Background: A shedding product of PTK7 was detected in the culture media from colon cancer cells.
Results: PTK7 is sequentially processed by ADAM17 and γ-secretase, and its cytosolic domain enhances oncogenic properties of colon cancer cells.
Conclusion: The cytosolic domain of PTK7 generated by sequential cleavage of ADAM17 and γ-secretase promotes tumorigenesis.
Significance: We provide a novel oncogenic mechanism of PTK7 upon its processing.
Keywords: Colon Cancer, Protein Processing, Shedding, Protein-tyrosine Kinase (Tyrosine Kinase), ADAM17, PTK7, Colon Cancer Cells, γ-Secretase, Tumorigenesis
Abstract
Protein-tyrosine kinase 7 (PTK7) is a member of the defective receptor protein-tyrosine kinases and is known to function as a regulator of planar cell polarity during development. Its expression is up-regulated in some cancers including colon carcinomas. A 100-kDa fragment of PTK7 was detected in the culture media from colon cancer cells and HEK293 cells. The shed fragment was named sPTK7-Ig1–7 because its molecular mass was very similar to that of the entire extracellular domain of PTK7 that contains immunoglobulin-like loops 1 to 7 (Ig1–7). The shedding of sPTK7-Ig1–7 was enhanced by treatment with phorbol 12-myristate 13-acetate. In addition to the sPTK7-Ig1–7 found in the culture medium, two C-terminal fragments of PTK7 were detected in the cell lysates: PTK7-CTF1, which includes a transmembrane segment and a cytoplasmic domain, and PTK7-CTF2, which lacks most of the transmembrane segment from PTK7-CTF1. Analysis of PTK7 processing in the presence of various protease inhibitors or after knockdown of potential proteases suggests that shedding of PTK7 into sPTK7-Ig1–7 and PTK7-CTF1 is catalyzed by ADAM17, and further cleavage of PTK7-CTF1 into PTK7-CTF2 is mediated by the γ-secretase complex. PTK7-CTF2 localizes to the nucleus and enhances proliferation, migration, and anchorage-independent colony formation. Our findings demonstrate a novel role for PTK7 in the tumorigenesis via generation of PTK7-CTF2 by sequential cleavage of ADAM17 and γ-secretase.
Introduction
Protein-tyrosine kinase 7 (PTK7)4 (also known as colon carcinoma kinase-4, CCK-4) is a receptor tyrosine kinase-like molecule containing an extracellular domain with seven immunoglobulin-like (Ig) loops, a transmembrane domain, and a catalytic domain lacking kinase activity (1–4). PTK7 is conserved across phylogenetically diverse groups from Hydra to human (5). Off-track (Dtrk/OTK), PTK7 ortholog in Drosophila was reported to be a hemophilic, Ca2+-independent cell adhesion molecule in the developing nervous system that regulates neuronal recognition and axon guidance (6). Later it was shown that Dtrk/OTK contributes to repulsive axon guidance signaling by associating with Plexins in response to semaphorin binding (7). In chickens, formation of a complex composed of Plexin-A1, KLG (PTK7 ortholog) and Sema6D is important for cardiac morphogenesis, especially the formation of the ventricle segment (8). In Xenopus, interaction between Plexin-A1 and PTK7 is required for neural crest migration (9).
A role of PTK7 as a regulator of planar cell polarity and noncanonical Wnt signaling was first identified in mice expressing a truncated form of PTK7. The mutation resulted in perinatal lethality, and the mice exhibited characteristic defects of planar cell polarity, such as open neural tubes and disorganized stereociliary bundle orientation (10). PTK7 regulates neural crest migration by recruiting Dishevelled to the membrane (11). PTK7 must interact with RACK1 to recruit Dishevelled for proper neural tube closure in Xenopus (12). Although a role for PTK7 in the canonical Wnt pathway has not been well defined, we have shown that Wnt3a-stimulated β-catenin/T cell factor transcriptional activity is weakened in PTK7-deficient cells (13). In contrast, Peradziryi et al. (14) reported that PTK7/Otk inhibits canonical Wnt signaling but activates noncanonical Wnt signaling by acting as a Frizzled co-receptor.
Up-regulation of PTK7 is observed in various cancers including colon cancer (2, 15), gastric cancer (16), lung cancer (17), acute myeloid leukemia (18), esophageal squamous cell carcinoma (19), and liposarcoma (20). Ectopic expression of PTK7 in leukemia cells promotes cell migration and survival, whereas knockdown of PTK7 shows the opposite effects (21). Knockdown of PTK7 in HCT-116 cells also inhibits cell proliferation and induces apoptosis (22). Similarly, knockdown of PTK7 in liposarcoma cells reduces cell proliferation and invasion and induces apoptosis (20). Interestingly, PTK7 was detected in an analysis of the secretome from pancreatic cancer cells (23) and colon cancer cells (24), suggesting the shedding of PTK7.
Shedding is an important regulatory mechanism for cellular signaling (25). Shedding of membrane proteins such as pro-TNF-α and heparin-binding EGF can release ligands inducing signal transduction (26). In contrast, shedding can down-regulate or terminate signaling by removing the signaling capability of proteins on the cell surface, like Ephrins, or by producing soluble decoy receptors that sequester cognate ligands, like sVEGFR-1 (27, 28). Sheddases that cleave extracellular domains are often members of a disintegrin and metalloprotease (ADAM) family or matrix metalloproteinase (MMP) family, which are Zn2+-dependent proteases. After cleavage of the extracellular domain by a sheddase, some cell surface receptors are further cleaved by intramembrane-cleaving proteases (I-CliPs) within the transmembrane domain in a process termed regulated intramembrane proteolysis. In some proteins such as Notch (29) and erythroblastic leukemia viral oncogene homolog 4 (ErbB4) (30), the cytosolic fragment generated by regulated intramembrane proteolysis can translocate to the nucleus and regulate transcription. In other proteins, the cytosolic fragments play various roles related to the function of the complete protein. The cytosolic domain of Ephrin-B2 activates Src by competing with Csk which phosphorylates and inhibits Src (31). Cleavage of E-cadherin by an I-Clip down-regulates cell adhesion and enhances Wnt signaling through the release of β-catenin (32).
In an attempt to understand the role of PTK7, we generated recombinant soluble PTK7 (sPTK7), which contains the entire extracellular domain consisting of Ig1–7 and acts as a decoy receptor to counteract PTK7 function. We previously demonstrated that treatment with sPTK7 induces an effect similar to PTK7 knockdown and inhibits VEGF-induced tube formation, migration, invasion of HUVECs, and VEGF-induced angiogenesis in vivo (33). ADAMs or MMPs that act as sheddases are often up-regulated during carcinogenesis. Thus, we hypothesized that sPTK7 may be generated in cancers by shedding of PTK7. In addition, MMP-14-dependent shedding of PTK7 was observed in breast cancer cells, and MMP-14 overexpression in fibrosarcoma HT-1080 cells induced the release of an extracellular domain of PTK7 containing the first six Ig loops (sPTK7-Ig1–6) (34). However, we found that, in colon cancer cells, PTK7 is shed into a form containing seven Ig loops, sPTK7-Ig1–7. Here, we report that in colon cancer cells PTK7 is sequentially processed in an extracellular cleavage event followed by intramembrane cleavage. We further demonstrate that the resulting cytosolic fragment of PTK7 has oncogenic properties.
EXPERIMENTAL PROCEDURES
Antibodies and Reagents
Mouse anti-human PTK7 monoclonal antibody against human sPTK7-His was generated and purified by Ab Frontier (Seoul, Korea). Anti-FLAG M2-agarose and the anti-FLAG M2 antibody were obtained from Sigma-Aldrich. Anti-GAPDH antibody was purchased from Ab Frontier. Anti-lamin A/C antibody was purchased from Santa Cruz Biotechnology (Santa Cruz, CA). Anti-TIMP-1 and anti-TIMP-2 were purchased from Calbiochem. Anti-TIMP-3, anti-TIMP-4, anti-ADAM10, and anti-ADAM17 antibodies were obtained from Millipore. GM6001 was from Santa Cruz Biotechnology and Chemicon. TAPI-1, pepstatin A, and E-64 were obtained from Calbiochem. DAPT and l-685458 were purchased from Sigma. Phorbol 12-myristate 13-acetate (PMA) was from A. G. Scientific (San Diego, CA). The hydroxamate-based ADAM inhibitors, GW280264X (selective for ADAM10 and ADAM17) and GI254023X (selective for ADAM10), were described previously (35, 36). siRNA pools for ADAM10 (M-004503-02), ADAM17(l-003453–00), and negative control (d-001810-10) were purchased from Dharmacon (Chicago, IL).
