.
2. Materials and methods
2.1 Cell lines and cell culture
Panc1 and Panc28 pancreatic cancer cell lines were obtained from the American Type Culture Collection (Manassas, VA) and maintained in Dulbecco’s modified Eagle’s medium (DMEM) nutrient mixture with Ham’s F-12 (DMEM/Ham’s F-12; Sigma-Aldrich, St Louis, MO) supplemented with 5% fetal bovine serum (FBS), 0.22% sodium bicarbonate and 10 mL/L 100× antibiotic antimycotic solution (Invitrogen, Carlsbad, CA). Cells were maintained at 37°C in the presence of 5% CO2 and the solvent [dimethyl sulfoxide (DMSO)] used in the experiments was ≤ 0.15%.
2.2 Plasmids
The GAL4-Nurr1 chimeras GAL4-Nurr1 (full length, amino acid 1 to 598), GAL4-Nurr1-AB (amino acid 1 to 259), and GAL4-Nurr1-(C-F) (amino acid 260 to 598) were constructed by inserting PCR-amplified each fragment into the BamHI/HindIII site of pM vector (Clontech, Mountain View, CA). The FLAG-tagged full-length Nurr1 (FLAG-Nurr1) were constructed by inserting PCR-amplified full-length Nurr1 fragment into the HindIII/BamHI site of p3XFLAG-CMV-10 expression vector (Sigma-Aldrich). The NBREx3-Luc was generously provided by Dr. Jacques Drouin (University of Montreal, Quebec, Canada). All other reporter constructs have been previously described [15].
2.3 Antibodies, chemicals, reagents, siRNA oligonucleotides and primers
The NR4A2 antibody (sc-991) was purchased from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA). The TR3/Nur77 antibody (IMG-528) was purchased from Imgenex (San Diego CA). The FLAG (F3165) and β-actin antibodies were purchased from Sigma-Aldrich. The methylene-substituted diindolylmethanes were synthesized in this laboratory by condensation of indole or substituted indoles with corresponding aromatic aldehyde (2:1 indole/aldehydes) at 80°C in a pH5 buffer essentially as described [16, 24, 25]. Compounds were crystallized from benzene/hexane (≥ 1 time) and purities were > 97% as determined by gas chromatography or gas chromatography-mass spectrometry [16, 24, 25]. Indole was purchased from Sigma-Aldrich and the following benzaldehyde derivatives were used to make a series of DIM-C-pPhX analogs where X represents the para-substituent: p-trifluoromethylbenzaldehyde, p-bromobenzaldehyde, p-fluorobenzaldehyde, p-t-butylbenzaldehyde, p-N-dimethylamino, benzaldehyde (no substituent), p-hydroxybenzaldehyde, p-phenylbenzaldehyde, p-cyanobenzaldehyde, p-tolualdehyde, p-chlorobenzaldehyde, p-iodobenzaldehyde, p-carboxymethylbenzaldehyde, p-methoxybenzaldehyde, p-butoxybenzaldehyde, and p-trifluromethoxybenzaldehyde. The meta- and ortho-bromo substituted isomers were prepared by condensing indole with meta- and ortho-bromobenzaldehye, respectively. The N-methyl and 2-methyl analogs of DIM-C-pPhBr were prepared by condensing p-bromobenzaldehyde with N-methylindole and 2-methylindole, respectively as described [25]. The C-DIM analogs containing heteroaromatic substituents were synthesized by condensing indole with the following aldehydes: 2-furaldehyde, 2-thiophene-carboxaldehyde, 3-thiophenecarboxaldehyde, pyrrole-2-carboxaldehyde, piperonal, 4-pyridine-carboxyaldehyde, N-O-4-pyridine-carboxaldehyde, and indole-3-carboxaldehyde. All indole derivatives, aldehydes and 6-mercaptopurine (MP) were purchased from Sigma-Aldrich. Reporter lysis buffer, luciferase and β-galactosidase (β-gal) reagents were purchased from Promega (Madison, WI) and Tropix (Applied Biosystems). Plasmid and total-RNA extraction kits were purchased from Qiagen (Valencia, CA). SYBR Green (Applied Biosystems, Foster City, CA) was used for triplicate real-time PCR reaction. All the primers and the small inhibitory RNAs were prepared by Sigma-Aldrich. Two siRNA oligonucleotides were used in combination to target NR4A2 (Nurr1): 5′- CAG UUA CCA CUC UUC GGG A dTdT -3′ and 5′- CGU GUG UUU AGC AAA UAA A dTdT -3′. The sequences of the primers used for real-time PCR were as follows: NR4A2 sense 5′- AGT CTG ATC AGT GCC CTC GT -3′, antisense 5′- TAT GCT GGG TGT CAT CTC CA -3′; VIP sense 5′- TCA GGT TCA TTT GCT CCC TC -3′, antisense 5′- TCT TCT CAC AGA CTT CGG CA -3′. SPP1 (osteopontin) sense 5′- TTG CAG TGA TTT GCT TTT GC -3′, antisense 5′- GCC ACA GCA TCT GGG TAT TT -3′; NRP1 sense 5′- AAG GTT TCT CAG CAA ACT ACA GTG - 3′, antisense 5′- GGG AAG AAG CTG TGA TCT GGT C -3′. NRP2 sense 5′- GAT TCG GGA TGG GGA CAG TGA -3′, antisense 5′- GGT GAA CTT GAT GTA GAG CAT GGA -3′.
