Abstract
The Deleted in Liver Cancer (DLC) protein family comprises proteins that exert their function mainly by the Rho GTPase-activating protein (GAP) domain and by regulation of the small GTPases. Since Rho GTPases are key factors in cell proliferation, polarity, cytoskeletal remodeling and migration, the aberrant function of their regulators may lead to cell transformation. One subgroup of these proteins is the DLC family. It was found that the first identified gene from this family, DLC1, is often lost in hepatocellular carcinoma and may be involved as a tumor suppressor in the liver. Subsequent studies evaluated the hypothesis that the DLC1 gene acts as a tumor suppressor, not only in liver cancer, but also in other types of cancer. Following DLC1, two other members of the DLC protein family, DLC2 and DLC3, were identified. However, limited published data are available concerning the role of these proteins in malignant transformation. This review focuses on the structure and the role of DLC1 and its relatives in physiological conditions and summarizes data published thus far regarding DLC function in the neoplastic process.
1. Introduction
In recent years, significant progress has been made in understanding the biological functions mediated by Rho GTPases. As key regulators of diverse cellular pathways GTPases affect such crucial processes as transcriptional regulation, cell cycle progression, apoptosis, and membrane trafficking (1,2). This family of small (20-30 kDa) signaling G proteins (guanine nucleotide-binding proteins) constitutes a major branch of the Ras superfamily (3). Membership in the superfamily of Ras proteins is determined by the presence of the GTPase domain. A total of 23 Rho proteins have been identified, among which RhoA, Rac1 and Cdc42 are the best characterized (4). Rho GTPases are also involved in the cytoskeleton formation of the cell via the regulation of actin dynamics (5). RhoA induces stress fiber formation and focal adhesion assembly, thereby regulating cell shape, attachment and motility. On the other hand, Rac1 promotes lamellipodium formation and membrane ruffling. Cdc42 has been shown to act in the formation of filopodia, finger-like actin-rich protrusions, thought to be involved in the sensing of the extracellular environment (2,6).
As with other small GTP-binding proteins of the Ras superfamily, the Rho GTPases act by switching between an inactive GDP-bound and an active GTP-bound conformation, with the latter form capable of interacting with a wide range of downstream effectors, thereby activating them. The cycling of Rho GTPases between these GTP- and GDP-bound states is modulated by the three classes of regulatory proteins, the guanine nucleotide exchange factors (GEFs) which catalyze the exchange of GDP for GTP, the GTPase-activating proteins (GAPs) that promote hydrolysis of GTP to GDP, and the guanine nucleotide dissociation inhibitors (GDIs), which bind to the GDP-bound form and not only prevent nucleotide exchange, but also sequester Rho GTPases in the cytoplasm (4).
The effect on the wide spectrum of biological functions suggests the involvement of Rho GTPases and their regulators in cancer progression. Increasing evidence obtained from numerous in vitro and in vivo studies shows that deregulated signaling of Rho proteins may lead to tumorigenesis (7). The fact that no constitutively active Rho mutants have been reported in human tumors suggests that aberrant Rho GTPase signaling in cancer is caused by the alterations of their regulators (4,7). Findings of a large number of studies revealed that regulators of Rho proteins are over- or down-expressed in various types of human cancer (8-11). The most common alteration reported for Rho regulators in cancer is inactivation of RhoGAPs. One branch of this protein family is a member of the deleted in liver cancer (DLC) family. In the late 80s, the identification of a gene found to be commonly deleted in liver tumors, the so-called DLC1, focused the attention on the role of this protein family in tumorigenesis. Subsequent studies revealed that this type of genetic loss is found in a number of other neoplasms (11-14).
To the best of our knowledge, no reports summarizing the nature of the DLC family proteins are currently available. This review focuses on the structure, function and expression of DLC1 and its two other homologs, DLC2 and DLC3, in physiological conditions and in human malignancies.
2. The DLC family consists of three structurally related proteins
The human genome encodes approximately 70 RhoGAPs that share a conserved GAP domain, whose functional role is to turn off Rho-mediated signaling (15). One subgroup of the human RhoGAPs contains DLC1 (also termed as STARD12 or ARHGAP7), a human homolog of the rat p122RhoGAP (14,16). The ArhGAP7/DLC1 gene is localized on chromosome 8p21-22 and encodes a 1091-amino acid protein with a predicted molecular mass of 122 kDa. By means of quantitative RT-PCR assay, it was determined that DLC1 is widely expressed in normal tissues, with high abundance in the lung and ovary, and moderately in the thyroid, spleen, intestine and kidney. The adrenal gland, liver and pancreas exhibit the lowest expression (17).