Constructs for Expression of PTK7 or Its Domains
pcDNA3-hPTK7-FLAG, which expresses FLAG-tagged human PTK7, and pcDNA3-hPTK7-Ext-His, which expresses the His-tagged extracellular domain of PTK7, were described previously (33). To construct pcDNA3.1-PTK7-Cyt-FLAG, which expresses human PTK7 cytosolic domain containing residues 726–1070 of PTK7 and a C-terminal FLAG tag, the cDNA was PCR-amplified using pcDNA3-hPTK7-FLAG as a template and 5′-GTCGCTAGCATGTGCAAGAAGCGCTGCAAAGC-3′, including an NheI site (italicized), a start codon (bold), and nucleotide positions of 2323–2342 of PTK7 cDNA (GenBank accession number U40271), and 5′-TAGAAGGCACAGTCGAGG-3′, nucleotide positions 1036–1053 of pcDNA3 vector (Invitrogen), as primer pairs. The amplified fragments were digested with NheI and EcoRI and ligated into pcDNA3.1. To generate pcDNA3.1-PTK7-CTF2-FLAG encoding residues 722–1070 of PTK7 and the C-terminal FLAG tag, nucleotides encoding residues 722–725 of PTK7 were inserted into pcDNA3.1-PTK7-Cyt-FLAG by DpnI-mediated site-directed mutagenesis method using a QuikChange kit (Stratagene) and 5′-GCAGTAGAACATGAGCATGGTGGCGCTAGCCAGCTTGGGTCTCCCTATAGTGA-3′ and 5′-GCGCCACCATGCTCATGTTCTACTGCAAGAAGCGCTGCAAAGCCA-3′, including a Kozak's consensus sequence (underlined), a start codon (bold), and nucleotide positions 2311–2325 of PTK7 cDNA (GenBank accession number U40271), as primers.
Cell Lines, Cell Culture, and Transfection
The colorectal cancer cell lines (HCT-8, HCT-15, HCT-116, SW480, DLD-1, LoVo, and HT-29) and HEK293 cells were cultured in DMEM supplemented with 10% FBS, 100 units/ml penicillin, and 100 μg/ml streptomycin. For transfection with plasmid DNA, subconfluent cells were transfected by the calcium phosphate method (37) followed by selection with 1.2 mg/ml G418. After 2 weeks, all colonies were cultured as a mixed population or individual clones. For transfection with siRNA (100 nm), subconfluent cells were transfected using Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions.
Immunoprecipitation and Western Blotting of Cell Lysates and Media
Subconfluent cells were incubated in serum-free medium either for 8 h without PMA or for 30 min with 100 ng/ml PMA. To collect proteins secreted into the medium, the medium was spun down at 2,000 rpm for 5 min and then precipitated with cold TCA (Sigma). The cells were lysed with RIPA lysis buffer (50 mm Tris-HCl, pH7.4, 150 mm NaCl, 1% Triton X-100, 0.5% deoxycholate, 0.1% SDS). For immunoprecipitation, cell lysates were incubated with mouse anti-FLAG M2-agarose and then washed in PBS three times. For Western blot analysis, cell lysates, TCA-precipitated medium, or immunoprecipitated proteins were resuspended in SDS sample buffer containing 100 mm β-mercaptoethanol and were resolved by SDS-PAGE and transferred to a PVDF membrane.
N-terminal Sequencing
SW480 cells expressing PTK7-FLAG were preincubated in serum-free medium with 10 μm GM6001 or dimethyl sulfoxide for 30 min and then treated with 100 ng/ml PMA for 30 min. The cells were lysed with RIPA lysis buffer and immunoprecipitated with anti-FLAG M2-agarose. The precipitated C-terminal fragments of PTK7 were resolved by SDS-PAGE, transferred to a PVDF membrane, and stained with Coomassie Brilliant Blue. N-terminal sequences of the C-terminal fragments were analyzed by the Edman degradation method at the Tufts Core Facility (Tufts University, Medford, MA).
Fractionation of Cytosolic and Nuclear Proteins
SW480 cells expressing FLAG-PTK7 were pretreated with 10 μm DAPT for 30 min and then treated with 100 ng/ml PMA for 30 min. The cells were collected by scraping in PBS, spun down at 2,000 rpm for 5 min, resuspended in a hypotonic buffer (10 mm HEPES, pH 7.4, 10 mm KCl, 1.5 mm MgCl2), and incubated on ice for 10 min. Nonidet P-40 was then added to a final concentration of 0.625%, and the lysate was incubated on ice for 20 min. The lysate was centrifuged at 2,000 rpm for 5 min to separate the cytosolic fraction from the nuclear pellet. The nuclear pellet was washed twice in hypotonic buffer, resuspended in RIPA lysis buffer, and incubated for 20 min. The nuclear fraction was obtained by centrifugation at 13,000 rpm for 15 min.
Immunofluorescence Staining and Confocal Microscopy
SW480 cells expressing PTK7-CTF2-FLAG or transfected with a control vector were grown on poly-l-lysine-coated coverslips. Immunofluorescence staining was performed with mouse anti-FLAG M2 antibody (1 μg/ml; Sigma), Alexa Fluor 488-conjugated goat anti-mouse IgG antibody (1 μg/ml; Invitrogen), and DAPI (0.25 μg/ml; Invitrogen). Immunofluorescence was visualized with a confocal microscope (Carl Zeiss).
Cell Proliferation Assay
Cell proliferation was assessed by an MTT assay. Subconfluent cells were seeded at a density of 4 × 103 cells in 96-well plates and incubated in DMEM containing 5% FBS. After incubation for the indicated time intervals, cells were stained with 0.5 mg/ml MTT in DMEM for 4 h and then disrupted using dimethyl sulfoxide. Absorbance of the extract at 565 nm was measured.
Cell Migration Assay
Cell migration was assayed using modified Boyden chambers (8-μm pore size, Costar Transwell filters; Corning-Costar, Lowell, MA) coated with 0.1% gelatin. Subconfluent cells were incubated in serum-free medium for 24 h. The 1 × 105 cells in 100 μl of serum-free medium were loaded on the upper chamber. DMEM (600 μl) containing 5% FBS served as a chemoattractant in the lower chamber. After 24 h, cells in the upper surface of the upper chamber were removed with cotton swabs. Cells that had migrated to the lower surface of the upper chamber were stained with hematoxylin and eosin and counted under a light microscope.
Anchorage-independent Colony Formation Assay
Anchorage-independent colony formation was measured as described previously (38). Cells (5 × 104/well) were suspended in DMEM-10% FBS containing 0.35% Bacto agar (Difco, Sparks, MD) incubated at 42 °C and overlaid onto the solidified 0.5% agar layer containing DMEM-10% FBS. After 3 weeks of incubation, the number of colonies formed was counted with a microscope.
RESULTS
sPTK7-Ig1–7 Is Shed in Colon Cancer Cells
To examine whether shedding of PTK7 occurs in colon cancer cell lines, Western blot analysis with an antibody against the extracellular domain of PTK7 in various colon cancer cell lines (HCT-8, HCT-15, HCT-116, DLD-1, LoVo, HT-29, and SW480) and HEK293 cells was performed. We detected a 100-kDa band in the conditioned medium from colon cancer cells and HEK293 cells (Fig. 1A). A band of similar molecular mass was observed in HEK293 cells ectopically expressing the sPTK7-His polypeptide. Considering that sPTK7-His contains all seven Ig loops in the extracellular domain of PTK7 and a His tag (33), this result indicates that the shed fragment of PTK7 in colon cancer cells should also contain all seven Ig loops in the extracellular domain. Thus, it was named sPTK7-Ig1–7.