2.4 Transfection, luciferase assay and quantitative real-time PCR
Cells were plated on 12-well plates at 7×104 per well in DMEM/F12 supplemented with 2.5% charcoal-stripped FBS and 0.22% sodium bicarbonate. After 24 hr growth, various amounts of DNA [i.e., UASx5-Luc (400 ng), GAL4-Nurr1 (40 ng) and β-gal (40 ng)] were cotransfected into each well by Lipofectamine 2000 reagent (Invitrogen, Carlsbad, CA) according to the manufacturer’s protocol. After 5-6 hr of transfection, cells were treated with plating media (as above) containing either solvent (DMSO) or the indicated concentration of compound for 18 hr. Cells were then lysed using a freeze-thaw protocol and 30 μL of cell extract was used for luciferase and β-gal assays. LumiCount (Packard, Meriden, CT) was used to quantify luciferase and β-gal activities. Luciferase activity values were normalized against corresponding β-gal activity values as well as protein concentrations determined by Bradford assay. For RNA interference experiment, cells were transfected with equal amount of both siRNA duplex (i.e. 75 pmol each/well for 6-well plate) using Lipofectamine 2000 reagent for 24 hr prior to treatment. Total RNA was extracted, reverse transcription and real-time PCR were carried out as described previously [15], and messenger RNA (mRNA) levels were normalized to the expression of TATA-binding protein (TBP).
2.5 Subcellular localization assay and western blot analysis
Cells were seeded on cover glass and transfected with adenovirus expressing FLAG-Nurr1 (5 MOI) for 4 hr. At 18 hr after transfection, cells were treated with DIM-C-pPhBr for 12 hr and immunostained with anti-FLAG antibody. Cells were then mounted in mounting medium including DAPI (Vector Laboratory, CA) and the fluorescent images were obtained using a Zeiss Axioplan2 fluorescence microscope (Carl Zeiss, Jena, Germany). For western blotting, cells (2×105) were plated on six-well plates in DMEM/Ham’s F-12 media containing 10% charcoal-stripped FBS for 16 hr and then treated with indicated concentrations of compounds. Cellular lysates and their subsequent separation by electrophoresis were carried out as described previously using β-actin as a loading control [15] and Sp1 as a representative nuclear protein [16].
2.6 Statistical analysis
Statistical significance of differences in luciferase activities and gene expression levels between groups was analyzed using unpaired Student’s t-test. A P value of <0.05 was considered statistically significant. Results are expressed as means ± standard deviations for at least three independent determinations for each treatment group.
3. Results
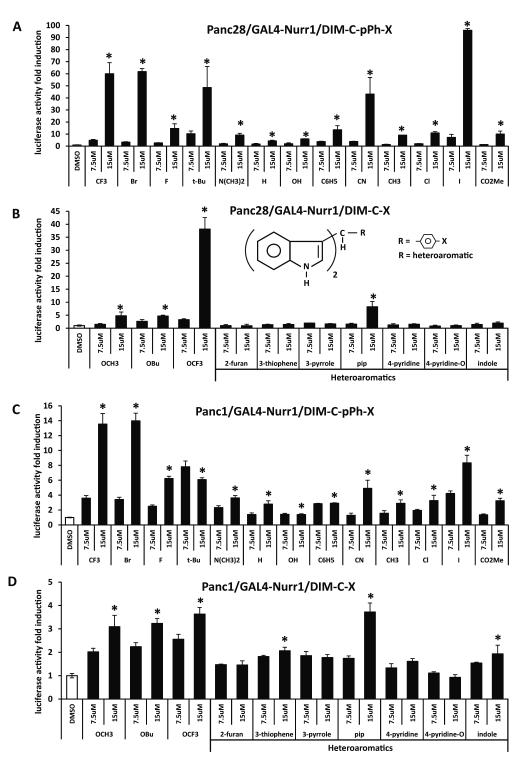
Panc1 and Panc28 pancreatic cancer cell lines were used in these studies for screening a series of C-DIM compounds as activators of NR4A2. Transfection of Panc28 cells with a UASx5-luc construct containing 5 tandem GAL4 response elements gave relatively low basal luciferase activity compared to Panc1 cells. For the C-DIM screening assay, Panc28 cells were transfected with UASx5-luc and the GAL4-Nurr1 chimera (containing the yeast GAL4 DNA binding domain fused to wild type NR4A2) and treated with DMSO, 7.5 or 15 μM concentrations of various C-DIMs which contained p-substituted phenyl or heteroaromatic groups (Figs. 1A and 1B). Compounds containing p-substituted CF3, Br, t-Bu, CN, I and OCF3 groups induced the highest activity at the 15 μM concentration. Among the heteroaromatics, only 1,1-bis(3′-indolyl)-1-[2,4-(methylendioxy)benzaldehyde]methane, (C-DIM-pip), the piperonal condensation product, induced activity > 5-fold. A similar approach was used in Panc1 cells and the fold-induction of luciferase activity by C-DIMs was significantly lower than observed in Panc28 cells (Figs. 1C and 1D) and this may be due, in part, to the relatively high basal luciferase activity in this cell line transfected with UASx5-luc. Despite the compression of induced luciferase activities, most of the p-substituted phenyl and heteroaromatic compounds that activated GAL4-Nurr1 in Panc28 cells were also active in Panc1 cells.
Figure 1.
Activation of GAL4-Nurr1 chimeras by C-DIMs. UASx5-Luc (400 ng) and GAL4-Nurr1 (40 ng) were cotransfected into Panc28 (A, B) and Panc1 (C, D) cells for 6 hr and then treated with 7.5 and 15 μM phenyl-substituted C-DIMs including trifluoromethyl (CF3), bromo (Br), fluoro (F), tert-butyl (t-Bu), dimethylamino (N(CH3)2), hydrogen (H), hydroxy (OH), phenyl (C6H5), cyano (CN), methyl (CH3), chloro (Cl), iodo (I), carboxymethyl (CO2Me) (A, C), methoxy (OCH3), tert-butoxy (OBu) or trifluoromethoxy (OCF3) (B, D) group on the para position, or the heterocyclic C-DIMs including 2-furan, 3-thiophene, 3-pyrrole, piperonal, 4-pyridine, 4-pyridine-N-oxide or indole ring (B, D) for 18 hr. Luciferase activity was determined as described in the Materials and Methods. Results are expressed as means ± SD for at least three separate determinations for each treatment. *, P < 0.05, high concentration treatment (15 μM) vs. solvent control (DMSO).
