Abstract
Riboswitches are structured mRNA elements involved in gene regulation that respond to the intracellular concentration of specific small molecules. Binding of their cognate ligand is thought to elicit a global conformational change of the riboswitch, in addition to modulating the fine structure of the binding site. X-ray crystallography has produced detailed descriptions of the three-dimensional structures of the ligand-bound conformations of several riboswitches. We have employed small-angle X-ray scattering (SAXS) to generate low-resolution reconstructions of the ligand-free states of the ligand-binding domains of riboswitches that respond to thiamine pyrophosphate (TPP), and cyclic diguanylate (c-di-GMP), a bacterial second messenger. Comparison of the SAXS reconstructions with the crystal structures of these two riboswitches demonstrates that the RNAs undergo dramatic ligand-induced global conformational changes. However, this is not an universal feature of riboswitches. SAXS analysis of the solution behavior of several other riboswitch ligand-binding domains demonstrates a broad spectrum of conformational switching behaviors, ranging from the unambiguous switching of the TPP and c-di-GMP riboswitches to complete lack of switching for the flavin mononucleotide (FMN) riboswitch. Moreover, the switching behavior varies between examples of the same riboswitch from different organisms. The range of observed behaviors suggests that in response to the evolutionary need for precise genetic regulation, riboswitches may be tuned to function more as dimmers or rheostats than binary on/off switches.
Introduction
Riboswitches are structured domains of the untranslated regions and introns of mRNAs that directly recognize small molecule metabolites and second messengers and regulate gene expression in cis. While some riboswitches execute genetic control through ligand-induced self-cleavage1, 2 or hybridization in trans,3 most riboswitches are thought to undergo a conformational change upon binding their cognate effector molecule, which subsequently translates into a genetic on/off switch.
Riboswitches can be thought of as being comprised of two distinct, separable domains, one responsible for small molecule binding (the 'aptamer domain') and another for transducing small molecule binding into a genetic regulatory signal (the 'expression platform').4 The former exhibits the majority of the sequence conservation in the riboswitch and it alone is responsible for recognition of the cellular small molecule. Because of this, many studies have focused on the aptamer domain in an effort to understand riboswitch function, including the role of conformational switching induced by cognate ligand binding. Three-dimensional structures have been reported for 11 of the approximately 20 different classes riboswitch aptamer domains discovered to date, all in their ligand-bound conformations (reviewed in refs. 5–7).
Modes of small molecule recognition
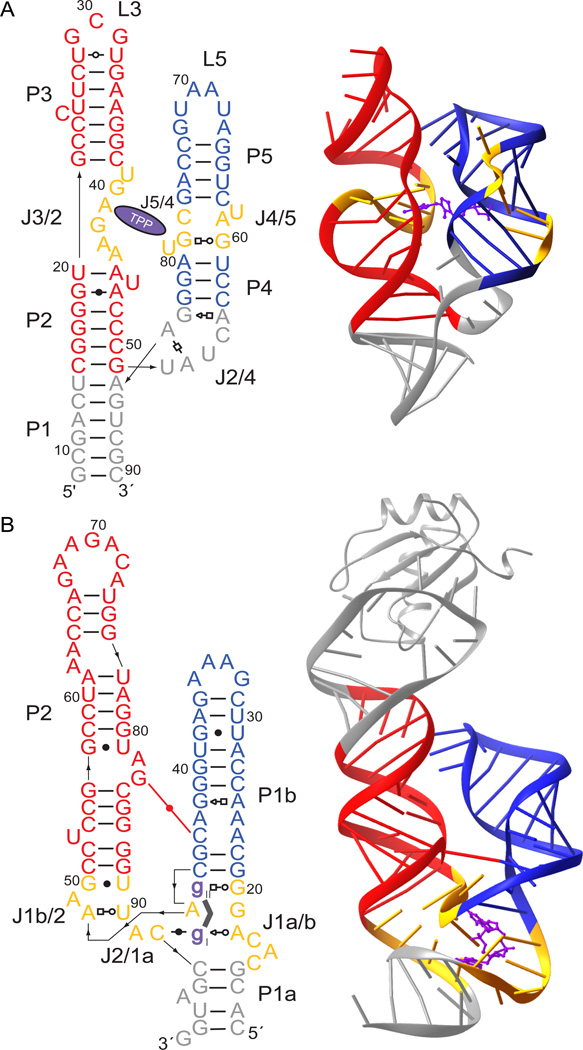
Crystallographic analyses in our laboratory and others has revealed the molecular details of small-molecule recognition by the TPP and c-di-GMP riboswitch aptamer domains, which are ~80 and ~90 nucleotides long, respectively.8–12 The ligand-bound conformations of these aptamer domains are globally similar, despite differences in the size, shape, and overall charge of their respective cognate small molecules (Figure 1). Both RNAs adopt h-shaped folds comprised of two parallel helical elements connected by a three-way junction to a helical 'handle' containing the 5'- and 3'- termini of the aptamer domain. The parallel helices contain irregular helical regions and loops that participate in long-range (tertiary) interactions important for stabilizing the overall fold of the RNA domain.