FIGURE 1.
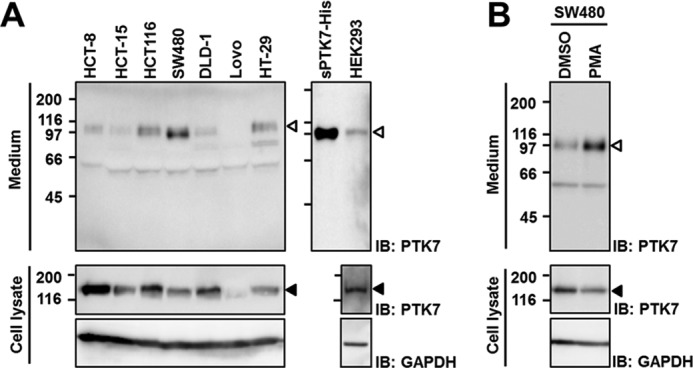
PTK7 is shed in colon cancer cells. A, constitutive shedding of PTK7. Human colon cancer HCT-8, HCT-15, HCT-116, SW480, DLD-1, LoVo, and HT-29 cells and HEK293 cells were incubated in serum-free medium for 8 h. Recombinant sPTK7-His (20 ng) was loaded as a positive control. B, PMA-induced PTK7 shedding. SW480 cells were replaced with serum-free medium and stimulated with PMA (100 ng/ml) for 30 min. Incubated media were concentrated by TCA precipitation. The concentrated media and cell lysates were analyzed by SDS-PAGE and Western blotting (IB). The positions of sPTK7-Ig1–7 and PTK7 are denoted by open arrowheads and closed arrowheads, respectively. Numbers on the left indicate positions of molecular mass markers (kDa).
Phorbol esters such as PMA are potent activators of PKC and have been reported to stimulate shedding of diverse proteins such as IL-6R, ErbB4, and TNF-α (39). We thus analyzed whether PMA increases shedding of sPTK7-Ig1–7 in SW480 cells. PMA treatment enhanced release of sPTK7-Ig1–7 into conditioned medium when compared with mock- or dimethyl sulfoxide-treated cells (Fig. 1B). The apparent molecular mass of sPTK7-Ig1–7 did not change in the presence of PMA (Fig. 1B).
A Metalloprotease Contributes to sPTK7-Ig1–7 Shedding
To address which protease mediates PTK7 shedding, we analyzed PTK7 shedding in the presence of various protease inhibitors in SW480 cells. A panmetalloprotease inhibitor (GM6001) and an MMP and ADAM17 inhibitor (TAPI-1) blocked constitutive shedding of PTK7, but inhibitors of serine proteases (AEBSF), cysteine and serine proteases (leupeptin), aspartic proteases (pepstatin), and cysteine proteases (E-64) did not (Fig. 2A). Like constitutive PTK7 shedding, PMA-induced PTK7 shedding was also inhibited by the metalloprotease inhibitors, GM6001 and TAPI-1, but not by other protease inhibitors (Fig. 2B). Thus, the shed fragment size and metalloprotease requirement were the same in constitutive or PMA-induced sPTK7-Ig1–7 shedding.
FIGURE 2.
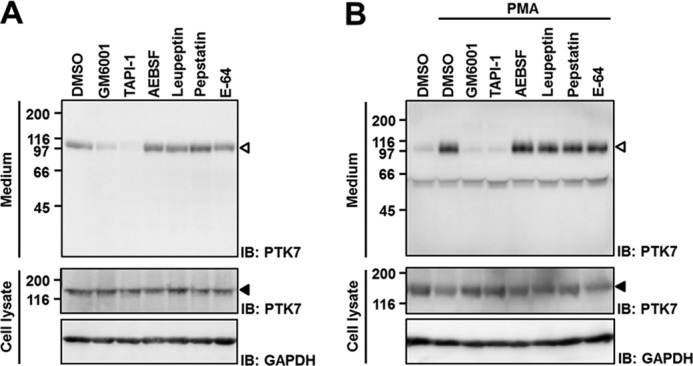
Effect of protease inhibitors on PTK7 shedding. A, effect of protease inhibitors on constitutive PTK7 shedding. SW480 cells were incubated in serum-free medium for 8 h in the presence of various protease inhibitors. B, effect of protease inhibitors on PMA-induced PTK7 shedding. SW480 cells were preincubated in serum-free medium in the presence of the protease inhibitors for 30 min and then treated with PMA (100 ng/ml) for 30 min. Concentrations of the inhibitors were 10 μm GM6001, 50 μm TAPI-1, 100 μm AEBSF, 10 μm leupeptin, 10 μm pepstatin, and 10 μm E-64. Incubated media were concentrated by TCA precipitation. The concentrated media and cell lysates were analyzed by SDS-PAGE and Western blotting (IB). The positions of sPTK7-Ig1–7 and PTK7 are denoted by open arrowheads and closed arrowheads, respectively. DMSO, dimethyl sulfoxide.
Both the Extracellular Domain and Transmembrane Domain of PTK7 Are Cleaved
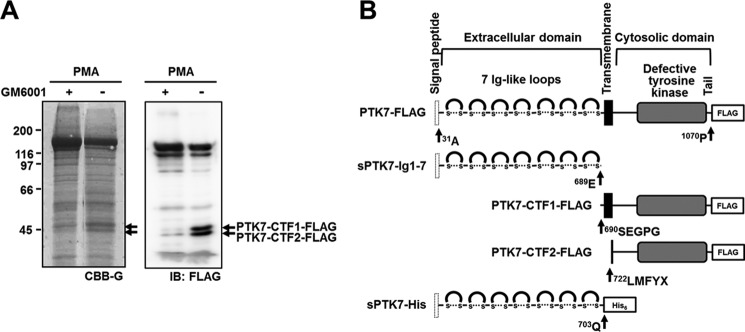
To identify the cleavage site of PTK7 shedding, PTK7-FLAG was stably expressed in SW480 cells. SW480 cells expressing PTK7-FLAG were stimulated with PMA in the presence or absence of GM6001, and PTK7 shedding was analyzed. Immunoprecipitation with an anti-FLAG antibody followed by Western blot analysis revealed the presence of two CTFs of PTK7, which we named PTK7-CTF1-FLAG and PTK7-CTF2-FLAG (Fig. 3A).
FIGURE 3.
PTK7 cleavage sites were identified by N-terminal sequencing. A, SW480 cells overexpressing PTK7-FLAG were preincubated in serum-free medium with GM6001 (10 μm) for 30 min and then treated with PMA (100 ng/ml) for 30 min. The lysate was immunoprecipitated using anti-FLAG M2-agarose and analyzed with SDS-PAGE and transferred to a PVDF membrane. The membrane was stained with Coomassie Brilliant Blue G-250 (CBB-G, left) and immunoblotted (IB) with a FLAG antibody (right). B, schematic diagrams for PTK7-FLAG, sPTK7-Ig1–7, PTK7-CTF1-FLAG, PTK7-CTF2-FLAG, and sPTK7-His are shown. N-terminal sequences of PTK7-CTF1-FLAG and PTK7-CTF2-FLAG which were analyzed by Edman degradation sequencing are indicated.
