Background: Cytokinesis is regulated by phosphorylation and dephosphorylation of proteins.
Results: MINK1 associated with STRN4, a regulatory subunit of PP2A, and depletion of either protein inhibited completion of cytokinesis.
Conclusion: MINK1 and STRN4 are required for abscission, the final stage of cytokinesis.
Significance: Our study reveals novel regulatory mechanisms for abscission.
Keywords: Cell Cycle, Cell Division, Cytokinesis, Serine Threonine Protein Kinase, Serine Threonine Protein Phosphatase
Abstract
Cytokinesis is initiated by constriction of the cleavage furrow and terminated by abscission of the intercellular bridge that connects two separating daughter cells. The complicated processes of cytokinesis are coordinated by phosphorylation and dephosphorylation mediated by protein kinases and phosphatases. Mammalian Misshapen-like kinase 1 (MINK1) is a member of the germinal center kinases and is known to regulate cytoskeletal organization and oncogene-induced cell senescence. To search for novel regulators of cytokinesis, we performed a screen using a library of siRNAs and found that MINK1 was essential for cytokinesis. Time-lapse analysis revealed that MINK1-depleted cells were able to initiate furrowing but that abscission was disrupted. STRN4 (Zinedin) is a regulatory subunit of protein phosphatase 2A (PP2A) and was recently shown to be a component of a novel protein complex called striatin-interacting phosphatase and kinase (STRIPAK). Mass spectrometry analysis showed that MINK1 was a component of STRIPAK and that MINK1 directly interacted with STRN4. Similar to MINK1 depletion, STRN4-knockdown induced multinucleated cells and inhibited the completion of abscission. In addition, STRN4 reduced MINK1 activity in the presence of catalytic and structural subunits of PP2A. Our study identifies a novel regulatory network of protein kinases and phosphatases that regulate the completion of abscission.
Introduction
Cytokinesis is the final step of cell division that physically disconnects two daughter cells by constriction of the cleavage furrow and membrane abscission. A complicated coordination of the dynamic remodeling of the cellular cytoskeleton and intertwined signal networks in time and space regulates the precise progression of cytokinesis. Phosphorylation by mitotic kinases, such as Polo-like kinase 1 (PLK1),2 Aurora-A, and Aurora-B, is one of the major regulatory mechanisms that control the dynamic cellular changes during cytokinesis (1). For example, PLK1 is localized to different subcellular structures, such as the centrosome, central spindle, and midbody, and phosphorylates numerous proteins to ensure the accurate distribution of chromosomes, constriction of the cleavage furrow and abscission (2, 3). In addition to these kinases, recent studies have used a library of siRNAs to identify other kinases required for cytokinesis (4). The elucidation of the physiological functions of these kinases is crucial to fully realize the complicated process of cytokinesis.
Accumulating evidence has demonstrated that mitotic phosphatases are important for the accurate progression of cell division (5–7). Protein phosphatase 1 (PP1) and protein phosphatase type 2A (PP2A) are major Ser/Thr phosphatase holoenzymes that account for over 90% of total Ser/Thr phosphatase activity in cells. PP1 is composed of catalytic and regulatory subunits, and PP2A is a heterotrimeric complex of catalytic, structural, and regulatory subunits (8). Although PP2A has only two catalytic subunits, there are a number of regulatory subunits that determine the specific functions of phosphatase holoenzymes (9). PP2A has four subgroups of regulatory subunits, B, B′, B″, and B‴. B‴ members are commonly known as striatins, which include STRN1 (striatin), STRN3 (SG2NA), and STRN4 (Zinedin) (10). Striatins are calmodulin- and caveolin-binding proteins with WD repeats and are involved in signaling and trafficking (11–15). These numerous regulatory subunits give phosphatases diverse cellular functions to control the dynamic changes during cell division (16). For example, the B55α regulatory subunit of PP2A regulates the postmitotic assembly of the nuclear envelope and Golgi apparatus (17). In addition, a recent study using a library of siRNAs revealed that regulatory subunits of PP2A regulate spindle formation, kinetochore functions, and progression into cytokinesis (18).
Misshapen-like kinase 1 (MINK1) is a member of the germinal center kinase (GCK) family and is known to regulate a variety of biological processes (19). The MINK1 Drosophila homolog Misshapen (Msn) controls embryonic dorsal closure, photoreceptor axon pathfinding, and planar cell polarity by modulating the activity of JNK and the phosphorylation of Bifocal and Prickle (20–24). In addition, Msn was reported to phosphorylate Smad1 to inhibit TGF-β/bone morphogenetic protein (BMP)-mediated signaling (25). Mammalian MINK1 is implicated in the activation of JNK, in Ras-mediated p38 MAP kinase activation, and regulates cellular senescence, cytoskeletal organization and cell motility (26–29). These studies indicate that MINK1 plays crucial roles in fundamental cellular processes, but the precise functions still remain largely unknown.
In this report, we searched for novel regulators of cytokinesis using a library of siRNAs and identified MINK1 as one of the essential factors for cytokinesis. Mass spectrometry analysis revealed that MINK1 is a component of the recently identified large protein assembly called STRIPAK (striatin interacting phosphatase and kinase). STRIPAK is composed of striatins, catalytic and structural subunits of PP2A, and other accessory proteins (30–32). Biochemical analysis demonstrated the direct interaction of MINK1 and a member of the striatin family, STRN4. We also report that STRN4 modulates MINK1 activity in the presence of catalytic and structural subunits of PP2A and is required for the completion of cytokinesis.
EXPERIMENTAL PROCEDURES
Cells, Antibodies, and Chemicals
HeLa and 293T cells were propagated in Dulbecco's modified Eagle's medium (Wako, Osaka, Japan) supplemented with 10% fetal bovine serum (Equitech-BIO, Kerrville, TX). Anti-MINK1 antibody was generated by injecting 200 μg of GST-MINK1 (aa346–431) mixed with Freud adjuvant (Sigma-Aldrich, Taufkirchen, Germany) into a rabbit four times every 2 weeks. Serum was obtained, and anti-MINK1 antibody was purified using an NHS-activated column (GE Healthcare BioSciences, Uppsala, Sweden) coupled with GST-MINK1. Anti-GST antibody was eliminated using recombinant GST. Anti-STRN4 antibody was generated using aa1–147 of STRN4 fused with GST. Anti-HA antibody was generated using HA peptide (YPYDVPDYA) fused with GST. Other antibodies were obtained from the following manufacturers: Anti-GFP antibody, NeuroMab, Davis, CA; anti-FLAG antibody, Wako; anti-cyclin B1 antibody, Cell Signaling, Danvers, MA; anti-α-tubulin and anti-beta-actin antibodies, Sigma-Aldrich. CDK1 inhibitor, RO3306, was obtained from Merck4Biosciences (Darmstadt, Germany), and PLK1 inhibitor, BI2536, from Axon Medchem BV (Groningen, Netherlands).
siRNA Screening
Forty proteins that are phosphorylated in mitosis and whose cellular functions have never been associated with mitosis were selected based on the previous report (33). siRNAs targeting these genes were purchased from Invitrogen. HeLa cells were cultured on 48-well plates and transfected with each siRNA. Seventy-two hours later, the cells were fixed with ice cold methanol/acetone (1:1) and immunostained with Hoechst (Dojindo, Kumamoto, Japan) and anti-α-tubulin antibody. Multiple pictures were taken using a fluorescence microscope (IX71, Olympus, Tokyo, Japan), and multinucleated cells were manually evaluated.
Plasmids
Human full-length STRN4 was obtained from Kazusa DNA Research Institute (Chiba, Japan). Human full-length MINK1, TNIK, MAP4K4, PPP2R1A, and PP2P2CA cDNAs were amplified by PCR from the HeLa cDNA library and ligated into the pQCXIP vector (Clontech, Mountain View, CA) with N-terminal GFP, FLAG, HA, or Myc tag. The deletion constructs for MINK1 were generated by PCR.
In Vitro Translation
cDNAs were cloned into the pcDNA3.1 vector (Invitrogen), and in vitro translation was performed using TnT Quick-coupled Transcription/Translation System (Promega, Madison, WI) according to the manufacturer's protocol.