Figure 1.
Secondary and schematic three-dimensional structures28 of the (A) TPP and (B) c-di-GMP riboswitch aptamer domains. The parallel helices involved in long-range interactions are colored red and blue. Regions involved in ligand binding are colored gold and the ligands are colored purple. The grey region atop of the c-di-GMP aptamer domain comprises the U1A protein and its binding site, introduced to facilitate crystallization.
The most dramatic difference between these two riboswitch aptamer domains is in their modes of small molecule recognition. In the TPP aptamer domain, the central, irregular regions of each of the two parallel helices directly participate in metabolite binding. One folds into a structure resembling the T-loop of tRNA and recognizes the pyrimidine moiety of TPP, while the other contacts the pyrophosphate of TPP, which is bound as a Mg2+ chelate. In contrast, the analogous parallel helices of the c-di-GMP aptamer domain utilize their non-A-form central regions for presentation of an interhelical Watson-Crick base pair, rather than for cognate small-molecule binding. In the c-di-GMP aptamer domain, small molecule recognition is achieved within the three-helix junction. This junction consists of 14 nt and binds to both nucleobases of c-di-GMP molecule through a combination of base pairing and stacking interactions. The three-helix junction of the TPP aptamer domain is comprised of only 7 nt, and makes no interactions with the metabolite.
The specifics of the ligand recognition strategies of these aptamer domains could not have been deduced a priori from the molecular properties of their respective cognate small molecules. For instance, that the c-di-GMP would adopt a ‘sandwiched’ conformation in complex with the aptamer domain was not necessarily expected, as some proteins are known to recognize this second messenger in an extended conformation, with the nucleobases splayed out in opposite directions.13 In light of the TPP aptamer domain structure, it might have seemed plausible for the c-di-GMP to be recognized in such an extended conformation, bridging the two parallel helices of the RNA. Alternatively, given the 2-fold symmetry of c-di-GMP, a dimer of aptamers could have recognized the second messenger. Indeed, many c-di-GMP binding proteins are symmetric oligomers.14, 15
Mode of recognition predicts nature of global response?
Can riboswitch aptamer domains be classified based on crystal structures of their ligand-bound states, and can such a classification predict the global structural response of the domains to ligand binding? Based on the six structures available at the time, Montange and Batey16 proposed that riboswitches could be classified into two distinct families. The aptamer domains of Type I riboswitches bind to their respective cognate small molecules at the junction of several helices or at a pseudoknot (e.g. the purine17 or the glmS18 riboswitches, respectively) while Type II aptamer domains utilize structural elements distant in secondary structure to bind their ligands (e.g. the TPP riboswitch). The two types can be described as ‘junction’ vs. ‘other’ modes of ligand recognition, because the aptamer domains that do not bind their metabolite in a multi-helical junction (or a pseudoknot) use a variety of distinct peripheral structural elements to achieve specific binding. Furthermore, Montange and Batey16 suggested that this structural classification would predict the global responses to small molecule binding of the aptamer domain. Specifically, because Type I riboswitches recognize their ligands at a central helical junction, their global architectures are predicted to be preformed and ligand binding is proposed to induce only local rearrangements of the binding pocket. In contrast, Type II riboswitches require distal structural elements to coalesce to form their ligand binding pocket; therefore, they must undergo a global structural rearrangement upon ligand binding.