To identify the cleavage sites, the CTF bands that were detected in Coomassie Brilliant Blue-stained SDS gels (Fig. 3A) were subjected to N-terminal sequencing by the Edman degradation method. The N-terminal sequences of PTK7-CTF1-FLAG and PTK7-CTF2-FLAG were 690SEGPG and 722LMFYX, respectively (Fig. 3B). The amino acid residue at the X position was Cys which could not be detected by this method. This finding suggests that PTK7 is cleaved at two different positions; between amino acids Glu689 and Ser690 near the C terminus of the extracellular domain (Ala31 to Gln703) and between amino acids Gly721 and Leu722 near the C terminus of a transmembrane segment (Thr704 to Tyr725). Therefore, sPTK7-Ig1–7 that spans Ala31 to Glu689 (Fig. 3B) is shed into the extracellular space, and PTK7-CTF2-FLAG which spans Leu722 to Pro1070 is likely released into the cytosol.
sPTK7-Ig1–7 Shedding Is Mediated by ADAM17
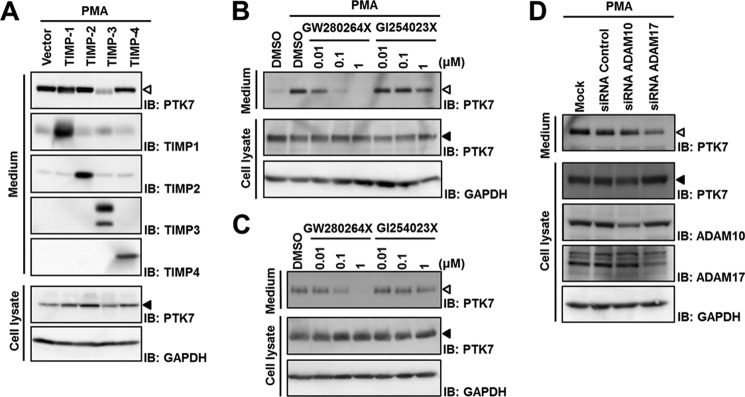
To characterize which metalloprotease is involved, sPTK7-Ig1–7 shedding was analyzed in the presence of TIMPs, which are major physiological metalloprotease inhibitors. SW480 cells, in which one of four TIMPs was stably expressed, were stimulated with PMA to enhance sPTK7-Ig1–7 shedding. Shedding of sPTK7-Ig1–7 was efficiently suppressed by TIMP-3, but not by TIMP-1, TIMP-2, or TIMP-4 (Fig. 4A). TIMP-3 inhibits various MMPs (MMP-1, MMP-2, MMP-3, MMP-7, MMP-9, MMP-13, MMP-14, and MMP-15) (40) and ADAM10 and ADMA17. However, the MMPs inhibited by TIMP-3 are also inhibited by at least one other TIMP that cannot block PTK7 shedding, suggesting that the sheddase is ADAM10 and/or ADAM17 and not an MMP. However, TIMP-1 inhibits ADAM10 but not ADAM17 and does not prevent PTK7 shedding (41, 42), thus indicating that ADAM17 is likely the PTK7 sheddase.
FIGURE 4.
PTK7 shedding is dependent on ADAM-17. A, SW480 cells stably expressing each TIMP were incubated in serum-free medium with PMA (100 ng/ml) for 24 h. B and C, SW480 cells were preincubated in serum-free medium with various concentrations of the ADAM-specific inhibitors GW280264X and GI254023X for 30 min and then incubated with PMA (100 ng/ml) for 30 min (B) or without PMA for 8 h (C). D, SW480 cells were transiently transfected with siRNA for negative control, ADAM10, and ADAM17. At 72 h after transfection, cells were replaced with serum-free medium and stimulated with PMA (100 ng/ml) for 30 min. Cell lysates and concentrated media were analyzed by SDS-PAGE and Western blotting (IB). The positions of sPTK7-Ig1–7 and PTK7 are denoted by open arrowheads and closed arrowheads, respectively. DMSO, dimethyl sulfoxide.
In addition, a synthetic ADAM inhibitor GW280264X, which inhibits both ADAM17 and ADAM10, inhibits sPTK7-Ig1–7 shedding in a dose-dependent manner in SW480 cells with or without PMA stimulation (Fig. 4, B and C). Another ADAM inhibitor GI254023X, which inhibits ADAM10 about 100 times more than ADMA17 (35, 36), is a poor inhibitor of sPTK7-Ig1–7 shedding (Fig. 4, B and C). Moreover, siRNA-mediated ADAM17 knockdown reduced PMA-induced sPTK7-Ig1–7 shedding in SW480 cells, but ADAM10 knockdown did not (Fig. 4D). These results demonstrate that ADAM17 contributes to PTK7 shedding.
Generation of PTK7-CTF2 Is Dependent on γ-Secretase Activity
As shown above, PTK7-CTF2 was generated by proteolytic cleavage within the transmembrane segment (Fig. 3B). Many receptor proteins that are cleaved in the extracellular domain by ADAMs are processed within a transmembrane segment by a γ-secretase complex (43). To analyze whether a γ-secretase complex is involved in PTK7 processing, generation of PTK7-CTF2 was analyzed in SW480 cells expressing PTK7-FLAG in the presence of the γ-secretase inhibitors, DAPT and l-685458, with or without GM6001. Treatment with DAPT or l-685458 completely blocked the generation of PTK7-CTF2-FLAG (Fig. 5). Interestingly, GM6001 alone prevented the generation of PTK7-CTF2 as well as production of PTK7-CTF1. These results indicate that sequential cleavage of PTK7 by ADAM17 and γ-secretase releases sPTK7-Ig1–7 into the extracellular space and PTK7-CTF2 into the cytoplasm.
FIGURE 5.
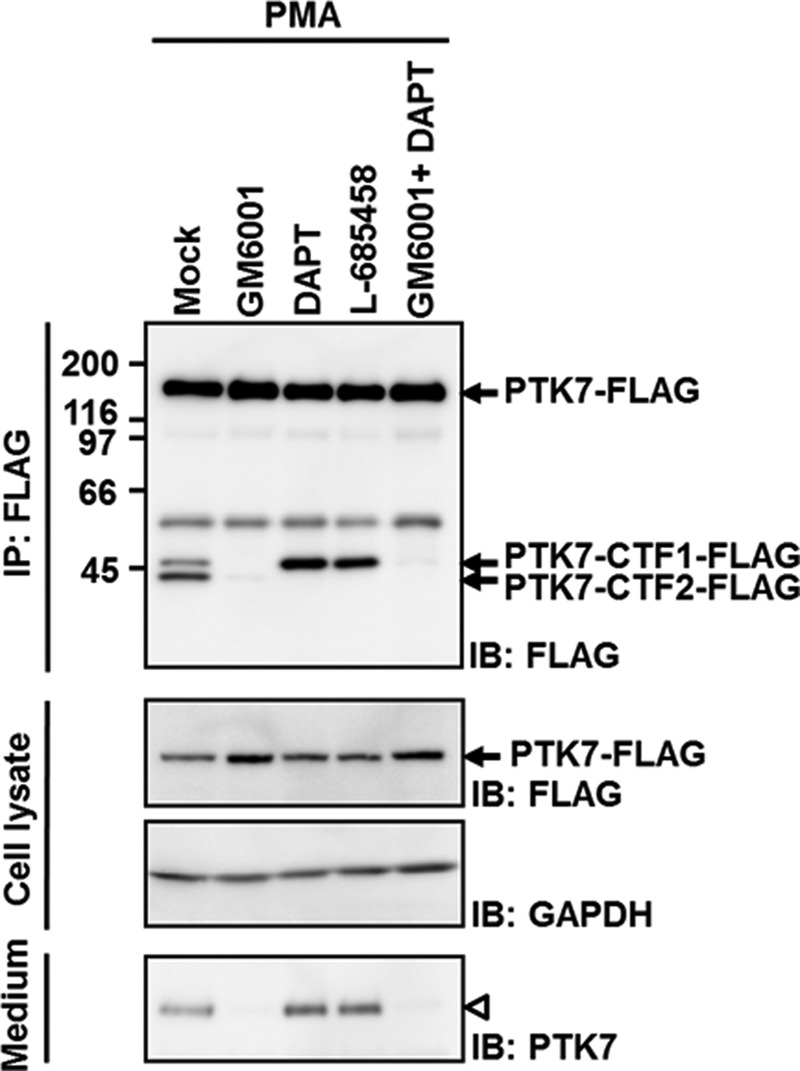
Generation of PTK7-CTF2 is dependent on γ-secretase. SW480 cells expressing PTK7-FLAG were incubated in serum-free medium with GM6001 (10 μm) and the γ-secretase inhibitors, DAPT (10 μm) and l-685458 (2 μm) for 30 min and were then incubated with PMA (100 ng/ml) for 30 min. The cells were lysed with RIPA lysis buffer and immunoprecipitated (IP) using anti-FLAG M2-agarose. The lysate were analyzed by SDS-PAGE and immunoblotting (IB) for FLAG. The positions of sPTK7-Ig1–7 and PTK7 are denoted by open arrowheads and closed arrowheads, respectively.
