Background: Considerable interest surrounds how 8-oxoguanine DNA glycosylase (hOGG1) distinguishes rare oxoG lesions from undamaged G residues.
Results: Even when G is forcibly inserted into the lesion-recognition pocket on the enzyme, it is not cleaved.
Conclusion: The hOGG1 active site can discriminate G from oxoG at the stage of catalysis.
Significance: HOGG1 has a catalytic checkpoint that prevents accidental cleavage of undamaged DNA.
Keywords: Base Excision Repair, DNA Repair, Enzyme Catalysis, Molecular Dynamics, X-ray Crystallography, 8-Oxoguanine, DNA Glycosylase
Abstract
A poorly understood aspect of DNA repair proteins is their ability to identify exceedingly rare sites of damage embedded in a large excess of nearly identical undamaged DNA, while catalyzing repair only at the damaged sites. Progress toward understanding this problem has been made by comparing the structures and biochemical behavior of these enzymes when they are presented with either a target lesion or a corresponding undamaged nucleobase. Trapping and analyzing such DNA-protein complexes is particularly difficult in the case of base extrusion DNA repair proteins because of the complexity of the repair reaction, which involves extrusion of the target base from DNA followed by its insertion into the active site where glycosidic bond cleavage is catalyzed. Here we report the structure of a human 8-oxoguanine (oxoG) DNA glycosylase, hOGG1, in which a normal guanine from DNA has been forcibly inserted into the enzyme active site. Although the interactions of the nucleobase with the active site are only subtly different for G versus oxoG, hOGG1 fails to catalyze excision of the normal nucleobase. This study demonstrates that even if hOGG1 mistakenly inserts a normal base into its active site, the enzyme can still reject it on the basis of catalytic incompatibility.
Introduction
Reactive oxygen species, produced primarily as by-products of oxidative phosphorylation, represent a chronic threat to the integrity and vitality of cells (1). These powerful electrophiles react with many components of the cell, but the consequences are particularly dangerous when DNA is the target, as the resulting oxidative damage fundamentally alters the covalent structure and therefore the information content of DNA, thus giving rise to mutations. The predominant nucleobase lesion arising from DNA oxidation, 8-oxoguanine (oxoG),2 is among the most mutagenic of all known endogenous nucleobase lesions (2, 3). Despite the fact that the oxoG lesion differs by only two atoms from its progenitor G (Fig. 1A) and bears no alteration in its Watson-Crick base pairing functionality, oxoG mispairs during DNA replication with A instead of C to initiate the transversional mutation of G:C pairs to T:A (2, 4, 5). This particular type of mutation is the second most prevalent mutation, after G:C to A:T, observed in human cancers (6). In human cells, repair of oxoG is initiated mainly by human 8-oxoguanine glycosylase 1 (hOGG1), which catalyzes excision of the oxidatively damaged nucleobase and degradation of its sugar moiety, leading to DNA strand nicks that are substrates for the restorative components of the base excision DNA repair machinery (7–9).
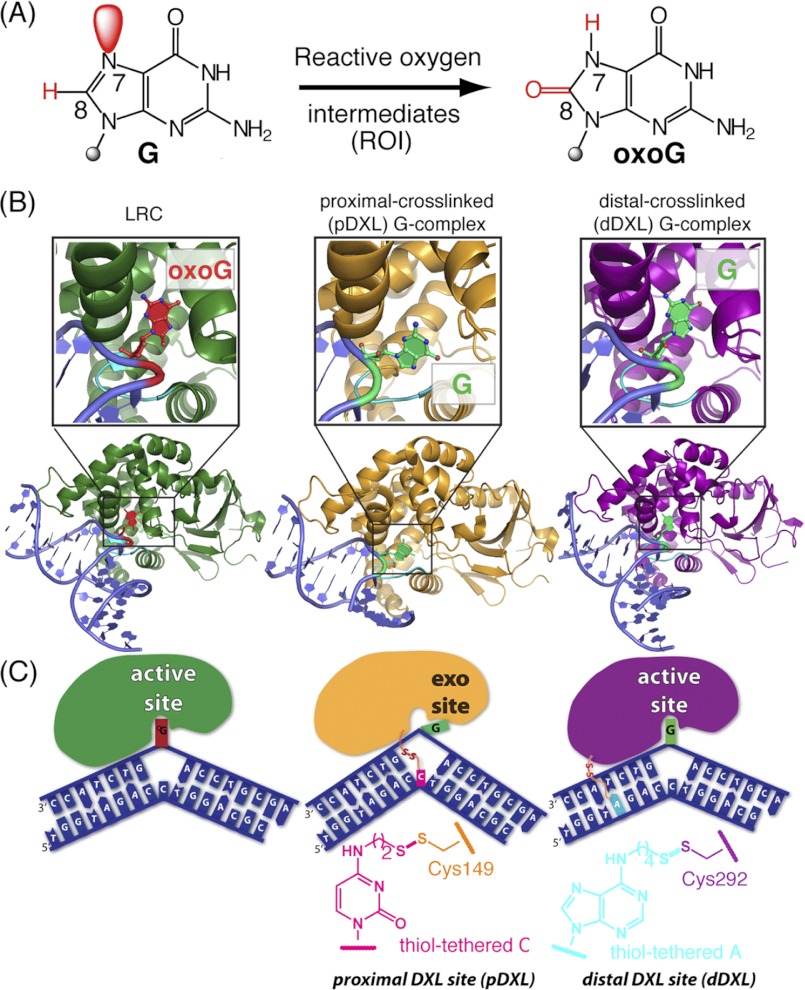
FIGURE 1.
Overview of hOGG1-DNA complexes and disulfide cross-linking strategies. A, chemical structures of guanine and 8-oxoguanine, with the constitutional differences at the 7 and 8 positions highlighted in red. B, schematic representations of: left, a lesion recognition complex (LRC, 1EBM (10)) obtained using a recognition-competent but catalytically inactive mutant of hOGG1 bound to DNA, with oxoG (red) fully inserted into the active site lesion-recognition pocket; middle, the proximally disulfide cross-linked (pDXL) G-complex (orange, 1YQR (34)), with the target G (green) in the exo-site; and right, distally dDXL G-complex (3IH7, present work) of hOGG1 bound to normal DNA, with the target G (green) in the active site. The DNA interrogation loop containing the conserved NNN motif is colored cyan. C, schematic representations of the structures in B. In the pDXL and dDXL structures, the protein is wild-type, except for Cys residues engineered for purposes of cross-linking (N149C and S292C, respectively). Chemical structures of the tethered nucleobases employed in cross-linking are shown.
In previous studies, we determined that oxoG excision by hOGG1 proceeds via a covalent catalysis mechanism in which the oxoG is subject to nucleophilic displacement by the ϵ-amino group of Lys-249 (10, 11). Mutation of this residue to the non-nucleophilic Gln generates a variant of the enzyme (K249Q hOGG1) that lacks catalytic activity but retains the ability to recognize oxoG-containing DNA specifically. Mutation of an active site aspartate residue, Asp-268, which is highly conserved among members of the protein superfamily to which hOGG1 belongs (12), similarly generates a catalytically incompetent but recognition-competent version of the protein (D268N hOGG1). The ability to trap otherwise transient lesion-recognition complexes (LRC) using these mutations has enabled the determination of high-resolution x-ray structures that have illuminated many important features of structure and function in this system (10, 13). In these LRC structures (Fig. 1B, left) the DNA is sharply bent at the site of the oxoG lesion, which is completely extruded from the helical stack and is inserted deeply into an extrahelical lesion-recognition pocket on the enzyme, where it is poised for catalysis of base excision. Amino acid residues lining this pocket make numerous specific contacts with the oxoG, and additional residues contact the DNA backbone and invade the duplex in and around the site vacated by oxoG. Although these structures have shed a great deal of light on recognition and cleavage of the oxoG lesion by hOGG1, they have left open the important question of how hOGG1 overcomes one of the most formidable needle in a haystack challenges in biology: that of efficiently locating and identifying one oxoG residue embedded in DNA containing, on average, a greater than million-fold excess of G, with only two atoms available to differentiate the pernicious oxoG from its innocuous G relative. This question is made all the more difficult by the fact that oxoG causes little if any structural distortion of DNA (14, 15) and has only modest effects on duplex stability and dynamics (16–20). Key to an understanding of lesion searching is to probe in depth how hOGG1 interrogates non-lesion-containing DNA while patrolling for lesions.
When proteins interact with DNA lacking any specific recognition feature, the two macromolecules form large ensembles of rapidly interconverting complexes, and this inherent heterogeneity has hampered attempts to characterize nonspecific protein-DNA interactions at atomic resolution (21–32). We have overcome this problem by developing a straightforward means by which to drastically limit the region a protein can explore while diffusing along DNA, through the introduction of an intermolecular disulfide cross-link (DXL) between protein and DNA. This modification has enabled the crystallization and structure determination of multiple protein-DNA complexes (33–44).
Of greatest relevance to the present work is our previous use of DXL technology to trap hOGG1 in the act of attempting to present an undamaged base, G, to the enzyme active site (34). In this previous example, we attached the disulfide-bearing tether to the cytosine (C) opposite the target G residue (the “estranged” C), such that extrusion of the target G would be required for DXL formation, and DXL formation would in turn prevent return of the target G to its intrahelical, base-paired state. The structure of this complex revealed that the target G was indeed extrahelical, however, instead of being inserted into the enzyme active site, it resided in an “exo-site,” a patch of protein surface adjacent to the enzyme active site (Fig. 1B, middle). This exo-site G-complex and an LRC were used as starting points for alchemical free energy simulations (34). They suggested that the rejection of G from the active site, and the acceptance of oxoG, resulted from repulsive interactions between G and active site functionality, which were replaced by attractive interactions in the case of oxoG.
