For more than a century, microbiologists studied pathogens in pure culture, whereas cell biologists studied mammalian cells in tissue culture. Both fields used rich media and conditions optimized for in vitro growth. A few years ago, researchers realized that these laboratory methods of growing pathogens and their hosts were quite artificial and had very little in common with real life, where pathogens and hosts coexist, interact, and compete in conditions that are often far from optimal. To better mimic what happens in real life, the study of the interaction between microbes and host cells was proposed, taking advantage of technological progress that allowed cocultures of microbes and cells to be handled together. A new discipline (cellular microbiology) was born (1). This discipline took over quite rapidly, a new journal named after this new discipline was started (Cellular Microbiology, Blackwell, Oxford), and textbooks were published (2, 3) describing in detail the techniques to be used in cellular microbiology and the scientific problems that could be addressed. These textbooks are just a few months old, when a powerful new approach to cellular microbiology is described (4). This novel approach pushes the limits that we had previously and suggests it is probably already time for a new edition of these textbooks. The new technique, reported in part by Belcher et al. in this issue of PNAS (4) and by other recent studies, describes how, instead of studying one parameter at a time, we can use microchips to study, within a single experiment, all of the host genes and those of the bacteria whose expression is modified during host–pathogen interaction: the global picture of the dialogue between the pathogen and the host in one experiment!
Microarrays to Study Host–Pathogen Interactions.
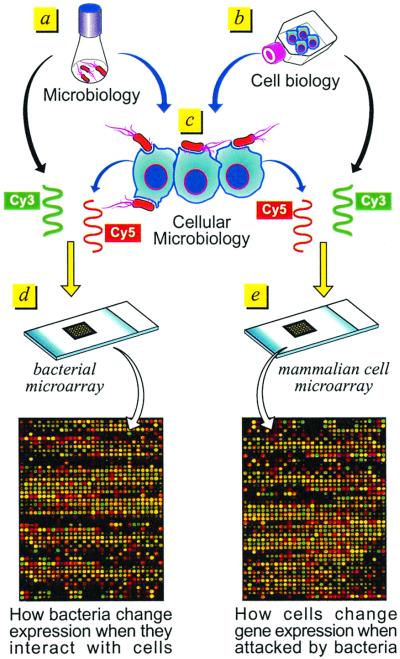
A typical microarray experiment is shown in Fig. 1. RNA is prepared from bacteria grown in standard laboratory conditions (Fig. 1a), from host cells grown under optimal conditions in tissue culture (b), and from cells that have been infected with bacteria (c). A probe is then prepared from each of the RNAs by reverse transcription, usually by using oligo(dT) to probe host-cell genes and random oligonucleotide primers to anneal to bacterial genes. A balanced mixture of oligos specific for each gene present in the microarray may also be used. (This is particularly valuable for preparing probes of bacterial RNA.) To probe printed microarrays, RNA is converted into a fluorescent cDNA probe by incorporation of different fluorochromes during the reverse-transcription reaction (typically by using Cy3- and Cy5-dCTP). The probes are then used to hybridize microarrays containing bacterial genes (Fig. 1d) or eukaryotic genes (e). High-density genome-wide microarrays of eukaryotic cells are commercially available. The Hu6800 Array (Affymetrix , Santa Clara, CA), which in a chip of 1.28 × 1.28 cm contains probes for approximately 6,800 human genes, is the one used in most published studies; however, today a set of 5 chips (Human Genome U95 set), each containing approximately 12,000 genes/ESTs per chip for a total of more than 60,000 human genes and ESTs, is available. Many other systems are also available [UniGem V (Incyte Pharmaceuticals, Palo Alto, CA), MassArray Spectrochip (Sequenom, Hamburg, Germany and San Diego, California), Universal Arrays (Genometrix, The Woodlands, TX), Hychip (Hyseq, Sunnyvale, CA), and the Atlas mouse cDNA expression array (CLONTECH)]. Homemade chips can also be printed when the genes are available. Pat Brown (Stanford University, Stanford, CA) is the leader in printing homemade high-density arrays. In collaboration with Lou Staudt of the National Cancer Institute, he has created the “Lymphochip,” which contains about 18,432 human gene elements (5), and he is now collaborating with David Botstein (Stanford University) on a separate array with >25,000 elements (genes) on a single glass microscope slide. Chips representing the whole genome of a pathogen usually become available as soon as the genomic sequence of the bacterium is published; sometimes they are available even before publication. Usually chips are printed by using each of the bacterial genes, which have been previously amplified by PCR. Microarrays containing all genes of the sequenced bacterial genomes are either available or are being prepared. These include Helicobacter pylori, Neisseria meningitidis, Mycobacterium tuberculosis, and Escherichia coli. Chips containing the genes of many other bacteria will be available within the next few months.