Given their structures in complex with their respective cognate ligands, the c-di-GMP riboswitch would be classified as a Type I or ‘junction’ riboswitch, whereas the TPP riboswitch would be classified as a Type II, or ‘other’. It follows that the Type I c-di-GMP riboswitch aptamer domain would be prefolded and not undergo global structural rearrangements upon binding the second messenger, while the Type II TPP riboswitch aptamer would be expected to undergo a global rearrangement upon binding its cognate metabolite. Based on the structure of the c-di-GMP riboswitch aptamer domain and in-line probing data, Strobel and colleagues12 predicted this RNA would be prefolded. In the case of the TPP riboswitch, Micura and colleagues19 inferred from fluorescence spectroscopic studies of aptamer domain constructs with 2-aminopurine substitutions that the RNA undergoes large-scale folding in response to metabolite binding. Calorimetric analysis of TPP binding by the latter riboswitch is consistent with this inference.20
The c-di-GMP and TPP riboswitches in solution
We employed small-angle X-ray scattering (SAXS) to investigate of the nature of the global ligand-induced structural responses of the c-di-GMP and TPP riboswitch aptamer domains. SAXS is a powerful tool for examining conformations of macromolecules in solution and has been used to monitor RNA folding for over a decade.21 From a SAXS experiment, the radius of gyration (Rg) and maximum dimension of the molecule are obtained, providing global measures of the shape of a macromolecule in solution. From a practical standpoint, one advantage of SAXS is that a variety of solution conditions (e.g. a riboswitch without and with saturating concentrations of its cognate ligand) can be readily probed.22 Since the mechanism of riboswitch function may involve a global ligand binding-induced conformational switch, SAXS is an appropriate experimental technique.
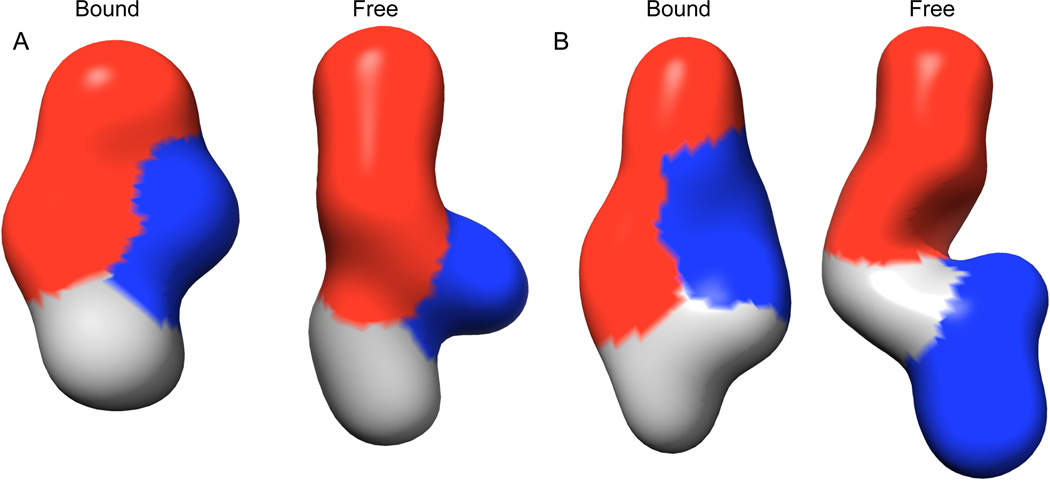
We examined the conformation of the TPP and c-di-GMP riboswitch aptamer domains under near-physiologic23 Mg2+ concentrations (1.5 – 2.5 mM) and under highly stabilizing Mg2+ conditions (10 mM), both in the absence and presence of their cognate ligands. In 2.5 mM Mg2+, the TPP aptamer domain undergoes considerable compaction upon metabolite binding as indicated by the change in Rg (ΔRg) of 3.8 Å (Table 1). The ΔRg is smaller (2.3 Å) in the presence of 10 mM Mg2+, indicating that highly stabilizing Mg2+ is capable of inducing a partially pre-organized conformation relative to physiologic Mg2+ (Table 1). Under all Mg2+ concentrations examined, this riboswitch aptamer domain undergoes metabolite-induced compaction, albeit to varying degrees (Table 1 and ref. 24) Ab initio reconstructions based on the SAXS data provide low-resolution views of the conformations of this RNA under various conditions (Figure 2).24, 25 Comparison of the free and TPP-bound reconstructions indicate that the helical elements involved in metabolite recognition are not pre-arranged in a side-by-side orientation prior to binding TPP (Figure 2).
Table 1.
Rg values in varying Mg2+ concentrations for TPP and c-di-GMP aptamer domains.
Figure 2.
