Background: MicroRNA-29-YY1 regulatory circuitry functions during skeletal myogenesis.
Results: A genome-wide search revealed Rybp as a novel target of miR-29, and it silences myogenic loci together with YY1.
Conclusion: Rybp functions as a repressor of myogenesis.
Significance: This study identifies a novel regulatory circuitry underlying muscle formation and highlights the intimate interplay among transcription factors, epigenetic regulators, and microRNAs.
Keywords: MicroRNA, Myogenesis, Polycomb, Skeletal Muscle, Transcription/Developmental Factors, Rybp, YY1, MicroRNA-29
Abstract
Skeletal muscle cell differentiation (myogenesis) is a process orchestrated by a complex network involving transcription factors, epigenetic regulators, and microRNAs. Previous studies identified miR-29 as a pro-myogenic factor that interacts with components of Polycomb repressive complex, YY1 and Ezh2. In a genome-wide survey of miR-29-mediated transcriptome changes in C2C12 myoblasts, many epigenetic factors were found to be down-regulated by miR-29. Among them, Rybp was shown to be a direct target of miR-29 through binding to its 3′ UTR. Functional studies demonstrated that Rybp is down-regulated during myogenesis and acts as a negative regulator of skeletal myogenesis both in vitro during C2C12 differentiation and in vivo in injury-induced muscle regeneration. Furthermore, we found that Rybp and YY1 co-occupy several myogenic loci, including miR-29 itself, to silence their expression, thus forming a Rybp-miR-29 feedback loop. Rybp overexpression was found to enhance the enrichment of Ezh2 and trimethylation of H3K27 at target loci, suggesting it may facilitate the recruitment or stabilization of the Polycomb repressive complex. Collectively, our results identify Rybp as a novel regulator of myogenesis that co-acts with YY1 to silence miR-29 and other myogenic loci.
Introduction
Skeletal muscle growth and regeneration are attributed to satellite cells (muscle stem cells), which are characterized by the expression of paired-box transcription factor 7 (Pax7) and when activated, become immature muscle cells or myoblasts that will proliferate and differentiate. The formation of mature muscle proceeds with the exit of myoblasts from the cell cycle, the expression of muscle-specific genes, and the suppression of genes that are specific to other cell lineages and tissues (1). A major portion of our understanding of myogenic differentiation is focused at the level of transcription, orchestrated by a complex network of muscle-specific transcription factors, including MyoD, myogenic factor 5 (Myf5), myogenin, myogenic regulatory factor 4 (MRF4), and myocyte enhancer factor 2 (Mef2). These factors activate muscle-specific genes to coordinate myoblasts to terminally withdraw from the cell cycle and subsequently fuse into multinucleated myotubes (2). In addition to transcription factors, epigenetic regulators exert an important layer of transcriptional control (3). The interplay between transcription factors and these regulators on muscle loci constitutes an important part of the signaling pathways.
Studies from recent years have also incorporated microRNAs (miRNAs)3 into the complex network of myogenic regulation. As noncoding single-stranded RNAs of 21 to 25 nucleotides and a novel class of gene regulators, miRNAs negatively regulate their targets at the post-transcriptional level through binding to their 3′ UTR leading to either mRNA cleavage or translational repression (4). A subset of these miRNAs, miR-1, miR-133, and miR-206, play central regulatory roles in muscle biology and are called muscle miRs. In addition to the above muscle miRs, emerging evidence also supports the involvement of non-muscle miRs in myogenesis (5). In an effort to elucidate the transcriptional and post-transcriptional regulation during skeletal myogenesis, we identified an NF-κB (NF-κB)-Yin Yang1 (YY1)-miR-29 signaling axis (6). YY1, a ubiquitously expressed transcription factor, upon activation by NF-κB signaling, functions to repress muscle differentiation by epigenetically silencing the transcription of miR-29 through recruiting histone methyltransferase Ezh2 as well as histone deacetylase 1 (6, 7). At the onset of myogenesis, YY1 is down-regulated by decreased NF-κB signaling, which results in the replacement of the YY1-Ezh2-histone deacetylase 1 repressive complex by an activator complex containing MyoD, serum response factor along with associated acetyltransferases cAMP-response element-binding protein and p300/cAMP-response element-binding protein-associated factor, leading to the activation of miR-29. MiR-29 in turn targets Yy1 resulting in its further decrease. These findings highlight the intimate interaction of the transcription factor, miRNA, and chromatin modulator.
In addition to Yy1, miR-29 directly targets DNMT3a and DNMT3b in lung cancer and acute myeloid leukemia (AML) (8, 9). MiR-29b can also indirectly silence DNMT1 by targeting SP1, a transactivator of DNMT1 (9). These findings together with ours support the ability of miR-29 in regulating epigenetic machinery. We thus speculated that in addition to Yy1, miR-29 may regulate many other epigenetic factors whose unveiling will be facilitated by a genome-wide search.
As members of Polycomb group proteins (PcGs), YY1 and Ezh2 together regulate a number of muscle loci including both muscle structure genes and muscle relevant miRNAs (5). The PcG genes encode a structurally diverse group of proteins, which form large complexes arising from their mutual interactions. Despite their well characterized functions in embryonic stem cells (ESCs) (10, 11), it is unknown if other PcG proteins are involved in myogenesis and whether they are part of the YY1/Ezh2 containing PRC.
In this work, a genome-wide transcriptome survey was employed to uncover that a number of epigenetic factors were down-regulated in miR-29 expressing C2C12 cells, with Rybp, an YY1 interacting PcG protein, being one of them. MiR-29 was able to target Rybp directly through a conserved binding site on its 3′ UTR. Both in vitro and in vivo functional studies showed that Rybp is a negative regulator of skeletal myogenesis, which functions as a co-repressor of YY1 to silence the myogenic loci including miR-29 itself. Our study thus identified a novel functional Rybp-miR-29 regulatory circuitry in skeletal muscle formation.
EXPERIMENTAL PROCEDURES
Cells
Mouse C2C12 myoblasts (CRL-1772) were obtained from ATCC and cultured in DMEM supplemented with 10% FBS, 2 mm l-glutamine, 100 units/ml of penicillin, and 100 μg of streptomycin at 37 °C in 5% CO2. For myogenic differentiation, cells were seeded in 60- or 100-mm plates and shifted to DMEM containing 2% horse serum when 90% confluence. Primary myoblasts were isolated from approximately 1-week-old mice muscles by the described procedures (12, 13). Briefly, total hind limb muscles (3 to 6 mice per group) were digested with type IV collagenase (Invitrogen, 5 mg/ml) and dispase II (Invitrogen, 1.4 mg/ml) for 0.5 h, and cell suspensions were further homogenized by pipetting before being filtered through 70- and 40-μm filters. The obtained cells were pre-plated on uncoated cell culture plates in F10 media (Invitrogen) to selectively enrich for myoblasts. After two rounds of pre-plating, the cell suspension was plated on gelatin-coated plates (Iwaki) in F10 medium (Invitrogen) supplemented with 20% FBS and basic fibroblast growth factor (Invitrogen, 25 ng/ml). Primary myoblasts were used at passages 3–5 after isolation.
Transfection and Infection
Transient transfection with miRNA precursor oligos, siRNA oligos, or DNA plasmids was performed on 60- or 100-mm dishes with Lipofectamine 2000 reagent as suggested by the manufacturer (Invitrogen). For luciferase experiments, C2C12 and primary myoblasts were transfected in 12-well plates. Cell extracts were prepared and luciferase activity was monitored as previously described (14) or using a dual-luciferase kit (Promega). To generate C2C12 cells stably expressing Rybp, a Rybp expressing plasmid (4 μg) was transfected into C2C12 cells using Lipofectamine 2000 (Invitrogen). 36 h after transfection, cells were placed in 400 μg/ml of G418 (Invitrogen) for stable selection. Stable clones were pooled together after ∼2 week selection.
Oligonucleotides
Precursor miRNA oligos were obtained from Ambion. miRCURY LNA microRNA inhibitor or control oligos were obtained from Exiqon. The 19-nucleotide siRNA duplexes against mouse Rybp coding region (siRNA, 5′-CGACATGTCAGCAGTGAAT-3′) or scrambled oligos were obtained from RiboBio. In each case 100 nm oligos were used for transient transfection into cells.