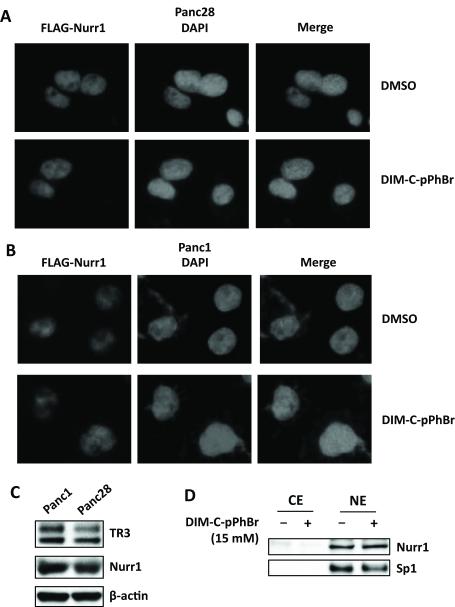
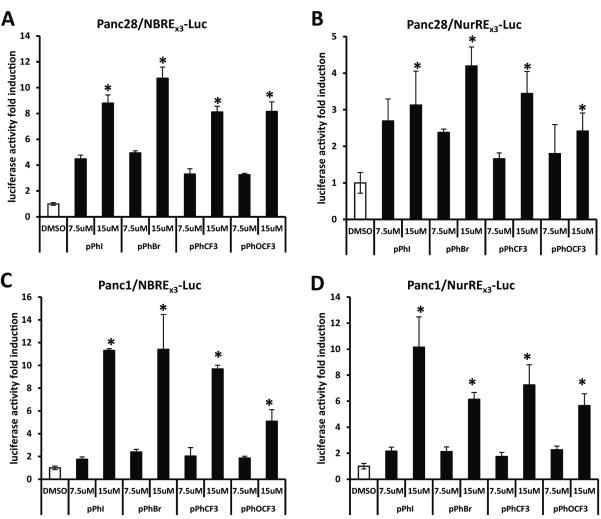
The subcellular location of NR4A2 was determined in Panc1 cells transfected with FLAG-Nurr1 (Fig. 2A). Immunostaining showed only nuclear FLAG, and treatment with 15 μM DIM-C-pPhBr for 12 hr did not induce any changes in nuclear NR4A2 staining. Immunostaining with NR4A2 antibodies also gave a nuclear signal which was weak (data not shown) and this necessitated the use of FLAG-tagged NR4A2. A similar immunostaining pattern was observed in Panc28 cells treated with solvent control (DMSO) or 15 μM DIM-C-pPhBr for 12 hr (Fig. 2B), and the western blot in Figure 2C shows that both NR4A2 and NR4A1 proteins were expressed in Panc1 and Panc28 cells. Moreover, after treatment of Panc1 cells with DMSO or 15 μM DIM-C-pPhBr for 24 hr, Nurr1 protein was isolated in nuclear (NE) but not cytosolic (CE) extracts, and expression was not changed by the treatment (Fig. 2D). Sp1 protein served as a nuclear protein control for this experiment. Preliminary studies showed that transfection of Panc1 and Panc28 cells with NBREx3-luc or NuREx3-luc constructs resulted in low basal activity and inducibility; however, higher activities and inducibility were observed only after transfection with FLAG-Nurr1 expression plasmid (data not shown), suggesting endogenous levels of NR4A2 were limiting in cells transfected with the response element constructs. Based on these results, 10 ng FLAG-Nurr1 construct was transfected for the structure-activity studies in Panc1 and Panc28 cells. Results in Figures 3A and 3B show that in Panc28 cells transfected with NBREX3-luc or NurREx3-luc constructs, respectively, the most active C-DIM compounds identified in the GAL4-Nurr1/UASx5-luc screening assays were also active in Panc28 cells, and similar results were observed in Panc1 cells (Figs. 3C and 3D). The compounds exhibited similar potencies and some cell context- and construct (NBREx3-luc vs. NuREx3-luc)-dependent differences.
Figure 2.
Expression and subcellular localization of NR4A2 in Panc28 and Panc1 cells. Panc28 (A) and Panc1 (B) cells were transfected with adenovirus expressing FLAG-Nurr1 for 4 hr; media was changed and after 18 hr, cells were treated with 15 μM of DIM-C-pPhBr for 12 hr and immunostained with anti-FLAG antibody. Fluorescent images were obtained as described in the Materials and Methods. (C) Whole cell lysates from Panc1 and Panc28 cells were analyzed by western blotting and β-actin was used as a loading control. (D) Cells were treated with DMSO or 15 μM DIM-C-pPhBr for 24 hr, and cytosolic (CE) or nuclear (NE) extracts were analyzed by western blots (Sp1 was a nuclear protein control).
Figure 3.
Selected NR4A2 activators increase activities of Nur response elements containing NBREx3-Luc and NurREx3-Luc luciferase genes. NBREx3-Luc (200 ng) (A, C) or NurREx3-Luc (200 ng) (B, D) was cotransfected with FLAG-Nurr1 (10 ng) into Panc28 (A, B) or Panc1 (C, D) cells for 6 hr and then treated with 7.5 and 15 μM p-substituted phenyl-C-DIMs DIM-C-pPhI, -pPhBr, -pPhCF3, and -pPhOCF3 for 18 hr. Luciferase activity was determined as described in the Materials and Methods. Results are expressed as means ± SD for at least three separate determinations for each treatment. *, P < 0.05, high concentration treatment (15 μM) vs. solvent control (DMSO).