There are two additional members of the DLC family; DLC2 (or STARD13), located on chromosome 13q12 (18), and DLC3 (also known as KIAA0189 or STARD8), located on the X chromosome at q13 brand (19). The DLC2 encodes a 1113-amino acid protein with a molecular weight of 125 kDa, whereas the protein product of the DLC3 transcript has 1103 aa with a calculated molecular mass of 121 kDa. DLC2 has a broad tissue distribution, with the highest levels in the brain, heart and liver (9,19). The third member of the deleted in liver cancer protein family is also detected in a broad spectrum of human tissues, with high abundance in the lung, kidney and placenta (19).
3. Typical DLC protein possesses three domains: SAM, RhoGAP and START
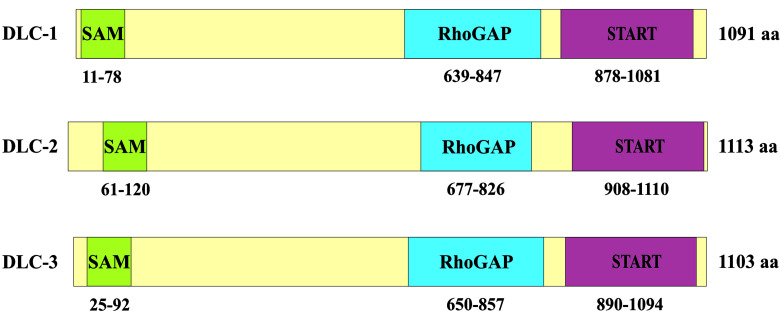
DLC proteins have three distinct domains: the sterile α motif (SAM) localized at its N-terminus, a conserved RhoGAP (GAP) domain found close to the middle of the protein and the steroidogenic acute regulatory-related lipid transfer (START) domain at the C-terminus (Fig. 1) (18-20).
Figure 1. Schematic representation of DLC family proteins showing three major functional domains (SAM, RhoGAP and START domain).
The biological activity of DLC1 is mainly executed by the RhoGAP domain (~150-200 aa), which promotes the hydrolysis of GTP bound to the Rho GTPases. This catalytic activity is mediated by an ‘arginine finger’ donated by GAP. It was reported that DLC1 and DLC3 contain a conserved ‘arginine finger’ at position 677 and 688, respectively (19,20). In in vitro assays both the full-length DLC1 and the isolated RhoGAP domain reveal activity on RhoA, RhoB and RhoC, to a lesser extent on Cdc42, and no activity on Rac1 (11,21). By inactivating these small GTPases, DLC1 affects cell morphology and control actin cytoskeletal remodeling (16,22). Similar to DLC1, both DLC2 and DLC3 contain a RhoGAP domain and exhibit GAP activity for RhoA and Cdc42 in vitro (18,23).
SAM domains (~70 aa) were found in nuclear and signaling proteins (e.g., p73 and p63 proteins, Eph-related tyrosine kinases and Ets transcription factors) (24,25). The SAM domain was thought to act as a protein interaction module through the formation of homo- and hetero-oligomers with other SAM domains, or even RNA and DNA (26). Results of structural studies of the DLC2 SAM domain have shown that it may also bind lipids (27).
However, much less is known regarding the exact role of the SAM domain in DLC function. One structure-function study has shown that the SAM domain may serve as an autoinhibitory domain for RhoGAP catalytic activity (28). Zhong et al have identified the eukaryotic elongation factor-1A1 (EF1A1) as a novel target of the SAM domain of DLC1 (29). EF1A1 is involved in protein synthesis (30) and transporting β-actin mRNA (31). It regulates the actin network through its G-actin-binding and F-actin-bundling activity, and also by stabilizing microtubules (32-34). The expression of EF1A1 is increased in a wide variety of human cancers, including pancreas, lung, prostate, breast, colon and melanomas (30). All of these activities support the hypothesis that EF1A1 has a significant regulatory role in cell growth and the cytoskeletal network. Zhong et al have demonstrated that the DLC1 SAM domain is capable of adopting a four-helix fold that creates a unique hydrophobic motif, allowing it to bind directly to EF1A1. Findings of these authors show that this interaction facilitates EF1A1 distribution to the membrane periphery and membrane ruffles and suppresses cell migration in a GAP-independent manner (29).
The START domain (~210 aa) is a well-conserved lipid‑binding domain, which is primarily found in proteins that transfer lipids between organelles and are involved in lipid metabolism as well as in modulation of signaling events involved in lipid processing. In mammals, 15 START domain‑containing proteins were identified (35). Among these proteins, steroidogenic acute regulatory protein (StAR) and metastatic lymph node 64 (MLN64) appear to be the most well-known representatives (36). Both StAR and MLN64 bind cholesterol (37), and are also known to play a role in lipid transport into mitochondria (38,39). In particular, StAR localizes to the mitochondria and stimulates the translocation of cholesterol from the outer to the inner mitochondrial membranes (40). DLC2 has been found to localize in mitochondria and was found in proximity to the lipid droplets through the START domain (41). Future research is required in order to establish the lipid ligand of DLC2-START and clarify whether the START domain of DLC2 and its homologs, DLC1 and DLC3, play a role in mitochondrial lipid transport.