DNA Constructs
To construct a Rybp-3′ UTR reporter plasmid, a 401-bp fragment encompassing the miR-29 binding site was amplified by primers (forward primer, 5′-AGTTCTAGAAGACTCCATCCTGTCAGCATATC-3′; reverse primer, 5′-AGTTCTAGAGGAAAAAGGCCGGAATTCGA-3′) and cloned into pGL3 vector (Promega) at the XbaI site. To construct a mutant reporter plasmid, the seed region was mutated from TGGTGCTT to TCATAAGT. To generate a Rybp expression plasmid, the full-length coding region of Rybp was amplified by primers (forward primer, 5′-AGTGCTAGCATGACCATGGGCGACAAGAAG-3′; reverse primer, 5′-AGTGGTACCTCAGAAAGATTCATCATTCACTGC-3′), digested, and cloned into pcDNA3.1(+) vector (Invitrogen) at NheI and KpnI sites. An YY1 expression plasmid was a gift from Y. Shi (Harvard University) and used as described (7). A miR-29-promoter luciferase reporter was created and used as described (6). Renilla luciferase reporter was obtained from Promega and used according to the manufacturer's instructions.
RT-PCR and Real-time RT-PCR
Total RNAs were extracted using TRIzol reagent (Invitrogen). Expression of mature miRNAs was determined using the miRNA-specific TaqMan microRNA assay kit (Applied Biosystem) on an ABI PRISM 7900HT Sequence Detection System (Applied Biosystem). U6 was used for normalization. Expression of mRNA analysis was performed with SYBR Green Master Mix (Bio-Rad) as described using GAPDH for normalization (7). Primers used are listed in supplemental Table S1.
Immunoblotting, Immunostaining, and Immunohistochemistry
For Western blotting analyses, total cell extracts were prepared and used as previously described (14). The following dilutions were used for each antibody: Myogenin (Santa Cruz Biotechnology; 1:2000), YY1 (Santa Cruz Biotechnology; 1:2000), Rybp (Millipore; 1:2000), Troponin (Sigma; 1:2000), MyHC (Sigma; 2,000), α-Tubulin (Sigma; 1:5000), Pax7 (Developmental Studies Hybridoma Bank; 1:2000), eMyHC (Leica, 1:2000), and GAPDH (Santa Cruz Biotechnology; 1:5000). Densimetric quantification of the Western bands was performed using Quantity One software (Bio-Rad). Immunofluorescence on cultured cells was performed using the following antibodies: Rybp (Millipore; 1:400), MyHC (Sigma; 1:350), and YY1 (Santa Cruz Biotechnology; 1:400). Frozen muscle sections were prepared and stained as previously described (17). Immunofluorescence staining on frozen muscle sections was performed using the following antibodies: MyoD (Santa Cruz, 1:100), Myogenin (Santa Cruz, 1:100), and Pax7 (DSHB, 1:100). H&E staining on frozen muscle sections was performed as previously described (17). Quantification of the number of fibers with centrally located nuclei (CLN) and IF positively stained cells was performed from a minimum of 20 randomly chosen fields, from 5 to 6 sections throughout the length of the muscle in 4–6 mice per group. All fluorescent images were captured with a confocal laser scanning microscope (FV1000, Olympus, Japan). All samples were imaged with ×20 or 40 objective lens. Pictures were captured in Kahlman frame giving an average of two scans using the Olympus microscope FV1000 and the accompanying software FV10-ASW (version 01.07.02.02, Olympus).
Co-immunoprecipitation Assays
Co-IP was performed as described before (18). Briefly, 10 μg of normal IgG (Santa Cruz Biotechnology), antibodies against YY1 (Santa Cruz Biotechnology), or Rybp (Millipore) was cross-linked to 50 μl (bed volume) of Protein A/G PLUS-agarose (Santa Cruz Biotechnology) by dimethyl pimelimidate (Sigma). C2C12 cells were first cross-linked with 200 μg/ml of 3,3′-dithiodipropionic acid di(N-hydroxysuccinimide ester) (Sigma) for 20 min and then harvested and lysed in radioimmunoprecipitation assay (RIPA) buffer (25 mm HEPES, pH 7.4, 1% Nonidet P-40, 0.1% SDS, 0.5% sodium deoxycholate, 1 × Protease Inhibitor Mixture). The antibody-conjugated beads were incubated with 500 μg of the above cell lysate overnight at 4 °C with rotation. After extensive washing with RIPA buffer, the bound proteins were eluted by boiling in 20 μl of 2× sample buffer (125 mm Tris-HCl, pH 6.8, with 4% SDS, 20% (v/v) glycerol) and subjected to Western blot analysis.
ChIP Assay
ChIP assays were performed as recommended by the manufacturer (Upstate) using 5 μg of antibodies against Rybp (Millipore), YY1 (Santa Cruz Biotechnology), Ezh2 (Cell signaling), trimethyl-histone H3-K27 (Millipore), or isotype IgG (Santa Cruz Biotechnology) used as a negative control. Genomic DNA pellets were resuspended in 20 μl of water. qRT-PCR was performed with 1 μl of immunoprecipitated material with SYBR Green Master Mix (Bio-Rad). Relative enrichment is calculated as the amount of amplified DNA normalized to input and relative to values obtained after normal IgG immunoprecipitation, which were set as 1. Primers used are listed in supplemental Table S1.
Animal Studies
C57B/L mice were housed in the animal facility of the Chinese University of Hong Kong (CUHK) under conventional conditions with constant temperature and humidity and fed a standard diet. Animal experimentation was approved by the CUHK Animal Experimentation Ethics Committee (Ref No. 10/027/MIS). For cardiotoxin (CTX) injection, approximately 5-week-old mice were injected with 60 μl of CTX at 10 μg/ml into the tibialis anterior (TA) muscles. Oligos were prepared by preincubating 10 μm siRNA oligos with Lipofectamine 2000 for 15 min and injections were made in a final volume of 60 μl in OPTI-EM (Invitrogen) at days 1/4, 2, and 4. Mice were sacrificed and TA muscles were harvested at days 1, 2, 4, and 6, and total RNAs and proteins were extracted for real-time RT-PCR and Western analyses. For immunofluorescene staining of MyoD, Myogenin, and Pax7, muscle sections were collected at day 3.
Statistical Analysis
Statistical significance was assessed by the Student's t test (*, p < 0.05; **, p < 0.01; ***, p < 0.001).
RESULTS
Rybp Is Down-regulated in miR-29 Expressing C2C12 Cells
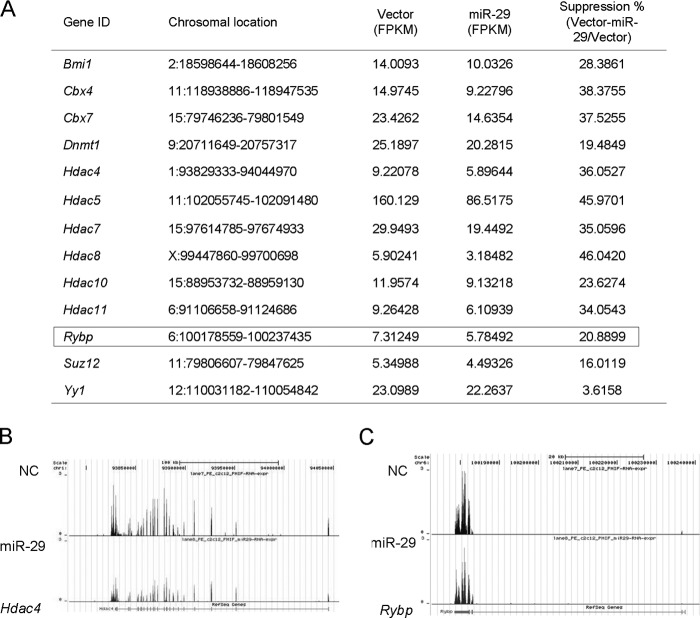
To gain insights into the miR-29-mediated events in muscle cells, we previously performed a genomewide transcriptome analysis by high throughput RNA sequencing (19). 739 genes were found to be down-regulated by miR-29 overexpression. Interestingly, many epigenetic factors were decreased, including previously identified targets of miR-29, Hdac4 and Dnmt1 (Fig. 1, A and B), suggesting miR-29 is an important epigenetic regulator. When compared with vector control cells, these genes were suppressed to a different extent with Cbx4 38 versus Suz12 16%. Yy1 RNAs, on the other hand, were not found to be significantly down-regulated. This is in line with our previous finding showing that miR-29 represses Yy1 translation but not transcription (6). Among the down-regulated genes, Rybp caught our attention because it was consistently predicted to be a direct target of miR-29 by three algorithms, TargetScan (20), miRanda (21), and PicTar (22). Moreover, as a known modifier of chromatins and a member of the PcG genes, its role in myogenesis has never been explored. As revealed by RNA-seq, it was suppressed about 21% by miR-29 overexpression (Fig. 1, A and C).