Immunoblotting and Immunoprecipitation
Cells were lysed with Laemmli sample buffer (20% glycerol, 135 mm Tris-HCl, pH 6.8, 4% SDS, 10% 2-mercaptoethanol, 0.003% BPB) and boiled for 5 min. Protein concentrations of lysates were measured using the RC-DC Protein Assay (Bio-Rad Laboratories). Equal protein quantities of cell lysates were separated on SDS-polyacrylamide electrophoresis (SDS-PAGE) gels and transferred to PVDF membranes (Millipore, Billerica, MA). The membranes were blocked with 1% nonfat skim milk, incubated with each primary antibody for 1 h, washed with TBS-T buffer (10 mm Tris-HCl, pH 7.4, 150 mm NaCl, 0.05% Tween 20) and incubated with secondary antibodies. The proteins were visualized by enhanced chemiluminescence (GE Healthcare). To immunoprecipitate proteins, cells were lysed with TNE buffer (25 mm Tris-HCl, pH 7.4, 150 mm NaCl, 0.1% Nonidet P-40 (Wako)) or RIPA buffer (50 mm Tris-HCl pH 7.4, 150 mm NaCl, 0.1% SDS, 0.5% DOC, 1% Nonidet P-40) and centrifuged at 15,000 rpm for 20 min to clear cell debris. Cell lysates containing equal amounts of protein were incubated with primary antibodies coupled to protein A- or protein G-agarose beads (Thermo Scientific, Waltham, MA) at 4 °C for 1 h. After extensive washing with TNE buffer, the proteins were eluted with Laemmli sample buffer, boiled, and then subjected to immunoblotting.
Phosphatase Treatment
Cells were lysed with RIPA buffer with a mixture of phosphatase inhibitors (Nacalai tesque, Kyoto, Japan) and immunoprecipitated with anti-MINK1 antibody. The immunoprecipitates were washed with phosphatase reaction buffer (50 mm HEPES pH 7.5, 100 mm NaCl, 0.1 mm EDTA pH 8.0, 2 mm MnCl2, 5 mm DTT), either treated or not treated with 10 units of λ phosphatase at room temperature for 20 min and then subjected to immunoblot analysis.
Protein Identification by Mass Spectrometry
293T cells were lysed with TNE buffer with phosphatase inhibitor cocktails and protease inhibitors and immunoprecipitated with either anti-MINK1 or control antibody. The immunoprecipitates were washed four times with TNE buffer and then suspended in Laemmli sample buffer. The samples were separated on a 10% SDS-PAGE gel and stained using a SilverQuest Silver Staining Kit (Invitrogen). The protein bands were excised, and the gel pieces were destained, reduced, alkylated, and digested using an In-Gel Tryptic Digestion Kit (Thermo Scientific). The peptides were sequenced using the LC-MS/MS system (Paradigm MS4, Michrom Bioresources, Sacramento, CA; HTS-PAL, CTC Analytics AG, Zwingen, Swiss; LTQ Orbitrap XL, Thermo Scientific), and the proteins were identified using the Mascot software package (Matrix Science, London, UK).
Time-lapse Analysis
HeLa cells cultured on glass-based dishes were transfected with siRNAs; 24 h later, the cells were monitored using a time-lapse microscope system (LCV110, Olympus) for 48 h. Images were acquired and analyzed using the MetaMorph Imaging System (Universal Imaging, Silicon Valley, CA).
Transfection
The sequences of the siRNAs used to suppress MINK1 expression were 5′-GGAACAAGAUUCUGCACAATT-3′ (MINK1–01), 5′-GCAGCCAAGUUUACUUCAUTT-3′ (MINK1–02). The sequences of the siRNAs for STRN4 depletion were 5′-GGGUCAAACUCCAAGGCAUTT-3′ (STRN4–01), and 5′-GCCUCUGUCUGUUUGCCAUTT-3′ (STRN4–02). The sequence of the control siRNA targeting luciferase was 5′-CUUACGCUGAGUACUUCGATT-3′. Cells were transfected with 20 nm siRNA using Lipofectamine RNAiMAX (Invitrogen) according to the manufacturer's instructions. Plasmids were introduced into cells using LipofectamineTM 2000 (Invitrogen) according to the manufacturer's protocol. To detect the interaction of MINK1 and STRN4 in 293T cells, cells cultured on 3.5-cm dishes were transfected with 1 μg of plasmid encoding each gene. Forty-eight hours later, the cells were lysed with TNE buffer and immunoprecipitated.
Immunofluorescence Analysis
Cells were grown on glass coverslips coated with fibronectin, fixed with ice cold methanol/acetone (1:1) for 10 min, and blocked with phosphate-buffered saline (PBS) containing 7% fetal bovine serum for 30 min. Cells were incubated with primary antibody in PBS for 1 h, washed three times with PBS, incubated with Alexa Fluor 488 or Alexa Fluor 594-labeled secondary antibody in PBS for 1 h, and then analyzed using a fluorescence microscope (BX60, Olympus).
Generation of Stable Cell Lines
siRNA-resistant GFP-MINK1 was generated by PCR and cloned into pQCXIP retrovirus vector. To produce kinase-inactive siRNA-resistant GFP-MINK1, the lysine at amino acid 54 was substituted to arginine by PCR. 293T cells were transfected with pQCXIP vector encoding wild-type or mutant GFP-MINK1 with pVPack-GP and pVPack-Ampho vectors (Stratagene, Tokyo, Japan). The culture supernatant was collected 48 h later and applied to HeLa cells with 2 μg/ml of polybrene (Sigma-Aldrich). Cells were cultured for 24 h, and then 1 μg/ml of puromycin (Sigma-Aldrich) was added to select for infected cells.
Cell Synchronization
Cells were treated with 40 ng/ml of nocodazole for 14.5 h, and mitotic cells were collected by shaking off. The collected cells were washed with PBS three times and then cultured in DMEM with 10% FBS for the indicated time periods. To synchronize cells in mitosis using thymidine, cells were cultured in the presence of 2 mm thymidine for 24 h and then released for 9 h. Mitotic cells were collected by shaking off.
In Vitro Kinase Assay
To determine the catalytic activity of MINK1 during mitosis, GFP-MINK1 HeLa cells synchronized in mitosis were lysed with RIPA buffer and immunoprecipitated with anti-GFP-antibody. The immunoprecipitates were washed three times with RIPA buffer, washed twice with kinase buffer (50 mm Tris-HCl, pH 7.4, 5 mm MgCl2, 5 mm MnCl2, 5 mm dithiothreitol, 0.01% Triton X-100), and then mixed with 2 μg of MBP (Sigma-Aldrich) and [γ32-P]ATP for 40 min at 30 °C. The reaction was terminated by adding Laemmli sample buffer, and then the samples were separated by SDS-PAGE and analyzed by autoradiography.
To examine the effects of STRN4 on the catalytic activity of MINK1, FLAG-tagged STRN4 was transiently expressed in 293T cells and purified. 293T cells were transfected with a plasmid encoding either FLAG or FLAG-STRN4; 24 h later, the cells were lysed with RIPA buffer and immunoprecipitated with anti-FLAG antibody. The immunoprecipitate was washed with RIPA buffer and kinase buffer and then eluted with FLAG-peptide (Wako). One-tenth of the immunoprecipitated GFP-MINK1 and FLAG-STRN4 was applied to SDS-PAGE gel with different amounts of the recombinant proteins of GST-fused MINK1 (aa346–431) and STRN4 (aa1–147), which were used to immunize rabbits. The approximate amount of GFP-MINK1 and FLAG-STRN4 was measured using ImageJ. Immunoprecipitated GFP-MINK1 was mixed with MBP, [γ-32P]ATP, and different amounts of STRN4 for 40 min at 30 °C. The reaction was terminated by adding Laemmli sample buffer, and then the samples were separated by SDS-PAGE and analyzed by autoradiography.