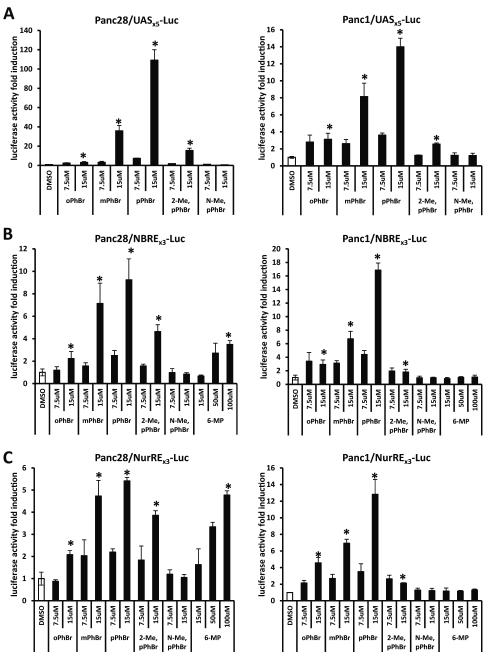
Among the Nurr1-active C-DIMs, previous studies showed that DIM-C-pPhBr exhibited minimal activation of PPARγ or TR3 [16, 24]. Therefore, this compound was used as a model for further investigating the structure-dependent activation of NR4A2, and results obtained for the C-DIM analogs were compared to 6-MP, an activator of Nurr1 in CV-1 and HEK293 cells [21]. Results summarized in Figures 4A - 4C compare the activity of DIM-C-pPhBr with the corresponding ortho- and meta-bromo-substituted analogs (DIM-C-oPhBr and DIM-C-mPhBr) on activation of GAL4-Nurr1/UASx5-luc, NRBEx3-luc and NuREx3-luc (cotransfected with 10 ng FLAG-Nurr1), respectively, and the results show that the order of potency was para-≥ meta- > ortho-bromo-substituted analogs for all three reporter constructs. The 2-methylindole-substituted derivative of DIM-C-pPhBr was also active; however, the 1-N-methyl indole analog was inactive in both cell lines. In contrast, 6-MP activated NBREX3-luc and NuREx3-luc in Panc28 cells but not Panc1 cells (Fig. 4C). The structure-activity study for activation of Nurr1 showed that among the bromophenyl analogs, DIM-C-pPhBr was the most potent compound and a free indole group was necessary for activation.
Figure 4.
Differential NR4A2 activation by DIM-C-PhBr analogs and 6-mercaptopurine. UASx5-Luc (400 ng) (A) was cotransfected with GAL4-Nurr1 (40 ng); NBREx3-Luc (200 ng) (B) or NurREx3-Luc (200 ng) (C) was cotransfected with FLAG-Nurr1 (10 ng) into Panc28 or Panc1 cells for 6 hr and then treated with 7.5 and 15 μM ortho-, meta-, para-substituted or indole ring-substituted bromo-phenyl-C-DIMs for 18 hr. A gradient of 15 μM, 50 μM and 100 μM 6-mercaptopurine was included in the treatment and compared with the DIM-C-PhBr analogs (B, C). Luciferase activity was determined as described in the Materials and Methods. Results are expressed as means ± SD for at least three separate determinations for each treatment. *, P < 0.05, high concentration treatment (15 μM or 100 μM) vs. solvent control (DMSO).
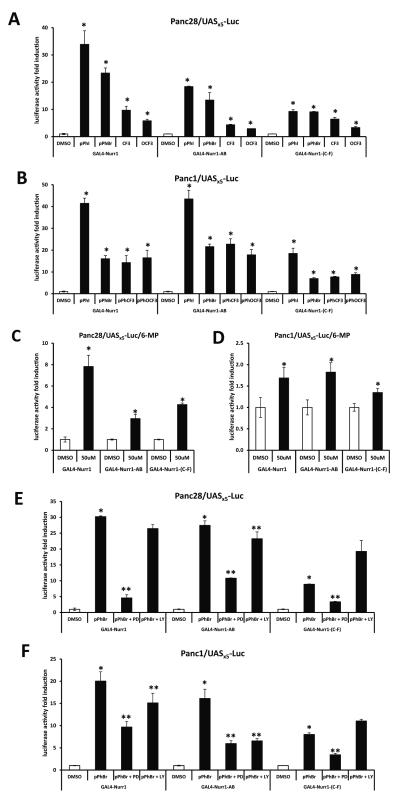
Activation of Nurr1 by 6-MP in CV-1 and HEK293 cells was dependent on the N-terminal A/B domain of the receptor [26] and therefore, we further investigated activation of wild-type GAL4-Nurr1 and truncated GAL4-Nurr1 (A/B) and GAL4-Nurr1 (C-F) chimeras expressing the N- and C-terminal regions of the receptor, respectively. In Panc28 and Panc1 cells (Figs. 5A and 5B), DIM-C-pPhBr and the p-iodo, p-trifluoromethyl and p-trifluoromethoxy analogs significantly induced transactivation in cells transfected with wild-type or variant GAL4-Nurr1 constructs. Similar results were observed for 6-MP (Figs. 5C and 5D); however, the fold induction in Panc1 cells was < 2-fold and this was consistent with the failure of 6-MP to activate NBREx3-luc or NuREx3-luc in this cell line. Previous studies showed that multiple kinase inhibitors block activation of NR4A2 in different cell lines [26-28], and preliminary inhibitor screening studies (Supplemental Fig. S1) in pancreatic cancer cells showed that inhibition of mitogen-activated protein kinase (MAPK) (PD98059) and phosphatidylinositol-3-kinase (PI3-K) (LY294002) were among the most active and least active inhibitors, respectively, using wild-type GAL4-Nurr1/UASx5-luc. In Panc28 cells transfected with wild-type or variant GAL4-Nurr1 chimeras, PD but not LY inhibited DIM-C-pPhBr-induced transactivation and, in cells transfected GAL4-Nurr1-(C-F), PD also decreased DIM-C-pPhBr-induced transactivation (Fig. 5E). The inhibitors alone had minimal effects on luciferase activity compared to DMSO (control). The pattern of inhibition by PD was similar in Panc1 cells (Fig. 5F); however, LY slightly inhibited and enhanced DIM-C-pPhBr-induced transactivation in cells transfected with GAL4-Nurr1 and GAL4-Nurr-(C-F), respectively, but exhibited inhibitory activity comparable to PD in cells transfected with GAL4-Nurr-AB. These results clearly demonstrate the complex effects of just two kinase inhibitors on activation of NR4A2 by DIM-C-pPhBr, suggesting that multiple phosphorylation sites in different domains of NR4A2 are involved.