FIGURE 1.
Overexpression of miR-29 in C2C12 cells down-regulates Rybp. A, down-regulated epigenetic factors in C2C12 cells stably expressing miR-29 as revealed by RNA-seq. The abundances were reported in FPKM (fragments per kilobase of transcript per million fragments mapped). Suppression ratio was obtained by calculating the percentage of the expression difference between vector and miR-29 cells relative to vector. B, coverage plot showing the ∼200 kb genomic region encompassing the Hdac4 gene on chromosome 1; the gene structure is shown below the graph. C, coverage plot showing the ∼60 kb region encompassing Rybp gene on chromosome (Chr) 6; the gene structure is shown below the graph.
miR-29 Directly Targets Rybp through Binding to Its 3′ UTR
To investigate whether the down-regulation of Rybp is due to direct targeting, A search for miR-29 binding sites within the Rybp 3′ UTR revealed that all three miR-29 family members, miR-29a, -29b, -29c, were predicted to hybridize to an evolutionarily conserved site among vertebrate species (Fig. 2A and supplemental Fig. S1). A perfect match exists between the seed region of miR-29 and the 3′ UTR of Rybp, suggesting that miR-29 can directly repress Rybp expression. This was tested by cloning a 401-bp fragment of the Rybp 3′ UTR encompassing the target site downstream of the firefly luciferase gene. Co-transfection of this reporter (WT) with each of the miR-29 family members caused similar repression of luciferase reporter (Fig. 2B). This was specific to miR-29 binding because the reporter activity was less affected when transfections were repeated with an irrelevant miRNA, miR-194, or with a mutant miR-29 binding site in the Rybp 3′ UTR. At the functional level, we predicted that miR-29 binding to the Rybp 3′ UTR would lead to Rybp repression. Indeed, ectopic expression of miR-29 by transfection of miR-29 oligos or transfection of a pMIF-miR-29 plasmid caused decreased expression of Rybp at both RNA and protein levels (Fig. 2, C and D, and supplemental Fig. S2, A–D), and knockdown of miR-29 by anti-miR oligos led to increased expression of Rybp (Fig. 2, C and D, and supplemental Fig. S2E). Interestingly, in addition to miR-29, Rybp is also predicted to be targeted by muscle miRNAs, miR-1 and miR-206 (supplemental Fig. S3A). Indeed, results from expression and luciferase reporter assays performed as above demonstrated that miR-1 and miR-206 can also directly target Rybp (supplemental Fig. S3, B–F). This is in consistent with the common idea that an mRNA can be targeted by multiple miRNAs. It also further indicates the relevancy of Rybp to myogenesis.
FIGURE 2.
miR-29 suppresses Rybp expression through direct binding to its 3′ UTR. A, a schematic illustration of base pairing between mmu-miR-29c with the 3′ UTR of the mouse Rybp gene. B, a wild type (WT) or mutant Rybp 3′ UTR luciferase reporter was transfected into C2C12 cells along with the indicated precursor miRNA oligos. Luciferase activities were determined at 48 h post-transfection and normalized to Renilla luciferase activity. Data represent the average of three independent experiments ± S.D. C, Rybp mRNA expression was measured in NC, miR-29, anti-NC, or anti-miR-29 transfected C2C12 cells by qRT-PCR and normalized with GAPDH. Expression folds are shown with respect to NC or anti-NC cells where normalized copy numbers were set to 1. D, Rybp protein expression was measured in the above transfected C2C12 cells by Western blotting using α-Tubulin as a loading control. Numbers below indicate densimetric quantification of the Western bands.
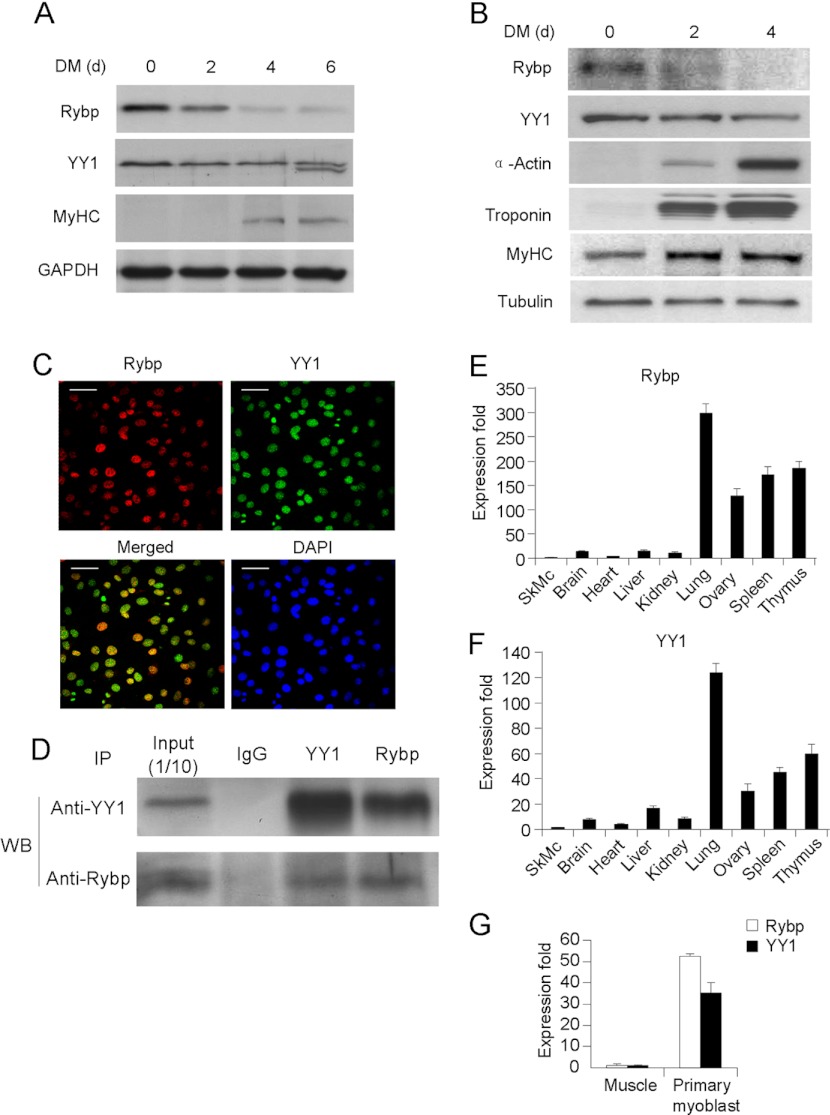
Rybp Is Down-regulated in C2C12 Myogenesis Concomitant with YY1
To probe into the function of Rybp in skeletal myogenesis, we examined the expression levels of Rybp during C2C12 myoblast differentiation. As shown in Fig. 3A, its expression gradually decreased during a time course of days 0, 2, 4, and 6, whereas the myogenic marker, myosin heavy chain (MyHC) increased. This was concomitant with gradually decreased YY1 expression as well as a gradual increase of miR-29, miR-1, and miR-206 (supplemental Fig. S4), suggesting a functional connection between Rybp and YY1. Consistently, in primary myoblasts isolated from mouse limb muscles, expression of Rybp decreased gradually along with YY1 during differentiation in contrast to the up-regulated expressions of myogenic markers, α-skeletal actin (α-Actin), Troponin, and MyHCIIB (MyHC) (Fig. 3B). Immunofluorescence (IF) staining also revealed that Rybp was co-localized with YY1 in the nuclei of C2C12 myoblasts (Fig. 3C). Furthermore, a physical interaction of Rybp and YY1 was detected in C2C12 myoblasts by co-immunoprecipitation (co-IP) (Fig. 3D). Together, the above findings suggest that Rybp may be an anti-myogenic factor that acts in concert with YY1. Additional evidence for their functional relationship came from the expression patterns of their transcripts across tissues. Expressions of Rybp and Yy1 were found to be highly coordinated with the highest expression levels in lung and the lowest in skeletal muscle (Fig. 3, E and F). Although Rybp is low in mature skeletal muscle, it was found to be highly enriched in freshly isolated myoblasts together with Yy1 (Fig. 3G), suggesting its role in regulating myoblast growth or differentiation.