Between the SAM and RhoGAP domains there is a long unstructured region that contains a phosphorylation-independent binding site, which Kawai et al termed as the FAT (focal adhesion targeting) domain, due to the fact that its presence determines of DLC focal adhesion localization (42). Independent studies have reported that DLC1, DLC2 and DLC3 are recruited to focal adhesion sites via their FAT sequence, which binds to the Src homology 2 (SH2) domains of focal adhesion proteins and interacts with tensins (43-45). However, the same region of DLC1 has been found to possess the ability to interact with the PTB domain of tensin2 (46-48) and tensin1 (44). The role of the interaction with the PTB domain in the localization of DLC1 to focal adhesions remains controversial and requires further investigation. Using pull-down assay, the interaction between the other members of the DLC family, DLC2 and DLC3, and tensin2 PTB was documented (46).
The central region of DLC1 was reported to target caveolae by interacting with caveolin-1 (48). Although the physiological function and significance of DLC1 in caveolar localization remain unclear, it appears that caveolae are one of the compartments whereby DLC1 may exhibit its tumor‑suppressive role and affect the cytoskeletal reorganization by its RhoGAP activity.
The C-terminal region of DLC1 is known to interact with the phospholipase C-δ1 (PLCδ1), as well as enhance its activity (16). DLC3 has also been shown to activate PLCδ1 in an in vitro assay (23). It is suggested that this interaction supports involvement of DLC proteins in modulating actin cytoskeleton.
4. Expression analysis of DLCs reveals their down-regulation in various types of human cancer
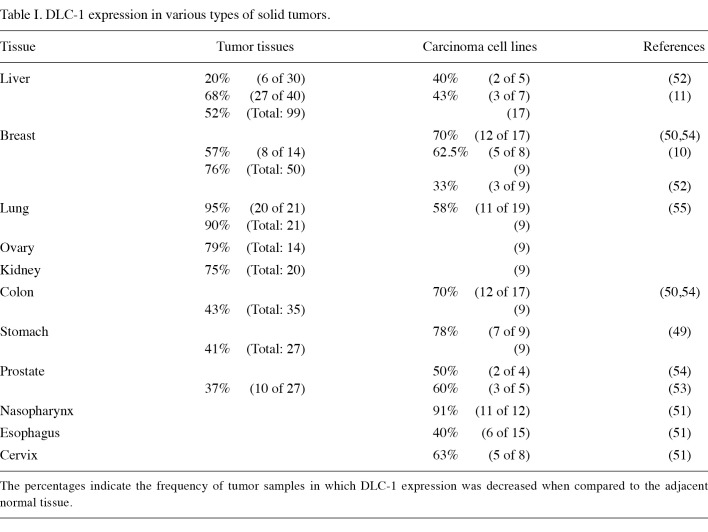
DLC1 was first identified as a tumor suppressor gene in hepatocellular carcinoma (14). Nevertheless, the accumulating data have shown its involvement in the pathogenesis of various types of human cancer. In accordance with this theory, downregulation of DLC1 was observed in a wide range of tumor tissues, including liver, breast, lung, ovarian, kidney, colon, stomach and prostate (Table I) (9-11,17,49-55). Underexpression of DLC1 was associated with either heterozygous deletions of the DLC1 gene or hypermethylation of the gene promoter region (11,50,51,53,56). The downregulation of DLC1 was also documented in breast cancer cell lines derived from aggressive metastatic tumors (57).
Table 1. DLC-1 expression in various types of solid tumors.