As described above, stalling the hOGG1 extrusion process with G occupying the exo-site was made possible by attachment of the DXL proximal to the lesion, at a site designed to enforce extrahelicality of the target G (proximal DXL site, hereafter designated pDXL). We envisioned it might be possible to trap hOGG1 interrogating an intact, intrahelical target G:C base pair, as has been done for the bacterial counterpart MutM (38), by shifting the tether to a site more distal to the lesion, one that did not directly enforce disruption of the target base pair; for this purpose, we selected an adenine (A) residue 4 base pairs removed from the target G:C (distal DXL site, hereafter dDXL) (Fig. 1C). We have previously reported the use of the dDXL site to study oxoG insertion into a hOGG1 active site mutationally encumbered by the presence of a bulky side chain (43).
Here we report the x-ray structure of hOGG1 bound to non-lesion-containing DNA, cross-linked at the distal site (Fig. 1B, right). This structure reveals that the target G:C base pair is disrupted, with the target guanine extruded from the DNA base stack and inserted almost completely into the extrahelical active site pocket of the enzyme. Our molecular dynamics (MD) simulations hint that the favorable insertion of G into the enzyme active site results from dDXL-associated suppression of structural fluctuations around the active site. This structure provides the first direct view of hOGG1 interrogating an undamaged nucleobase in a fully wild-type active site. Remarkably, the position of guanine overlays almost exactly with that of oxoG in the LRCs. Moreover, despite the high conformational similarity between this complex and the LRC, and although the protein in this structure bears a fully functional active site whose catalytic power is not compromised by distal cross-linking, the target G in this complex remains uncleaved during a period of weeks. These findings suggest that insertion of a nucleobase into the hOGG1 active site is necessary but not sufficient for catalysis of base excision, implying a checkpoint mechanism that discriminates a cognate oxoG lesion from the far more prevalent G on the basis of catalytic competence.
MATERIALS AND METHODS
Cross-linked Complex Formation and Crystallization
A fragment of hOGG1 (amino acids 12–327) bearing the S292C mutation (43) was expressed in Escherichia coli BL21(DE3)pLysS cells and purified essentially as previously described (13, 34), except nickel-nitrilotriacetic acid-agarose resin (Qiagen) was substituted for Talon resin. DNA oligomers 5′-ACGTCCAGGTCTACC-3′ and 5′-TGGTXGACCTGGACG-3′ (in which X denotes O6-phenyldeoxyinosine (ΦdI, Glen Research, Sterling, VA) were synthesized on the Expedite 8900 using standard solid phase synthesis methods and purified by denaturing PAGE. The thiol-bearing tether was installed by treatment of the ΦdI-containing oligonucleotide with (H2N-CH2-CH2-CH2-CH2-S-)2 as described (43). Single-stranded thiol-tethered oligonucleotides were incubated at 10 μm with 20 μm hOGG1 S292C (final reaction concentrations) in 10 mm Tris-HCl, pH 7.5, 100 mm NaCl, 1 mm EDTA for at least 18 h before 11 μm of the complementary DNA strand was added and the duplex allowed to anneal on ice for at least 40 min. The cross-linked double-stranded complex was purified by anion exchange chromatography (MonoQ or HiTrapQ, GE Healthcare) with a 35-ml gradient from 180–600 mm NaCl. Fractions were analyzed by nonreducing SDS-PAGE, combined, buffer exchanged to crystallization buffer (10 mm Tris, pH 7.5, 50 mm NaCl, 1 mm EDTA), and concentrated to 230–300 μm. A small number of crystals yielding x-ray diffraction of reasonable quality were obtained using an OptiSalts screen (Qiagen) after extensive efforts. These were collected in a nylon loop (Hampton Research), cryoprotected by brief (<10 s) dipping in 25% glycerol in reservoir solution, and flash frozen in liquid nitrogen. Only one of these crystals yielded diffraction of sufficient quality for structure determination. This crystal was obtained using hanging drop vapor diffusion by equilibrating 1 μl of 300 μm protein-DNA complex and 1 μl of reservoir solution against reservoir solution containing 63 mm magnesium acetate, 12.6% PEG-8K, 90 mm sodium cacodylate, 10 mm sodium acetate, pH 4.6, 60 mm sodium fluoride.
Structure Determination
X-ray diffraction data were collected under cryogenic conditions on beamline 24-ID at the Advanced Photon Source, Argonne National Laboratory. These data were integrated with HKL2000 and merged and scaled using SCALEPACK (45). The crystals have one protein-DNA complex in the asymmetric unit and belong to the hexagonal space group P6522. The protein component alone from a hOGG1-oxoG DNA complex (PDB code 2NOH (43)) was used to calculate the initial phases for structural refinement. After rigid body refinement followed by simulated annealing and B-factor refinement, the partial model was used to calculate σA-weighted (46) 2Fo − Fc and difference maps in CNS (47). The presence of strong electron density for the omitted DNA nucleotides guided their inclusion in subsequent models using Coot (48), followed by refinement with CNS and PHENIX (49). Crystallographic statistics for the distally cross-linked G-complex of hOGG1 are presented in Table 1. All depictions of the structure were created with PyMol (50).
TABLE 1.
Data collection and refinement statistics
The hOGG1 dDXL G-complex was space group, P6522; cell parameters, a = b = 90.91 Å, c = 211.63 Å, α = β = 90.0°, and γ = 120°.
| Data collectiona | |
| Source | APS 24ID |
| Wavelength (Å) | 0.97921 |
| Resolution (Å) | 50.0-3.1 (3.21-3.1) |
| Rsym (%)a | 11.3 (54.5) |
| Total No. of obs. | 131,064 |
| No. of unique obs. | 10,056 (974) |
| Completeness (%) | 100.0 (99.7) |
| 〈I〉/σ〈I〉 | 21.2 (2.6) |
| Refinement | |
| Resolution (Å) | 32.19-3.1 |
| No. reflections | 9,362 |
| No. of non-hydrogen atoms | 3023 |
| Rwork (%)b | 22.84 |
| Rfree (%)c | 26.73 |
| Root mean square deviation bond (Å)d | 0.002 |
| Root mean square deviation angle (°)d | 0.579 |
| Ramachandran plot analysise (% favored, allowed, outlier) | 95.1, 4.5, 0.3 |
| Average B-factors | Protein, 67.5; DNA, 130.1 |
Catalytic Activity
Human OGG1 S292C was incubated at 4 °C overnight with oligonucleotide 1 (2:1 at 20 μm protein) 5′-TGGTAGACCTGGACGC, where A is the convertible adenine nucleoside (51) (Glen Research) functionalized with a four-carbon mixed disulfide tether. The single-stranded complex was purified by MonoQ (GE Healthcare), buffer exchanged, and diluted to 10 μm in buffer containing 10 mm Tris-HCl, pH 7.5, 100 mm sodium chloride, 1 mm EDTA. The 10-μl 5 μm DNA cleavage reaction was initiated by addition of 1 molar eq of complementary 5′-32P-labeled single-stranded DNA containing either a G or an oxoG at the target position. For comparison, oxoG-containing duplex DNA was added in a 1:1 molar ratio to hOGG1 S292C for a 10-μl reaction at 5 μm. Aliquots of 0.5 μl were removed from the reaction at regular intervals, quenched with 25 μl of stop solution containing 0.5 m piperidine, 90% formamide, 1× TBE (100 mm Tris-HCl, 90 mm boric acid, and 1 mm EDTA) and heated at 95 °C for 30 min. Approximately 10 pmol of the DNA were resolved by 20% urea-PAGE, run at 325 volts for 1.5–2 h with 1× TBE as the running buffer. Gels were exposed to a storage phosphorimaging plate (Fuji BAS 1000) and the plates were scanned (Typhoon, GE) at maximum sensitivity to visualize the bands. Bands were quantified using ImageQuant software (GE Healthcare) and the percent cleavage at each time point was determined by dividing the intensity of the cleavage product(s) by the total intensity of DNA in each lane.
System Preparation for hOGG1 LRC Structures Used for Molecular Dynamics Simulations
The crystal structure of the hOGG1-DNA complex with an oxoG bound in the active site (PDB code 1YQR (34)) was used to build the structure presented in Fig. 2. To obtain a wild-type structure, Gln-249 was changed to Lys by removing side chain oxygen and nitrogen atoms and introducing terminal amine and methylene groups. Cys-149 was also modeled back to the wild-type Asn residue. For the systems with G, we removed the O8 atom from oxoG. Then, the coordinates of missing atoms, including hydrogen atoms, were generated based on the CHARMM force field parameters (52). Protonation states for all ionizable residues were chosen corresponding to pH 7, except the protonation states of His residues, which were deduced on the basis of possible hydrogen bonding interactions. The active site Cys-253 was deprotonated based on the predictions of previous alchemical free energy simulations (34). The resulting protein and DNA duplex was neutralized by placing 24 Na+ ions 4.5 Å away from the phosphorus atom along the line passing through the phosphorus atom and the midpoint of the two nonbridging oxygens. All ordered waters from the crystal structure were included and the resulting systems were further solvated with a rhombic dodecahedron box of 11,712 water molecules, followed by the removal of water molecules that were within 2.5 Å of any nonhydrogen atoms of the protein, nucleic acid, Na+ ions, and crystal waters. The final model contains 35,174 atoms: 5,864 protein and DNA atoms, 24 Na+ ions, and 9,762 water molecules.