PTK7-CTF2 Is Localized Mainly in the Nucleus
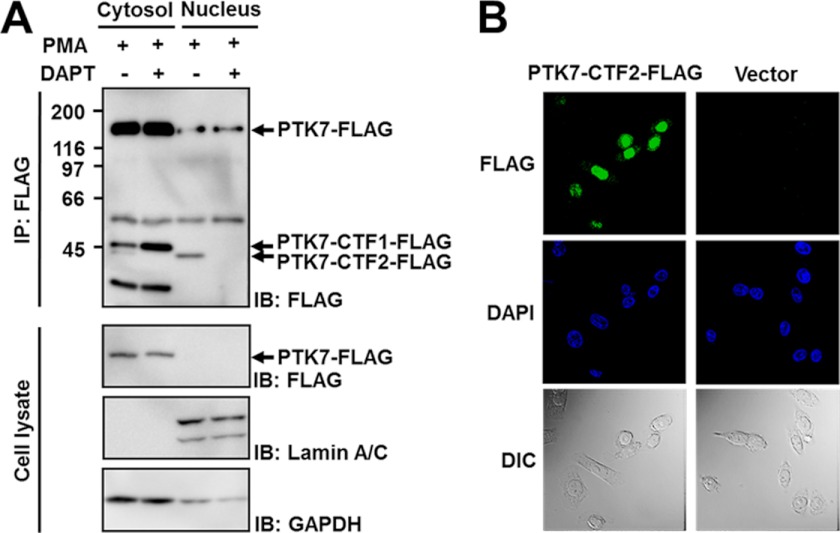
In an attempt to determine a role for PTK7-CTF2, we analyzed its intracellular localization of PTK7-CTF2-FLAG in SW480 cells using two different methods. First, SW480 cells expressing PTK7-FLAG were separated into cytosolic and nuclear fractions. PTK7-CTF1-FLAG was found in the cytosolic fraction, but PTK7-CTF2-FLAG was mainly in the nuclear fraction (Fig. 6A). Second, immunofluorescence demonstrated that PTK7-CTF2-FLAG expressed in SW480 cells is mainly localized to the nucleus (Fig. 6B).
FIGURE 6.
PTK7-CTF2 displays a nuclear localization. A, SW480 cells stably expressing PTK7-FLAG were incubated in serum-free medium with DAPT (10 μm) for 30 min and then with PMA for 30 min. The cells were lysed with hypotonic buffer, and the cytosolic fraction and nuclei were separated. The nuclei were lysed with RIPA lysis buffer. FLAG-tagged polypeptides were immunoprecipitated (IP) from cytosolic and nuclear fractions using anti-FLAG M2-agarose. The immunoprecipitates and lysates were analyzed by SDS-PAGE and immunoblotting (IB) for PTK7. The positions of sPTK7-Ig1–7 and PTK7 are denoted by open arrowheads and closed arrowheads, respectively. B, SW480 cells stably expressing PTK7-CTF2-FLAG or transfected with control vector were fixed with 3.7% formaldehyde. PTK7-CTF2-FLAG cells were labeled with anti-FLAG M2 antibody and rabbit anti-mouse IgG conjugated with Alexa Fluor 488. Nucleus was stained with DAPI. Immunofluorescence was observed by a confocal microscope. DIC, differential interference contrast.
PTK7-CTF2 Expression Increases Cell Proliferation and Migration
To investigate further the function of nuclear PTK7-CTF2, we carried out several assays to monitor biological functions. An MTT assay showed that SW480 cells expressing PTK7-CTF2-FLAG (Fig. 7A) grow up to two times faster than SW480 cells transfected with an empty vector (Fig. 7B). A migration assay using a Boyden chamber showed that SW480 cells expressing PTK7-CTF2-FLAG migrate approximately four times faster than control SW480 cells (Fig. 7C). Further, anchorage-independent colony formation assays indicated that expression of PTK7-CTF2-FLAG enhanced colony formation in soft agar, with almost twice as many colonies as the vector control (Fig. 7D). Together, these findings suggest that PTK7-CTF2 translocation to the nucleus will enhance the oncogenic potential of SW480 cells.
FIGURE 7.
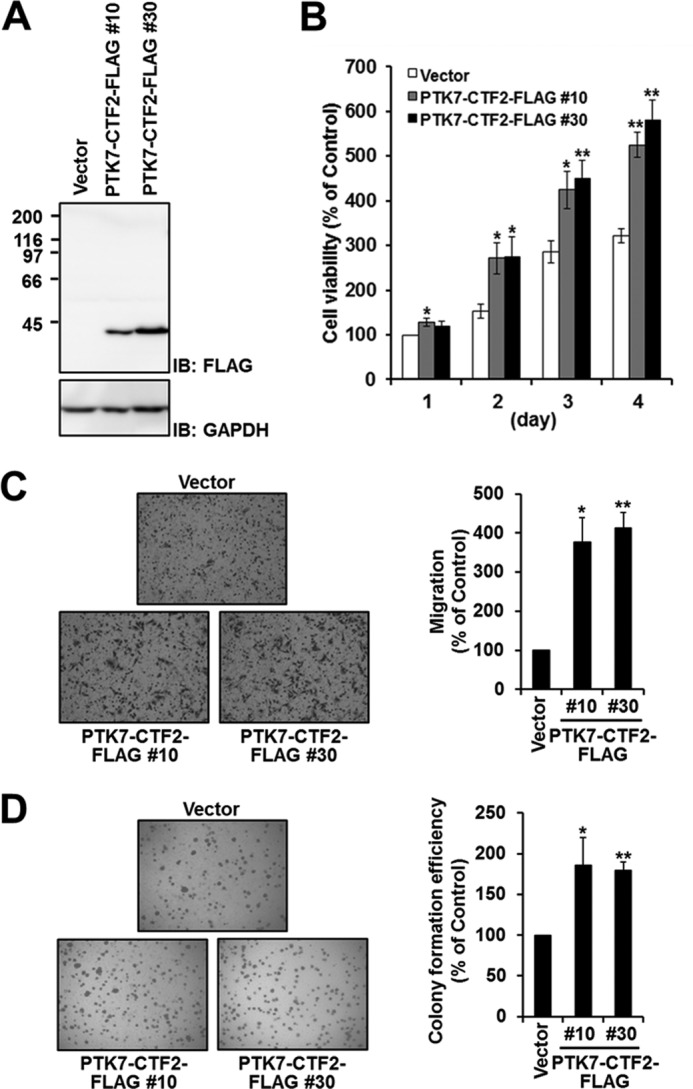
PTK7-CTF2 increases cell proliferation, migration, and colony formation. A, expression of PTK7-CTF2-FLAG in SW480 cells. SW480 cells stably expressing PTK7-CTF2-FLAG (#10 and #30 clones) or control vector were lysed with RIPA lysis buffer. The lysates were analyzed by SDS-PAGE and immunoblotting (IB) for FLAG. B, cell proliferation assay. The cells were incubated in DMEM containing 5% FBS in 96-well plates. After incubation for the indicated time, viable cells were measured by MTT assay. C, cell migration assay. The cells that migrated across the Transwell chamber were stained with hematoxylin and eosin and were counted under a light microscope. D, colony formation assay. The cells suspended in DMEM-10% FBS containing 0.35% Bacto agar were overlaid onto a solidified 0.5% agar layer containing DMEM-10% FBS. After 3 weeks of incubation, the number of colonies that formed was counted with a microscope. Each value is the mean ± S.D. (error bars) of three independent experiments. *, p < 0.05 versus vector; **, p < 0.01 versus vector.
DISCUSSION
In this study, we found shed PTK7 in the medium from multiple colon cancer cell lines. The shed polypeptide is ∼100 kDa and includes all seven Ig loops of the extracellular domain of PTK7. Treatment with PMA enhances shedding, but does not alter the size of the shed fragment in SW480 cells. Shedding was inhibited by a panmetalloprotease inhibitor, GM6001, and the MMP/ADAM inhibitors, TAPI-1 and TIMP-3, but not by other TIMPs or nonmetalloprotease inhibitors. In addition, an ADAM10/17 inhibitor, GW280264X, inhibited PTK7 shedding whereas another ADAM inhibitor that is 100 times more selective for ADAM10 than ADAM17, GI254023X, poorly inhibited, in SW480 cells with or without PMA stimulation (Fig. 4) and in other colon cancer cells (data not shown). Furthermore, ADAM17 knockdown reduced PMA-induced sPTK7-Ig1–7 shedding. These data demonstrate that ADAM17 should be a sheddase of PTK7.