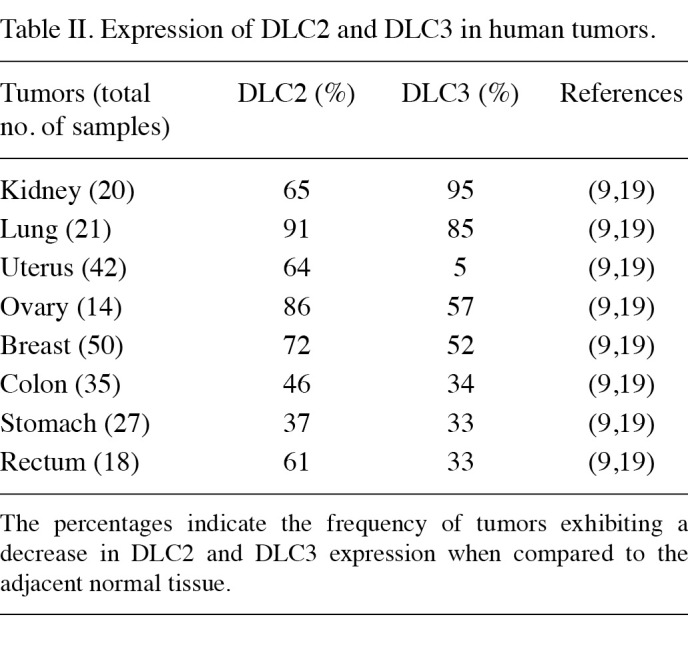
Following the findings of dysregulation of the DLC1 gene function in a variety of solid tumors, downregulation of DLC2 and DLC3 genes was also shown to be involved in human neoplasm development. DLC2 was found to be downregulated in breast, lung, ovarian, renal, uterine, gastric, colon, rectal and liver tumors (Table II) (9,19,58). Notably, the comparison of DLC1 and DLC2 gene expression in the same cell lines revealed that DLC2 is more frequently downregulated than DLC1 in HCC cell lines (9). The expression of the third member of the deleted in liver cancer protein family was shown to be significantly low or undetectable in the majority of the human breast, prostate and ovarian cancer cell lines. A cancer profiling array has revealed a decreased level of DLC3 in primary tumors originating from kidney, lung, uterine, ovarian and breast tissues (Table II) (19).
Table 2. Expression of DLC2 and DLC3 in human tumors.
5. DLCs act as tumor suppressor genes
In recent years extensive studies have revealed DLC1 to be a bona fide tumor suppressor gene not only in HCC, but also in other types of cancer. Accumulating evidence has attributed the suppressive function of DLC1 to RhoGAP activity. As mentioned above, DLC1 was shown to exhibit in vitro GAP activity for RhoA and Cdc42 (11). Negative regulation of the Rho/ROCK/MLC pathway in HCC cell lines by DLC1 was shown to be RhoGAP-dependent (59). DLC1 RhoGAP defective mutant failed to inhibit stress fiber formation in HCC lines (60), whereas the overexpression of DLC1 resulted in morphological change with disruption of actin stress fibers (22,59). Using various cancer cell models, DLC1 was shown to inhibit cell proliferation, suppress cell migration and invasion, and induce apoptosis (44,52,54,60,61). Additionally, DLC1 was reported to act as a suppressor of metastatic foci formation in breast cancer cells. Restoration of DLC1 expression in metastatic cell lines has been shown to result in the inhibition of cell migration and invasion as well as a significant reduction in metastases in athymic mice (57).
DLC2 has been identified as the second tumor suppressor protein of the DLC family and is considered to be equally important. The specific GAP activity on RhoA and Cdc42 suggested that DLC2 inhibits Rho-mediated cytoskeletal reorganization and counteracts Ras-mediated cell transformation in mouse fibroblasts (18). Further research has shown that DLC2 inactivates RhoA, leading to the inhibition of the chemically-induced stress fiber formation. This inactivation may be followed subsequently by a change of the cell morphology, from angular and spindle-shaped to round-shaped with dendritic protrusion formation, as was shown in mouse fibroblasts and hepatoma cells. Moreover, the overexpression of DLC2 suppresses cell proliferation, motility and anchorage‑independent growth in the human hepatoma cell line, HepG2 (62). DLC2 was also reported to have an inhibitory effect on the growth of breast cancer cells in vitro (63).
Supporting the assertion that DLC family members play a significant role in human malignancies, the findings of three reports indicate that DLC3 acts as a potential tumor suppressor (19,23,44). Kawai et al have demonstrated that DLC3 serves as a GAP for both RhoA and Cdc42 in in vitro assays. Furthermore, the overexpression of DLC3 in HeLa cells disrupts actin stress fibers and changes cell morphology in a GAP-dependent manner (23). The introduction of DLC3 into human breast and prostate cancer cell lines has been shown to inhibit cell proliferation, colony formation and growth in soft agar (19).
With reference to the above-mentioned studies, the DLC family proteins may fulfill their suppressive function by decreasing the levels of active, GTP-bound Rho proteins or inhibiting the GDP-GTP cycling process, affecting cytoskeletal remodeling, cell shape, proliferation and migration, as well as induction of apoptosis. However, little is known about the molecular mechanisms through which DLC family proteins are capable of suppressing cell motility and cell growth. In their study, Holeiter et al have shown that enhanced migration of cells lacking DLC1 is dependent on the Rho effector protein Dia1 and does not require the activity of Rho kinase (ROCK) (64). Leung et al provide evidence for a key mechanism through which DLC proteins act as tumor suppressors (65). This study demonstrated that the expression of DLC2 protein is involved in the inactivation of the Raf-1-ERK1/2-p70S6K signaling pathway, which is crucial for cell proliferation. This inhibitory activity of DLC2 is attributed to RhoGAP function (65).