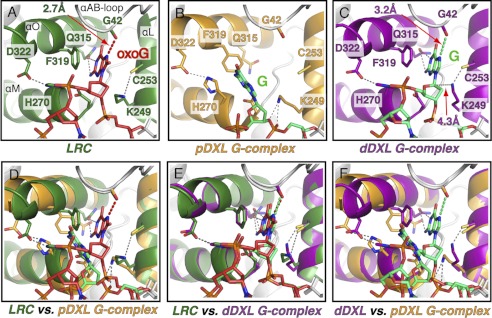
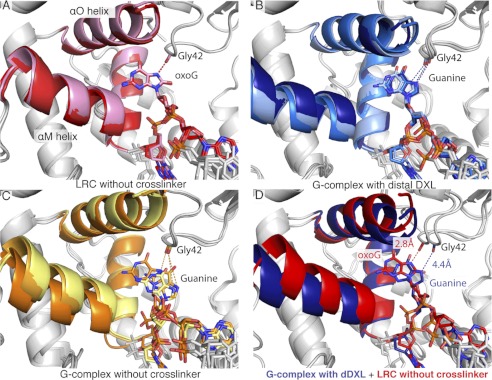
FIGURE 2.
Interaction of the hOGG1 active site with target nucleobases. Close-up views of the active site region in a hOGG1 LRC (green, A, see below for reference), the pDXL G-complex (orange, B, 1YQK, (34)), and the dDXL G-complex (purple, C, 3IH7, this work). In all views, the proteins are shown as schematics with the side chains of relevant active site residues displayed as stick models and colored by atom. The DNA is also shown as a stick model with oxoG colored red and G shown in green. Important interactions between the active site residues and the DNA are shown by dotted lines. For those structures in which the target base is inserted into the active site, the distance from Gly-42 O to the N7 of the target base is highlighted (distance indicated by a red arrow in A and C). Movement of the αO-helix upon insertion of guanine in the active site is highlighted in the overlays of the pDXL G-complex with either the LRC (D) or the dDXL G-complex (F). Several side chains labeled in the figure similarly shift upon G or oxoG insertion into the active site and stabilize the base. Structural alignment of the LRC with the dDXL G-complex (E) reveals the overall similarity between these two complexes (protein all atom alignment, root mean square deviation = 0.5 Å, Pymol (50)). Overlay of the LRC with the dDXL G-complex (E) illustrates the similarities in both the position of the inserted base and the active site residues that recognize the bases. The LRC structure used here is based on the structure of K249Q hOGG1 bound to oxoG-containing DNA (10), but energy minimized after the active site nucleophile, Lys-249, was restored in silico (details in ”Materials and Methods“).
Energy Minimization and Molecular Dynamics Simulations
Following the preparation of each structure, a total of 5000 steps of energy minimizations were performed with a series of decreasing constraints and restraints, by using conjugate gradient energy minimizations and Adopted Basis Newton-Raphson energy minimizations. Each system was then equilibrated at constant temperature and pressure for 0.91 ns, followed by MD simulations at constant volume and temperature for 30 ns. During the MD simulations, coordinates were saved at 1-ps intervals and later used to determine the average structures. The leapfrog Verlet algorithm (53) was used in the simulation with a 2-fs integration time step, with SHAKE being used for bonds involving hydrogens (54). The temperature was maintained at 298 K by coupling the system to an external thermal bath (55). The volume of the system was held constant with a rhombic dodecahedron lattice length parameter of 78.9 Å. We used the CHARMM27 force fields (52, 56) to represent the protein, ions, and nucleic acid, and the TIP3P model (57) to represent water molecules. For the protein backbone dihedral angles, the CMAP correction was applied (58). The force field parameters for oxoG were the same as presented previously (37). For the electrostatics, the particle mesh Ewald summation method (59) was used with the Ewald κ value of 0.340 Å−1 and the approximate grid spacing of 1.0 Å. The real space terms in the particle mesh Ewald summation method were evaluated with a cutoff distance of 9.0 Å. The van der Waals interactions were evaluated with the same cutoff distance, but were smoothly turned off at the cutoff distance (with a shift function). All calculations were carried out with the CHARMM program (version c33a2) (60).
RESULTS
Experimental Strategy
As previously described, the dDXL configuration employs a 4-carbon tether attached to an A residue located 4 base pairs away from the target G:C pair in the DNA, and this undergoes cross-linking to an engineered cysteine at position 292 in hOGG1 (Fig. 1C, right). Relocation of the DXL from the proximal (34) to the distal site (this work) significantly altered crystallization behavior, making it necessary to re-screen various DNA sequences and crystallization conditions (“Materials and Methods”) to obtain crystals of the dDXL complex. Cross-linking at the distal site also gave poorer yields of product than at the proximal site, but we found that the yield could be improved by resorting to a two-step procedure. The single DNA strand containing the thiol tether at the distal site was first cross-linked to the Cys-engineered hOGG1 protein, and then the complementary strand was annealed to this covalent hOGG1-ssDNA complex. The double-stranded cross-linked complex thus formed was purified, concentrated, and used in crystallization trials. To validate that this two-step strategy yielded a functional complex, we annealed a complementary strand containing an oxoG lesion to the single-stranded cross-linked complex and assayed for catalysis of DNA strand cleavage using denaturing gel electrophoresis (supplemental Fig. 1A). The expected 8-nt DNA cleavage product was observed (supplemental Fig. 1B, lanes 11-20), and it was formed at a rate similar to that observed for S292C hOGG1 in the absence of cross-linking (supplemental Fig. 1B, lanes 1–10). On the other hand, the nonlesion-containing complex produced by annealing the corresponding G-containing oligonucleotide gave no cleavage product (supplemental Fig. 1B, lanes 21–29) over a 1-h time frame and even after 5 to 7 days, no specific cleavage product is detected (supplemental Fig. 1B, lanes 30–31). We only observed the expected nonspecific nuclease bands that are very difficult to avoid in any protein-DNA sample incubated over such a long time period. These data indicate that (i) the procedure for assembly of the distally cross-linked complex yields a catalytically active species, and (ii) in the dDXL G-complex, the target G is not a substrate for hOGG1.
Structure of the dDXL G-complex
Despite extensive attempts to optimize crystallization conditions through variation of the DNA component and buffer conditions, most of the crystals we obtained of the dDXL G-complex were severely branched, fragile, and gave unsatisfactory diffraction data. The most promising hits were obtained using the sequence shown in Fig. 1C, right, and exhaustive buffer and additive screening with this complex yielded a single crystal that gave diffraction data suitable for structural analysis. Using these data, the structure of the dDXL G-complex was solved to 3.1 Å (Table 1) starting with the phases calculated from a high resolution hOGG1 structure (2.01 Å, PDB code 2NOH (43)), followed by cycles of refinement.
The overall structure of the dDXL G-complex (Fig. 1B, right) is very similar to that of the LRC and the pDXL G-complex (Fig. 1B, left and middle, respectively). The protein straddles the DNA strand containing the target nucleoside, inducing a sharp bend in the duplex at the site of the target G:C pair; the flanking duplex arms that project outward from the target site are unbent. The αEF-loop on hOGG1 uses its conserved NNN motif (residues 149–151) to invade the duplex at the target site. The target nucleoside, G in the case of the dDXL G-complex, is fully extruded from the helical stack, whereas the estranged C remains intrahelical. Although the target G is clearly extrahelical in both the pDXL and dDXL G-complexes, its precise disposition is markedly different in the two structures (compare middle and right blow-ups in Fig. 1B). Whereas the target G in the pDXL G-complex is completely disengaged from the lesion-recognition pocket on hOGG1 and resides in the exo-site, the target G in the dDXL G-complex is instead engaged in the active site pocket, adopting a conformation nearly identical to that of an oxoG lesion poised for repair (compare right and left blow-ups in Fig. 1B; see below). Although the target G is inserted into the active site pocket in the dDXL G-complex, and hOGG1 in this complex bears a wild-type active site, the electron density maps clearly show that the target G remains covalently bonded to its own sugar moiety (supplemental Fig. S2), consistent with the behavior of the same complex in solution (supplemental Fig. S1B).