To examine the activity of MINK1 in the presence of PPP2R1A, PPP2CA, and STRN4, a complex of FLAG-PPP2R1A and GFP-PPP2CA was transiently expressed in 293T cells and purified. 293T cells were transfected with plasmids encoding FLAG-PPP2R1A and GFP-PPP2CA; 24 h later, the cells were lysed with TNE buffer and immunoprecipitated with anti-FLAG antibody. The immunoprecipitate was washed with TNE buffer, and the complex was eluted with FLAG-peptide. The approximate amount of the complex was estimated by immunoblot with anti-FLAG antibody using FLAG-STRN4 as a standard. GFP-MINK1 was immunoprecipitated and incubated with FLAG-STRN4 and different amounts of the FLAG-PPP2R1A/GFP-PPP2CA complex for 40 min at 30 °C. Laemmli sample buffer was added to terminate the reaction, which was then separated by SDS-PAGE and analyzed by autoradiography.
To examine whether CDK1/cyclin B1 and PLK1 can phosphorylate MINK1, kinase-inactive MINK1 (MINK1 K54R) was produced by PCR and used as a substrate. One microgram of CDK1/cyclin B1 or PLK1 (Millipore) was incubated with GFP-MINK1 K54R in the presence of [γ-32P]ATP for 40 min at 30 °C and subjected to autoradiography after SDS-PAGE gel separation.
RESULTS
The Depletion of MINK1 Induces Multinucleated Cells
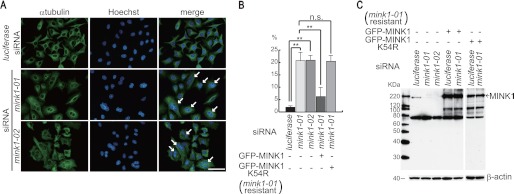
Cytokinesis-related proteins are often phosphorylated during mitosis. A previous study identified a large number of proteins that are phosphorylated in mitosis (33). Based on their findings, we manually selected 40 genes whose products are phosphorylated in mitosis and whose physiological functions have not been associated with cytokinesis. To determine whether these genes are required for cytokinesis, HeLa cells cultured on 48-well plates were transfected with siRNAs targeting these genes and fixed 72 h later to visualize the cells with Hoechst and anti-α-tubulin antibody. Silencing of the proteins essential for cytokinesis is known to induce multinucleated cells. Multiple pictures for each group of siRNA-transfected cells were taken, and the number of multinucleated cells was evaluated. In this screen, we found that transfection of two different siRNAs targeting MINK1 significantly increased the number of multinucleated cells (Fig. 1A). The quantification of these results revealed that ∼20% of cells were multinucleated (Fig. 1B). To confirm that MINK1 suppression, but not an off-target effect of the siRNAs, resulted in the increase of multinucleated cells, we established HeLa cells that constitutively expressed GFP-tagged siRNA-resistant MINK1 by retroviral infection. The number of multinucleated cells induced by MINK1 siRNA transfection was dramatically reduced by the expression of siRNA-resistant MINK1 (Fig. 1B). Next, we determined if catalytic activity was required to rescue the phenotype induced by the siRNA transfection. The lysine at amino acid 54 is essential for the catalytic activity of MINK1. GFP-tagged siRNA-resistant MINK1 with the substitution of Lys-54 to arginine was expressed in HeLa cells by retroviral infection and transfected with MINK1 siRNA to deplete the endogenous protein. Catalytically inactive MINK1 was unable to reduce the ratio of multinucleated cells by MINK1 siRNA transfection (Fig. 1B). We generated an affinity-purified polyclonal antibody using a MINK1 fragment of aa346–431 to confirm the depletion of MINK1 by the siRNAs. Immunoblot analysis demonstrated that endogenous MINK1 was depleted by both of the siRNAs, whereas expression of the siRNA-resistant MINK1 was not affected by either siRNA (Fig. 1C). These results suggest that loss of MINK1 function compromises cytokinesis. We performed immunofluorescence analysis to determine the localization of MINK1 during mitosis; however, MINK1 did not localize to specific sites, such as the mitotic spindle, kinetochore, cleavage furrow, and midbody (supplemental Fig. S1).
FIGURE 1.
The depletion of MINK1 induces multinucleated cells. A, HeLa cells were cultured on glass coverslips and transfected with the indicated siRNAs. Seventy-two hours later, cells were fixed and immunostained with anti-α-tubulin antibody and Hoechst. The arrows indicate multinucleated cells (scale bar, 50 μm). B, HeLa cells that constitutively expressed GFP-tagged siRNA-resistant wild-type or kinase inactive MINK1 were established by retroviral infection. The cells were treated as in A, and the ratio of multinucleated cells was evaluated. Three independent experiments were performed, and 200 cells were evaluated for each experiment (the data are represented as the mean ± S.D.; **, p < 0.01; n.s., not significant). C, cells were transfected with the indicated siRNA; 72 h later, the cells were lysed and immunoblotted with anti-MINK1 antibody.
MINK1 Is Required for Abscission
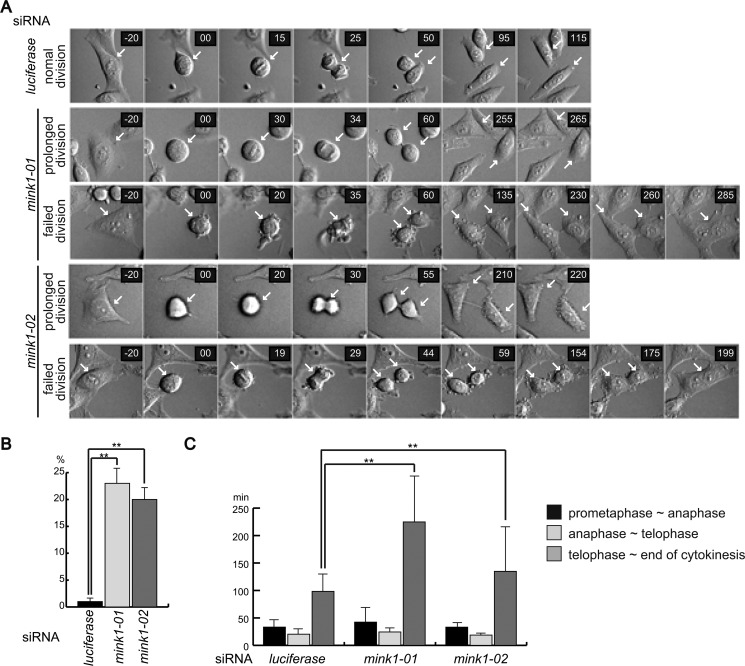
To further characterize the cytokinesis phenotype caused by MINK1 depletion, time-lapse imaging of cell division after siRNA treatment was performed. Luciferase siRNA-transfected cells showed normal kinetics of cell division, and all of the cells we observed completed cytokinesis to produce two separated daughter cells (Fig. 2A). Although all the MINK1 siRNA-transfected cells formed a cleavage furrow, and ingression proceeded to form intercellular bridge between two daughter cells, some MINK1-knockdown cells were unable to abscise and fused back to form binucleated cells (Fig. 2A). The quantification of cells with failed cytokinesis revealed that ∼20% of MINK1-depleted cells became binucleated after incomplete cytokinesis (Fig. 2B). In addition, we noticed that abscission of the intercellular bridge was significantly delayed in most of the MINK1-knockdown cells. We evaluated the time period of each stage of cell division. MINK1 depletion did not affect the progression from prometaphase to telophase, but the time from telophase to the completion of abscission was longer compared with luciferase siRNA-transfected cells (Fig. 2C). These results indicate that MINK1 is required to abscise the intercellular bridge to complete cytokinesis.
FIGURE 2.
MINK1 is required for the completion of abscission. A, HeLa cells were transfected with control or MINK1 siRNAs; 48 h later, the cells were observed using time-lapse microscopy. Representative images of two types of cell divisions of MINK1 siRNA-transfected cells are shown. prolonged division indicates cells that completed cytokinesis with extended time period. failed division indicates cells that were unable to complete cytokinesis. The arrows point dividing cells. B, graph shows the ratio of cells that failed cytokinesis. Forty cells for each group of siRNA-transfected cells were evaluated, and three independent experiments were performed (the data are represented as the mean ± S.D.; **, p < 0.01). C, time period of each stage of mitosis was evaluated. The graph shows the average time period of 30 cells for each siRNA transfection (data represented as the mean ± S.D.; **, p < 0.01).