By binding to phospholipase C-δ1, DLC1 enhances the hydrolysis of PIP2, which interacts with various actin‑regulatory proteins that affect the architecture of the actin cytoskeleton (16,22,66). In addition to the PLCδ1-enhancing and RhoGAP activities, proper focal adhesion localization and interaction with tensins have been demonstrated to be essential for the growth-suppression function of DLC1 (44,45,47,48). DLC1 mutants deficient in tensin binding disrupt the focal adhesion localization and lose their ability to suppress colony formation (44,45). Additional investigation is required to establish whether this localization is crucial for the tumor suppression activity of other members of the DLC family.
The DLC protein family contribution in normal tissue development was also documented (67). Although mice hetero-zygous for the DLC1 gene showed no physical abnormalities, mouse embryos, which were homozygous, died before day 10.5 postcoitus with severe defects in a number of organs, including the neural tube, brain, heart and placenta. In addition, mouse embryonic fibroblasts isolated from DLC1 mutant embryos revealed disruption of the actin filament and altered focal adhesions (67).
6. Conclusion
Deleted in liver cancer family proteins are a group of Rho GTPases whose aberrant function suggests dysregulation of numerous cell processes that can promote tumorigenesis. Accumulating evidence indicates that DLC proteins are regulated in various ways: at a genetic level by gene deletion, at an epigenetic level by the aberrant promoter methylation, or at a cellular level by the regulation of protein stability and localization. Future studies are required to establish which form of DLC regulation occurs most commonly in cancer and whether restoration of the proper function of the DLCs is capable of restraining cell transformation. Few studies regarding the function of the two family members, DLC2 and DLC3 are currently available. Additional studies may reveal which member of this family plays a key role in individual types of neoplasia, leading to identification of the protein that is due to become the subject of subsequent studies as a potential drug for targeted therapy.
Contributor Information
D. Lukasik, Department of Pathology, Medical University of Warsaw, Warsaw 02-106, Poland
E. Wilczek, Department of Pathology, Medical University of Warsaw, Warsaw 02-106, Poland
A. Wasiutynski, Department of Pathology, Medical University of Warsaw, Warsaw 02-106, Poland
B. Gornicka, Department of Pathology, Medical University of Warsaw, Warsaw 02-106, Poland
References
- Jaffe AB and Hall A: Rho GTPases: biochemistry and biology. Journal 21: 247-269, 2005. [DOI] [PubMed]
- Heasman SJ and Ridley AJ: Mammalian Rho GTPases: new insights into their functions from in vivo studies. Nat Rev Mol Cell Biol 9: 690-701, 2008. [DOI] [PubMed]
- Corbett KD and Alber T: The many faces of Ras: recognition of small GTP-binding proteins. Trends Biochem Sci 26: 710-716, 2001. [DOI] [PubMed]
- Grise F, Bidaud A and Moreau V: Rho GTPases in hepatocellular carcinoma. Biochim Biophys Acta 1795: 137-151, 2009. [DOI] [PubMed]
- Ridley AJ: Rho GTPases and actin dynamics in membrane protrusions and vesicle trafficking. Trends Cell Biol 16: 522-529, 2006. [DOI] [PubMed]
- Wennerberg K and Der CJ: Rho-family GTPases: it's not only Rac and Rho (and I like it). J Cell Sci 117: 1301-1312, 2004. [DOI] [PubMed]
- Ellenbroek SI and Collard JG: Rho GTPases: functions and association with cancer. Clin Exp Metastasis 24: 657-672, 2007. [DOI] [PubMed]
- Xu XR, Huang J, Xu ZG, et al: Insight into hepatocellular carcinogenesis at transcriptome level by comparing gene expression profiles of hepatocellular carcinoma with those of corresponding noncancerous liver. Proc Natl Acad Sci USA 98: 15089-15094, 2001. [DOI] [PMC free article] [PubMed]
- Ullmannova V and Popescu NC: Expression profile of the tumor suppressor genes DLC-1 and DLC-2 in solid tumors. Int J Oncol 29: 1127-1132, 2006. [PubMed]