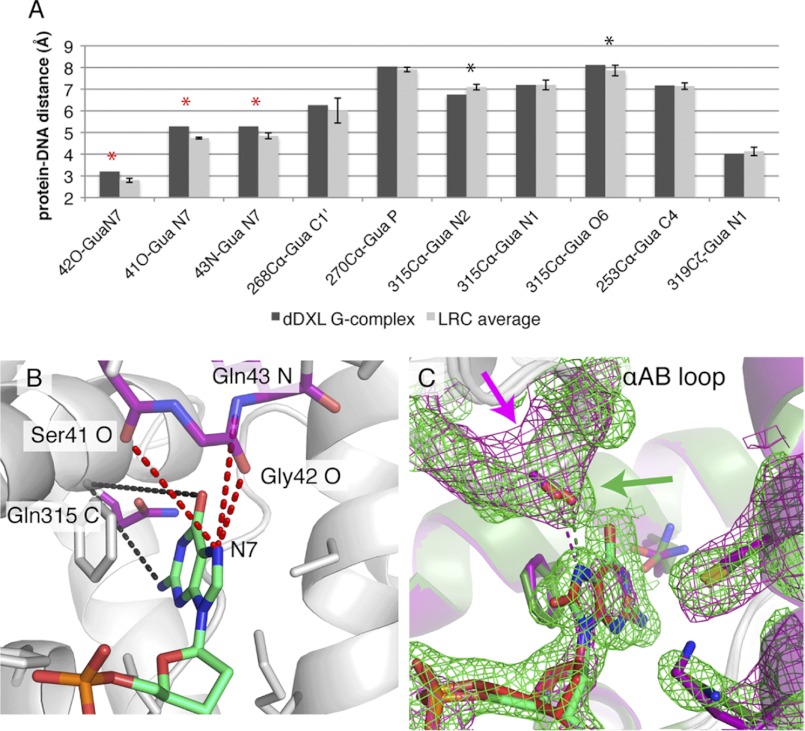
The hOGG1 lesion-recognition pocket is formed by the convergence of three α-helices, αL, αM, and αO, and the αAB-loop, residues of which contact the oxoG nucleobase and its 5′-phosphate (Fig. 2A). The αAB-loop contributes the main chain carbonyl of Gly-42, the oxygen atom of which hydrogen bonds to the oxoG N7-H. The αL-helix contributes Lys-249, the catalytic nucleophile, and Cys-253, the side chains of these residues were proposed to form an (RNH3+/RS−) ion pair that interacts favorably with the oxoG 5-membered ring through dipole/dipole forces (34). The αM-helix contributes His-270, the side chain of which hydrogen bonds to a nonbridging phosphate oxygen on the oxoG 5′-phosphate. Finally, the αO-helix contributes Gln-315, which hydrogen bonds to the Watson-Crick face of oxoG; Phe-319, the aryl ring makes a face-to-face π/π interaction with the oxoG nucleobase; and Asp-322, the carboxylate of which serves to orient His270 through hydrogen bonding. In structures having the active site vacated and the exo-site occupied, the αO-helix retracts to open up the active site, and this helix movement is associated with disengagement of His-270 from the 5′-phosphate and engagement in an edge to face interaction with the Phe-319 side chain (Fig. 2, B, D, and F). Exo-site occupancy is also associated with rupture of the Lys-249/Cys-253 interaction and establishment of a hydrogen bonding interaction between Lys-249 and the oxoG 3′-phosphate (Fig. 2, B, D, and F). The dDXL G-complex shows all the hallmarks of the “closed” active site conformation, with the same repertoire of contacts between the resident G and the protein as seen with oxoG in an LRC (compare Fig. 2, A and C, see E for an overlay). We were quite intrigued by this seemingly perfect superposition between the dDXL G-complex and the LRC, given the significant calculated energy difference between inserting a G versus an oxoG into the active site (34). However, although precise determinations of atomic positions are not possible at the resolution of the dDXL G-complex, our best interpretation of the data indicates slight repositioning of active site functionality and the DNA target in the dDXL complex relative to the corresponding elements in an LRC. For example, the key Gly-42 interaction appears to be somewhat longer in the former than in the latter (3.2 versus 2.7 Å, respectively, Fig. 2, C versus A), and the entire DNA strand in the dDXL G-complex is accordingly shifted slightly away from the active site, relative to its position in an LRC. Because all of the LRC structures bear some alteration, a mutated amino acid residue or a substrate analog in the DNA relative to the native DNA-protein complex, no single LRC structure can be unambiguously considered to represent the true native, pre-catalytic complex. Therefore, we compared the dDXL G-complex with the entire series of available hOGG1 LRC structures. Shown in Fig. 3A are several DNA/protein contact distances for the dDXL G-complex and averaged for the six hOGG1/oxoG LRCs. Almost all of the contact distances in the dDXL G-complex do not deviate significantly from those in the six LRC complexes (fall within 1 S.D. of the averaged LRC values). However, the guanine nucleobase does appear to be slightly mispositioned in the active site by virtue of a small retrograde tilt away from Gln315 at the back of the active site (black asterisks in Fig. 3A, and black distances in Fig. 3B). The other more notable perturbation encompasses the three contacts involving the αAB-loop (red asterisks in Fig. 3A, red distances in Fig. 3B): i.e. the three dDXL G-complex distances fall outside of three times the S.D. from the corresponding distance averaged for the LRC complexes. Thus, the structural perturbations in the active site as a result of G appear to be localized to the αAB-loop region, an intriguing result considering that the flexibility (as measured by α-carbon B factors) of this loop is lower than the average.
FIGURE 3.
Effect of G versus oxoG insertion on the hOGG1 active site. A, at several positions in the protein-DNA interface, distances were measured between active site atoms and the target G or oxoG, and the distances are presented as bar graphs. The distances within several LRCs were averaged (gray columns) and compared with the dDXL G-complex (black columns), with key distances being highlighted with asterisks (see text). Error bars are 1 S.D. The LRC complexes used in this average are as follows: K249Q LRC (1EBM (10)), pDXL LRC (1YQK (34)), dDXL LRC (2NOL (43)), the K249Q LRC with Lys-249 restored in silico (as in Fig. 2, A, D, and E); an LRC generated using a completely wild-type enzyme with DNA containing a target oxoG having a 2′-α-fluoro substituent (3KTU, S. Lee and G. L. Verdine, unpublished data); the D268N LRC (1N3C (13)). B, atomic coordinate points for the columns with asterisks in A are illustrated using the dDXL G-complex. C, a σA-weighted 2Fo − Fc map of the dDXL G-complex was generated (purple mesh, contoured at 1σ) via refinement by a rigid body fit initiated with the K249Q LRC model (1EBM minus the oxoG nucleoside, Gln249, and nonbonded molecules/ions). The map was then superposed on the final σA-weighted 2Fo − Fc map of the K249Q LRC in Phenix (green mesh, contoured at 1σ, phenix.superpose_maps utility (49)), to correct for the unit cell differences between the two structures. Arrows point to the nonoverlapping density that indicates a shift has occurred. Final models for the dDXL G-complex (purple schematic) and the K249Q LRC (green schematic) are also superposed to match their corresponding maps.
The low B factors of the αAB-loop region attest to the significance of the small structural differences observed here. We nevertheless, sought to further establish the validity of these results, in particular through the application of methods to minimize the uncertainties in structure determination. Related structures are generally compared by superimposing the models so that conformational changes at various sites of interest can be observed. When the magnitude of these changes is not much larger than the coordinate error of the model, that is, the positional uncertainty of each atom, one can argue that this method is not sufficiently robust. In cases involving more subtle conformational shifts, it is preferable to compare the electron density maps themselves. (The dDXL G-complex structure was not sufficiently isomorphous with LRC to enable interpretable isomorphous difference maps to be generated.) Due to the “phase problem” of crystallography, these maps will always bear model bias to some extent. However, we used this fact to our advantage to validate the shifted position of the αAB-loop in the dDXL G-complex structure, as we indicate below.
Specifically, we first generated a “control” 2Fo − Fc electron density map using the K249Q LRC structure factors and model deposited in the PDB (PDB code 1EBM, chosen for having the highest resolution, 2.1 Å, among available LRC structures). Next, we used the same model to calculate a 2Fo − Fc map using the structure factors of the dDXL G-complex, the only model adjustments allowed during the limited refinement being a rigid body fit, which accounted for the minor positional difference between these two structures within the unit cell (detailed in Fig. 3, legend). Superposition of these 2Fo − Fc maps reveals a clear outward (away from the active site) shift of the αAB-loop in the dDXL G-complex (Fig. 3C, purple mesh), despite the bias of the dDXL G-complex 2Fo − Fc map toward the original conformation of the loop (green mesh).
Further validation of our final structural model of the αAB-loop (specifically, residues Leu-39 to Arg-46) in the dDXL G-complex is evident from the increase in both the working (from 0.228 to 0.250) and free (from 0.263 to 0.276) R factors upon transposing this loop segment to its original position in the K249Q LRC. Our analyses indicate that the positional shifts in the Gly-42 loop are significant. These shifts appear to represent an important impediment to proper alignment of G with respect to the hOGG1 active site.
Guanine Is Misaligned in hOGG1 Lesion-recognition Pocket
Hints as to the precise nature of this impediment were previously obtained from calculations based on the pDXL G-complex and a computationally generated wild-type LRC (34). The simulations indicated that repulsive interactions between G and the hOGG1 active site are a major factor in thermodynamic discrimination of G versus oxoG by the hOGG1 lesion-recognition pocket. The modest retraction of G from the active site loop in the dDXL G-complex reported here is certainly consistent with avoidance of repulsive interactions. Specifically, the calculations suggested that lone-pair/lone-pair repulsion between the G N7 atom and the Gly-42 carbonyl are an important contributor to rejection of G by the enzyme active site. However, although these atoms are farther apart in the dDXL G-complex than in the clustered LRCs, their close distance and the lack of any more dramatic structural adjustment in the protein at Gly-42 in the dDXL G-complex still strikes us as unexpected. Because the strength of dipole-dipole interactions diminishes with the cubic of the distance between two dipole moments, it is conceivable that the modest retraction seen here provides some relief of repulsive strain between Gly-42 and guanine N7 (by 1.3 kcal/mol of the interaction energy based on the CHARMM potential (52, 56) as G retracts its position from LRC to G-complex) to allow G to reside in the active site pocket. On the other hand, the dipole-dipole interaction between the Lys-249/Cys-253 pair and the base bound in the active site was suggested computationally to be substantially weaker, for G than for oxoG, perhaps destabilizing the interactions with G, yet we observe this interaction to be retained in the dDXL G-complex. The Lys-249/Cys-253 salt bridge interaction in our structure might experience a relative reduction in stability due to the presence of G (supplemental Fig. S2), but such a parameter cannot be accurately determined from x-ray diffraction data alone.