MINK1 Is Phosphorylated in Mitosis
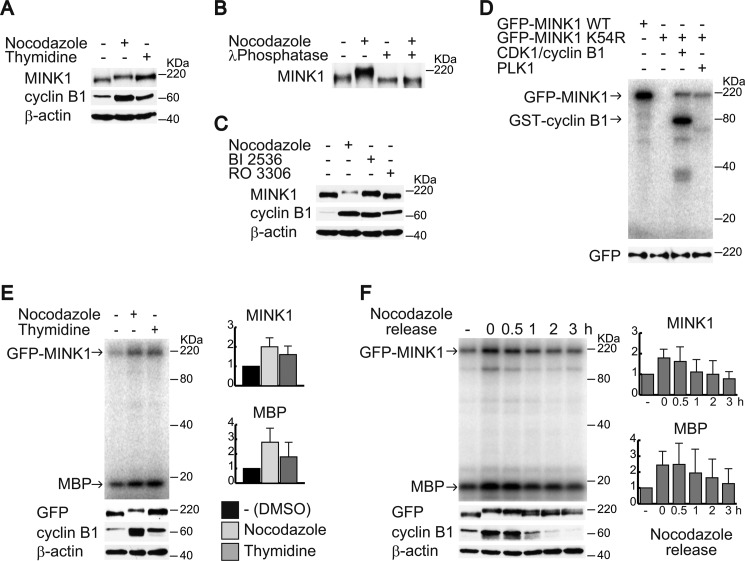
We examined the phosphorylation of MINK1 in mitosis. Cells were synchronized in mitosis by either nocodazole arrest or thymidine release. Cell lysates were then subjected to immunoblot analysis to detect the electrophilic mobility shift of MINK1. As shown in Fig. 3A, the mobility shift of MINK1 was observed in cells synchronized in mitosis. To confirm that the mobility shift is due to phosphorylation, MINK1 was immunoprecipitated from lysates of cells in mitosis and then treated with λ-phosphatase. The phosphatase treatment clearly eliminated the mobility shift of MINK1, indicating that MINK1 is phosphorylated in mitosis (Fig. 3B). To determine the kinase responsible for the MINK1 gel shift, we used inhibitors for CDK1 and PLK1; both kinases are essential for the progression of mitosis, and the addition of these inhibitors arrest cells in the G2/M phase. The mobility shift was eliminated by the addition of the CDK1 inhibitor, but not the PLK1 inhibitor (Fig. 3C). We then performed an in vitro kinase assay to determine whether these kinases can directly phosphorylate MINK1. Catalytically inactive GFP-tagged MINK1 was used as a substrate for the assay to avoid detection of autophosphorylation. As shown in Fig. 3D, both CDK1 and PLK1 could phosphorylate MINK1 in vitro. Although the mobility shift of MINK1 is mediated by CDK1-dependent phosphorylation, PLK1 may also phosphorylate MINK1 in mitosis.
FIGURE 3.
MINK1 is phosphorylated in mitosis. A, HeLa cells were synchronized in mitosis by nocodazole arrest or thymidine release. The cells were lysed, and the lysates were immunoblotted with the indicated antibodies. B, cells were treated or not treated with nocodazole for 16 h, lysed, and MINK1 was immunoprecipitated. The immunoprecipitates were incubated with or without lambda phosphatase and then immunoblotted with anti-MINK1 antibody. C, cells were treated with nocodazole, BI2536 (PLK1 inhibitor), or RO3306 (CDK1 inhibitor), and then the cell lysates were immunoblotted with the indicated antibodies. D, phosphorylation of GFP-MINK1 by PLK1 and CDK1/cyclin B1 was examined by an in vitro kinase assay. Catalytically inactive GFP-MINK1 (K54R) was used as a substrate. An immunoblot of anti-GFP antibody shows equal amounts of substrates were used for the assay. E, HeLa cells that constitutively expressed GFP-MINK1 were synchronized in mitosis by nocodazole arrest or thymidine release. The cells were lysed, immunoprecipitated with anti-GFP antibody, and then subjected to the in vitro kinase assay. A portion of cell lysates (cyclin B1 and β-actin) and immunoprecipitated GFP-MINK1 were subjected to immunoblot with the indicated antibodies. Note that phosphorylated MINK1 in the in vitro kinase assay does not show a mobility shift because a 12.5% acrylamide gel was used. The graphs show relative band intensities of phosphorylated GFP-MINK1 and MBP normalized by the amount of immunoprecipitated GFP-MINK1. The data are represented as mean ± S.D. from three independent experiments. F, HeLa cells that constitutively expressed GFP-MINK1 were synchronized in mitosis by nocodazole and released. The cells were lysed at the indicated time point, and immunoprecipitated GFP-MINK1 was subjected to the in vitro kinase assay. A portion of cell lysates (cyclin B1 and β-actin) and immunoprecipitated GFP-MINK1 were subjected to immunoblot with the indicated antibodies. The graphs show relative band intensities of phosphorylated GFP-MINK1 and MBP normalized by the amount of immunoprecipitated GFP-MINK1. The data are represented as mean ± S.D. from three independent experiments.
We next investigated the catalytic activity of MINK1 during mitosis. To eliminate a possibility that the anti-MINK1 antibody inhibits the catalytic activity of MINK1, we used HeLa cells that constitutively expressed GFP-MINK1 and used anti-GFP antibody for immunoprecipitation. GFP-MINK1 was immunoprecipitated from the lysates of asynchronized cells and synchronized cells in mitosis and subjected to an in vitro kinase assay using myelin basic protein (MBP) as a substrate. Catalytic activity and autophosphorylation of MINK1 was increased in mitosis (Fig. 3E). To further investigate the activity of MINK1 during mitosis, GFP-MINK1 cells were nocodazole arrested and released. Cells at 0, 30, 60, 120, and 180 min after nocodazole release were collected and immunoprecipitated with anti-GFP antibody and subjected to an in vitro kinase assay. Morphological analysis of cells with tubulin and nuclear staining indicated that cells at 30, 60, 120, and 180 after nocodazole release represent cells in metaphase, metaphase-anaphase, telophase-cytokinesis, and G1 phase, respectively (data not shown). The catalytic activity and autophosphorylation of MINK1 started to decrease 1 h after nocodazole release, when the cyclin B1 expression was reduced. In addition, the mobility shift of MINK1 was decreased 1 h after nocodazole release. These results suggest that the catalytic activity of MINK1 in mitosis is associated with phosphorylation.