FIGURE 3.
Rybp is down-regulated during C2C12 myogenesis concomitant with YY1. A, C2C12 myoblasts were induced to differentiate in differentiation medium (DM) for 0, 2, 4, or 6 days. Total proteins were extracted and used for Western blotting for Rybp, YY1, and MyHC. GAPDH was used as a loading control. B, primary myoblasts were induced to differentiate in DM for 0, 2, and 4 days. Total proteins were extracted and used for Western blotting analysis for Rybp, YY1, α-Actin, Troponin, and MyHC. α-Tubulin was used as a loading control. C, C2C12 cells were stained for Rybp (red) and YY1 (green) by IF. The merged signals of Rybp and YY1 are yellow. DAPI staining indicates the nuclei. Bars = 50 μm. D, protein lysates were extracted from C2C12 myoblasts and subjected to co-IP assay using antibodies against YY1 or Rybp. E and F, total RNAs were extracted from the indicated mouse tissues, and the expression levels of Rybp and Yy1 mRNAs were measured by qRT-PCR and normalized with GAPDH. Data are represented as mean ± S.D. G, expression levels of Rybp or Yy1 in muscle tissue or primary myoblasts were measured by qRT-PCR.
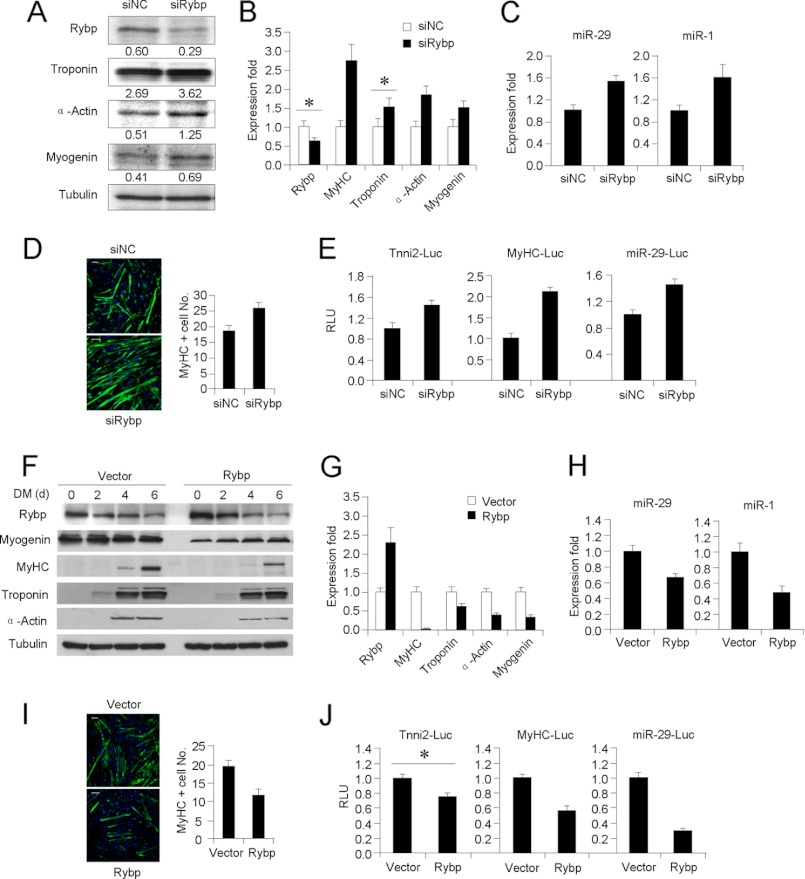
Rybp Functions as a Negative Regulator of Skeletal Myogenesis
To test the functional relevance of Rybp during myogenesis, we knocked-down Rybp in C2C12 cells with a siRNA oligo (siRybp). The decreased expression of Rybp was found to enhance C2C12 myogenic differentiation as evidenced by the up-regulated expressions of MyHC, Troponin, α-Actin, and Myogenin at both protein and RNA levels (Fig. 4, A and B). Specifically, miR-29 and miR-1, two other direct transcriptional targets of YY1, were up-regulated by Rybp decrease (Fig. 4C). IF staining also revealed an increased number of MyHC-positive myotubes upon decrease of Rybp (Fig. 4D). Reporter assay with luciferase reporter plasmids containing Troponin, MyHC, miR-29, and promoter/enhancer upstream of luciferase gene, demonstrated an increase of luciferase activity in siRybp cells as compared with cells transfected with negative control oligos (siNC) (Fig. 4E). Collectively, the above findings suggest that Rybp inhibits C2C12 myogenic differentiation. To strengthen the above findings, overexpressing Rybp stably in C2C12 was found to delay differentiation compared with cells expressing vector control as revealed by an expression assay, IF, and reporter assay performed as described above (Fig. 4, F–J).
FIGURE 4.
Rybp suppresses myogenic differentiation. A, C2C12 cells were transfected with siNC or siRybp oligos, and differentiated for 2 days. Total proteins were isolated and Western blotting was performed to probe for Rybp, Troponin, α-Actin, and Myogenin expressions using α-Tubulin as a loading control. Numbers below indicate densimetric quantification of the Western bands. B, total RNAs were extracted from the above transfected cells and used for qRT-PCR analysis of Rybp, MyHC, Troponin, α-Actin, and Myogenin normalized with GAPDH. Expression folds are shown with respect to siNC cells where normalized copy numbers were set to 1. Data are plotted as mean ± S.D. C, qRT-PCR was performed in the above transfected cells to detect the expressions of miR-29 and miR-1 using U6 as normalization control. D, the above transfected cells were stained for MyHC by IF (left). Bars = 50 μm. Positively stained cells were quantified by counting 10 randomly chosen fields and are represented as mean ± S.D. (right). E, C2C12 cells were transfected with Tnni2-Luc, MyHC-Luc, or miR-29-Luc reporters along with pCMV-LacZ and siNC or siRybp oligos. Cells were differentiated for 48 h, and luciferase activities were determined and normalized to activities of β-glactosidase. Relative activity is shown with respect to control cells, where normalized luciferase values were set to 1. Data represent the average of three independent experiments ± S.D. F, C2C12 cells stably expressing Rybp or vector control were differentiated and harvested at the indicated time points. Total proteins were isolated and Western blotting was performed to probe for Rybp, MyHC, Troponin, and α-Actin expressions using α-Tubulin as loading control. G, C2C12 cells were transfected with Rybp expressing plasmid or vector control, and then differentiated for 2 days. Total RNAs were extracted from the above transfected cells and used for qRT-PCR analysis of Rybp, MyHC, Troponin, α-Actin, and Myogenin as in B. H, qRT-PCR was performed in the above transfected cells to detect the expression of miR-29 and miR-1 using U6 as normalization control. I, the above transfected cells were stained for MyHC by IF (left). Bars = 50 μm. Positively stained cells were quantified by counting 10 randomly chosen fields and are represented as mean ± S.D. (right). J, C2C12 cells were transfected with Tnni2-Luc, MyHC-Luc, or miR-29-Luc reporters along with pCMV-LacZ and vector or Rybp expressing plasmids. Cells were differentiated for 48 h, and luciferase activities were determined as in E.