Structural Influence of Cross-linking
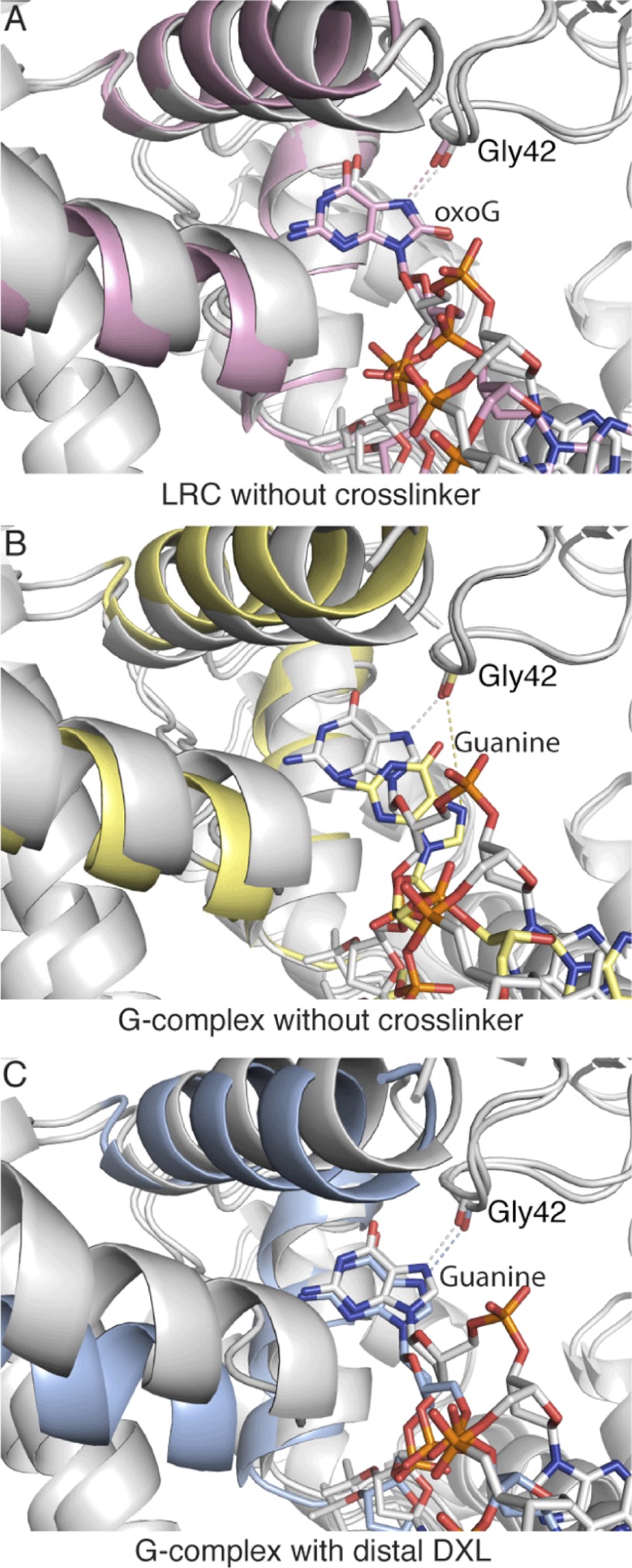
In previous studies, we observed that cross-linking at the distal site seems to bias hOGG1-DNA complexes toward insertion into the active site pocket. Specifically, we observed that introduction of a bulky mutation (Q315F) into the active site pocket caused the enzyme to reject an oxoG substrate when DXLed at the proximal site, but when DXLed at the distal site, the oxoG was partially inserted into the active site, and certain amino acid side chains of the active site reorganized themselves to accommodate the target base (43). Might it be possible that this same bias lowers the thermodynamic penalty of inserting G into the enzyme active site? Ideally, we could answer this question by comparing the present crystal structure to the crystal structure of hOGG1 bound to normal DNA in the absence of a cross-linker and noting if/how the conformation of the target base changes when the influence of the distal cross-linker is removed. Because such a structure is inaccessible experimentally, molecular dynamics (MD) simulations were used to generate an in silico model of the structure of wild-type hOGG1 extrahelically interrogating a normal guanine base. Three simulations were carried out, each using a crystal structure of hOGG1 bound to oxoG-containing DNA as a starting point (PDB code 1YQR). In the first simulation, any mutated residues (i.e. N149C and K249Q) were restored to wild-type in silico to yield the “LRC without cross-linking” (gray structure, Fig. 4A). The starting model was further modified by exchanging the oxoG for a G to yield the “G-complex without cross-linking” (gray structure, Fig. 4B). Finally, the distal cross-linker and the S292C mutation were introduced into the model to produce the “G-complex with distal DXL” (gray structure, Fig. 4C). For each system, MD simulations were run for 30 ns and averaged structures were determined over three 10-ns intervals.
FIGURE 4.
Comparison of the MD simulations and their initial input models. Starting models used for each molecular dynamics simulation are shown in gray and aligned (protein all atom alignment) with the averaged structure from the first 10 ns of the simulation, shown in pink (LRC without cross-linking) (A), yellow (G-complex without the distal cross-linker) (B), or powder blue (C) (G-complex with distal cross-linker).
Overlays of the averaged structure from the first 10 ns with the initial model used to start the simulation reveal that whereas the target DNA base conformations in the uncross-linked oxoG complex and the dDXLed G-complex remain close to those of their respective starting models, the guanine in the uncross-linked G-complex shifts significantly away from the active site (compare Fig. 4, panels A and C with B). OxoG maintains practically the same orientation throughout the 30-ns simulation (Fig. 5A), as does G in the dDXLed G-complex (Fig. 5B). When the distal DXL is removed, guanine begins to move considerably and in the end, is ejected from the active site (Fig. 5C). The cross-linking notably suppresses the motion of the αO and αM helices that interact with the region of the enzyme containing the cross-linked residue. As described above, these helices contain residues (His-270, Phe-319, and Gln-315) that recognize the base in the active site. The suppressed fluctuations of the helices appear to be correlated with, or even to promote, the stable occupancy of the G in the active site (compare supplemental Movies S1, S2, and S3). In addition, the average structures from the dDXLed G and uncross-linked oxoG system show that the cross-linked G is inserted less deeply into the active site than the oxoG, which is in accord with the crystal structure counterparts of these systems (Fig. 2, A and C, red arrows); specifically, the separation between the Gly-42 O and guanine N7 in the dDXLed G-complex system is larger than it is in the uncross-linked oxoG system (Fig. 5D). As mentioned above, the shift of G is likely due to the electrostatic repulsion between G and Gly-42. The present crystal structure, MD simulations, and DNA cleavage assay data clearly demonstrate that the interaction with Gly-42 is critical in the discrimination of oxoG in the active site of hOGG1 and that G is unstable in the active site without the cross-link. This is in accord with previous structures and simulation results (34).
FIGURE 5.
Averages of MD simulation structures. The following structures were obtained by averaging the coordinates saved during MD simulations over three 10-ns intervals (represented in order from early to late in the 30-ns simulation by lighter to darker hues) for the lesion recognition complex in the absence of any cross-linking (A, red structures) or the distally cross-linked G-complex in the presence (B, blue structures) or absence (C, orange structures) of the cross-linker moiety. The overlays of the three averaged structures reveal the comparative mobility of the DNA conformation in the G-complex that lacks the distal cross-linker. Panel D shows the relative displacement of the G in the distally cross-linked complex (blue structure) with respect to the position of the oxoG in the LRC (red structure). Structural averages from the final 10 ns of the simulations were used for the overlay.