Low-resolution SAXS reconstructions of the TPP and c-di-GMP riboswitches in the ‘bound’ and ‘free’ conformations. (A) The low-resolution reconstruction of the TPP ‘bound’ aptamer domain is colored as in Figure 1. The location of the red helix in the ‘free’ reconstruction is assigned based on experiments described in ref. 24. The assignment of regions corresponding to the blue and grey helices is somewhat ambiguous.25 (B) The ‘bound’ and ‘free’ reconstructions of the c-di-GMP aptamer domain are colored according to Figure 1 and assigned as described in ref. 11.
SAXS investigation of the c-di-GMP riboswitch aptamer domain indicates that it also undergoes a pronounced global conformational change upon binding its cognate ligand.11 The observed ΔRg between the free and c-di-GMP-bound conformations in 2.5 mM Mg2+ is 4.6 Å, a change even larger than that observed for the TPP riboswitch aptamer domain (Table 1). The magnitude of this ligand binding-induced conformational change is surprising given the prediction of a pre-formed structure that undergoes only local changes upon c-di-GMP binding for this Type I riboswitch.16 Low-resolution reconstructions of the aptamer domain in the free and bound states are dramatically different from each other (Figure 2). The SAXS reconstruction of the ligand-bound state is in excellent agreement with the cocrystal structure. The reconstruction of the free state of the aptamer domain has two arms whose dimensions are strikingly similar to those of helices P1b and P2. This suggests that in the absence of c-di-GMP, these helices are not oriented side-by-side, but are splayed out in opposite directions, in a manner reminiscent of the ligand-free state of the TPP riboswitch. Back-calculation of scattering profiles from such a model of the ligand-free c-di-GMP riboswitch, as well as nuclease probing experiments, support this conclusion.11
The cocrystal structure and the SAXS reconstructions were obtained with an RNA construct whose sequence corresponds to that of a c-di-GMP riboswitch from Vibrio cholerae.11 The unexpectedly large ligand binding-induced global conformational change of this RNA is probably not an anomaly of this specific riboswitch sequence, because SAXS analysis of the aptamer domain of a second c-di-GMP riboswitch (from Thiomicrospira crunogena) also reveals a ligand-binding induced compaction (Table 1). In 2.5 mM Mg2+ and the absence of c-di-GMP, this RNA exhibits an Rg = 28.0 Å. Upon the addition of saturating c-di-GMP, the aptamer domain compacts to an Rg = 25.2 Å.
Diversity of riboswitch responses to ligand
It appears that the structural responses of riboswitch aptamer domains to ligand binding are far more nuanced than implied by the binary classification of Montange and Batey16. For example, independently from our studies of an Escherichia coli TPP riboswitch, Doniach and colleagues examined by SAXS the aptamer domain of a TPP riboswitch from Arabidopsis thaliana.25 The TPP riboswitch aptamer domains from the two organisms have contrasting behaviors. In highly stabilizing Mg2+ conditions (10 mM) and in the absence of metabolite, the TPP riboswitch aptamer domain from E. coli exhibits an Rg = 24.4 Å. In contrast, a much more expanded conformation (Rg = 28 Å) is observed for the RNA from A. thaliana under the same conditions.25 In the presence of 10 mM Mg2+ and saturating concentrations of TPP, the Rg for both of these aptamers compacts to 22 Å, which is consistent with the observation that the cocrystal structures of the two RNAs are very similar.9 The distinctly different metabolite-free conformations of these two TPP riboswitch aptamer domains demonstrate that small molecule-driven structural responses are not comparable even between orthologous riboswitches from different organisms.
The idiosyncratically tuned nature of the global metabolite binding-induced responses of riboswitch aptamer domains extends across riboswitch classes as well. We employed SAXS to investigate riboswitch aptamer domains that bind S-adenosyl methionine (SAM-I), lysine, and FMN.24 We found that the SAM-I aptamer domain behaves similarly to the TPP riboswitch aptamer domain in 2.5 mM Mg2+ in that it displays a ΔRg of ~ 3 Å between its free and bound conformations. However, in the absence of SAM, more stabilizing Mg2+ concentrations (10 mM) compact the RNA to within 1 Å of its bound conformation, demonstrating a global response that is highly dependent on the environmental conditions. The global conformation of the lysine riboswitch aptamer domain is insensitive to the presence of its cognate ligand under all Mg2+ concentrations examined. Instead, a significant conformational change is induced by varying Mg2+ concentrations. Finally, the FMN riboswitch aptamer domain is globally insensitive to metabolite binding as well as Mg2+ concentrations: this RNA is already maximally compact in 1.5 mM Mg2+ in the absence of metabolite. Altogether, our SAXS experiments show that the global conformational response of riboswitch aptamer domains does not neatly follow the classification proposed by Montange and Batey.16 At least one Type I aptamer domain (c-di-GMP) undergoes a global switch, and Type II aptamer domains (SAM, TPP) display a range of behaviors in response to ligand binding and solution conditions.24
Conclusion: riboswitches, digital or analog?