Rybp Functions as a Co-repressor of YY1 to Silence Myogenic Loci in Myoblasts
To gain insights into the molecular mechanism on how Rybp repressed myogenesis, we speculated that Rybp may function in the same repressive complex containing YY1 and Ezh2 to epigenetically silence muscle genes and muscle relevant miRNA targets in myoblasts. To test this notion, ChIP assays were performed against Rybp and YY1 using chromatins isolated from C2C12 cells stably expressing vector or Rybp. Both proliferating myoblasts and differentiated myotubes were included in the study. Promoters/enhancers of three known YY1 targets, miR-29, miR-1, and Tnni2, were examined for their association with Rybp and YY1. As expected, in vector myoblasts, strong binding of Rybp was detected on all three target regions along with YY1, and the association dramatically reduced in myotubes (Fig. 5, A and B), suggesting that Rybp functions together with YY1 to silence target expression in proliferating myoblasts and removed together upon myogenic differentiation. Rybp overexpression led to an increase of Rybp binding on these loci but not YY1 enrichment (Fig. 5, A and B), implying that Rybp may not have impact on YY1 targeting. Knowing that Ezh2 is a co-repressor of YY1 on these loci, we further examined Ezh2 binding. Interestingly, Rybp overexpression was found to enhance Ezh2 association (Fig. 5C). Consistently, levels of H3K27me3 displayed the same pattern as Ezh2 (Fig. 5D). Collectively, these data suggest that Rybp functions to suppress muscle genes and miRNAs transcription together with YY1 and Ezh2. This notion was further strengthened by transfecting YY1 or/and Rybp expression plasmids along with a miR-29 promoter reporter in C2C12 cells. Rybp or YY1 alone was found to inhibit the reporter activity to a similar extent, whereas Rybp expression increased YY1 inhibition at a dose-dependent manner (Fig. 5E).
FIGURE 5.
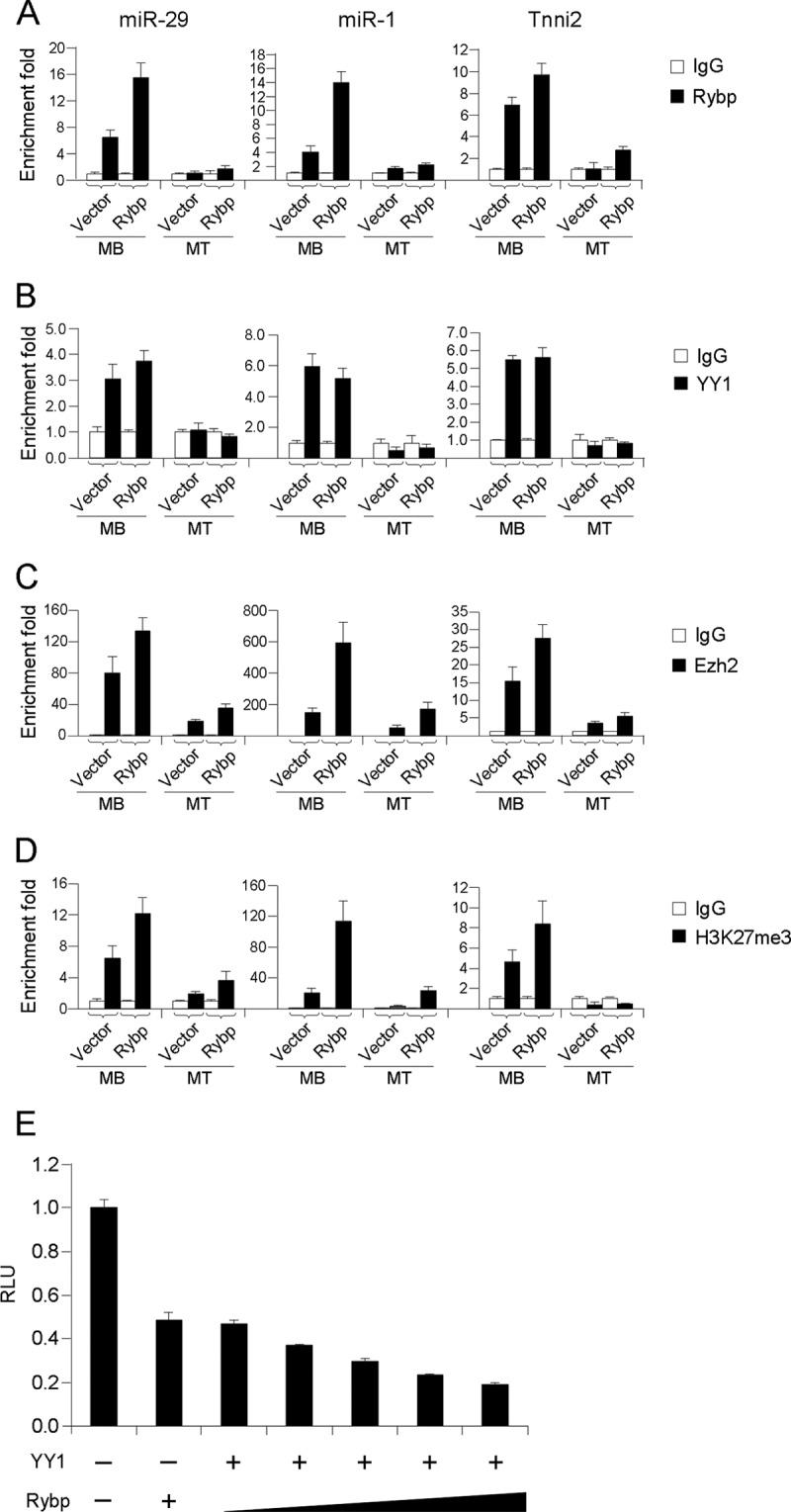
Rybp functions as a co-repressor of YY1 to silence myogenic loci. A–D, C2C12 cells expressing vector control or Rybp were cultured as myoblasts or differentiated into myotubes, at which stages chromatins were collected for ChIP-PCR assays using antibodies against Rybp, YY1, Ezh2, H3K27me3, or IgG as controls. Primers for amplifying promoters/enhancers of miR-29, miR-1, and Tnni2 were used for qRT-PCR analysis. Enrichment folds are shown with respect to IgG control where normalized PCR values were set to 1. Data are plotted as mean ± S.D. E, C2C12 cells were transfected with 0.25 μg of the miR-29-promoter-luc reporter plasmid along with 0.5 μg of Rybp, or 0.5 μg of YY1 plasmid combined with Rybp plasmids (0, 0.2, 0.3, 0.4, or 0.5 μg). Relative activities were shown with respect to control cells, where normalized luciferase values were set to 1. Data represent the average of three independent experiments ± S.D.