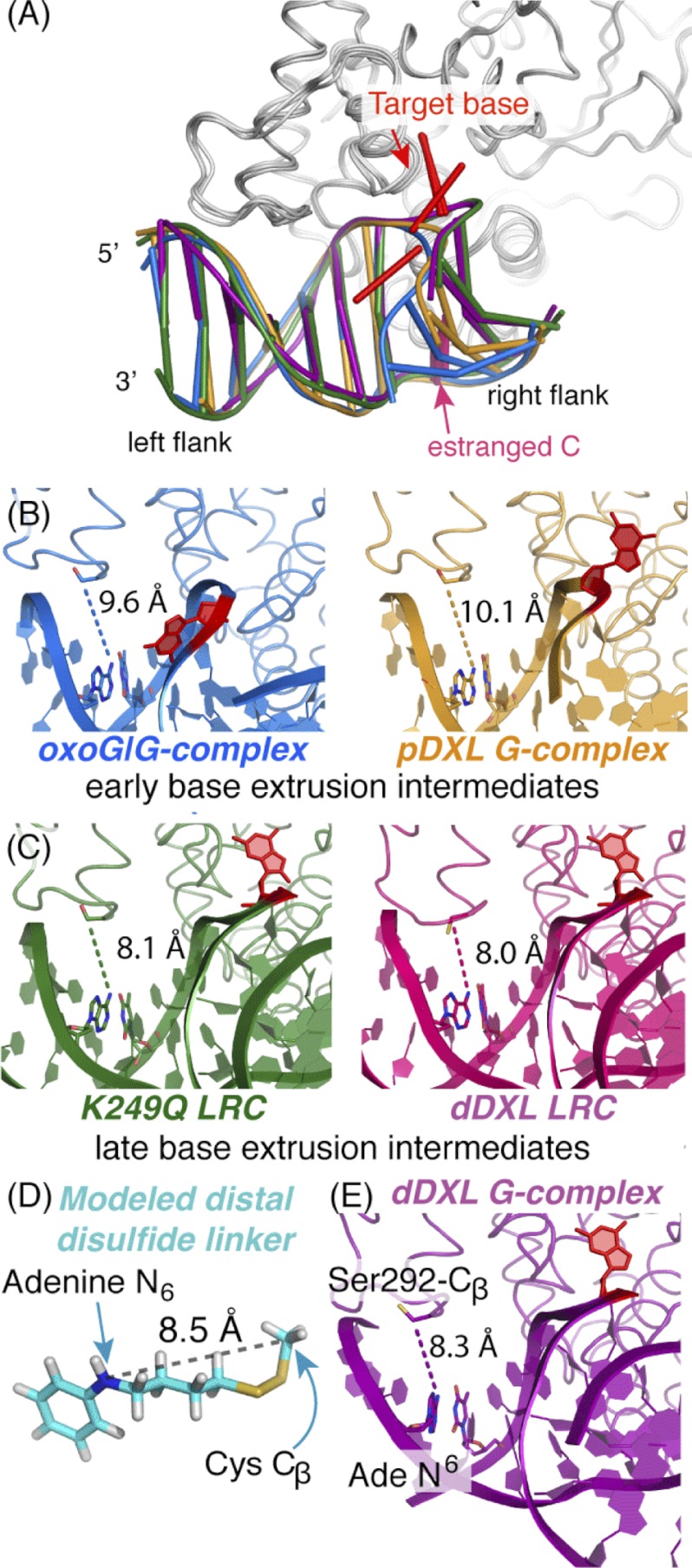
Although the MD simulations clearly identify the influence of the distal cross-linker on the conformation and dynamics of G in the active site of hOGG1, the mechanism of action remains unclear. One potential mechanism, however, is evident from the structural record. We have compared the distance separating the tether attachment points for distal site disulfide cross-linking, namely the N6 of adenine 5 and the β-carbon of residue 292, between the reported structures of hOGG1 that illuminate many of the base extrusion intermediates, from those that occur “early” to “late” in the pathway (Fig. 6A) (18, 22, 24, 26). From this comparison we found that this distance varies as a function of progression along the nucleobase extrusion pathway. Specifically, the N6-A5/β-CCys-292 distance is 9.6 Å or greater for early extrusion intermediates and 8.5 Å on average for fully extruded intermediates (Fig. 6, B and C, and supplemental Fig. S3). Molecular modeling shows that the most extended conformation of the particular cross-link system used in the dDXL, which also happens to be the lowest in energy, has an N6-A5/β-CCys-292 span of 8.46 Å (Fig. 6D and supplemental Fig. S4; see the caption of supplemental Fig. S4 for a detailed description of the modeling protocol). This distance is too short to traverse the N6-A5/β-CCys-292 span found in early extrusion intermediates but is compatible with the span in fully extruded intermediates, including the dDXL G-complex (Fig. 6E). Based on this analysis and MD simulation results, we propose that distal site cross-linking using a 4-carbon tether applies tension locally on the DNA at the tether attachment point, thereby suppressing local conformational fluctuations of the protein, specifically the αM and αO helices. Therefore, the previously calculated 3.5 kcal/mol repulsion between the G NH and the Gly-42 O is overcome by both the cross-linking, which biases toward active site binding, and the modest retraction of G, which locally relieves the repulsion with Gly-42.
FIGURE 6.
Structural snapshots along the hOGG1 nucleobase extrusion pathway. Shown are representations of hOGG1-DNA complexes representing various stages along the nucleobase extrusion pathway: blue, the oxoG·G-complex (2I5W (42)), representing an early intermediate; orange, the pDXL G-complex (1YQK (34)), a later intermediate; the K249Q LRC in the absence (green, 1EBM (13)) and presence (hot pink, 2NOL (43)) of distal site cross-linking, representing the final state preceding catalysis; and purple, the dDXL G-complex (3IH7, this work). A, comparison of adjustments in the DNA component of the protein-DNA complexes. This structural comparison was generated by least-squares superposition of the protein component (residues 12–300) using the program Coot (48). The left flank of the DNA bears the distal cross-linking site; note the shift in position as a function of progression along the nucleobase extrusion pathway. The target base is colored red and the estranged C is light magenta. B, model of the lowest energy conformation for the four-carbon disulfide-bearing tether used in distal cross-linking of hOGG1 to DNA. Denoted explicitly is the distance (8.5 Å) between the atoms corresponding to the N6 atom of the tethered adenine and the β-carbon of Cys-292 (the adenine ring is replaced by benzene in the model. See supplemental Fig. S4, legend for details of the minimization procedure and the entire ensemble of conformers. Note that the lowest energy conformation extends the longest distance of all the stable conformers.) C–E, a comparison of distances between the adenine N6 to the Cβ of residue 292 in structures of hOGG1-DNA complexes at various stages of the nucleobase extrusion pathway. In all cases, the protein is displayed as a ribbon and the DNA is shown in schematic representations. The side chain of residue 292, the tethered adenine, and its thymine partner are colored by atom and shown as stick models, as is the target base, in which all atoms are colored red. D, complexes of hOGG1 in which the target base is partially extruded are termed early base extrusion intermediates. The distal site distances tend to be longer on average than those of LRCs (E), which represent late base extrusion intermediates. The DNA ladder representation was generated using X3DNA (66), nuccyl, and PyMol (50).
DISCUSSION
We have shown that hOGG1 in the dDXL G-complex bears a fully intact, wild-type catalytic apparatus, which, nonetheless, is ineffective at promoting base excision of a target G. This failure to catalyze base excision is clearly due to rejection of G as a substrate, and not to disulfide cross-linking, because mere replacement of G by oxoG in this 46-kDa protein-DNA complex results in efficient base excision. The present data therefore point to the existence of a mechanism at a late stage in the hOGG1 base extrusion pathway that serves to prevent the enzyme from accidentally excising the normal G nucleobase. The mechanism would come into play as a catalytic checkpoint only in the rare instances in which the target G has surmounted the energy barrier to transition from the exo-site to the active site pocket. It remains unclear by what means the catalytic checkpoint prevents G cleavage. We note that the N-glycosidic bond of G has been shown to be more labile toward hydrolysis at neutral pH than oxoG (61). Therefore, a discrimination mechanism that relies on the intrinsic differences in lability between the glycosidic bond of G versus oxoG cannot be in operation in hOGG1. Indeed, hOGG1 rejecting G as a substrate in the structure reported here presents even more of a mystery and intensifies the search problem hOGG1 faces: not only is oxoG extremely rare and structurally similar to G, but it is also inherently more difficult to cleave than G. So by what mechanism might this catalytic checkpoint be working? We envision that the transition state leading to glycosidic bond cleavage of G could inter alia be elevated relative to oxoG by displacement of key active site elements, leading to a suboptimal reaction trajectory, or by deprivation of the transition state stabilization afforded oxoG through hydrogen bonding with Gly-42. Also, as discussed above, the target G in the dDXL G-complex is slightly retracted from the active site pocket, relative to LRC structures, and this may prevent attainment of the optimal conformational state for the attack by Lys-249. Consistent with this notion, introduction of active site mutations that even modestly perturb the active site disposition of oxoG, namely D268N (13) and Q315F (43), have extremely deleterious effects on the rate of oxoG cleavage by the enzyme. Not all base excision DNA repair enzymes are as discriminating as hOGG1; for example, 3-methyladenine DNA glycosylases (AlkA and AAG) cleave normal nucleobases from DNA at an appreciable frequency (62, 63). The same factors of substrate recognition and catalysis that cause hOGG1 to be much more restricted than AlkA and AAG in substrate scope for DNA lesions may also lead hOGG1 to be less prone to making mistakes in distinguishing normal from damaged DNA.
We have employed disulfide cross-linking to stabilize and capture an ordinarily unstable, fleeting species in which hOGG1 has inserted a normal guanine base into the enzyme active site. Efforts are currently underway to crystallize the dDXL G-complex with a longer tether,3 so as to gauge the effect of reducing tether-dependent torsional stress. Because the tether length employed in disulfide cross-linking can easily be varied, we expect that such tunable DXL-dependent manipulation of protein conformational dynamics will find use in structural studies of a variety of dynamic DNA/protein interaction systems. Indeed, we have already captured a similar base extrusion intermediate of MutM, the bacterial counterpart of hOGG1, using a cross-linker-mediated DNA tugging strategy (64).
The remarkable observation reported here is that a relatively subtle structural perturbation due to introduction of a covalent cross-link between hOGG1 and DNA, 4 base pairs distal to the extruded base pair can nevertheless shift the energetic landscape of the complex to favor extrusion of the target G and insertion into the enzyme active site. This enforced presentation produces a complex having a structure very similar to that produced with an oxoG lesion, where the differences appear to arise from divergence in how the active site interacts with a G nucleobase versus oxoG. Based on these observations and on previous calculations, it is likely that during routine interrogation of DNA, hOGG1 occasionally exposes guanine to its active site. The present results suggest such accidental presentation does not give rise to spurious base excision, because of the ability of the hOGG1 active site to discriminate between oxoG and G at the level of catalysis.
Supplementary Material
Acknowledgments
We thank the staff at the NE-CAT at the Advanced Photon Source, especially C. Narayanasami Sukumar, for assistance during data collection, the National Energy Research Scientific Computing Center and FAS Science Division Research Computing Group at Harvard University for providing computer resources. We also thank Drs. Lourdes Gude-Rodriguez, Seongmin Lee, Danaya Pakotiprapha, Marie Spong, and especially Gordon Louie for critical reading of the manuscript, and data analysis advice. We also thank Rachelle Gaudet and David Jeruzalmi and other laboratory members for useful discussions and guidance on data processing. Use of the Advanced Photon Source was supported by the United States Dept. of Energy, Office of Science, Office of Basic Energy Sciences, under Contract No. DE-AC02-06CH11357.