Figure 5.
Activation of NR4A2 and different domains of NR4A2 by C-DIMs and 6-MP. Full length (GAL4-Nurr1, 20 ng) or truncated GAL4-Nurr1 containing the A/B domain (GAL4-Nurr1-AB, 20 ng) or C to F domains [GAL4-Nurr1-(C-F), 20 ng] was cotransfected with UASx5-Luc (200 ng) into Panc28 (A, C) or Panc1 (B, D) cells for 6 hr and then treated with 10 μM DIM-C-pPhI, -pPhBr, -pPhCF3, -pPhOCF3 or 50 μM 6-mercaptopurine (C, D) for 18 hr. Panc28 (E) or Panc1 (F) cells were transfected with 10 ng full/truncated GAL4-Nurr1 and 200 ng UASx5-Luc for 6 hr and pre-incubated with 20 μM kinase inhibitors PD98059 or LY294002 for 45 min and treated with 10 μM DIM-C-pPhBr for 18 hr. Luciferase activity was determined as described in the Materials and Methods. Results are expressed as means ± SD for at least three separate determinations for each treatment. *, P < 0.05, treatment vs. solvent control (DMSO). **, P < 0.05, kinase inhibitor and DIM-C-pPhBr cotreatment vs. DIM-C-pPhBr treatment.
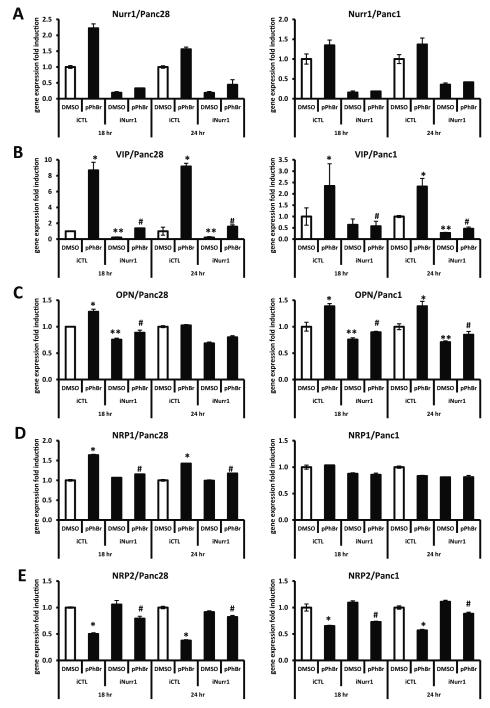
Previous studies have identified several NR4A2-regulated genes in different cell lines and these include vasoactive intestinal peptide (VIP), osteopontin (OPN) and neuropilin 1 (NRP1) [23, 24, 26-31]. The effects of DIM-C-pPhBr on NR4A2-dependent expression of these genes was investigated in Panc28 and Panc1 cells treated with 12 μM DIM-C-pPhBr and transfected with iNurr1 or iCTL (non-specific) oligonucleotides for RNA knockdown. Figure 6A summarizes the effects of iNurr1 vs. iCTL on knockdown of NR4A2 which was highly efficient in both cell lines. Treatment with DIM-C-pPhBr for 18 or 24 hr induced VIP in both Panc28 and Panc1 cells, and knockdown of NR4A2 significantly decreased both basal and induced activity (Fig. 6B). OPN induction after treatment of Panc28 (18 hr) or Panc1 (18 and 24) cells with DIM-C-pPhBr was also Nurr1-dependent (Fig. 6C), whereas neuropilin 1 (NRP1) was induced by DIM-C-pPhBr in Panc28 but not in Panc1 cells (Fig. 6D). NRP2 is also coexpressed with NRP1 in pancreatic cancer cells [32, 33], and Figure 6E shows that DIM-C-pPhBr decreased NRP2 expression in Panc28 and Panc1 cells; however, based on knockdown studies, the effects were NRR4A2-dependent and -independent, respectively. Supplemental Figure 2 shows the effectiveness of NR4A2 knockdown on the expression of Nurr1 protein in Panc1 cells, and the results also show that VIP protein was induced by DIM-C-pPhBr (NR4A2-dependent). These results demonstrate that DIM-C-pPhBr not only activates NR4A2 in pancreatic cancer cells but also induces NR4A2-dependent genes, and current studies are focused on the functional role of activated NR4A2 in mediating gene/protein expression and the anticancer activities of NR4A2-active C-DIMs.
Figure 6.
Effects of DIM-C-pPhBr on expression of several genes with or without NR4A2 knockdown. Panc28 and Panc1 cells were transfected with siRNAs targeting NR4A2 transcripts (iNurr1) or non-specific control oligonucleotides (iCTL). At 24 hr after transfection, cells were treated with 12 μM DIM-C-pPhBr for 18 and 24 hr. Relative expression levels of NR4A2 (A) and NR4A2-dependent genes vasoactive intestinal peptide (B), osteopontin (C), neuropilin-1 (D) and -2 (E) were determined by real-time PCR analysis as described in the Materials and Methods. Results are expressed as means ± SD for at least three separate determinations for each treatment. *, P < 0.05, treatment vs. solvent control (DMSO). **, P < 0.05, iNurr1 vs. iCTL. #, P < 0.05, knockdown and treatment combination vs. treatment only.