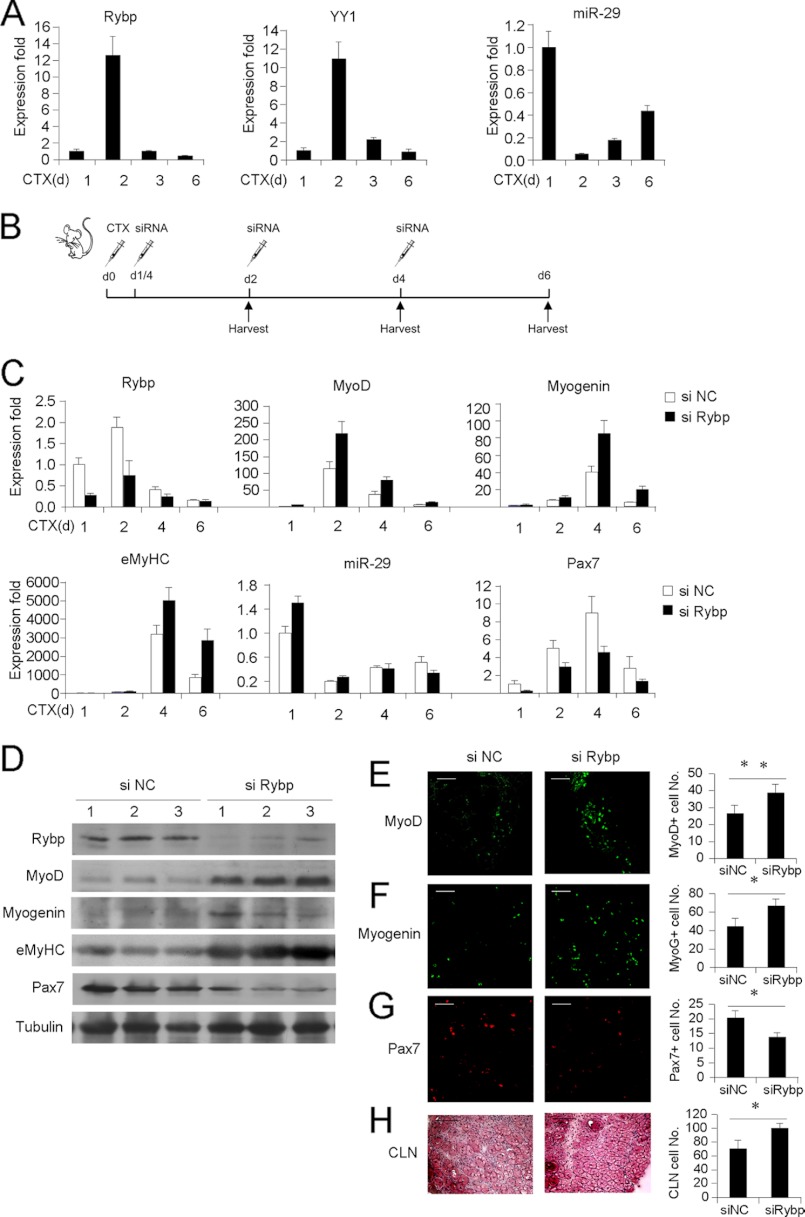
Knockdown of Rybp in Vivo Enhances Injury-induced Muscle Regeneration
Last, to extend our in vitro findings to in vivo muscle formation, we explored the function of Rybp in a widely used muscle regeneration model in which the injection of CTX results in muscle injury and in turn induces muscle regeneration. After injection with CTX, TA muscle displays the typical degeneration-regeneration process: fiber degeneration and immune cell infiltration were immediately evident within 1 to 2 days, meanwhile satellite cells start proliferating followed by activation and myogenic differentiation 3 to 4 days afterward, newly formed fibers with CLN are evident within 5 to 6 days, and muscle architecture is largely restored within 10 days (12).
The expression profile of Rybp mRNAs was examined at days 1, 2, 3, and 6 days post-injection. A sharp increase of more than 10-fold in the first 2 days post-injury was observed, after which its expression level gradually fell down until day 6 (Fig. 6A, left panel). This was temporally correlated with Yy1 expression (middle panel), suggesting that a dynamic and coordinated change of Rybp and Yy1 expressions occurred during in vivo muscle degeneration-regeneration. On the contrary, miR-29 expression was inversely correlated with Rybp and Yy1 (right panel): it sharply decreased in the degeneration stage, and then gradually increased in the regeneration stage. This was in keeping with the in vitro finding that Rybp and YY1 together inhibit miR-29 transcription.
FIGURE 6.
Knockdown of Rybp improves muscle regeneration in vivo. A, TA muscles from 6-week-old C57/BL6 background mice were injected with CTX. RNAs were then extracted from injected muscles at the indicated days postinjection, and qRT-PCR was performed to measure the expressions of Rybp and Yy1 normalized with GAPDH or miR-29 normalized with U6. Expression folds are shown with respect to day 1 where Rybp, Yy1, and miR-29 levels were set to a value of 1. Quantitative values are represented as mean ± S.D. B, TA muscles were injected with CTX at day 0, followed by injection with siNC (left leg) and siRybp1 oligos (right leg) 6 h later. Reinjection of siRNA oligos was performed every other day for two more times. The injected muscles were harvested at the indicated days. n = 4 for each group. C, total RNAs were extracted from the above injected muscles. Expression levels of Rybp, MyoD, Myogenin, eMyHC, miR-29, and Pax7 RNAs were detected by qRT-PCR. Expression folds are shown with respect to siNC where expression levels were set to a value of 1. Data are represented as mean ± S.D. D, proteins were extracted from the above injected muscles and probed for Rybp, MyoD, Myogenin, eMyHC, or Pax7 using α-tubulin as a loading control. E–G, frozen muscle sections were prepared from the above injected muscles on day 3 post-injection and IF staining was performed for MyoD, Myogenin, or Pax7. H, H&E staining was used to visualize fibers with CLN on day 6 post-CTX injection. Positively stained cells or fibers with CLN were quantified. *, p < 0.05; **, p < 0.01.
To ascertain the functional significance of Rybp regulation, siNC or siRybp oligos were injected into CTX-injured TA muscles following the administration scheme outlined in Fig. 6B. Six hours after CTX injection into TA muscles, siNC or siRybp oligos were administrated into the left and right hind limbs, respectively. The injections were made every 2 days for a total of three times. The treated muscles were collected at days 1, 2, 4, and 6. Results from qRT-PCR and Western analyses revealed that the expressions of Rybp were decreased at all the time points (Fig. 6, C and D), suggesting a successful knockdown of Rybp by this approach. Expression levels of MyoD, Myogenin, and embryonic MyHC (eMyHC, a marker for regenerating fibers) were found to be up-regulated by siRybp injection (Fig. 6, C and D), suggesting an acceleration of the myogenic program as discovered in vitro. Consistently, miR-29 expression was found to be up-regulated at days 1 and 2 during the regeneration peak (Fig. 6C, day 1 and 2). Interestingly, the expression of Pax7 was found to be down-regulated (Fig. 6, C and D). Further evidence came from IF staining of the above markers on frozen muscle sections. MyoD and Myogenin positively stained cells were significantly increased by siRybp treatment (Fig. 6, E and F). Pax7 positively stained cells, on the other hand, were decreased (Fig. 6G). Additionally, siRybp treatment led to an increased number of newly formed fibers characterized by CLN (Fig. 6H). Together, these findings imply that Rybp is a functional anti-myogenic factor during muscle regeneration in vivo.
DISCUSSION
This study was initiated to investigate the genome-wide impact of miR-29 on epigenetic regulators in skeletal muscle cells. Unexpectedly, Rybp, a PcG member was discovered to be a likely target of miR-29. Subsequent study validated that Rybp is a novel target of miR-29. Further investigation identified Rybp as a negative regulator of skeletal myogenesis, which acts in concert with YY1. Rybp and YY1 were found to co-occupy several target promoters/enhancers of YY1 including miR-29 itself to silence their expression in myoblasts, thus forming a miR-29-Rybp feedback regulatory loop. Increased Rybp led to an enhancement of Ezh2 association on the target loci, suggesting that Rybp may facilitate PRC recruitment or stabilization. Moreover, we demonstrated that knockdown of Rybp accelerated injury-induced muscle regeneration in vivo, strengthening the argument that Rybp is a relevant regulator of skeletal muscle formation (Fig. 7).
FIGURE 7.
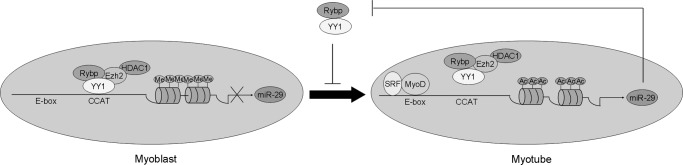
A model of Rybp/YY1-miR-29 regulatory circuitry in skeletal myogenesis. The model depicts the role of the Rybp/YY1-miR-29 regulatory circuitry in myogenic differentiation. In myoblast, a repressive complex containing YY1/Rybp/Ezh2/HDAC4 binds to the promoter region of miR-29, leading to the trimethylation of histone lysine 27 and subsequent silencing of miR-29 transcription. Upon myogenesis starts, the recruitment of the MyoD/serum response factor activating complex and the displacement of the repressive complex lead to the activation of miR-29, which in turns targets both Rybp and YY1 to ensure the progress of myogenic differentiation. Straight line, promoter/enhancer region of mmu-miR-29b/c with arrow denotes TSS; CCAT, YY1 binding element; E-box, MyoD binding site; Me, methylation of histone lysine 27; Ac, acetylation of histones.