MINK1 Is a Component of STRIPAK and Directly Associates with STRN4
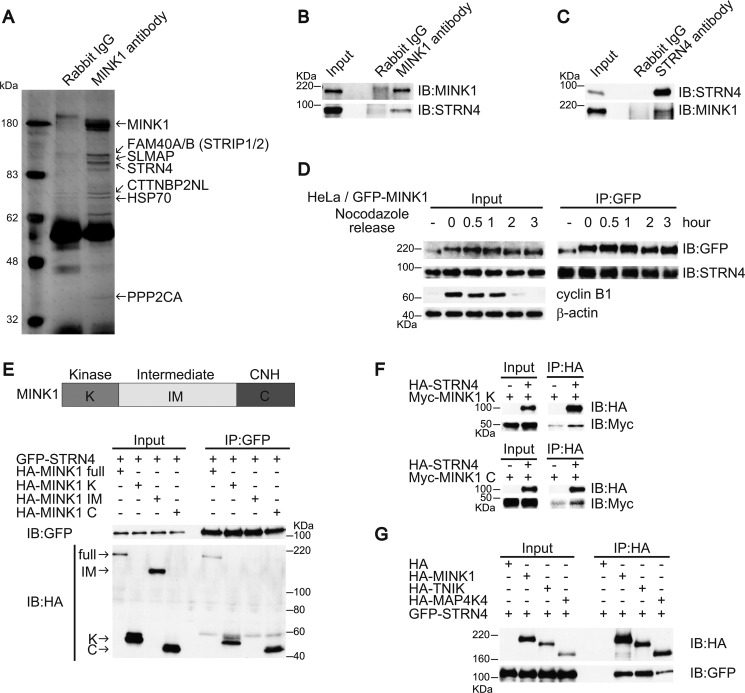
To gain further insight into the role of MINK1 in cytokinesis, we aimed to identify associated proteins. 293T cells were lysed, immunoprecipitated by anti-MINK1 antibody, and separated by SDS-PAGE. After the gel was silver stained, specific bands were excised and identified using mass spectrometry. Interestingly, STRN4 (Zinedin) and PPP2CA, which are regulatory and catalytic subunits of PP2A phosphatase holoenzyme, were found to coprecipitate with MINK1. In addition, FAM40A/B (STRIP1/2), SLMAP, and CTTNBP2NL were in complex with MINK1 (Fig. 4A). The physiological functions of these proteins have not yet been determined, but recent proteomics analysis has demonstrated that these proteins are components of a large protein complex called STRIPAK (30). Regulatory subunits are known to mediate the association of substrates and phosphatase holoenzymes; thus, we examined the interaction of MINK1 and STRN4. A plasmid encoding GFP-STRN4 was transfected into 293T cells with a plasmid encoding either HA-MINK1 or HA alone and then immunoprecipitated with anti-HA antibody. GFP-STRN4 coprecipitated with HA-MINK1 but not with HA (supplemental Fig. S2A). Immunoprecipitation with anti-GFP antibody also revealed the specific coprecipitation of HA-MINK1 and GFP-STRN4 (supplemental Fig. S2B). We next examined the association of endogenous proteins. To immunoprecipitate STRN4, we generated affinity-purified polyclonal antibody using 1–147aa of STRN4 as an antigen. The immunoprecipitation of endogenous MINK1 or STRN4 clearly demonstrated the association of these two proteins (Fig. 4, B and C). The association of these proteins during mitosis was also investigated by immunoprecipitation. GFP-MINK1 HeLa cells were arrested with nocodazole and released after different periods of time. The cells were lysed, immunoprecipitated with anti-GFP antibody, and subjected to an immunoblot to probe for STRN4. As shown in Fig. 4D, both proteins were stably associated during mitosis. MINK1 is composed of an N-terminal catalytic domain, C-terminal citron homology (CNH) domain, and an intermediate region. We investigated which region of MINK1 was essential to bind to STRN4 and found that both catalytic and CNH domains could independently interact with STNR4 (Fig. 4E). To confirm the direct interaction, HA-STRN4 and the kinase or CNH region of MINK1 that was Myc-tagged was produced by in vitro translation, and their interaction was examined. As shown in Fig. 4F, STRN4 directly interacted with both kinase and CNH regions of MINK1. TNIK and MAP4K4 are MINK1 homologs and contain an N-terminal kinase domain and C-terminal CNH domain. We tested if these proteins could also interact with STRN4. Immunoprecipitation analysis demonstrated that not only MINK1 but also TNIK and MAP4K4 could bind to STRN4 (Fig. 4G).
FIGURE 4.
MINK1 associates with STRN4. A, 293T cells were lysed, and the lysates were immunoprecipitated with anti-MINK1 antibody. The immunoprecipitates were separated by SDS-PAGE gel and silver stained. B, 293T cells were lysed, and the lysates were immunoprecipitated with either control or anti-MINK1 antibody; the immunoprecipitates were blotted with anti-MINK1 or anti-STRN4 antibodies. C, 293T cells were lysed, and the lysates were immunoprecipitated with either control or anti-STRN4 antibody; the immunoprecipitates were blotted with anti-MINK1 or anti-STRN4 antibodies. D, GFP-MINK1 HeLa cells were nocodazole arrested and released. The cells were lysed at the indicated time points, and the lysates were immunoprecipitated with anti-GFP antibody; the immunoprecipitates were subjected to immunoblot with the indicated antibodies. E, GFP-STRN4 was transiently expressed in 293T cells with HA-tagged deletion constructs of MINK1. The cells were lysed, and the lysates were immunoprecipitated with anti-GFP antibody; the immunoprecipitates were blotted with anti-HA and anti-GFP antibodies. F, HA-STRN4 and Myc-tagged deletion constructs of MINK1 were translated in vitro. The proteins were mixed and then immunoprecipitated with anti-HA antibody; the immunoprecipitates were immunoblotted with anti-HA and anti-Myc antibodies. G, GFP-STRN4 was transiently expressed in 293T cells with HA-MINK1, HA-TNIK, and HA-MAP4K4. The cells were lysed, and the lysates were immunoprecipitated with anti-HA antibody; the immunoprecipitates were immunoblotted with anti-HA and anti-GFP antibodies.
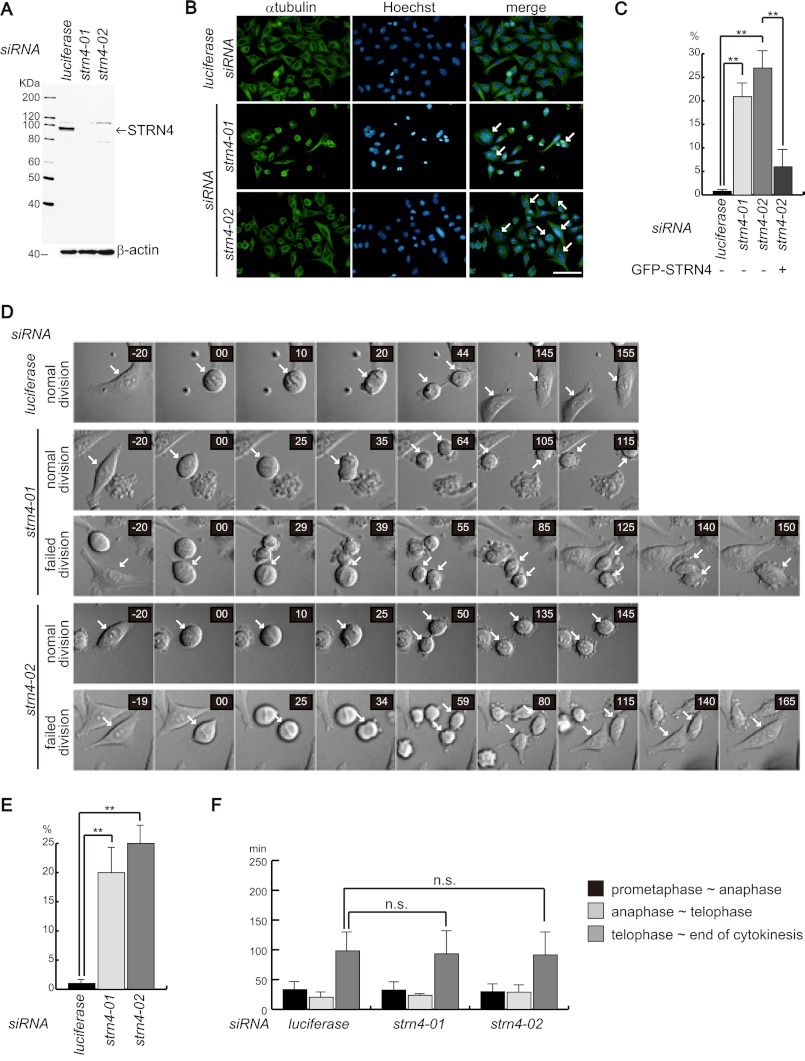
STRN4 Was Required for Abscission
We next determined whether STRN4 was required for the progression of cytokinesis. To deplete STRN4, two siRNAs that efficiently suppressed STNR4 expression were used (Fig. 5A). HeLa cells were transfected with these siRNAs; the cells were fixed 72 h later and stained with anti-α-tubulin antibody and Hoechst. As shown in Fig. 5B, the depletion of STRN4 expression by either siRNA induced multinucleated cells. The quantification of multinucleated cells revealed that more than 20% of the siRNA-transfected cells became multinucleated (Fig. 5C). We also performed rescue experiment. HeLa cells were transfected with expression vector encoding GFP-STRN4 together with STRN4 siRNA (strn4–02) that targeted 3′-UTR of STNR4. Seventy-two hours later, cells were fixed and the ratio of multinucleated cells in GFP-positive cells was evaluated. Exogenous expression of STRN4 reduced the ratio of multinucleated cells induced by STRN4 depletion (Fig. 5C). These results indicate that STRN4 is required for the completion of cytokinesis. We analyzed the subcellular localization of STRN4 during cell division. Similar to MINK1, STRN4 was localized in the cytoplasm at every stage of mitosis, and no specific subcellular localization was observed (supplemental Fig. S3). To further investigate the role of STRN4 in cytokinesis, we observed STRN4-depeleted cells using time-lapse microscopy. STRN4-knockdown cells showed the formation and ingression of the cleavage furrow; however, some cells were unable to abscise and eventually became binucleated cells (Fig. 5D). The examination of these cells revealed that ∼20–25% of STRN4-depleted cells failed to complete abscission (Fig. 5E). We measured the time periods of each stage of cell division in STRN4-knockdown cells. Although MINK1-depleted cells took a longer time to complete cytokinesis, silencing of STRN4 did not affect the length of cytokinesis (Fig. 5F).