4. Discussion
Although endogenous ligands for the NR4A orphan receptors have not been identified, there is increasing evidence that several agents can modulate nuclear NR4A-dependent transactivation [8, 9, 11-20]. Cytosporone B and related analogs have been extensively investigated as NR4A1 (TR3) agonists and they activate nuclear NR4A1 and induce nuclear export of this receptor which acts directly on mitochondria to induced apoptosis [14, 20]. The cytosporone B analogs activate both wild-type and the LBD of NR4A1 (using GAL4-receptor chimeras), and direct binding of cytosporone B and related compounds to NR4A1 was confirmed using a BIAcore surface plasmon resonance-based instrument. DIM-C-pPhOCH3 was initially identified as a NR4A1-active compound that induces apoptosis and inhibits cancer cell growth through activation of nuclear NR4A1, and this has been linked to induction of several genes associated with these cellular responses [9, 15-18]. However, the effects of DIM-C-pPhOCH3 were due to activation of nuclear NR4A1 and nuclear export of NR4A1 was not induced.
Another C-DIM analog, namely DIM-C-pPhCI, was characterized as an activator of Nurr1 in bladder cancer cells, and knockdown of NR4A2 by RNA interference inhibited DIM-C-pPhCI-induced apoptosis [22]. In this study, we investigated the structure-activity relationships among a series of triarylmethane C-DIM analogs that contain a bis(3′-indolyl) moiety and either p-substituted phenyl or heteroaromatic groups in Panc28 and Panc1 pancreatic cell lines. Several p-substituted phenyl (CF3, Br, t-Bu, CN, I and OCF3) and the piperonal (heteroaromatic) compounds were potent activators of wild-type GAL4-Nurr1 in Panc28 and Panc1 cells with fold-inducibility higher in the former cell line. Interestingly, DIM-C-pPhCI which was characterized as a Nurr1 activator in bladder cancer cells [29] was also active in pancreatic cancer cells but was much less active than several other C-DIM analogs.
Previous studies show that DIM-C-pPhBr did not activate PPARγ or NR4A1 [16, 24] and this compound was used as a model to determine the role of ortho-, meta- and para-bromophenyl ring substitution and the free indole NH group on activation of NR4A2 (Fig. 4). There were some cell context-dependent differences in Panc28 and Panc1 cells; however, DIM-C-pPhBr (p-bromo substituent) was more active that the corresponding ortho- and meta-bromo isomers and methylation of the indole NH group resulted in complete loss of activity, and similar results were observed for activation of TR3 [16]. In contrast, the N-methyl derivative of DIM-C-pPhCI activated GAL4-NR4A2 in bladder cancer cells [22]; however, most other reports demonstrate the importance of a free NH group for induction of apoptosis or for activation of other nuclear receptors by C-DIM compounds [16, 24].
6-MP was previously identified as an activator of NR4A2 and NR4A3 (but not NR4A1) in CV1 and HEK293 cells [21, 26] and, using various GAL4-chimeras, it was shown that activation of NR4A2 by 6-MP in these cells was dependent on the N-terminal A/B domain of the receptor. In this study, 6-MP induced transactivation in Panc28 cells but exhibited minimal to non-detectable activation of NuREx3-luc or NBREx3-luc in Panc1 cells (Figs. 4B and 4C). 6-MP also activated all domains of NR4A2 using the GAL-Nurr1, GAL4-Nurr1-(AB) and GAL4-Nurr-(C-F) constructs in Panc28 cells and only exhibited minimal (but significant) activity in Panc1 cells (Fig. 5C). These data indicate that, in contrast to previous results indicating that 6-MP activates Nurr1 through the N-terminal domain, the effects of this compound on Nurr1 and domains of Nurr1 are more complex and highly dependent on cell context. DIM-C-pPhBr also induced transactivation in cells transfected wild-type and variant Gal4-Nurr1 constructs, indicating that both N- and C-terminal domains of NR4A2 were activated by DIM-C-pPhBr. Preliminary studies showed that several kinase inhibitors blocked activation of Nurr1 by DIM-C-pPhBr (Suppl. Fig. S1); however, a direct comparison of MAPK and PI3-K inhibitors shows that MAPK played an important role in DIM-C-pPhBr-dependent activation of NR4A2 through multiple domains (Figs. 5C and 5D). Ongoing studies are investigating specific regions and amino acids that are important for kinase-mediated activation of NR4A2.
NR4A2-regulated genes are variable among different cell lines but include OPN and NRP1 in cancer cell lines and VIP in dopaminergic cells [29-31]. We also observed induction of VIP, NRP1 and OPN mRNA levels in Panc28 cells and VIP and OPN in Panc 1 cells treated with DIM-C-pPhBr, and induction was abrogated after knockdown of NR4A2 by RNA interference (Fig. 6). In contrast, DIM-C-pPhBr decreased NRP2 mRNA levels both cell lines (Fig. 6E) and, since NRP1 and NRP2 may be involved in the pathogenesis of pancreatic cancer [32, 33], it is possible that differential modulation of NRP1 and NRP2 by DIM-C-pPhBr may be important for the anti-carcinogenic activity of this compound and is currently being investigated.
In summary, this study shows that specific C-DIMs activate NR4A2, and using DIM-C-pPhBr as a model, it was shown that activation of NR4A2 was enhanced using a p-bromo substituent and a free indole NH group was required for activity. DIM-C-pPhBr and related compounds activated nuclear NR4A2 and both N- and C-terminal domains of the receptor were involved. The identification of NR4A2-active C-DIMs that activate the nuclear receptor will be important for future studies on identification of specific NR4A2 sites and pathways required for receptor activation and for determining the role of NR4A2 in mediating the anticancer activities of these agents. Previous studies also reported that DIM-C-pPhBr induces apoptosis and endoplasmic reticulum stress in pancreatic and colon cancer cells [34, 35], and these and other Nurr1-independent responses also contribute to the activity of this compound. Relative contributions of receptor-mediated and -independent activities are being investigated.