- Plaumann M, Seitz S, Frege R, Estevez-Schwarz L and Scherneck S: Analysis of DLC-1 expression in human breast cancer. J Cancer Res Clin Oncol 129: 349-354, 2003. [DOI] [PMC free article] [PubMed]
- Wong CM, Lee JM, Ching YP, Jin DY and Ng IO: Genetic and epigenetic alterations of DLC-1 gene in hepatocellular carcinoma. Cancer Res 63: 7646-7651, 2003. [PubMed]
- Wistuba, II, Behrens C, Virmani AK, et al: Allelic losses at chromosome 8p21-23 are early and frequent events in the pathogenesis of lung cancer. Cancer Res 59: 1973-1979, 1999. [PubMed]
- Chinen K, Isomura M, Izawa K, et al: Isolation of 45 exon-like fragments from 8p22→p21.3, a region that is commonly deleted in hepatocellular, colorectal, and non-small cell lung carcinomas. Cytogenet Cell Genet 75: 190-196, 1996. [DOI] [PubMed]
- Yuan BZ, Miller MJ, Keck CL, Zimonjic DB, Thorgeirsson SS and Popescu NC: Cloning, characterization, and chromosomal localization of a gene frequently deleted in human liver cancer (DLC-1) homologous to rat RhoGAP. Cancer Res 58: 2196‑2199, 1998. [PubMed]
- Tcherkezian J and Lamarche-Vane N: Current knowledge of the large RhoGAP family of proteins. Biol Cell 99: 67-86, 2007. [DOI] [PubMed]
- Homma Y and Emori Y: A dual functional signal mediator showing RhoGAP and phospholipase C-delta stimulating activities. Embo J 14: 286-291, 1995. [DOI] [PMC free article] [PubMed]
- Ko FC, Yeung YS, Wong CM, et al: Deleted in liver cancer 1 isoforms are distinctly expressed in human tissues, functionally different and under differential transcriptional regulation in hepatocellular carcinoma. Liver Int 30: 139-148, 2009. [DOI] [PubMed]
- Ching YP, Wong CM, Chan SF, et al: Deleted in liver cancer (DLC) 2 encodes a RhoGAP protein with growth suppressor function and is underexpressed in hepatocellular carcinoma. J Biol Chem 278: 10824-10830, 2003. [DOI] [PubMed]
- Durkin ME, Ullmannova V, Guan M and Popescu NC: Deleted in liver cancer 3 (DLC-3), a novel Rho GTPase-activating protein, is downregulated in cancer and inhibits tumor cell growth. Oncogene 26: 4580-4589, 2007. [DOI] [PubMed]
- Durkin ME, Yuan BZ, Zhou X, et al: DLC-1: a Rho GTPase-activating protein and tumour suppressor. J Cell Mol Med 11: 1185-1207, 2007. [DOI] [PMC free article] [PubMed]
- Healy KD, Hodgson L, Kim TY, et al: DLC-1 suppresses non- small cell lung cancer growth and invasion by RhoGAP-dependent and independent mechanisms. Mol Carcinog 47: 326-337, 2008. [DOI] [PMC free article] [PubMed]
- Sekimata M, Kabuyama Y, Emori Y and Homma Y: Morphological changes and detachment of adherent cells induced by p122, a GTPase-activating protein for Rho. J Biol Chem 274: 17757-17762, 1999. [DOI] [PubMed]
- Kawai K, Kiyota M, Seike J, Deki Y and Yagisawa H: START‑GAP3/DLC3 is a GAP for RhoA and Cdc42 and is localized in focal adhesions regulating cell morphology. Biochem Biophys Res Commun 364: 783-789, 2007. [DOI] [PubMed]
- Stapleton D, Balan I, Pawson T and Sicheri F: The crystal structure of an Eph receptor SAM domain reveals a mechanism for modular dimerization. Nat Struct Biol 6: 44-49, 1999. [DOI] [PubMed]
- Chi SW, Ayed A and Arrowsmith CH: Solution structure of a conserved C-terminal domain of p73 with structural homology to the SAM domain. Embo J 18: 4438-4445, 1999. [DOI] [PMC free article] [PubMed]
- Qiao F and Bowie JU: The many faces of SAM. Sci STKE 2005: re7, 2005. [DOI] [PubMed]
- Li H, Fung KL, Jin DY, et al: Solution structures, dynamics, and lipid-binding of the sterile alpha-motif domain of the deleted in liver cancer 2. Proteins 67: 1154-1166, 2007. [DOI] [PubMed]
- Kim TY, Healy KD, Der CJ, Sciaky N, Bang YJ and Juliano RL: Effects of structure of Rho GTPase-activating protein DLC-1 on cell morphology and migration. J Biol Chem 283: 32762-32770, 2008. [DOI] [PMC free article] [PubMed]
- Zhong D, Zhang J, Yang S, et al: The SAM domain of the RhoGAP DLC1 binds EF1A1 to regulate cell migration. J Cell Sci 122: 414-424, 2009. [DOI] [PubMed]