FIGURE 5.
STRN4 is required for abscission. A, HeLa cells were transfected with the indicated siRNA, and 72 h later, the cells were lysed and blotted with anti-STRN4 antibody. B, HeLa cells were cultured on glass coverslips and transfected with the indicated siRNAs. Seventy-two hours later, the cells were fixed and immunostained with anti-α-tubulin antibody and Hoechst. The arrows indicate multinucleated cells. C, HeLa cells were treated as in B, and the ratio of multinucleated cells was evaluated. To perform rescue experiment, HeLa cells were transfected with expression plasmid encoding GFP-STRN4 together with STRN4 siRNA (strn4–02) that targeted 3′-UTR of STRN4. Seventy-two hours later, ratio of multinucleated cells in GFP-positive cells were evaluated. Three independent experiments were performed, and 200 cells were evaluated for each experiment (the data are represented as the mean ± S.D.; **, p < 0.01). D, HeLa cells were transfected with control or STRN4 siRNAs; 48 h later, the cells were observed using time-lapse microscopy. Representative images of two types of cell divisions of STRN4 siRNA-transfected cells are shown. normal division indicates cells that completed cytokinesis. failed division indicates cells that were unable to complete cytokinesis. The arrows point dividing cells. E, graph shows the ratio of cells that failed cytokinesis. Forty cells for each group of siRNA-transfected cells were evaluated, and three independent experiments were performed (the data are represented as the mean ± S.D.; **, p < 0.01). F, time period of each stage of mitosis was evaluated. The graph shows the average time period of 30 cells for each group of siRNA-transfected cells (the data are represented as the mean ± S.D.; n.s., not significant).
STRN4 Reduces MINK1 Activity in the Presence of Catalytic and Structural Subunits of PP2A
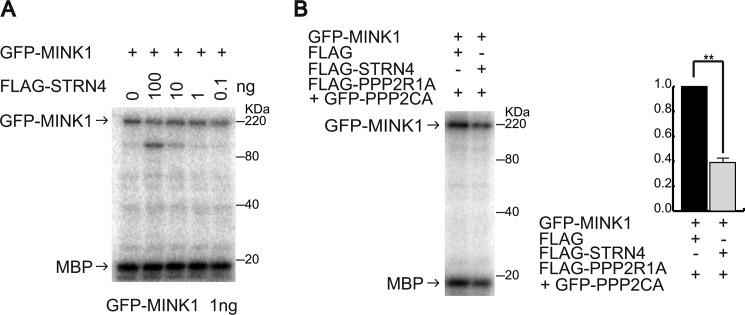
Dephosphorylation by phosphatases can regulate the activity of protein kinases; thus, we tested if STRN4 in complex with PP2A could modulate MINK1 activity. We first evaluated if STRN4 was sufficient to decrease the activity of MINK1 by an in vitro kinase assay. GFP-MINK1 was immunoprecipitated with anti-GFP antibody and subjected to an in vitro kinase assay with varying concentrations of FLAG-STRN4. The amounts of GFP-MINK1 and FLAG-STRN4 were approximated by Western blot using varying concentrations of recombinant proteins of MINK1 and STRN4 (supplemental Fig. S4, A and B). Although the amount of STRN4 we used was ∼100 times the amount GFP-MINK1 used, the activity of MINK1 was not reduced, which indicates that STRN4 is not sufficient to inhibit MINK1 activity (Fig. 6A). PPP2R1A and PPP2CA are structural and catalytic subunits of PP2A, respectively, and PPP2R1A associates with STRN4 (supplemental Fig. S4C). We examined the activity of GFP-MINK1 with STRN4 and different amounts of the PPP2R1A/PPP2CA complex. The PPP2R1A/PPP2CA complex decreased the catalytic activity of GFP-MINK1 in the absence of STRN4, but when GFP-MINK1, STRN4, and the PPP2R1A/PPP2CA complex were incubated at the ratio of 1:10:10, a clear decrease of GFP-MINK1 activity was observed in a STRN4-dependent manner (supplemental Fig. S4D). A STRN4-mediated decrease of MINK1 activity was repeatedly observed under the same conditions (Fig. 6B). These results suggest that STRN4 mediates the inactivation of MINK1 by PP2A.
FIGURE 6.
STRN4 induces a decrease of MINK1 activity in the presence of catalytic and structural subunits of PP2A. A, immunoprecipitated GFP-MINK1 (1 ng) was subjected to an in vitro kinase assay in the presence of different amounts of FLAG-STRN4. B, immunoprecipitated GFP-MINK1 (1 ng) was subjected to an in vitro kinase assay with or without the FLAG-PPP2R1A/GFP-PPP2CA complex (10 ng) in the presence of FLAG-STRN4 (10 ng). The graph shows the relative intensity of phosphorylated MBP from three independent experiments (the data are represented as the mean ± S.D.; **, p < 0.01).
DISCUSSION
Previous studies have demonstrated that mammalian MINK1 is essential for cytoskeletal organization and oncogene-induced cell senescence (27 29). In this study, we described a novel physiological function of MINK1 in mammalian cells. The silencing of MINK1 by two different siRNAs induced multinucleated cells, and exogenous expression of siRNA-resistant MINK1 significantly reduced the ratio of multinucleated cells induced by the siRNA transfection. Using time-lapse microscopy, we observed that all of the MINK1-knockdown cells initiated furrowing, but they displayed an extended time period of abscission. In addition, ∼20% of the cells were unable to complete abscission. These results clearly indicate that MINK1 is required for abscission. In addition, the catalytic activity of MINK1 is required for cytokinesis. Our findings suggest that MINK1 is a potential mitotic kinase that regulates abscission. Recent studies have shown that protein kinases, such as PLK1, Aurora-B, and WNK1, are required to complete abscission (34–36). However, detailed mechanisms of how these kinases regulate abscission are largely unknown. Identification of the physiological substrates of these mitotic kinases, including MINK1, is important to fully understand the regulatory mechanisms of abscission. A recent study has shown that p38 MAP kinase is associated with abscission. p38-mediated phosphorylation induces the interaction of PKC with 14-3-3, which inactivates RhoA to promote abscission in the late stage of cytokinesis (37). MINK1 is known to activate p38 (29); therefore, MINK1-dependent activation of p38 may play a role in the completion of abscission.