The diversity in the solution behavior of the aptamer domains revealed by SAXS analysis suggests that riboswitch response to ligand binding is better described as analog than digital. The degree of metabolite-induced aptamer switching is likely variably tuned, similar to the precise control offered by a dimmer switch, in contrast to the binary open or closed states of a single pole switch. Analysis of the regulation of gene expression by riboswitches suggest that they rarely function as simple digital “on/off” switches. In vivo and in vitro studies of the transcriptional regulation of gene expression by SAM-I and guanine riboswitches from different loci in the Bacillus subtilis genome revealed a wide range of transcript levels, transcriptional read-through efficiencies, and binding affinities for twelve SAM-I riboswitches26 and five guanine riboswitches.27 These studies suggest that the response of these riboswitches is tuned by sequence changes outside the near-invariant nucleotides directly involved in small molecule recognition. These sequence changes may reflect adaptation of each riboswitch for efficient control of gene expression within its own genomic locus.
Acknowledgements
The authors thank L. Guo for assistance with data collection at BioCAT. This work was supported by an NIH grant to A.R.F. (GM63576) and by the Howard Hughes Medical Institute. A.R.F.-D. is an Investigator of the Howard Hughes Medical Institute.
Footnotes
The authors declare no conflicts of interest.
References
- 1.Collins JA, Irnov I, Baker S, Winkler WC. Mechanism of mRNA destabilization by the glmS ribozyme. Genes Dev. 2007;21:3356–3368. doi: 10.1101/gad.1605307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Klein DJ, Been MD, Ferré-D'Amaré AR. Essential role of an active-site guanine in glmS ribozyme catalysis. J Am Chem Soc. 2007;129:14858–14859. doi: 10.1021/ja0768441. [DOI] [PubMed] [Google Scholar]
- 3.Loh E, Dussurget O, Gripenland J, Vaitkevicius K, Tiensuu T, Mandin P, et al. A transacting riboswitch controls expression of the virulence regulator PrfA in listeria monocytogenes. Cell. 2010;139:770–779. doi: 10.1016/j.cell.2009.08.046. [DOI] [PubMed] [Google Scholar]
- 4.Winkler W, Nahvi A, Breaker RR. Thiamine derivatives bind messenger RNAs directly to regulate bacterial gene expression. Nature. 2002;419:952–956. doi: 10.1038/nature01145. [DOI] [PubMed] [Google Scholar]
- 5.Edwards TE, Klein DJ, Ferré-D'Amaré AR. Riboswitches: small-molecule recognition by gene regulatory RNAs. Curr Op Struct Biol. 2007;17:273–279. doi: 10.1016/j.sbi.2007.05.004. [DOI] [PubMed] [Google Scholar]
- 6.Serganov A. The long and the short of riboswitches. Curr Op Struct Biol. 2009;19:251–259. doi: 10.1016/j.sbi.2009.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Henkin T. Riboswitch RNAs: using RNA to sense cellular metabolism. Genes Dev. 2008;22:3383–3390. doi: 10.1101/gad.1747308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Serganov A, Polonskaia A, Phan AT, Breaker RR, Patel DJ. Structural basis for gene regulation by a thiamine pyrophosphate-sensing riboswitch. Nature. 2006;441:1167–1171. doi: 10.1038/nature04740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Edwards TE, Ferré-D'Amaré AR. Crystal structures of the thi-box riboswitch bound to thiamine pyrophosphate analogs reveal adaptive RNA-small molecule recognition. Structure. 2006;14:1459–1468. doi: 10.1016/j.str.2006.07.008. [DOI] [PubMed] [Google Scholar]
- 10.Thore S, Frick C, Ban N. Structural basis of thiamine pyrophosphate analogues binding to the eukaryotic riboswitch. J Am Chem Soc. 2008;130:8116–8117. doi: 10.1021/ja801708e. [DOI] [PubMed] [Google Scholar]