Among the 21 ADAMs identified in the human genome, 13 members are proteolytically active. ADAM17 is one of the proteolytically active members (26). Expression of ADAM17 is increased in various cancers including breast cancer (44), colorectal cancer (45), pancreatic cancer (46), prostate cancer (47), renal cancer (48), and hepatocellular cancer (49). Overexpression of ADAM17 in breast cancer cells increased in vitro invasion and proliferation (50). Also, silencing of ADAM17 suppressed tumor formation by inhibiting ADAM17-mediated TGF-α release, a ligand for the EGF receptor, in renal carcinoma cells (51). Therefore, ADAM17 contributes to tumorigenesis and metastasis by promoting cell proliferation, adhesion, migration, and invasion (52–54).
During our study, Golubkov et al. (34) reported that PTK7 levels were reduced in breast cancer cells overexpressing MMP-14 and that sPTK7-Ig1–6 shed from PTK7 was observed in HT1080 cells overexpressing MMP-14. We observed a 73-kDa band, which may correspond to sPTK7-Ig1–6, at low level in culture medium of some colon cancer cells such as HT-29 cells (Fig. 1A). However, the major PTK7 shed product detected was 100-kDa band corresponding to sPTK7-Ig1–7 rather than 73-kDa band. These results strongly suggest that at least in colon cancer cells ADAM17 is a main sheddase of PTK7.
In this study, we found that PTK7 was processed not only at the extracellular domain to shed sPTK7-Ig1–7 but also within a transmembrane segment to generate PTK7-CTF2. Because the cleavage to generate PTK7-CTF2 occurs between amino acids Gly721 and Leu722 near the C terminus of a transmembrane segment (Thr704 to Tyr725), it is likely that the enzyme is a member of the I-CliP family. I-CliP members can be classified into three groups; site-2 proteases which are metalloproteases, γ-secretase complexes and signal peptide peptidases which are aspartyl proteases, and rhomboids which are serine proteases (55, 56). Among these, γ-secretase cleaves mainly type I transmembrane proteins (43), and therefore PTK7 which has the same membrane topology could represent a potential substrate. It was also reported that C-terminal fragments of many transmembrane receptors that are shed by ADAMs are further processed by γ-secretase in the transmembrane domain (43). As expected, we found that the γ-secretase inhibitors, DAPT and l-685458, blocked PTK7-CTF2 generation. Moreover, a metalloprotease inhibitor, GM6001 blocks generation of both PTK7-CTF1 and PTK7-CTF2. These results demonstrate that cleavage by ADAM17 is required for the γ-secretase cleavage, and thus that PTK7 is sequentially cleaved by ADAM17 and γ-secretase.
Intracellular domains of type I transmembrane receptors cleaved by γ-secretases are known to exert biological effects at other sites within the cell. The most well characterized function of such intracellular domains is transcriptional regulation in the nucleus. For example, the intracellular domain of ErbB4 complexed with the signaling protein TAB2, and the co-repressor N-CoR translocates to the nucleus and represses GFAP and S100β promoters (30). Likewise, PTK7-CTF2 is localized mainly in the nucleus. Ectopic expression of PTK7-CTF2 increases proliferation, migration, and anchorage-independent colony formation of SW480 cells. Thus, PTK7-CTF2 translocation from the plasma membrane to the nucleus likely regulates transcription, thus enhancing tumor generation and progression.
Accumulated data suggest that PTK7 is up-regulated in various cancers (2, 15–20) and that its expression promotes proliferation, survival, migration, and invasion (15, 20–22). We have also shown that treatment with sPTK7-Ig1–7 or PTK7 knockdown inhibits VEGF-induced migration, invasion, tube formation of HUVECs, and VEGF-induced angiogenesis in vivo (33). However, it has also been reported that PTK7 down-regulates actin cytoskeleton organization and actomyosin contraction and that sPTK7-Ig1–6 enhances cell locomotion and invasion in HT-1080 cells by reversing PTK7 functions (34). The functional differences between sPTK7-Ig1–7 and sPTK7-Ig1–6 could result from the presence of the seventh Ig loop in sPTK7-Ig1–7 or may be dependent on the cell type studied. Nonetheless, sPTK7-Ig1–7 is thought to act as a tumor suppressor by counteracting PTK7. However, to neutralize PTK7 function, high concentrations (>53 μm) of sPTK7 were required (33). Moreover, shed sPTK7 would diffuse out into extracellular space, and so we assume that the effect of shed sPTK7 would not be significant in cancer tissues. In contrast, another processing product of PTK7, PTK7-CTF2, is able to be effectively concentrated in the nucleus and thus activate signaling pathways to promote tumorigenesis and metastasis. Therefore, although it seems that PTK7 is oncogenic as an intact molecule, it maintains its oncogenic properties through the generation of PTK7-CTF2 in the malignant environment.
Here, we have shown that PTK7 is sequentially processed by ADAM17 at the C-terminal region of the extracellular domain and then by γ-secretase within the transmembrane segment and that the resulting fragments sPTK7-Ig1–7 and PTK7-CTF2 are released into the extracellular space and nucleus, respectively. We also showed that PTK7-CTF2 localized to the nucleus can increase proliferation, migration, and anchorage-independent colony formation. Therefore, in addition to the known oncogenic role of the intact PTK7 molecule, we add a novel role of PTK7 upon its shedding: PTK7-CTF2 translocates into nucleus and enhances oncogenic properties of the cell.
This work was supported by grants from the Basic Research Program Grant 2009-0076146, Studies on Ubiquitome Functions Grant 2005-2001143, and the Mid-Career Researcher Program Grant 2010-0026103 of the National Research Foundation, Ministry of Education, Science, and Technology, Republic of Korea and by the National R&D Program for Cancer Control Grant 1120110, Ministry of Health and Welfare, Republic of Korea (to S.-T. L.).
- PTK7
- protein-tyrosine kinase 7
- ADAM
- a disintegrin and metalloprotease
- CTF
- C-terminal fragment
- ErbB
- erythroblastic leukemia viral oncogene homolog
- I-Clip
- intramembrane domain-cleaving protease
- Ig
- immunoglobulin-like
- MMP
- matrix metalloproteinase
- MTT
- 3-(4,5-dimethyl thiazol-2-yl)-2,5-diphenyltetrazolium bromide
- OTK
- Off-track
- PMA
- phorbol 12-myristate 13-acetate
- RIPA
- radioimmuneprecipitation assay
- sPTK7
- soluble PTK7
- TIMP
- tissue inhibitor of metalloproteinases.