Affinity purification of MINK1-associating proteins coupled with mass spectrometry analysis identified that MINK1 is in complex with PP2A holoenzyme. STRN4, a regulatory subunit of PP2A, directly associated with MINK1, and depletion of STRN4-induced multinucleated cells and inhibited abscission. Striatins-associated PP2A has been reported to dephosphorylate and inactivate serine/threonine kinase 24 (STK24), a member of the germinal center kinases that is known to complex with striatins (31). Similarly, we found that STRN4 could reduce the catalytic activity of MINK1 in the presence of structural and catalytic subunits of PP2A. Although it remains unknown whether the STRN4-mediated decrease of MINK1 activity is associated with abscission, recent studies showed inactivation of mitotic kinases in the late stage of cytokinesis is essential for the promotion of abscission (34, 35). For example, inactivation of PLK1 is necessary for the dephosphorylation and proper localization of Cep55 to promote abscission (34). Although further analysis is needed, the decrease of MINK1 activity by PP2A may be required for the dephosphorylation of abscission factors to promote abscission. In addition to modulating MINK1 activity, STRN4 may have some additional roles in the control of abscission. Cells depleted of MINK1 adhered to the surface in the late stage of cytokinesis, but most of the STRN4-knockdown cells were defective in attachment and spreading during abscission (Fig. 5D). The organization of focal adhesions and adhesion to the surface during abscission generate traction forces to physically disconnect two daughter cells. The molecular mechanisms of how focal adhesions are formed to initiate cell adhesion during abscission are not well understood; STRN4 may be involved in these processes.
Recently performed proteomics analysis has revealed that striatins are core components of a large multiprotein complex known as striatin-interacting phosphatase and kinase (STRIPAK) (30). The STRIPAK complex contains catalytic and structural subunits of PP2A, striatins, Mob3, STRIP1/2, and other weakly associating proteins such as CCM3/PDCD10, members of germinal center kinases, CTTNBP2NL, and SLMAP. Some of the STRIPAK proteins are conserved in a wide range of species. In Drosophila melanogaster, the STRIPAK complex associates with Hpo (MST1/2 in mammals), a protein kinase involved in cell proliferation, and inactivates the Hpo-dependent signaling pathway (38). Thus, it seems likely the STRIPAK complex can modulate the cellular functions of associating kinases. It has been reported that MINK1 is involved in the MAP kinase pathway (26), TNIK is required for activation of Wnt target genes (39, 40), and MAP4K4 negatively attenuates mTOR signaling (41). Our findings suggest that the STRIPAK complex is also involved in the regulation of these signaling pathways. It remains unknown whether the STRIPAK complex is involved in the progression of cytokinesis. Large scale proteomics analysis has revealed that a number of STRIPAK components, including striatins, STRIP1/2, and CTTNBP2NL, associate with dynein (42). Dynein is a motor protein that localizes to the mitotic spindle to control the transport of proteins necessary for mitosis (43). Thus, the STRIPAK complex may play some roles in the progression of cell division. Although STRN4 was required for abscission, it remains unknown whether STRN4 needs to be associated with STRIPAK to control abscission. Further detailed analysis is needed to fully realize the cellular functions of this intriguing protein complex.
Supplementary Material
Acknowledgments
We thank the members of the Division of Cancer Biology for helpful discussions. We gratefully acknowledge Ikuyo Mizuguchi and Kentaro Taki (Division for Medical Research Engineering, Nagoya University Graduate School of Medicine) for technical assistance.
This research was supported in part by a grant from the Ministry of Education, Culture, Sports, Science, and Technology of Japan.

This article contains supplemental Figs. S1–S4.
- PLK1
- polo-like kinase
- MINK
- Misshapen-like kinase
- STRIPAK
- striatin-interacting phosphatase and kinase
- PP2A
- protein phosphatase 2A.
REFERENCES
- 1. Salaun P., Rannou Y., Prigent C. (2008) Cdk1, Plks, Auroras, and Neks: the mitotic bodyguards. Adv. Exp. Med. Biol. 617, 41–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Petronczki M., Lénárt P., Peters J. M. (2008) Polo on the rise-from mitotic entry to cytokinesis with Plk1. Dev. Cell. 14, 646–659 [DOI] [PubMed] [Google Scholar]
- 3. van de Weerdt B. C., Medema R. H. (2006) Polo-like kinases: a team in control of the division. Cell Cycle 5, 853–864 [DOI] [PubMed] [Google Scholar]
- 4. Bettencourt-Dias M., Giet R., Sinka R., Mazumdar A., Lock W. G., Balloux F., Zafiropoulos P. J., Yamaguchi S., Winter S., Carthew R. W., Cooper M., Jones D., Frenz L., Glover D. M. (2004) Genome-wide survey of protein kinases required for cell cycle progression. Nature 432, 980–987 [DOI] [PubMed] [Google Scholar]
- 5. Trinkle-Mulcahy L., Lamond A. I. (2006) Mitotic phosphatases: no longer silent partners. Curr. Opin. Cell Biol. 18, 623–631 [DOI] [PubMed] [Google Scholar]
- 6. De Wulf P., Montani F., Visintin R. (2009) Protein phosphatases take the mitotic stage. Curr. Opin. Cell Biol. 21, 806–815 [DOI] [PubMed] [Google Scholar]
- 7. Barr F. A., Elliott P. R., Gruneberg U. (2011) Protein phosphatases and the regulation of mitosis. J. Cell Sci. 124, 2323–2334 [DOI] [PubMed] [Google Scholar]
- 8. Eichhorn P. J., Creyghton M. P., Bernards R. (2009) Protein phosphatase 2A regulatory subunits and cancer. Biochim. Biophys. Acta 1795, 1–15 [DOI] [PubMed] [Google Scholar]
- 9. Virshup D. M., Shenolikar S. (2009) From promiscuity to precision: protein phosphatases get a makeover. Mol. Cell 33, 537–545 [DOI] [PubMed] [Google Scholar]
- 10. Benoist M., Gaillard S., Castets F. (2006) The striatin family: a new signaling platform in dendritic spines. J. Physiol. 99, 146–153 [DOI] [PubMed] [Google Scholar]
- 11. Moreno C. S., Park S., Nelson K., Ashby D., Hubalek F., Lane W. S., Pallas D. C. (2000) WD40 repeat proteins striatin and S/G(2) nuclear autoantigen are members of a novel family of calmodulin-binding proteins that associate with protein phosphatase 2A. J. Biol. Chem. 275, 5257–5263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Moreno C. S., Lane W. S., Pallas D. C. (2001) A mammalian homolog of yeast MOB1 is both a member and a putative substrate of striatin family-protein phosphatase 2A complexes. J. Biol. Chem. 276, 24253–24260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Gaillard S., Bartoli M., Castets F., Monneron A. (2001) Striatin, a calmodulin-dependent scaffolding protein, directly binds caveolin-1. FEBS Lett. 508, 49–52 [DOI] [PubMed] [Google Scholar]
- 14. Lu Q., Pallas D. C., Surks H. K., Baur W. E., Mendelsohn M. E., Karas R. H. (2004) Striatin assembles a membrane signaling complex necessary for rapid, nongenomic activation of endothelial NO synthase by estrogen receptor α. Proc. Natl. Acad. Sci. U.S.A. 101, 17126–17131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Gaillard S., Bailly Y., Benoist M., Rakitina T., Kessler J. P., Fronzaroli-Molinières L., Dargent B., Castets F. (2006) Targeting of proteins of the striatin family to dendritic spines: role of the coiled-coil domain. Traffic 7, 74–84 [DOI] [PubMed] [Google Scholar]
- 16. Wurzenberger C., Gerlich D. W. (2011) Phosphatases: providing safe passage through mitotic exit. Nat. Rev. Mol. Cell Biol. 12, 469–482 [DOI] [PubMed] [Google Scholar]