- Thornton S, Anand N, Purcell D and Lee J: Not just for housekeeping: protein initiation and elongation factors in cell growth and tumorigenesis. J Mol Med 81: 536-548, 2003. [DOI] [PubMed]
- Liu G, Grant WM, Persky D, Latham VM, Jr, Singer RH and Condeelis J: Interactions of elongation factor 1alpha with F-actin and beta-actin mRNA: implications for anchoring mRNA in cell protrusions. Mol Biol Cell 13: 579-592, 2002. [DOI] [PMC free article] [PubMed]
- Moore RC, Durso NA and Cyr RJ: Elongation factor-1alpha stabilizes microtubules in a calcium/calmodulin-dependent manner. Cell Motil Cytoskeleton 41: 168-180, 1998. [DOI] [PubMed]
- Gross SR and Kinzy TG: Translation elongation factor 1A is essential for regulation of the actin cytoskeleton and cell morphology. Nat Struct Mol Biol 12: 772-778, 2005. [DOI] [PubMed]
- Murray JW, Edmonds BT, Liu G and Condeelis J: Bundling of actin filaments by elongation factor 1 alpha inhibits polymerization at filament ends. J Cell Biol 135: 1309-1321, 1996. [DOI] [PMC free article] [PubMed]
- Alpy F, Legueux F, Bianchetti L and Tomasetto C: [START domain-containing proteins: a review of their role in lipid transport and exchange] (in French). Med Sci 25: 181-191, 2009. [DOI] [PubMed]
- Iyer LM, Koonin EV and Aravind L: Adaptations of the helix-grip fold for ligand binding and catalysis in the START domain superfamily. Proteins 43: 134-144, 2001. [DOI] [PubMed]
- Soccio RE, Adams RM, Romanowski MJ, Sehayek E, Burley SK and Breslow JL: The cholesterol-regulated StarD4 gene encodes a StAR-related lipid transfer protein with two closely related homologues, StarD5 and StarD6. Proc Natl Acad Sci USA 99: 6943-6948, 2002. [DOI] [PMC free article] [PubMed]
- Strauss JF III, Kishida T, Christenson LK, Fujimoto T and Hiroi H: START domain proteins and the intracellular trafficking of cholesterol in steroidogenic cells. Mol Cell Endocrinol 202: 59-65, 2003. [DOI] [PubMed]
- Zhang M, Liu P, Dwyer NK, et al: MLN64 mediates mobilization of lysosomal cholesterol to steroidogenic mitochondria. J Biol Chem 277: 33300-33310, 2002. [DOI] [PubMed]
- Soccio RE and Breslow JL: StAR-related lipid transfer (START) proteins: mediators of intracellular lipid metabolism. J Biol Chem 278: 22183-22186, 2003. [DOI] [PubMed]
- Ng DC, Chan SF, Kok KH, et al: Mitochondrial targeting of growth suppressor protein DLC2 through the START domain. FEBS Lett 580: 191-198, 2006. [DOI] [PubMed]
- Kawai K, Yamaga M, Iwamae Y, et al: A PLCdelta1-binding protein, p122RhoGAP, is localized in focal adhesions. Biochem Soc Trans 32: 1107-1109, 2004. [DOI] [PubMed]
- Kawai K, Seike J, Iino T, et al: START-GAP2/DLC2 is localized in focal adhesions via its N-terminal region. Biochem Biophys Res Commun 380: 736-741, 2009. [DOI] [PubMed]
- Qian X, Li G, Asmussen HK, et al: Oncogenic inhibition by a deleted in liver cancer gene requires cooperation between tensin binding and Rho-specific GTPase-activating protein activities. Proc Natl Acad Sci USA 104: 9012-9017, 2007. [DOI] [PMC free article] [PubMed]
- Liao YC, Si L, deVere White RW and Lo SH: The phosphotyrosine-independent interaction of DLC-1 and the SH2 domain of cten regulates focal adhesion localization and growth suppression activity of DLC-1. J Cell Biol 176: 43-49, 2007. [DOI] [PMC free article] [PubMed]
- Kawai K, Kitamura SY, Maehira K, Seike J and Yagisawa H: START-GAP1/DLC1 is localized in focal adhesions through interaction with the PTB domain of tensin2. Adv Enzyme Regul 50: 202-215, 2010. [DOI] [PubMed]
- Chan LK, Ko FC, Ng IO and Yam JW: Deleted in liver cancer 1 (DLC1) utilizes a novel binding site for Tensin2 PTB domain interaction and is required for tumor-suppressive function. PLoS One 4: e5572, 2009. [DOI] [PMC free article] [PubMed]
- Yam JW, Ko FC, Chan CY, Jin DY and Ng IO: Interaction of deleted in liver cancer 1 with tensin2 in caveolae and implications in tumor suppression. Cancer Res 66: 8367-8372, 2006. [DOI] [PubMed]