This work was supported, in whole or in part, by National Institutes of Health Grants GM044853 and CA100742 (to G. L. V.) and GM030804 (to M. K.), predoctoral fellowships from the National Science Foundation (2004) (to C. M. C. and P. S. K.), a postdoctoral fellowship from the National Cancer Center (to K. N.), the Harvard College Program for Research in Science and Engineering (2007) (to K. O.), Herchel Smith Harvard Summer Undergraduate Research Fellowship (2007) (to K. O.), the Harvard College Research Program (2008) (to K. O.), and Ruth Kirschstein National Research Service Award Fellowship F32 GM077935-01 (to B. R. B.).
The atomic coordinates and structure factors (code 3IH7) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).

This article contains supplemental Figs. S1–S4 and Movies S1–S3.
U. Shigdel and G. L. Verdine, unpublished data.
- oxoG
- 8-oxoguanine
- hOGG1
- human 8-oxoguanine glycosylase 1
- dDXL
- distally disulfide cross-linked
- pDXL
- proximally disulfide cross-linked
- LRC
- lesion recognition complex
- MD
- molecular dynamics
- PDB
- Protein Data Bank.
REFERENCES
- 1. Boonstra J., Post J. A. (2004) Molecular events associated with reactive oxygen species and cell cycle progression in mammalian cells. Gene 337, 1–13 [DOI] [PubMed] [Google Scholar]
- 2. Wood M. L., Esteve A., Morningstar M. L., Kuziemko G. M., Essigmann J. M. (1992) Genetic effects of oxidative DNA damage. Comparative mutagenesis of 7,8-dihydro-8-oxoguanine and 7,8-dihydro-8-oxoadenine in Escherichia coli. Nucleic Acids Res. 20, 6023–6032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Lindahl T. (1993) Instability and decay of the primary structure of DNA. Nature 362, 709–715 [DOI] [PubMed] [Google Scholar]
- 4. Shibutani S., Takeshita M., Grollman A. P. (1991) Insertion of specific bases during DNA synthesis past the oxidation-damaged base 8- oxodG. Nature 349, 431–434 [DOI] [PubMed] [Google Scholar]
- 5. Cheng K. C., Cahill D. S., Kasai H., Nishimura S., Loeb L. A. (1992) 8-Hydroxyguanine, an abundant form of oxidative DNA damage, causes G–T and A–C substitutions. J. Biol. Chem. 267, 166–172 [PubMed] [Google Scholar]
- 6. Abu-Amero K. K., Alzahrani A. S., Zou M., Shi Y. (2006) Association of mitochondrial DNA transversion mutations with familial medullary thyroid carcinoma/multiple endocrine neoplasia type 2 syndrome. Oncogene 25, 677–684 [DOI] [PubMed] [Google Scholar]
- 7. Michaels M. L., Tchou J., Grollman A. P., Miller J. H. (1992) A repair system for 8-oxo-7,8-dihydrodeoxyguanine. Biochemistry 31, 10964–10968 [DOI] [PubMed] [Google Scholar]
- 8. Michaels M. L., Miller J. H. (1992) The GO system protects organisms from the mutagenic effect of the spontaneous lesion 8-hydroxyguanine (7,8-dihydro-8-oxoguanine). J. Bacteriol. 174, 6321–6325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Mitra S., Boldogh I., Izumi T., Hazra T. K. (2001) Complexities of the DNA base excision repair pathway for repair of oxidative DNA damage. Environ. Mol. Mutagen. 38, 180–190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bruner S. D., Norman D. P., Verdine G. L. (2000) Structural basis for recognition and repair of the endogenous mutagen 8-oxoguanine in DNA. Nature 403, 859–866 [DOI] [PubMed] [Google Scholar]
- 11. Nash H. M., Lu R., Lane W. S., Verdine G. L. (1997) The critical active-site amine of the human 8-oxoguanine DNA glycosylase, hOGG1. Direct identification, ablation, and chemical reconstitution. Chem. Biol. 4, 693–702 [DOI] [PubMed] [Google Scholar]
- 12. Nash H. M., Bruner S. D., Schärer O. D., Kawate T., Addona T. A., Spooner E., Lane W. S., Verdine G. L. (1996) Cloning of a yeast 8-oxoguanine DNA glycosylase reveals the existence of a base-excision DNA-repair protein superfamily. Curr. Biol. 6, 968–980 [DOI] [PubMed] [Google Scholar]
- 13. Norman D. P., Chung S. J., Verdine G. L. (2003) Structural and biochemical exploration of a critical amino acid in human 8-oxoguanine glycosylase. Biochemistry 42, 1564–1572 [DOI] [PubMed] [Google Scholar]
- 14. Lipscomb L. A., Peek M. E., Morningstar M. L., Verghis S. M., Miller E. M., Rich A., Essigmann J. M., Williams L. D. (1995) X-ray structure of a DNA decamer containing 7,8-dihydro-8-oxoguanine. Proc. Natl. Acad. Sci. U.S.A. 92, 719–723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Bowman B. R., Lee S., Wang S., Verdine G. L. (2008) Structure of the E. coli DNA glycosylase AlkA bound to the ends of duplex DNA. A system for the structure determination of lesion-containing DNA. Structure 16, 1166–1174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Oda Y., Uesugi S., Ikehara M., Nishimura S., Kawase Y., Ishikawa H., Inoue H., Ohtsuka E. (1991) NMR studies of a DNA containing 8-hydroxydeoxyguanosine. Nucleic Acids Res. 19, 1407–1412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Plum G. E., Grollman A. P., Johnson F., Breslauer K. J. (1995) Influence of the oxidatively damaged adduct 8-oxodeoxyguanosine on the conformation, energetics, and thermodynamic stability of a DNA duplex. Biochemistry 34, 16148–16160 [DOI] [PubMed] [Google Scholar]
- 18. Koizume S., Kamiya H., Inoue H., Ohtsuka E. (1994) Synthesis and thermodynamic stabilities of damaged DNA involving 8-hydroxyguanine (7,8-dihydro-8-oxoguanin) in ras gene fragments. Nucleosides Nucleotides Nucleic Acids 13, 1517–1534 [Google Scholar]
- 19. McAuley-Hecht K. E., Leonard G. A., Gibson N. J., Thomson J. B., Watson W. P., Hunter W. N., Brown T. (1994) Crystal structure of a DNA duplex containing 8-hydroxydeoxyguanine-adenine base pairs. Biochemistry 33, 10266–10270 [DOI] [PubMed] [Google Scholar]
- 20. Crenshaw C. M., Wade J. E., Arthanari H., Frueh D., Lane B. F., Núñez M. E. (2011) Hidden in plain sight. Subtle effects of the 8-oxoguanine lesion on the structure, dynamics, and thermodynamics of a 15-base pair oligodeoxynucleotide duplex. Biochemistry 50, 8463–8477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Luger K., Mäder A. W., Richmond R. K., Sargent D. F., Richmond T. J. (1997) Crystal structure of the nucleosome core particle at 2.8-Å resolution. Nature 389, 251–260 [DOI] [PubMed] [Google Scholar]
- 22. Gao Y. G., Su S. Y., Robinson H., Padmanabhan S., Lim L., McCrary B. S., Edmondson S. P., Shriver J. W., Wang A. H. (1998) The crystal structure of the hyperthermophile chromosomal protein Sso7d bound to DNA. Nat. Struct. Biol. 5, 782–786 [DOI] [PubMed] [Google Scholar]
- 23. Robinson H., Gao Y. G., McCrary B. S., Edmondson S. P., Shriver J. W., Wang A. H. (1998) The hyperthermophile chromosomal protein Sac7d sharply kinks DNA. Nature 392, 202–205 [DOI] [PubMed] [Google Scholar]
- 24. Viadiu H., Aggarwal A. K. (2000) Structure of BamHI bound to nonspecific DNA. A model for DNA sliding. Mol. Cell 5, 889–895 [DOI] [PubMed] [Google Scholar]
- 25. Horton J. R., Liebert K., Hattman S., Jeltsch A., Cheng X. (2005) Transition from nonspecific to specific DNA interactions along the substrate-recognition pathway of DAM methyltransferase. Cell 121, 349–361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Townson S. A., Samuelson J. C., Bao Y., Xu S. Y., Aggarwal A. K. (2007) BstYI bound to noncognate DNA reveals a ”hemispecific“ complex. Implications for DNA scanning. Structure 15, 449–459 [DOI] [PubMed] [Google Scholar]
- 27. Winkler F. K., Banner D. W., Oefner C., Tsernoglou D., Brown R. S., Heathman S. P., Bryan R. K., Martin P. D., Petratos K., Wilson K. S. (1993) The crystal structure of EcoRV endonuclease and its complexes with cognate and non-cognate DNA fragments. EMBO J. 12, 1781–1795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Agback P., Baumann H., Knapp S., Ladenstein R., Härd T. (1998) Architecture of nonspecific protein-DNA interactions in the Sso7d-DNA complex. Nat. Struct. Biol. 5, 579–584 [DOI] [PubMed] [Google Scholar]
- 29. Kalodimos C. G., Biris N., Bonvin A. M., Levandoski M. M., Guennuegues M., Boelens R., Kaptein R. (2004) Structure and flexibility adaptation in nonspecific and specific protein-DNA complexes. Science 305, 386–389 [DOI] [PubMed] [Google Scholar]