Supplementary Material
Acknowledgements
This research was supported by the National Institutes of Health (R01CA124998) and Texas AgriLife Research.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Disclosure: There are no conflicts of interest to disclose.
6. References
- [1].Gronemeyer H, Gustafsson JA, Laudet V. Principles for modulation of the nuclear receptor superfamily. Nat Rev Drug Discov. 2004;3:950–64. doi: 10.1038/nrd1551. [DOI] [PubMed] [Google Scholar]
- [2].McKenna NJ, Cooney AJ, DeMayo FJ, Downes M, Glass CK, Lanz RB, et al. Minireview: Evolution of NURSA, the Nuclear Receptor Signaling Atlas. Mol Endocrinol. 2009;23:740–6. doi: 10.1210/me.2009-0135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Warnmark A, Treuter E, Wright AP, Gustafsson JA. Activation functions 1 and 2 of nuclear receptors: molecular strategies for transcriptional activation. Mol Endocrinol. 2003;17:1901–9. doi: 10.1210/me.2002-0384. [DOI] [PubMed] [Google Scholar]
- [4].Benoit G, Cooney A, Giguere V, Ingraham H, Lazar M, Muscat G, et al. International Union of Pharmacology. LXVI. Orphan nuclear receptors. Pharmacol Rev. 2006;58:798–836. doi: 10.1124/pr.58.4.10. [DOI] [PubMed] [Google Scholar]
- [5].Maxwell MA, Muscat GE. The NR4A subgroup: immediate early response genes with pleiotropic physiological roles. Nucl Recept Signal. 2006;4:e002. doi: 10.1621/nrs.04002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Pearen MA, Muscat GE. Minireview: Nuclear hormone receptor 4A signaling: implications for metabolic disease. Mol Endocrinol. 2010;24:1891–903. doi: 10.1210/me.2010-0015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Shi Y. Orphan nuclear receptors in drug discovery. Drug Discov Today. 2007;12:440–5. doi: 10.1016/j.drudis.2007.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Lee SO, Li X, Khan S, Safe S. Targeting NR4A1 (TR3) in cancer cells and tumors. Expert Opin Ther Targets. 2011;15:195–206. doi: 10.1517/14728222.2011.547481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Wang Z, Benoit G, Liu J, Prasad S, Aarnisalo P, Liu X, et al. Structure and function of Nurr1 identifies a class of ligand-independent nuclear receptors. Nature. 2003;423:555–60. doi: 10.1038/nature01645. [DOI] [PubMed] [Google Scholar]
- [10].Ke N, Claassen G, Yu DH, Albers A, Fan W, Tan P, et al. Nuclear hormone receptor NR4A2 is involved in cell transformation and apoptosis. Cancer Res. 2004;64:8208–12. doi: 10.1158/0008-5472.CAN-04-2134. [DOI] [PubMed] [Google Scholar]
- [11].Inamoto T, Czerniak BA, Dinney CP, Kamat AM. Cytoplasmic mislocalization of the orphan nuclear receptor Nurr1 is a prognostic factor in bladder cancer. Cancer. 2010;116:340–6. doi: 10.1002/cncr.24737. [DOI] [PubMed] [Google Scholar]
- [12].Li QX, Ke N, Sundaram R, Wong-Staal F. NR4A1, 2, 3--an orphan nuclear hormone receptor family involved in cell apoptosis and carcinogenesis. Histol Histopathol. 2006;21:533–40. doi: 10.14670/HH-21.533. [DOI] [PubMed] [Google Scholar]
- [13].Li H, Kolluri SK, Gu J, Dawson MI, Cao X, Hobbs PD, et al. Cytochrome c release and apoptosis induced by mitochondrial targeting of nuclear orphan receptor TR3. Science. 2000;289:1159–64. doi: 10.1126/science.289.5482.1159. [DOI] [PubMed] [Google Scholar]
- [14].Lin B, Kolluri SK, Lin F, Liu W, Han YH, Cao X, et al. Conversion of Bcl-2 from protector to killer by interaction with nuclear orphan receptor Nur77/TR3. Cell. 2004;116:527–40. doi: 10.1016/s0092-8674(04)00162-x. [DOI] [PubMed] [Google Scholar]
- [15].Lee SO, Abdelrahim M, Yoon K, Chintharlapalli S, Papineni S, Kim K, et al. Inactivation of the orphan nuclear receptor TR3/Nur77 inhibits pancreatic cancer cell and tumor growth. Cancer Res. 2010;70:6824–36. doi: 10.1158/0008-5472.CAN-10-1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Chintharlapalli S, Burghardt R, Papineni S, Ramaiah S, Yoon K, Safe S. Activation of Nur77 by selected 1,1-Bis(3′-indolyl)-1-(p-substituted phenyl)methanes induces apoptosis through nuclear pathways. J Biol Chem. 2005;280:24903–14. doi: 10.1074/jbc.M500107200. [DOI] [PubMed] [Google Scholar]
- [17].Cho SD, Lee SO, Chintharlapalli S, Abdelrahim M, Khan S, Yoon K, et al. Activation of nerve growth factor-induced B alpha by methylene-substituted diindolylmethanes in bladder cancer cells induces apoptosis and inhibits tumor growth. Mol Pharmacol. 2010;77:396–404. doi: 10.1124/mol.109.061143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Lee SO, Chintharlapalli S, Liu S, Papineni S, Cho SD, Yoon K, et al. p21 expression is induced by activation of nuclear nerve growth factor-induced Balpha (Nur77) in pancreatic cancer cells. Mol Cancer Res. 2009;7:1169–78. doi: 10.1158/1541-7786.MCR-08-0473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Zhan Y, Du X, Chen H, Liu J, Zhao B, Huang D, et al. Cytosporone B is an agonist for nuclear orphan receptor Nur77. Nat Chem Biol. 2008;4:548–56. doi: 10.1038/nchembio.106. [DOI] [PubMed] [Google Scholar]