- Kim TY, Jong HS, Song SH, et al: Transcriptional silencing of the DLC-1 tumor suppressor gene by epigenetic mechanism in gastric cancer cells. Oncogene 22: 3943-3951, 2003. [DOI] [PubMed]
- Yuan BZ, Durkin ME and Popescu NC: Promoter hypermethylation of DLC-1, a candidate tumor suppressor gene, in several common human cancers. Cancer Genet Cytogenet 140: 113-117, 2003. [DOI] [PubMed]
- Seng TJ, Low JS, Li H, et al: The major 8p22 tumor suppressor DLC1 is frequently silenced by methylation in both endemic and sporadic nasopharyngeal, esophageal, and cervical carcinomas, and inhibits tumor cell colony formation. Oncogene 26: 934-944, 2007. [DOI] [PubMed]
- Ng IO, Liang ZD, Cao L and Lee TK: DLC-1 is deleted in primary hepatocellular carcinoma and exerts inhibitory effects on the proliferation of hepatoma cell lines with deleted DLC-1. Cancer Res 60: 6581-6584, 2000. [PubMed]
- Guan M, Zhou X, Soulitzis N, Spandidos DA and Popescu NC: Aberrant methylation and deacetylation of deleted in liver cancer-1 gene in prostate cancer: potential clinical applications. Clin Cancer Res 12: 1412-1419, 2006. [DOI] [PubMed]
- Yuan BZ, Zhou X, Durkin ME, et al: DLC-1 gene inhibits human breast cancer cell growth and in vivo tumorigenicity. Oncogene 22: 445-450, 2003. [DOI] [PubMed]
- Yuan BZ, Jefferson AM, Baldwin KT, Thorgeirsson SS, Popescu NC and Reynolds SH: DLC-1 operates as a tumor suppressor gene in human non-small cell lung carcinomas. Oncogene 23: 1405-1411, 2004. [DOI] [PubMed]
- Zhang Q, Ying J, Zhang K, et al: Aberrant methylation of the 8p22 tumor suppressor gene DLC1 in renal cell carcinoma. Cancer Lett 249: 220-226, 2007. [DOI] [PubMed]
- Goodison S, Yuan J, Sloan D, et al: The RhoGAP protein DLC-1 functions as a metastasis suppressor in breast cancer cells. Cancer Res 65: 6042-6053, 2005. [DOI] [PMC free article] [PubMed]
- Xiaorong L, Wei W, Liyuan Q and Kaiyan Y: Underexpression of deleted in liver cancer 2 (DLC2) is associated with overexpression of RhoA and poor prognosis in hepatocellular carcinoma. BMC Cancer 8: 205, 2008. [DOI] [PMC free article] [PubMed]
- Wong CC, Wong CM, Ko FC, et al: Deleted in liver cancer 1 (DLC1) negatively regulates Rho/ROCK/MLC pathway in hepatocellular carcinoma. PLoS One 3: e2779, 2008. [DOI] [PMC free article] [PubMed]
- Wong CM, Yam JW, Ching YP, et al: Rho GTPase-activating protein deleted in liver cancer suppresses cell proliferation and invasion in hepatocellular carcinoma. Cancer Res 65: 8861‑8868, 2005. [DOI] [PubMed]
- Zhou X, Thorgeirsson SS and Popescu NC: Restoration of DLC-1 gene expression induces apoptosis and inhibits both cell growth and tumorigenicity in human hepatocellular carcinoma cells. Oncogene 23: 1308-1313, 2004. [DOI] [PubMed]
- Leung TH, Ching YP, Yam JW, et al: Deleted in liver cancer 2 (DLC2) suppresses cell transformation by means of inhibition of RhoA activity. Proc Natl Acad Sci USA 102: 15207-15212, 2005. [DOI] [PMC free article] [PubMed]
- Nagaraja GM and Kandpal RP: Chromosome 13q12 encoded Rho GTPase activating protein suppresses growth of breast carcinoma cells, and yeast two-hybrid screen shows its interaction with several proteins. Biochem Biophys Res Commun 313: 654-665, 2004. [DOI] [PubMed]
- Holeiter G, Heering J, Erlmann P, Schmid S, Jahne R and Olayioye MA: Deleted in liver cancer 1 controls cell migration through a Dia1-dependent signaling pathway. Cancer Res 68: 8743-8751, 2008. [DOI] [PubMed]
- Leung TH, Yam JW, Chan LK, Ching YP and Ng IO: Deleted in liver cancer 2 suppresses cell growth via the regulation of the Raf-1-ERK1/2-p70S6K signalling pathway. Liver Int 30: 1315‑1323, 2010. [DOI] [PubMed]
- Yin HL and Janmey PA: Phosphoinositide regulation of the actin cytoskeleton. Annu Rev Physiol 65: 761-789, 2003. [DOI] [PubMed]
- Durkin ME, Avner MR, Huh CG, Yuan BZ, Thorgeirsson SS and Popescu NC: DLC-1, a Rho GTPase-activating protein with tumor suppressor function, is essential for embryonic development. FEBS Lett 579: 1191-1196, 2005. [DOI] [PubMed]