Figure 1.
Schematic description of how microarrays can be used to study host-cell–pathogen interactions.
In the experiment shown in Fig. 1e, the RNA prepared in b and labeled with Cy3 (green) is mixed with the RNA prepared in c and labeled with Cy5 (red). The two fluorescent cDNAs are then mixed together and constitute the probe, which is hybridized to the chip. Sometimes RNA prepared from a pool of control conditions/cells is used to prepare a Cy3-labeled reproducible control, which is then mixed with Cy5-labeled cDNA prepared from the experimental condition, including uninfected cells. This allows an invariant comparator, so that any experimental profile can be compared with any other, even months apart, by normalizing all data to the standard Cy3 control. The ability to compare different studies from the same laboratory and from different laboratories is one of the major problems we face today. Therefore, particular care should be used to standardize conditions. After hybridization, laser scanning, image detection, and analysis of each chip, the ratio of the probes in Fig. 1 b and c provides a precise indication of the relative expression of the genes of the mammalian cells grown in the conditions in Fig. 1 b and c. More specifically, for the labeling reaction [RNA in Fig. 1b (Cy3)/RNA in Fig. 1c (Cy5)], a red dot indicates gene activation after bacterial addition to the cells, and a green dot means down-regulation of the gene, whereas a yellow dot indicates no change in gene expression. The final results of the analysis are the expression ratios of Fig. 1 b/c obtained from the mean value of two labeling conditions (direct and reverse labeling).
Similarly, genome-wide changes in bacterial gene expression after contact with eukaryotic cells can be obtained (Fig. 1d) by hybridizing bacterial chips with a mixture of the probes in Fig. 1 a and c. It should be kept in mind that DNA arrays provide a measure of the average mRNA present (steady-state abundance) and are not just a measure of the rate of transcription.
The Inflammatory Response Is the First Reaction to Bacterial Infection.
Several studies describing changes in host-cell transcription after infection with bacteria were published during the last few months. Cohen et al. (6) published the changes in gene expression of the human promyelocytic cell THP1 after infection with the pathogenic microorganism Listeria monocytogenes; Rosenberger et al. (7) and Eckmann et al. (8) reported the response of macrophages and epithelial cells respectively to Salmonella infection and Ichikawa et al. (9) reported the response of the A549 pneumocyte cell line exposed to Pseudomonas aeruginosa infection. In this issue of PNAS, Belcher et al. (4) report the change in gene expression of the simian virus 40-transformed human bronchial epithelial cell line BEAS-2B after infection with Bordetella pertussis. All studies report genes that are up-regulated and those that are down-regulated after incubation with bacterial pathogens [for instance, 74 were up and 23 down in Cohen et al. (6) and 33 up and 65 down in Belcher et al. (4)]. Up-regulated and down-regulated genes belong to different functional clusters (inflammation, chemotaxis, transcription, apoptosis, transduction, extracellular matrix, cell cycle, metabolism, and genes with unknown function). Many studies and improved computer algorithms will be needed before we can draw a general picture of which genes are induced in mammalian cells by most pathogens and which are specifically induced by individual pathogens. However, from these initial studies we can get a preliminary idea of the main reaction of a mammalian cell to bacterial attack. The inflammatory response is the first and main reaction of mammalian cells to infection. The neutrophil chemoattractant chemokines IL8 and GROβ, which are responsible for chemotaxis and activation of effector inflammatory cells, are activated in most systems. Similarly, chemokines and cytokines, which are directly responsible for inflammations such as IL1, IL6, IFNβ, MCP1, and tumor necrosis factor α, and transcription factors involved in the inflammatory response such as NFκB, are affected.
Pathology, Pathogenesis, and Pharmacogenomics.
In addition to the simple observation of host-gene up- and down-regulation, microarrays can also be used to ask very specific questions about the clinical manifestation of a disease and the role in pathogenesis of individual virulence factors and to predict the clinical outcome of specific drugs. This is the novel concept introduced by Belcher et al. (4).
Links to pathology have already been made in this work. Infiltrates of monocytes, neutrophils, and lymphocytes observed in the lungs of B. pertussis-infected animals are explained by the activation of the chemotactic and proinflammatory response induced in host cells. Diffuse bronchopneumonia with increased secretion of mucus observed in autopsies of pertussis victims can be explained by the increased secretion of mucins induced by B. pertussis infection. The hypothesis generated by the microarray experiment that increased mucin expression could play a role in pathogenesis was then experimentally confirmed by showing that B. pertussis binds mucin in vitro.