REFERENCES
- 1. Lee S. T., Strunk K. M., Spritz R. A. (1993) A survey of protein-tyrosine kinase mRNAs expressed in normal human melanocytes. Oncogene 8, 3403–3410 [PubMed] [Google Scholar]
- 2. Mossie K., Jallal B., Alves F., Sures I., Plowman G. D., Ullrich A. (1995) Colon carcinoma kinase-4 defines a new subclass of the receptor tyrosine kinase family. Oncogene 11, 2179–2184 [PubMed] [Google Scholar]
- 3. Park S. K., Lee H. S., Lee S. T. (1996) Characterization of the human full-length PTK7 cDNA encoding a receptor protein tyrosine kinase-like molecule closely related to chick KLG. J. Biochem. 119, 235–239 [DOI] [PubMed] [Google Scholar]
- 4. Jung J. W., Shin W. S., Song J., Lee S. T. (2004) Cloning and characterization of the full-length mouse Ptk7 cDNA encoding a defective receptor protein tyrosine kinase. Gene 328, 75–84 [DOI] [PubMed] [Google Scholar]
- 5. Peradziryi H., Tolwinski N. S., Borchers A. (2012) The many roles of PTK7: a versatile regulator of cell-cell communication. Arch. Biochem. Biophys. 524, 71–76 [DOI] [PubMed] [Google Scholar]
- 6. Pulido D., Campuzano S., Koda T., Modolell J., Barbacid M. (1992) Dtrk, a Drosophila gene related to the trk family of neurotrophin receptors, encodes a novel class of neural cell adhesion molecule. EMBO J. 11, 391–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Winberg M. L., Tamagnone L., Bai J., Comoglio P. M., Montell D., Goodman C. S. (2001) The transmembrane protein off-track associates with Plexins and functions downstream of semaphorin signaling during axon guidance. Neuron 32, 53–62 [DOI] [PubMed] [Google Scholar]
- 8. Toyofuku T., Zhang H., Kumanogoh A., Takegahara N., Suto F., Kamei J., Aoki K., Yabuki M., Hori M., Fujisawa H., Kikutani H. (2004) Dual roles of Sema6D in cardiac morphogenesis through region-specific association of its receptor, Plexin-A1, with off-track and vascular endothelial growth factor receptor type 2. Genes Dev. 18, 435–447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Wagner G., Peradziryi H., Wehner P., Borchers A. (2010) Plexin-A1 interacts with PTK7 and is required for neural crest migration. Biochem. Biophys. Res. Commun. 402, 402–407 [DOI] [PubMed] [Google Scholar]
- 10. Lu X., Borchers A. G., Jolicoeur C., Rayburn H., Baker J. C., Tessier-Lavigne M. (2004) PTK7/CCK-4 is a novel regulator of planar cell polarity in vertebrates. Nature 430, 93–98 [DOI] [PubMed] [Google Scholar]
- 11. Shnitsar I., Borchers A. (2008) PTK7 recruits dsh to regulate neural crest migration. Development 135, 4015–4024 [DOI] [PubMed] [Google Scholar]
- 12. Wehner P., Shnitsar I., Urlaub H., Borchers A. (2011) RACK1 is a novel interaction partner of PTK7 that is required for neural tube closure. Development 138, 1321–1327 [DOI] [PubMed] [Google Scholar]
- 13. Puppo F., Thomé V., Lhoumeau A. C., Cibois M., Gangar A., Lembo F., Belotti E., Marchetto S., Lécine P., Prébet T., Sebbagh M., Shin W. S., Lee S. T., Kodjabachian L., Borg J. P. (2011) Protein tyrosine kinase 7 has a conserved role in Wnt/β-catenin canonical signalling. EMBO Rep. 12, 43–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Peradziryi H., Kaplan N. A., Podleschny M., Liu X., Wehner P., Borchers A., Tolwinski N. S. (2011) PTK7/Otk interacts with Wnts and inhibits canonical Wnt signalling. EMBO J. 30, 3729–3740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Saha S., Bardelli A., Buckhaults P., Velculescu V. E., Rago C., St Croix B., Romans K. E., Choti M. A., Lengauer C., Kinzler K. W., Vogelstein B. (2001) A phosphatase associated with metastasis of colorectal cancer. Science 294, 1343–1346 [DOI] [PubMed] [Google Scholar]
- 16. Gorringe K. L., Boussioutas A., Bowtell D. D. (2005) Novel regions of chromosomal amplification at 6p21, 5p13, and 12q14 in gastric cancer identified by array comparative genomic hybridization. Genes Chromosomes Cancer 42, 247–259 [DOI] [PubMed] [Google Scholar]
- 17. Endoh H., Tomida S., Yatabe Y., Konishi H., Osada H., Tajima K., Kuwano H., Takahashi T., Mitsudomi T. (2004) Prognostic model of pulmonary adenocarcinoma by expression profiling of eight genes as determined by quantitative real-time reverse transcriptase polymerase chain reaction. J. Clin. Oncol. 22, 811–819 [DOI] [PubMed] [Google Scholar]
- 18. Müller-Tidow C., Schwäble J., Steffen B., Tidow N., Brandt B., Becker K., Schulze-Bahr E., Halfter H., Vogt U., Metzger R., Schneider P. M., Büchner T., Brandts C., Berdel W. E., Serve H. (2004) High-throughput analysis of genome-wide receptor tyrosine kinase expression in human cancers identifies potential novel drug targets. Clin. Cancer Res. 10, 1241–1249 [DOI] [PubMed] [Google Scholar]
- 19. Su H., Hu N., Yang H. H., Wang C., Takikita M., Wang Q. H., Giffen C., Clifford R., Hewitt S. M., Shou J. Z., Goldstein A. M., Lee M. P., Taylor P. R. (2011) Global gene expression profiling and validation in esophageal squamous cell carcinoma and its association with clinical phenotypes. Clin. Cancer Res. 17, 2955–2966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Gobble R. M., Qin L. X., Brill E. R., Angeles C. V., Ugras S., O'Connor R. B., Moraco N. H., Decarolis P. L., Antonescu C., Singer S. (2011) Expression profiling of liposarcoma yields a multigene predictor of patient outcome and identifies genes that contribute to liposarcomagenesis. Cancer Res. 71, 2697–2705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Prebet T., Lhoumeau A. C., Arnoulet C., Aulas A., Marchetto S., Audebert S., Puppo F., Chabannon C., Sainty D., Santoni M. J., Sebbagh M., Summerour V., Huon Y., Shin W. S., Lee S. T., Esterni B., Vey N., Borg J. P. (2010) The cell polarity PTK7 receptor acts as a modulator of the chemotherapeutic response in acute myeloid leukemia and impairs clinical outcome. Blood 116, 2315–2323 [DOI] [PubMed] [Google Scholar]
- 22. Meng L., Sefah K., O'Donoghue M. B., Zhu G., Shangguan D., Noorali A., Chen Y., Zhou L., Tan W. (2010) Silencing of PTK7 in colon cancer cells: caspase-10-dependent apoptosis via mitochondrial pathway. PLoS One 5, e14018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Grønborg M., Kristiansen T. Z., Iwahori A., Chang R., Reddy R., Sato N., Molina H., Jensen O. N., Hruban R. H., Goggins M. G., Maitra A., Pandey A. (2006) Biomarker discovery from pancreatic cancer secretome using a differential proteomic approach. Mol. Cell Proteomics 5, 157–171 [DOI] [PubMed] [Google Scholar]
- 24. Xue H., Lü B., Zhang J., Wu M., Huang Q., Wu Q., Sheng H., Wu D., Hu J., Lai M. (2010) Identification of serum biomarkers for colorectal cancer metastasis using a differential secretome approach. J. Proteome Res. 9, 545–555 [DOI] [PubMed] [Google Scholar]
- 25. Dello Sbarba P., Rovida E. (2002) Transmodulation of cell surface regulatory molecules via ectodomain shedding. Biol. Chem. 383, 69–83 [DOI] [PubMed] [Google Scholar]
- 26. Edwards D. R., Handsley M. M., Pennington C. J. (2008) The ADAM metalloproteinases. Mol. Aspects Med. 29, 258–289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Reiss K., Saftig P. (2009) The “a disintegrin and metalloprotease” (ADAM) family of sheddases: physiological and cellular functions. Semin. Cell Dev. Biol. 20, 126–137 [DOI] [PubMed] [Google Scholar]
- 28. Rahimi N., Golde T. E., Meyer R. D. (2009) Identification of ligand-induced proteolytic cleavage and ectodomain shedding of VEGFR-1/FLT1 in leukemic cancer cells. Cancer Res. 69, 2607–2614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Schroeter E. H., Kisslinger J. A., Kopan R. (1998) Notch-1 signalling requires ligand-induced proteolytic release of intracellular domain. Nature 393, 382–386 [DOI] [PubMed] [Google Scholar]