- 30. Parker J. B., Bianchet M. A., Krosky D. J., Friedman J. I., Amzel L. M., Stivers J. T. (2007) Enzymatic capture of an extrahelical thymine in the search for uracil in DNA. Nature 449, 433–437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Maiti A., Morgan M. T., Pozharski E., Drohat A. C. (2008) Crystal structure of human thymine DNA glycosylase bound to DNA elucidates sequence-specific mismatch recognition. Proc. Natl. Acad. Sci. U.S.A. 105, 8890–8895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Albright R. A., Mossing M. C., Matthews B. W. (1998) Crystal structure of an engineered Cro monomer bound nonspecifically to DNA. Possible implications for nonspecific binding by the wild-type protein. Protein Sci. 7, 1485–1494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Huang H., Chopra R., Verdine G. L., Harrison S. C. (1998) Structure of a covalently trapped catalytic complex of HIV-1 reverse transcriptase. Implications for drug resistance. Science 282, 1669–1675 [DOI] [PubMed] [Google Scholar]
- 34. Banerjee A., Yang W., Karplus M., Verdine G. L. (2005) Structure of a repair enzyme interrogating undamaged DNA elucidates recognition of damaged DNA. Nature 434, 612–618 [DOI] [PubMed] [Google Scholar]
- 35. Fromme J. C., Banerjee A., Huang S. J., Verdine G. L. (2004) Structural basis for removal of adenine mispaired with 8-oxoguanine by MutY adenine DNA glycosylase. Nature 427, 652–656 [DOI] [PubMed] [Google Scholar]
- 36. Corn J. E., Berger J. M. (2007) FASTDXL. A generalized screen to trap disulfide-stabilized complexes for use in structural studies. Structure 15, 773–780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Qi Y., Spong M. C., Nam K., Banerjee A., Jiralerspong S., Karplus M., Verdine G. L. (2009) Encounter and extrusion of an intrahelical lesion by a DNA repair enzyme. Nature 462, 762–766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Banerjee A., Santos W. L., Verdine G. L. (2006) Structure of a DNA glycosylase searching for lesions. Science 311, 1153–1157 [DOI] [PubMed] [Google Scholar]
- 39. Yang C. G., Yi C., Duguid E. M., Sullivan C. T., Jian X., Rice P. A., He C. (2008) Crystal structures of DNA/RNA repair enzymes AlkB and ABH2 bound to dsDNA. Nature 452, 961–965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Corn J. E., Pelton J. G., Berger J. M. (2008) Identification of a DNA primase template tracking site redefines the geometry of primer synthesis. Nat. Struct. Mol. Biol. 15, 163–169 [DOI] [PubMed] [Google Scholar]
- 41. Duguid E. M., Rice P. A., He C. (2005) The structure of the human AGT protein bound to DNA and its implications for damage detection. J. Mol. Biol. 350, 657–666 [DOI] [PubMed] [Google Scholar]
- 42. Banerjee A., Verdine G. L. (2006) A nucleobase lesion remodels the interaction of its normal neighbor in a DNA glycosylase complex. Proc. Natl. Acad. Sci. U.S.A. 103, 15020–15025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Radom C. T., Banerjee A., Verdine G. L. (2007) Structural characterization of human 8-oxoguanine DNA glycosylase variants bearing active site mutations. J. Biol. Chem. 282, 9182–9194 [DOI] [PubMed] [Google Scholar]
- 44. Bowman B. R., Lee S., Wang S., Verdine G. L. (2010) Structure of Escherichia coli AlkA in complex with undamaged DNA. J. Biol. Chem. 285, 35783–35791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Zbyszek Otwinowski W. M. (1997) Processing of x-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 46. Read R. J. (1986) Improved Fourier coefficients for maps using phases from partial structures with errors. Acta Crystallogr. A 42, 140–149 [Google Scholar]
- 47. Brunger A. T. (2007) Version 1.2 of the Crystallography and NMR system. Nat. Protoc. 2, 2728–2733 [DOI] [PubMed] [Google Scholar]
- 48. Emsley P., Cowtan K. (2004) Coot, model-building tools for molecular graphics. Acta Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 49. Adams P. D., Afonine P. V., Bunkoczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L. W., Kapral G. J., Grosse-Kunstleve R. W., McCoy A. J., Moriarty N. W., Oeffner R., Read R. J., Richardson D. C., Richardson J. S., Terwilliger T. C., Zwart P. H. (2010) PHENIX, a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. 66, 213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. DeLano W. L. (2002) The PyMOL Molecular Graphics System (Scientific D., ed) Schrödinger, LLC, New York [Google Scholar]
- 51. MacMillan A. M., Verdine G. L. (1991) Engineering tethered DNA molecules by the convertible nucleoside approach. Tetrahedron 14, 2603–2616 [Google Scholar]
- 52. MacKerell A. D., Jr., Bashford D., Bellott M., Dunbrack R. L., Jr., Evanseck J., Field M. J., Fischer S., Gao J., Guo H., Ha S., Joseph D., Kuchnir L., Kuczera K., Lau F. T. K., Mattos C., Michnick S., Ngo T., Nguyen D. T., Prodhom B., Reiher W. E., 3rd, Roux B., Schlenkrich M., Smith J., Stote R., Straub J., Watanabe M., Wiorkiewicz-Kuczera J., Yin D., Karplus M. (1998) All-atom empirical potential for molecular modeling and dynamics studies of protein. J. Phys. Chem. B 102, 3586–3616 [DOI] [PubMed] [Google Scholar]
- 53. Allen M. P., Tildesley D. J. (1989) Computer Simulations of Liquids, Oxford University Press, New York [Google Scholar]
- 54. Ryckaert J. P., Ciccotti G., Berendsen H. J. (1977) Numerical Integration of the Cartesian equations of motion of a system with constraints. Molecular dynamics of n-alkanes. J. Comput. Phys. 23, 327–341 [Google Scholar]
- 55. Berendsen H. J., Postma J. P., van Gunsteren W. F., DiNola A., Haak J. R. (1984) Molecular dynamics with coupling to an external bath. J. Chem. Phys. 81, 3684–3690 [Google Scholar]
- 56. Foloppe N., MacKerell A. D., Jr. (2000) All-atom empirical force field for nucleic acids. I. Parameter optimization based on small molecule and condensed phase macromolecular target data. J. Comput. Chem. 21, 86–104 [Google Scholar]
- 57. Jorgensen W., Chandrasekhar J., Madura J. D., Impey R. W., Klein M. L. (1983) Comparison of simple potential functions for simulating liquid water. J. Chem. Physics 79, 926–935 [Google Scholar]
- 58. MacKerell A. D., Jr., Feig M., Brooks C. L., 3rd (2004) Improved treatment of the protein backbone in empirical force fields. J. Am. Chem. Soc. 126, 698–699 [DOI] [PubMed] [Google Scholar]
- 59. Darden T., York D., Pedersen L. (1993) Particle mesh Ewald. An N log(N) method for Ewald sums in large systems. J. Chem. Phys. 98, 10089–10092 [Google Scholar]
- 60. Brooks B. R., Brooks C. L., 3rd, Mackerell A. D., Jr., Nilsson L., Petrella R. J., Roux B., Won Y., Archontis G., Bartels C., Boresch S., Caflisch A., Caves L., Cui Q., Dinner A. R., Feig M., Fischer S., Gao J., Hodoscek M., Im W., Kuczera K., Lazaridis T., Ma J., Ovchinnikov V., Paci E., Pastor R. W., Post C. B., Pu J. Z., Schaefer M., Tidor B., Venable R. M., Woodcock H. L., Wu X., Yang W., York D. M., Karplus M. (2009) CHARMM. The biomolecular simulation program. J Comput. Chem. 30, 1545–1614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Bialkowski K., Cysewski P., Olinski R. (1996) Effect of 2′-deoxyguanosine oxidation at C8 position on N-glycosidic bond stability. Z. Naturforsch. C 51, 119–122 [DOI] [PubMed] [Google Scholar]
- 62. Berdal K. G., Johansen R. F., Seeberg E. (1998) Release of normal bases from intact DNA by a native DNA repair enzyme. EMBO J. 17, 363–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Wyatt M. D., Allan J. M., Lau A. Y., Ellenberger T. E., Samson L. D. (1999) 3-Methyladenine DNA glycosylases. Structure, function, and biological importance. Bioessays 21, 668–676 [DOI] [PubMed] [Google Scholar]
- 64. Qi Y., Spong M. C., Nam K., Karplus M., Verdine G. L. (2010) Entrapment and structure of an extrahelical guanine attempting to enter the active site of a bacterial DNA glycosylase, MutM. J. Biol. Chem. 285, 1468–1478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Lovell S. C., Davis I. W., Arendall W. B., III, de Bakker P. I. W., Word J. M., Prisant M. G., Richardson J. S., Richardson D. C. (2003) Structure validation by Cα geometry: ϕ, ψ and Cβ deviation. Proteins Struct. Funct. Genet. 50, 437–450 [DOI] [PubMed] [Google Scholar]
- 66. Lu X. J., Olson W. K. (2003) 3DNA. A software package for the analysis, rebuilding, and visualization of three-dimensional nucleic acid structures. Nucleic Acids Res. 31, 5108–5121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Collaborative Computational Project Number 4 (1994) The CCP4 Suite: Programs for Protein Crystallography. Acta Cryst. D50, 760–763 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.