- 11.Kulshina N, Baird NJ, Ferré-D'amaré AR. Recognition of the bacterial second messenger cyclic diguanylate by its cognate riboswitch. Nat Struct Mol Biol. 2009;16:1212–1217. doi: 10.1038/nsmb.1701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Smith KD, Lipchock SV, Ames TD, Wang J, Breaker RR, Strobel SA. Structural basis of ligand binding by a c-di-GMP riboswitch. Nat Struct Mol Biol. 2009;16:1218–1223. doi: 10.1038/nsmb.1702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Barends TRM, Hartmann E, Griese JJ, Beitlich T, Kirienko NV, Ryjenkov DA, et al. Structure and mechanism of a bacterial light-regulated cyclic nucleotide phosphodiesterase. Nature. 2009;459:1015–1018. doi: 10.1038/nature07966. [DOI] [PubMed] [Google Scholar]
- 14.Benach J, Swaminathan S, Tamayo R, Handelman S, Folta-Stogniew E, Ramos J, et al. The structural basis of cyclic diguanylate signal transduction by PilZ domains. EMBO J. 2007;26:5153–5166. doi: 10.1038/sj.emboj.7601918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.De N, Pirruccello M, Krasteva P, Bae N, Raghavan R, Sondermann H. Phosphorylation-independent regulation of the diguanylate cyclase WspR. Plos Biol. 2008;6:e67. doi: 10.1371/journal.pbio.0060067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Montange RK, Batey RT. Riboswitches: emerging themes in RNA structure and function. Annual Review of Biophysics. 2008;37:117–133. doi: 10.1146/annurev.biophys.37.032807.130000. [DOI] [PubMed] [Google Scholar]
- 17.Batey RT, Gilbert SD, Montange RK. Structure of a natural guanine-responsive riboswitch complexed with the metabolite hypoxanthine. Nature. 2004;432:411–415. doi: 10.1038/nature03037. [DOI] [PubMed] [Google Scholar]
- 18.Klein DJ, Ferré-D'Amaré AR. Structural basis of glmS ribozyme activation by glucosamine-6-phosphate. Science. 2006;313:1752–1756. doi: 10.1126/science.1129666. [DOI] [PubMed] [Google Scholar]
- 19.Lang K, Rieder R, Micura R. Ligand-induced folding of the thiM TPP riboswitch investigated by a structure-based fluorescence spectroscopic approach. Nucleic Acids Res. 2007;35:5370–5378. doi: 10.1093/nar/gkm580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kulshina N, Edwards TE, Ferre-D'amare AR. Thermodynamic analysis of ligand binding and ligand binding-induced tertiary structure formation by the thiamine pyrophosphate riboswitch. RNA. 2010;16:186–196. doi: 10.1261/rna.1847310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lipfert J, Doniach S. Small-angle X-ray scattering from RNA, proteins, and protein complexes. Annual Rev Biophys Biomol Struct. 2007;36:307–327. doi: 10.1146/annurev.biophys.36.040306.132655. [DOI] [PubMed] [Google Scholar]
- 22.Lipfert J, Das R, Chu VB, Kudaravalli M, Boyd N, Herschlag D, et al. Structural transitions and thermodynamics of a glycine-dependent riboswitch from Vibrio cholerae. J Mol Biol. 2007;365:1393–1406. doi: 10.1016/j.jmb.2006.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Romani A, Scarpa A. Regulation of cell magnesium. Arch Biochem Biophys. 1992;298:1–12. doi: 10.1016/0003-9861(92)90086-c. [DOI] [PubMed] [Google Scholar]
- 24.Baird NJ, Ferré-D'Amaré AR. Idiosyncratically tuned switching behavior of riboswitch aptamer domains revealed by comparative small-angle X-ray scattering analysis. RNA. 2010;16:598–609. doi: 10.1261/rna.1852310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ali M, Lipfert J, Seifert S, Herschlag D, Doniach S. The Ligand-Free State of the TPP Riboswitch: A Partially Folded RNA Structure. J Mol Biol. 2010;396:153–165. doi: 10.1016/j.jmb.2009.11.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tomsic J, Mcdaniel B, Grundy F, Henkin T. Natural variability in S-adenosylmethionine (SAM)-dependent riboswitches: S-box elements in Bacillus subtilis exhibit differential sensitivity to SAM in vivo and in vitro. J Bacteriol. 2007;190:823–833. doi: 10.1128/JB.01034-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mulhbacher J, Lafontaine DA. Ligand recognition determinants of guanine riboswitches. Nucleic Acids Res. 2007;35:5568–5580. doi: 10.1093/nar/gkm572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Carson M. Ribbons. Methods Enzymol. 1997;277:493–505. [PubMed] [Google Scholar]