- 30. Sardi S. P., Murtie J., Koirala S., Patten B. A., Corfas G. (2006) Presenilin-dependent ErbB4 nuclear signaling regulates the timing of astrogenesis in the developing brain. Cell 127, 185–197 [DOI] [PubMed] [Google Scholar]
- 31. Georgakopoulos A., Litterst C., Ghersi E., Baki L., Xu C., Serban G., Robakis N. K. (2006) Metalloproteinase/Presenilin1 processing of ephrinB regulates EphB-induced Src phosphorylation and signaling. EMBO J. 25, 1242–1252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Marambaud P., Shioi J., Serban G., Georgakopoulos A., Sarner S., Nagy V., Baki L., Wen P., Efthimiopoulos S., Shao Z., Wisniewski T., Robakis N. K. (2002) A presenilin-1/γ-secretase cleavage releases the E-cadherin intracellular domain and regulates disassembly of adherens junctions. EMBO J. 21, 1948–1956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Shin W. S., Maeng Y. S., Jung J. W., Min J. K., Kwon Y. G., Lee S. T. (2008) Soluble PTK7 inhibits tube formation, migration, and invasion of endothelial cells and angiogenesis. Biochem. Biophys. Res. Commun. 371, 793–798 [DOI] [PubMed] [Google Scholar]
- 34. Golubkov V. S., Chekanov A. V., Cieplak P., Aleshin A. E., Chernov A. V., Zhu W., Radichev I. A., Zhang D., Dong P. D., Strongin A. Y. (2010) The Wnt/planar cell polarity protein-tyrosine kinase-7 (PTK7) is a highly efficient proteolytic target of membrane type-1 matrix metalloproteinase: implications in cancer and embryogenesis. J. Biol. Chem. 285, 35740–35749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Hundhausen C., Misztela D., Berkhout T. A., Broadway N., Saftig P., Reiss K., Hartmann D., Fahrenholz F., Postina R., Matthews V., Kallen K. J., Rose-John S., Ludwig A. (2003) The disintegrin-like metalloproteinase ADAM10 is involved in constitutive cleavage of CX3CL1 (fractalkine) and regulates CX3CL1-mediated cell-cell adhesion. Blood 102, 1186–1195 [DOI] [PubMed] [Google Scholar]
- 36. Ludwig A., Hundhausen C., Lambert M. H., Broadway N., Andrews R. C., Bickett D. M., Leesnitzer M. A., Becherer J. D. (2005) Metalloproteinase inhibitors for the disintegrin-like metalloproteinases ADAM10 and ADAM17 that differentially block constitutive and phorbol ester-inducible shedding of cell surface molecules. Comb. Chem. High Throughput Screen 8, 161–171 [DOI] [PubMed] [Google Scholar]
- 37. Wigler M., Sweet R., Sim G. K., Wold B., Pellicer A., Lacy E., Maniatis T., Silverstein S., Axel R. (1992) Transformation of mammalian cells with genes from prokaryotes and eukaryotes. 1979. BioTechnology 24, 444–452 [PubMed] [Google Scholar]
- 38. Kim H. Ie., Lee S. T. (2005) An intramolecular interaction between SH2-kinase linker and kinase domain is essential for the catalytic activity of protein-tyrosine kinase-6. J. Biol. Chem. 280, 28973–28980 [DOI] [PubMed] [Google Scholar]
- 39. Huovila A. P., Turner A. J., Pelto-Huikko M., Kärkkäinen I., Ortiz R. M. (2005) Shedding light on ADAM metalloproteinases. Trends Biochem. Sci. 30, 413–422 [DOI] [PubMed] [Google Scholar]
- 40. Zhao H., Bernardo M. M., Osenkowski P., Sohail A., Pei D., Nagase H., Kashiwagi M., Soloway P. D., DeClerck Y. A., Fridman R. (2004) Differential inhibition of membrane type 3 (MT3)-matrix metalloproteinase (MMP) and MT1-MMP by tissue inhibitor of metalloproteinase (TIMP)-2 and TIMP-3 rgulates pro-MMP-2 activation. J. Biol. Chem. 279, 8592–8601 [DOI] [PubMed] [Google Scholar]
- 41. Amour A., Slocombe P. M., Webster A., Butler M., Knight C. G., Smith B. J., Stephens P. E., Shelley C., Hutton M., Knäuper V., Docherty A. J., Murphy G. (1998) TNF-alpha converting enzyme (TACE) is inhibited by TIMP-3. FEBS Lett. 435, 39–44 [DOI] [PubMed] [Google Scholar]
- 42. Amour A., Knight C. G., English W. R., Webster A., Slocombe P. M., Knäuper V., Docherty A. J., Becherer J. D., Blobel C. P., Murphy G. (2002) The enzymatic activity of ADAM8 and ADAM9 is not regulated by TIMPs. FEBS Lett. 524, 154–158 [DOI] [PubMed] [Google Scholar]
- 43. Haapasalo A., Kovacs D. M. (2011) The many substrates of presenilin/γ-secretase. J. Alzheimers Dis. 25, 3–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Lendeckel U., Kohl J., Arndt M., Carl-McGrath S., Donat H., Röcken C. (2005) Increased expression of ADAM family members in human breast cancer and breast cancer cell lines. J. Cancer Res. Clin. Oncol. 131, 41–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Blanchot-Jossic F., Jarry A., Masson D., Bach-Ngohou K., Paineau J., Denis M. G., Laboisse C. L., Mosnier J. F. (2005) Up-regulated expression of ADAM17 in human colon carcinoma: co-expression with EGFR in neoplastic and endothelial cells. J. Pathol. 207, 156–163 [DOI] [PubMed] [Google Scholar]
- 46. Ringel J., Jesnowski R., Moniaux N., Lüttges J., Choudhury A., Batra S. K., Kloppel G., Lohr M. (2006) Aberrant expression of a disintegrin and metalloproteinase 17/tumor necrosis factor-α-converting enzyme increases the malignant potential in human pancreatic ductal adenocarcinoma. Cancer Res. 66, 9045–9053 [DOI] [PubMed] [Google Scholar]
- 47. Karan D., Lin F. C., Bryan M., Ringel J., Moniaux N., Lin M. F., Batra S. K. (2003) Expression of ADAMs (a disintegrin and metalloproteases) and TIMP-3 (tissue inhibitor of metalloproteinase-3) in human prostatic adenocarcinomas. Int. J. Oncol. 23, 1365–1371 [PubMed] [Google Scholar]
- 48. Roemer A., Schwettmann L., Jung M., Stephan C., Roigas J., Kristiansen G., Loening S. A., Lichtinghagen R., Jung K. (2004) The membrane proteases ADAMs and hepsin are differentially expressed in renal cell carcinoma: are they potential tumor markers? J. Urol. 172, 2162–2166 [DOI] [PubMed] [Google Scholar]
- 49. Ding X., Yang L. Y., Huang G. W., Wang W., Lu W. Q. (2004) ADAM17 mRNA expression and pathological features of hepatocellular carcinoma. World J. Gastroenterol. 10, 2735–2739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. McGowan P. M., Ryan B. M., Hill A. D., McDermott E., O'Higgins N., Duffy M. J. (2007) ADAM-17 expression in breast cancer correlates with variables of tumor progression. Clin. Cancer Res. 13, 2335–2343 [DOI] [PubMed] [Google Scholar]
- 51. Franovic A., Robert I., Smith K., Kurban G., Pause A., Gunaratnam L., Lee S. (2006) Multiple acquired renal carcinoma tumor capabilities abolished upon silencing of ADAM17. Cancer Res. 66, 8083–8090 [DOI] [PubMed] [Google Scholar]
- 52. Murphy G. (2008) The ADAMs: signalling scissors in the tumour microenvironment. Nat. Rev. Cancer 8, 929–941 [DOI] [PubMed] [Google Scholar]
- 53. Roy R., Yang J., Moses M. A. (2009) Matrix metalloproteinases as novel biomarkers and potential therapeutic targets in human cancer. J. Clin. Oncol. 27, 5287–5297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Duffy M. J., Mullooly M., O'Donovan N., Sukor S., Crown J., Pierce A., McGowan P. M. (2011) The ADAMs family of proteases: new biomarkers and therapeutic targets for cancer? Clin. Proteomics 8, 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Beel A. J., Sanders C. R. (2008) Substrate specificity of γ-secretase and other intramembrane proteases. Cell Mol. Life Sci. 65, 1311–1334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Weihofen A., Martoglio B. (2003) Intramembrane-cleaving proteases: controlled liberation of proteins and bioactive peptides. Trends Cell Biol. 13, 71–78 [DOI] [PubMed] [Google Scholar]