B. pertussis is an ideal bacterial pathogen to study pathogenesis. The bacterium produces many virulence factors that are well characterized from the biochemical and genetic point of view: toxins such as pertussis toxin (PT), adenylate cyclase, dermonecrotic toxin, tracheal cytotoxin, and adhesins such as filamentous hemagglutinin, pertactin, fimbriae, etc. Furthermore, knockout isogenic mutants are available for these factors. Comparing the host-cell response to wild-type B. pertussis and each of these mutants will help understand how each contributes to the clinical outcome. Belcher et al. (4) compared the variation in gene expression of BEAS-2B cells infected with wild-type B. pertussis and with a mutant strain producing an enzymatically inactive form of PT, which is one of the major virulence factors. The results provide a list of genes, the transcript abundance of which is changed by the PT enzymatic activity (ADP ribosylation of G proteins). To further dissect the role of PT enzymatic activity in eukaryotic cells, Belcher et al. (4) also compare changes in transcript abundance of cells incubated with purified wild-type and mutant toxins. Many genes previously not known to be regulated by G proteins were identified.
Finally, microarrays can be used in pharmacogenomic studies to determine how a drug can modify the transcriptional response to infection. By adding the antiinflammatory drug dexamethasone to the experiment in Fig. 1e, these microarrays could monitor how a drug recommended for clinical use can modify the response of a single cell.
So far, published studies describe only the host response to infection. Many studies of the bacterial response to host-cell contact are in progress, and I would not be surprised if some of them were published by the time this commentary goes to print. It will be interesting and instructive to follow the bidirectional molecular dialogue that takes place when a pathogen and host meet.
Toward Studies of Infected Tissues and Whole Organisms.
The possibility of studying pure cultures of pathogens while they interact with pure cultures of mammalian cells is exciting and represents a big step forward from traditional studies, where pathogens and host cells were studied separately. However, even the in vitro infection of host cells is far from a real-life scenario where pathogens infect animals and their tissues. Can we study host-cell and pathogen-gene expression in whole organisms? There is no doubt that the technology is available for this step. For instance, bacteria recovered from infected tissues (blood, cerebrospinal fluid, etc.) should be suitable for probe preparation. Similarly, macrophages and lymphocytes from infected organisms and tissue from patients with chronic infections can be recovered and used for probe preparation.
Acknowledgments
I am grateful to Renata Grifantini for useful advice and discussions and to Giorgio Corsi for artwork.
Footnotes
See companion article on page 13847.
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.010505497.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.010505497
References
- 1.Cossart P, Boquet P, Normark S, Rappuoli R. Science. 1996;271:315–316. doi: 10.1126/science.271.5247.315. [DOI] [PubMed] [Google Scholar]
- 2.Cossart P, Boquet P, Normark S, Rappuoli R, editors. Cellular Microbiology. Washington, DC: Am. Soc. Microbiol.; 2000. [Google Scholar]
- 3.Henderson B, Wilson M, McNab R, Lax A J, editors. Cellular Microbiology: Bacteria–Host Interactions in Health and Disease. West Sussex, U.K.: Wiley; 1999. [Google Scholar]
- 4.Belcher C E, Drenkow J, Kehoe B, Gingeras T R, McNamara N, Lemjabbar H, Basbaum C, Relman D A. Proc Natl Acad Sci USA. 2000;97:13847–13852. doi: 10.1073/pnas.230262797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Alizadeh A A, Eisen M B, Davis R E, Ma C, Lossos I S, Rosenwald A, Boldrick J C, Sabet H, Tran T, Yu X, et al. Nature (London) 2000;403:503–511. doi: 10.1038/35000501. [DOI] [PubMed] [Google Scholar]
- 6.Cohen P, Bouaboula M, Bellis M, Baron V, Jbilo O, Poinot-Chazel C, Galiegue S, Hadibi E H, Casellas P. J Biol Chem. 2000;275:11181–11190. doi: 10.1074/jbc.275.15.11181. [DOI] [PubMed] [Google Scholar]
- 7.Rosenberger C M, Scott M G, Gold M R, Hancock R E, Finlay B B. J Immunol. 2000;164:5894–5904. doi: 10.4049/jimmunol.164.11.5894. [DOI] [PubMed] [Google Scholar]
- 8.Eckmann L, Smith J R, Housley M P, Dwinell M B, Kagnoff M F. J Biol Chem. 2000;275:14084–14094. doi: 10.1074/jbc.275.19.14084. [DOI] [PubMed] [Google Scholar]
- 9.Ichikawa J K, Norris A, Bangers M G, Geiss G K, van't Wout A B, Bumgarner R E, Lory S. Proc Natl Acad Sci USA. 2000;97:9659–9664. doi: 10.1073/pnas.160140297. [DOI] [PMC free article] [PubMed] [Google Scholar]