- 17. Schmitz M. H., Held M., Janssens V., Hutchins J. R., Hudecz O., Ivanova E., Goris J., Trinkle-Mulcahy L., Lamond A. I., Poser I., Hyman A. A., Mechtler K., Peters J. M., Gerlich D. W. (2010) Live-cell imaging RNAi screen identifies PP2A-B55alpha and importin-beta1 as key mitotic exit regulators in human cells. Nat. Cell Biol. 12, 886–893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Chen F., Archambault V., Kar A., Lio' P., D'Avino P. P., Sinka R., Lilley K., Laue E. D., Deak P., Capalbo L., Glover D. M. (2007) Multiple protein phosphatases are required for mitosis in Drosophila. Curr. Biol. 17, 293–303 [DOI] [PubMed] [Google Scholar]
- 19. Dan I., Watanabe N. M., Kusumi A. (2001) The Ste20 group kinases as regulators of MAP kinase cascades. Trends. Cell Biol. 11, 220–230 [DOI] [PubMed] [Google Scholar]
- 20. Liu H., Su Y. C., Becker E., Treisman J., Skolnik E. Y. (1999) A Drosophila TNF-receptor-associated factor (TRAF) binds the ste20 kinase Misshapen and activates Jun kinase. Curr. Biol. 9, 101–104 [DOI] [PubMed] [Google Scholar]
- 21. Ruan W., Long H., Vuong D. H., Rao Y. (2002) Bifocal is a downstream target of the Ste20-like serine/threonine kinase misshapen in regulating photoreceptor growth cone targeting in Drosophila. Neuron 36, 831–842 [DOI] [PubMed] [Google Scholar]
- 22. Su Y. C., Maurel-Zaffran C., Treisman J. E., Skolnik E. Y. (2000) The Ste20 kinase misshapen regulates both photoreceptor axon targeting and dorsal closure, acting downstream of distinct signals. Mol. Cell. Biol. 20, 4736–4744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Cobreros-Reguera L., Fernández-Miñán A., Fernández-Espartero C. H., López-Schier H., González-Reyes A., Martín-Bermudo M. D. (2010) The Ste20 kinase misshapen is essential for the invasive behaviour of ovarian epithelial cells in Drosophila. EMBO Rep. 11, 943–949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Daulat A. M., Luu O., Sing A., Zhang L., Wrana J. L., McNeill H., Winklbauer R., Angers S. (2012) Mink1 regulates β-catenin-independent Wnt signaling via Prickle phosphorylation. Mol. Cell. Biol. 32, 173–185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kaneko S., Chen X., Lu P., Yao X., Wright T. G., Rajurkar M., Kariya K., Mao J., Ip Y. T., Xu L. (2011) Smad inhibition by the Ste20 kinase Misshapen. Proc. Natl. Acad. Sci. U.S.A. 108, 11127–11132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Dan I., Watanabe N. M., Kobayashi T., Yamashita-Suzuki K., Fukagaya Y., Kajikawa E., Kimura W. K., Nakashima T. M., Matsumoto K., Ninomiya-Tsuji J., Kusumi A. (2000) Molecular cloning of MINK, a novel member of mammalian GCK family kinases, which is up-regulated during postnatal mouse cerebral development. FEBS Lett. 469, 19–23 [DOI] [PubMed] [Google Scholar]
- 27. Hu Y., Leo C., Yu S., Huang B. C., Wang H., Shen M., Luo Y., Daniel-Issakani S., Payan D. G., Xu X. (2004) Identification and functional characterization of a novel human misshapen/Nck interacting kinase-related kinase, hMINK β. J. Biol. Chem. 279, 54387–54397 [DOI] [PubMed] [Google Scholar]
- 28. Nonaka H., Takei K., Umikawa M., Oshiro M., Kuninaka K., Bayarjargal M., Asato T., Yamashiro Y., Uechi Y., Endo S., Suzuki T., Kariya K. (2008) MINK is a Rap2 effector for phosphorylation of the postsynaptic scaffold protein TANC1. Biochem. Biophys. Res. Commun. 377, 573–578 [DOI] [PubMed] [Google Scholar]
- 29. Nicke B., Bastien J., Khanna S. J., Warne P. H., Cowling V., Cook S. J., Peters G., Delpuech O., Schulze A., Berns K., Mullenders J., Beijersbergen R. L., Bernards R., Ganesan T. S., Downward J., Hancock D. C. (2005) Involvement of MINK, a Ste20 family kinase, in Ras oncogene-induced growth arrest in human ovarian surface epithelial cells. Mol. Cell. 20, 673–685 [DOI] [PubMed] [Google Scholar]
- 30. Goudreault M., D'Ambrosio L. M., Kean M. J., Mullin M. J., Larsen B. G., Sanchez A., Chaudhry S., Chen G. I., Sicheri F., Nesvizhskii A. I., Aebersold R., Raught B., Gingras A. C. (2009) A PP2A phosphatase high density interaction network identifies a novel striatin-interacting phosphatase and kinase complex linked to the cerebral cavernous malformation 3 (CCM3) protein. Mol. Cell Proteomics 8, 157–171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Gordon J., Hwang J., Carrier K. J., Jones C. A., Kern Q. L., Moreno C. S., Karas R. H., Pallas D. C. (2011) Protein phosphatase 2a (PP2A) binds within the oligomerization domain of striatin and regulates the phosphorylation and activation of the mammalian Ste20-Like kinase Mst3. BMC Biochem. 12, 54–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kean M. J., Ceccarelli D. F., Goudreault M., Sanches M., Tate S., Larsen B., Gibson L. C., Derry W. B., Scott I. C., Pelletier L., Baillie G. S., Sicheri F., Gingras A. C. (2011) Structure-function analysis of core STRIPAK Proteins: a signaling complex implicated in Golgi polarization. J. Biol. Chem. 286, 25065–25075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Dephoure N., Zhou C., Villén J., Beausoleil S. A., Bakalarski C. E., Elledge S. J., Gygi S. P. (2008) A quantitative atlas of mitotic phosphorylation. Proc. Natl. Acad. Sci. U.S.A. 105, 10762–10767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Bastos R. N., Barr F. A. (2010) Plk1 negatively regulates Cep55 recruitment to the midbody to ensure orderly abscission. J. Cell Biol. 191, 751–760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Steigemann P., Wurzenberger C., Schmitz M. H., Held M., Guizetti J., Maar S., Gerlich D. W. (2009) Aurora B-mediated abscission checkpoint protects against tetraploidization. Cell 136, 473–484 [DOI] [PubMed] [Google Scholar]
- 36. Tu S. W., Bugde A., Luby-Phelps K., Cobb M. H. (2011) WNK1 is required for mitosis and abscission. Proc. Natl. Acad. Sci. U.S.A. 108 1385–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Saurin A. T., Durgan J., Cameron A. J., Faisal A., Marber M. S., Parker P. J. (2008) The regulated assembly of a PKCepsilon complex controls the completion of cytokinesis. Nat. Cell Biol. 10, 891–901 [DOI] [PubMed] [Google Scholar]
- 38. Ribeiro P. S., Josué F., Wepf A., Wehr M. C., Rinner O., Kelly G., Tapon N., Gstaiger M. (2010) Combined functional genomic and proteomic approaches identify a PP2A complex as a negative regulator of Hippo signaling. Mol. Cell. 39, 521–534 [DOI] [PubMed] [Google Scholar]
- 39. Mahmoudi T., Li V. S., Ng S. S., Taouatas N., Vries R. G., Mohammed S., Heck A. J., Clevers H. (2009) The kinase TNIK is an essential activator of Wnt target genes. EMBO J. 28, 3329–3340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Shitashige M., Satow R., Jigami T., Aoki K., Honda K., Shibata T., Ono M., Hirohashi S., Yamada T. (2010) Traf2- and Nck-interacting kinase is essential for Wnt signaling and colorectal cancer growth. Cancer Res. 70, 5024–5033 [DOI] [PubMed] [Google Scholar]
- 41. Guntur K. V., Guilherme A., Xue L., Chawla A., Czech M. P. (2010) Map4k4 negatively regulates peroxisome proliferator-activated receptor (PPAR) γ protein translation by suppressing the mammalian target of rapamycin (mTOR) signaling pathway in cultured adipocytes. J. Biol. Chem. 285, 6595–6603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Hutchins J. R., Toyoda Y., Hegemann B., Poser I., Hériché J. K., Sykora M. M., Augsburg M., Hudecz O., Buschhorn B. A., Bulkescher J., Conrad C., Comartin D., Schleiffer A., Sarov M., Pozniakovsky A., Slabicki M. M., Schloissnig S., Steinmacher I., Leuschner M., Ssykor A., Lawo S., Pelletier L., Stark H., Nasmyth K., Ellenberg J., Durbin R., Buchholz F., Mechtler K., Hyman A. A., Peters J. M. (2010) Systematic analysis of human protein complexes identifies chromosome segregation proteins. Science 328, 593–599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Karki S., Holzbaur E. L. (1999) Cytoplasmic dynein and dynactin in cell division and intracellular transport. Curr. Opin. Cell Biol. 11, 45–53 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.