- [20].Liu JJ, Zeng HN, Zhang LR, Zhan YY, Chen Y, Wang Y, et al. A unique pharmacophore for activation of the nuclear orphan receptor Nur77 in vivo and in vitro. Cancer Res. 2010;70:3628–37. doi: 10.1158/0008-5472.CAN-09-3160. [DOI] [PubMed] [Google Scholar]
- [21].Ordentlich P, Yan Y, Zhou S, Heyman RA. Identification of the antineoplastic agent 6-mercaptopurine as an activator of the orphan nuclear hormone receptor Nurr1. J Biol Chem. 2003;278:24791–9. doi: 10.1074/jbc.M302167200. [DOI] [PubMed] [Google Scholar]
- [22].Inamoto T, Papineni S, Chintharlapalli S, Cho SD, Safe S, Kamat AM. 1,1-Bis(3′-indolyl)-1-(p-chlorophenyl)methane activates the orphan nuclear receptor Nurr1 and inhibits bladder cancer growth. Mol Cancer Ther. 2008;7:3825–33. doi: 10.1158/1535-7163.MCT-08-0730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Dubois C, Hengerer B, Mattes H. Identification of a potent agonist of the orphan nuclear receptor Nurr1. ChemMedChem. 2006;1:955–8. doi: 10.1002/cmdc.200600078. [DOI] [PubMed] [Google Scholar]
- [24].Chintharlapalli S, Smith R, 3rd, Samudio I, Zhang W, Safe S. 1,1-Bis(3′-indolyl)-1-(p-substitutedphenyl)methanes induce peroxisome proliferator-activated receptor gamma-mediated growth inhibition, transactivation, and differentiation markers in colon cancer cells. Cancer Res. 2004;64:5994–6001. doi: 10.1158/0008-5472.CAN-04-0399. [DOI] [PubMed] [Google Scholar]
- [25].Qin C, Morrow D, Stewart J, Spencer K, Porter W, Smith R, 3rd, et al. A new class of peroxisome proliferator-activated receptor gamma (PPARgamma) agonists that inhibit growth of breast cancer cells: 1,1-Bis(3′-indolyl)-1-(p-substituted phenyl)methanes. Mol Cancer Ther. 2004;3:247–60. [PubMed] [Google Scholar]
- [26].Kim SY, Choi KC, Chang MS, Kim MH, Na YS, Lee JE, et al. The dopamine D2 receptor regulates the development of dopaminergic neurons via extracellular signal-regulated kinase and Nurr1 activation. J Neurosci. 2006;26:4567–76. doi: 10.1523/JNEUROSCI.5236-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Nordzell M, Aarnisalo P, Benoit G, Castro DS, Perlmann T. Defining an N-terminal activation domain of the orphan nuclear receptor Nurr1. Biochem Biophys Res Commun. 2004;313:205–11. doi: 10.1016/j.bbrc.2003.11.079. [DOI] [PubMed] [Google Scholar]
- [28].Kovalovsky D, Refojo D, Liberman AC, Hochbaum D, Pereda MP, Coso OA, et al. Activation and induction of NUR77/NURR1 in corticotrophs by CRH/cAMP: involvement of calcium, protein kinase A, and MAPK pathways. Mol Endocrinol. 2002;16:1638–51. doi: 10.1210/mend.16.7.0863. [DOI] [PubMed] [Google Scholar]
- [29].Lammi J, Huppunen J, Aarnisalo P. Regulation of the osteopontin gene by the orphan nuclear receptor NURR1 in osteoblasts. Mol Endocrinol. 2004;18:1546–57. doi: 10.1210/me.2003-0247. [DOI] [PubMed] [Google Scholar]
- [30].Luo Y, Henricksen LA, Giuliano RE, Prifti L, Callahan LM, Federoff HJ. VIP is a transcriptional target of Nurr1 in dopaminergic cells. Exp Neurol. 2007;203:221–32. doi: 10.1016/j.expneurol.2006.08.005. [DOI] [PubMed] [Google Scholar]
- [31].Hermanson E, Borgius L, Bergsland M, Joodmardi E, Perlmann T. Neuropilin1 is a direct downstream target of Nurr1 in the developing brain stem. J Neurochem. 2006;97:1403–11. doi: 10.1111/j.1471-4159.2006.03829.x. [DOI] [PubMed] [Google Scholar]
- [32].Zhang XK. Targeting Nur77 translocation. Expert Opin Ther Targets. 2007;11:69–79. doi: 10.1517/14728222.11.1.69. [DOI] [PubMed] [Google Scholar]
- [33].Li M, Yang H, Chai H, Fisher WE, Wang X, Brunicardi FC, et al. Pancreatic carcinoma cells express neuropilins and vascular endothelial growth factor, but not vascular endothelial growth factor receptors. Cancer. 2004;101:2341–50. doi: 10.1002/cncr.20634. [DOI] [PubMed] [Google Scholar]
- [34].Lei P, Abdelrahim M, Cho SD, Liu S, Chintharlapalli S, Safe S. 1,1-Bis(3′-indolyl)-1-(p-substituted phenyl)methanes inhibit colon cancer cell and tumor growth through activation of c-jun N-terminal kinase. Carcinogenesis. 2008;29:1139–47. doi: 10.1093/carcin/bgn103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Lei P, Abdelrahim M, Cho SD, Liu X, Safe S. Structure-dependent activation of endoplasmic reticulum stress-mediated apoptosis in pancreatic cancer by 1,1-bis(3′-indoly)-1-(p-substituted phenyl)methanes. Mol Cancer Ther. 2008;7:3363–72. doi: 10.1158/1535-7163.MCT-08-0439. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.