Rybp Is a Novel PcG Repressor of Skeletal Myogenesis
PcG repressor proteins play key roles in establishing and maintaining gene expression patterns during cellular differentiation and development. There are two major biochemical complexes, PRC1 and PRC2, which have inherent histone-modifying activity critical for their function in gene repression. PcG target genes are often co-regulated by PRC1 and PRC2, which are hierarchically recruited to target loci (23). It is commonly accepted that Ezh2-containing PRC2 promotes the trimethylation of H3K27, which then leads to recruitment of PRC1. Being able to interact with both YY1 and PRC1 protein, Ring1, Rybp, and its homolog YAF2 is proposed to recruit PRC1 through interacting with RING1 (24). Previous studies have identified Rybp as a transcription silencer in cancer (25, 26), embryogenesis (27), and central nervous system development (28). Our current study is the first to demonstrate a role of Rybp in skeletal myogenesis. In line with the other studies, our results suggested Rybp as a transcriptional repressor and a repressor of myogenesis. Knockdown of Rybp was sufficient to accelerate C2C12 myogenesis, suggesting it as an indispensable factor. This is in contrast to a recent report showing Rybp is dispensable for gene repression in ESCs (29). Moreover, in vivo injection of siRybp oligos accelerated CTX-induced muscle regeneration, demonstrating that the role of Rybp is relevant in muscle formation. In contrast to increasing the number of cells stained with MyoD, Myogenin, and eMyHC, siRybp injection down-regulated the number of cells with Pax7 staining, suggesting it may positively regulate satellite cell proliferation. This will be an interesting direction for future investigation. Previously, we showed that YY1 is relevant in DMD pathogenesis and treatment of dystrophic muscles with miR-29 can increase muscle regeneration through down-regulating YY1 levels (17). Considering their intimate interaction, it will be interestingly to find out whether Rybp is also involved in this type of muscle disease.
Rybp Functions as a Co-repressor of YY1
The majority of studies on Rybp characterized it as a recruiter of PRC1. Rybp can bind both Ring1B and its substrate, ubiquitinated histone H2A, and thus participate in the long term stable repression established by the PRC1 complex (30). A very latest finding unveiled a H3K27me3 independent pathway of PRC1 recruitment, which is mediated directly through Rybp (31). However, it was not know how Rybp is recruited to target loci. Even though the physical interaction of YY1 and Rybp has been discovered long ago in an in vitro study (27), the biological significance of this interaction was hardly explored. In one study, Rybp and YY1 were found to form a transcriptional complex with E2F2/E2F3 controlling Cdc6 promoter activation (32). It was also proposed that YY1/Rybp acts as a recruiter of PcG complexes to their targets. However, despite some evidence for such an activity, chromatin association studies in ESCs failed to show YY1 co-localization with PcG targets (33). Our results, on the other hand, provided clear evidence for an in vivo association of YY1/Rybp on target loci. Co-occupancy of YY1, Rybp, Ezh2, and H3K27me3 at the same genomic region was observed on three known YY1 target loci, miR-29 and Tnni2 promoter (6, 7) as well as miR-1 enhancer (12). This is distinct to the recent observation showing Rybp binds to genomic regions with no or very low H3K27me3 in ESCs (29), suggesting a distinct function of Rybp in skeletal muscle cells versus ESCs. Furthermore, the fact that our ChIP assays detected co-occupancy of Ezh2, H3K27me3, and Rybp on the same genomic location suggest that Rybp may be somehow involved in recruitment or stabilization of PRC2. This notion is in line with a recent study of PcG regulation on the HOXD cluster in hESC differentiation (34). Knockdown of RYBP resulted in a loss of both BMI1 (PRC1) and SUZ12 (PRC2) from the promoter and a complete loss of H3K27me3. Thus, the YY1/Rybp interaction may be involved in PRC2 recruitment/stabilization in addition to PRC1 recruitment. Nevertheless, we cannot exclude the possibility that such a mechanism only occurs on the limited genomic loci. Rybp may exert diverse functions in a target and cell-type dependent manner. The coordinated function of YY1 and Rybp was further evidenced from their correlated expression patterns in both in vitro C2C12 cell differentiation and in vivo CTX-induced muscle regeneration. Interestingly, this expression pattern was also observed in multiple tissues, suggesting that the interaction of YY1 and Rybp may not be limited to muscle only.
miR-29 Functions as an Epigenetic Modulator
Our results reinforced the idea that miR-29 is a critical regulator of epigenetic machinery. In addition to Rybp and known targets, Hdac4 (35) and Dnmt1 (9), a dozen epigenetic modulators were down-regulated in miR-29 expressing cells. Among them Bmi1, Cbx4, Cbx7, and Suz12 are all PcG genes, suggesting miR-29 has strong impact on PcG-mediated chromatin modification. A number of Hdac genes, Hdac4, Hdac5, Hdac7, and Hdac8, were also down-regulated, suggesting a likely regulation of miR-29 on HDAC-mediated chromatin remodeling. Not only was Rybp directly targeted, its own expression is under transcriptional suppression by miR-29, thus forming a negative feedback loop. This regulation ensures that in myoblasts miR-29 is silenced; once differentiation ensued, NF-κB-decreased Yy1 relieves miR-29 suppression. Increasing levels of miR-29 in turn down-regulate both YY1 and Rybp thereby promoting the myogenic program. Although both YY1 and Rybp are directly targeted by miR-29, the former is at the translational level only (Fig. 1, RNA-seq data, and Ref. 6) but the latter is at both transcriptional and translational levels. Our studies thus demonstrate the prevalence of miRNA-involved regulatory loops.
Other than miR-29, miR-1 was found to form a direct regulatory circuitry with YY1 (12) and Rybp; miR-26 (16) and miR-214 (15) regulate Ezh2. These miRs thus constitute a growing list of epi-miRs in skeletal muscle. Our current study highlights the fact that skeletal myogenesis is a sophisticated process involving interactions of epi-miRs, epigenetic modulators, and transcription factors. Understanding these interactions will not only deepen our knowledge in the field of transcriptional/post-transcriptional regulation of myogenesis but also offer novel insights into the mechanisms behind various muscle diseases. For example, disruption of NF-κB-YY1-miR-29 circuitry leads to the formation of Rhabdomyosarcoma (6) and also contributes to muscle fibrogenesis in Duchenne muscular dystrophy (17). Our current study showed that knockdown of Rybp in vivo up-regulates miR-29 and accelerates injury-induced muscle regeneration (Fig. 6), suggesting this regulatory axis could be relevant in muscle diseases.
Supplementary Material
The work was supported by General Research Funds from the Research Grants Council (RGC) of the Hong Kong Special Administrative Region, China, Chinese University of Hong Kong (CUHK) 476309 and 476310 (to H. W.) and 473211 (to H. S.), CUHK Grants 2041492 and 2041662 (to H. W.), and CUHK Grant 2041474 (to H. S.).

This article contains supplemental Figs. S1–S4 and Table S1.
- miRNA
- microRNA
- ESC
- embryonic stem cell
- CTX
- cardiotoxin
- TA
- tibialis anterior
- IF
- immunofluorescence
- CLN
- centrally located nuclei
- PcG
- Polycomb group protein
- qRT
- quantitative reverse transcription
- HDAC
- histone deacetylase
- MyHC
- myosin heavy chain.
REFERENCES
- 1. Buckingham M. (2006) Myogenic progenitor cells and skeletal myogenesis in vertebrates. Curr. Opin. Genet. Dev. 16, 525–532 [DOI] [PubMed] [Google Scholar]
- 2. Sabourin L. A., Rudnicki M. A. (2000) The molecular regulation of myogenesis. Clin. Genet. 57, 16–25 [DOI] [PubMed] [Google Scholar]
- 3. Perdiguero E., Sousa-Victor P., Ballestar E., Muñoz-Cánoves P. (2009) Epigenetic regulation of myogenesis. Epigenetics 4, 541–550 [DOI] [PubMed] [Google Scholar]
- 4. Bartel D. P. (2004) MicroRNAs. Genomics, biogenesis, mechanism, and function. Cell 116, 281–297 [DOI] [PubMed] [Google Scholar]
- 5. Wang H., Sun H., Guttridge D. C. (2009) MicroRNAs, novel components in a muscle gene regulatory network. Cell Cycle 8, 1833–1837 [DOI] [PubMed] [Google Scholar]
- 6. Wang H., Garzon R., Sun H., Ladner K. J., Singh R., Dahlman J., Cheng A., Hall B. M., Qualman S. J., Chandler D. S. (2008) NF-[kappa] B-YY1-miR-29 regulatory circuitry in skeletal myogenesis and rhabdomyosarcoma. Cancer Cell 14, 369–381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Wang H., Hertlein E., Bakkar N., Sun H., Acharyya S., Wang J., Carathers M., Davuluri R., Guttridge D. C. (2007) NF-κB regulation of YY1 inhibits skeletal myogenesis through transcriptional silencing of myofibrillar genes. Mol. Cell. Biol. 27, 4374–4387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Fabbri M., Garzon R., Cimmino A., Liu Z., Zanesi N., Callegari E., Liu S., Alder H., Costinean S., Fernandez-Cymering C. (2007) MicroRNA-29 family reverts aberrant methylation in lung cancer by targeting DNA methyltransferases 3A and 3B. Proc. Natl. Acad. Sci. U.S.A. 104, 15805–15810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Garzon R., Liu S., Fabbri M., Liu Z., Heaphy C. E., Callegari E., Schwind S., Pang J., Yu J., Muthusamy N., Havelange V., Volinia S., Blum W., Rush L. J., Perrotti D., Andreeff M., Bloomfield C. D., Byrd J. C., Chan K., Wu L. C., Croce C. M., Marcucci G. (2009) MicroRNA-29b induces global DNA hypomethylation and tumor suppressor gene reexpression in acute myeloid leukemia by targeting directly DNMT3A and -3B and indirectly DNMT1. Blood 113, 6411–6418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Boyer L. A., Plath K., Zeitlinger J., Brambrink T., Medeiros L. A., Lee T. I., Levine S. S., Wernig M., Tajonar A., Ray M. K. (2006) Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature 441, 349–353 [DOI] [PubMed] [Google Scholar]
- 11. Lee T. I., Jenner R. G., Boyer L. A., Guenther M. G., Levine S. S., Kumar R. M., Chevalier B., Johnstone S. E., Cole M. F., Isono K. (2006) Control of developmental regulators by Polycomb in human embryonic stem cells. Cell 125, 301–313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Lu L., Zhou L., Chen E. Z., Sun K., Jiang P., Wang L., Su X., Sun H., Wang H. (2012) A novel YY1-miR-1 regulatory circuit in skeletal myogenesis revealed by genome-wide prediction of YY1-miRNA network. PLoS One 7, e27596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Rando T. A., Blau H. M. (1994) Primary mouse myoblast purification, characterization, and transplantation for cell-mediated gene therapy. J. Cell Biol. 125, 1275–1287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Guttridge D. C., Albanese C., Reuther J. Y., Pestell R. G., Baldwin A. S., Jr. (1999) NF-κB controls cell growth and differentiation through transcriptional regulation of cyclin D1. Mol. Cell. Biol. 19, 5785–5799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Juan A. H., Kumar R. M., Marx J. G., Young R. A., Sartorelli V. (2009) Mir-214-dependent regulation of the polycomb protein Ezh2 in skeletal muscle and embryonic stem cells. Mol. Cell 36, 61–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wong C. F., Tellam R. L. (2008) MicroRNA-26a targets the histone methyltransferase Enhancer of Zeste homolog 2 during myogenesis. J. Biol. Chem. 283, 9836–9843 [DOI] [PubMed] [Google Scholar]
- 17. Wang L., Zhou L., Jiang P., Lu L., Chen X., Lan H., Guttridge D. C., Sun H., Wang H. (2012) Loss of miR-29 in myoblasts contributes to dystrophic muscle pathogenesis. Mol. Ther. 20, 1222–1233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Diao Y., Wang X., Wu Z. (2009) SOCS1, SOCS3, and PIAS1 promote myogenic differentiation by inhibiting the leukemia inhibitory factor-induced JAK1/STAT1/STAT3 pathway. Mol. Cell. Biol. 29, 5084–5093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Zhou L., Wang L., Lu L., Jiang P., Sun H., Wang H. (2012) Inhibition of miR-29 by TGF-β-Smad3 signaling through dual mechanisms promotes transdifferentiation of mouse myoblasts into myofibroblasts. PLoS One 7, e33766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lewis B. P., Shih I. H., Jones-Rhoades M. W., Bartel D. P., Burge C. B. (2003) Prediction of mammalian microRNA targets. Cell 115, 787–798 [DOI] [PubMed] [Google Scholar]
- 21. Enright A. J., John B., Gaul U., Tuschl T., Sander C., Marks D. S. (2003) MicroRNA targets in Drosophila. Genome Biol. 5, R1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Krek A., Grün D., Poy M. N., Wolf R., Rosenberg L., Epstein E. J., MacMenamin P., da Piedade I., Gunsalus K. C., Stoffel M., Rajewsky N. (2005) Combinatorial microRNA target predictions. Nat. Genet. 37, 495–500 [DOI] [PubMed] [Google Scholar]
- 23. Müller J., Verrijzer P. (2009) Biochemical mechanisms of gene regulation by polycomb group protein complexes. Curr. Opin. Genet. Dev. 19, 150–158 [DOI] [PubMed] [Google Scholar]
- 24. Wilkinson F., Pratt H., Atchison M. L. (2010) PcG recruitment by the YY1 REPO domain can be mediated by Yaf2. J. Cell. Biochem. 109, 478–486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Zheng L., Schickling O., Peter M. E., Lenardo M. J. (2001) The death effector domain-associated factor plays distinct regulatory roles in the nucleus and cytoplasm. J. Biol. Chem. 276, 31945–31952 [DOI] [PubMed] [Google Scholar]
- 26. Danen-van Oorschot A. A., Voskamp P., Seelen M. C., van Miltenburg M. H., Bolk M. W., Tait S. W., Boesen-de Cock J. G., Rohn J. L., Borst J., Noteborn M. H. (2004) Human death effector domain-associated factor interacts with the viral apoptosis agonist Apoptin and exerts tumor-preferential cell killing. Cell Death Differ. 11, 564–573 [DOI] [PubMed] [Google Scholar]
- 27. García E., Marcos-Gutiérrez C., del Mar Lorente M., Moreno J. C., Vidal M. (1999) RYBP, a new repressor protein that interacts with components of the mammalian Polycomb complex, and with the transcription factor YY1. EMBO J. 18, 3404–3418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Pirity M. K., Locker J., Schreiber-Agus N. (2005) Rybp/DEDAF is required for early postimplantation and for central nervous system development. Mol. Cell. Biol. 25, 7193–7202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Hisada K., Sánchez C., Endo T. A., Endoh M., Román-Trufero M., Sharif J., Koseki H., Vidal M. (2012) RYBP represses endogenous retroviruses, preimplantation- and germline-specific genes in mouse embryonic stem cells. Mol. Cell. Biol. 32, 1139–1149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Arrigoni R., Alam S. L., Wamstad J. A., Bardwell V. J., Sundquist W. I., Schreiber-Agus N. (2006) The polycomb-associated protein Rybp is a ubiquitin-binding protein. FEBS Lett. 580, 6233–6241 [DOI] [PubMed] [Google Scholar]
- 31. Tavares L., Dimitrova E., Oxley D., Webster J., Poot R., Demmers J., Bezstarosti K., Taylor S., Ura H., Koide H., Wutz A., Vidal M., Elderkin S., Brockdorff N. (2012) RYBP-PRC1 complexes mediate H2A ubiquitylation at polycomb target sites independently of PRC2 and H3K27me3. Cell 148, 664–678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Schlisio S., Halperin T., Vidal M., Nevins J. R. (2002) Interaction of YY1 with E2Fs, mediated by RYBP, provides a mechanism for specificity of E2F function. EMBO J. 21, 5775–5786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Mendenhall E. M., Koche R. P., Truong T., Zhou V. W., Issac B., Chi A. S., Ku M., Bernstein B. E. (2010) GC-rich sequence elements recruit PRC2 in mammalian ES cells. PLoS Genet. 6, e1001244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Woo C. J., Kharchenko P. V., Daheron L., Park P. J., Kingston R. E. (2010) A region of the human HOXD cluster that confers polycomb-group responsiveness. Cell 140, 99–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Winbanks C. E., Wang B., Beyer C., Koh P., White L., Kantharidis P., Gregorevic P. (2011) TGF-β Regulates miR-206 and miR-29 to control myogenic differentiation through regulation of HDAC4. J. Biol. Chem. 286, 13805–13814 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.