Background: Serum amyloid A (SAA) is normally associated with the high-density lipoprotein (HDL).
Results: Heparan sulfate (HS) dissociates SAA from HDL, leading to AA amyloidosis. The activity requires a minimum length of 12–14 sugar units.
Conclusion: The result proposes an explanation for the findings that short HS precludes AA amyloidosis.
Significance: This study defines a novel role for HS in AA amyloidosis.
Keywords: Amyloid, Apolipoproteins, HDL, Heparan Sulfate, Heparin, AA Amyloidosis, Serum Amyloid A
Abstract
Inflammation-related (AA) amyloidosis is a severe clinical disorder characterized by the systemic deposition of the acute-phase reactant serum amyloid A (SAA). SAA is normally associated with the high-density lipoprotein (HDL) fraction in plasma, but under yet unclear circumstances, the apolipoprotein is converted into amyloid fibrils. AA amyloid and heparan sulfate (HS) display an intimate relationship in situ, suggesting a role for HS in the pathogenic process. This study reports that HS dissociates SAA from HDLs isolated from inflamed mouse plasma. Application of surface plasmon resonance spectroscopy and molecular modeling suggests that HS simultaneously binds to two apolipoproteins of HDL, SAA and ApoA-I, and thereby induce SAA dissociation. The activity requires a minimum chain length of 12–14 sugar units, proposing an explanation to previous findings that short HS fragments preclude AA amyloidosis. The results address the initial events in the pathogenesis of AA amyloidosis.
Introduction
The acute-phase protein serum amyloid A (SAA)2 is an apolipoprotein associated with the high-density lipoprotein (HDL) fraction in plasma (1). Although SAA normally constitutes a minor part of HDL, during the acute-phase response, SAA is substantially up-regulated (0.001–1 mg/ml) and thus becomes a major component of the lipoprotein (2). Concomitantly, the level of apolipoprotein A-I (ApoA-I), which is normally the most abundant apolipoprotein on HDL, is substantially reduced, and the inflammation-associated lipoprotein is referred to as HDL-SAA.
During longstanding inflammatory conditions, SAA can accumulate as deposits in organs, primarily in the spleen, liver, and kidneys, causing inflammation-related (AA) amyloidosis (3, 4). The development of the disease is a progressive process, often associated with persistent or reoccurring acute inflammation such as rheumatoid arthritis and familial Mediterranean fever (5). When compared with other amyloid conditions, such as Alzheimer disease and islet amyloid in patients with type II diabetes, AA amyloidosis is relatively rare, but often severe due to functional failure of the affected organs. AA amyloidosis may have varying etiology, but consistently high circulating SAA levels are a prerequisite for development of the disease. However, only a proportion of the patients with persistent inflammatory conditions develop this form of amyloidosis. Thus, factors other than circulating SAA concentration must be critical for the generation of amyloid.
AA amyloidosis can be experimentally induced in mice by intravenous injection of amyloid fibrils (also known as amyloid-enhancing factor) in combination with an inflammatory challenge (6). The procedure causes amyloid formation in affected organs within 24 h (7). Experiments with this model have revealed an intimate structural and temporal relationship between AA amyloidosis and heparan sulfate (HS) (8), a sulfated polysaccharide expressed on the cell surfaces and in the extracellular matrix, suggesting a role for HS in SAA amyloidogenesis. Further, cell surface HS converts HDL-associated SAA into AA amyloid fibrils in cell culture, a process that can be inhibited by a C-terminal SAA peptide selectively binding to HS (9). A pH-sensitive motif on SAA has also been identified. This motif facilitates the SAA-HS interaction, leading to aggregation of the polypeptide under mild acidic conditions (10). Collectively, these findings indicate that a direct interaction between SAA and HS is essential for the amyloid formation.
Further, we have previously demonstrated that a transgenic mouse overexpressing heparanase, an HS-degrading enzyme, is resistant to experimental induction of AA amyloid (11). The overexpression of heparanase resulted in a drastic shortening of the HS chain length, demonstrating a decisive role for the polysaccharide chain length in SAA aggregation.
In this study, we investigated the molecular basis of this finding. The results reveal that HS/heparin-SAA interaction causes SAA to dissociate from the HDL-SAA complex at mild acidic conditions. This process is critically dependent on the molecular size of the polysaccharide. The results offer an explanation for earlier observations and, most importantly, provide information for the potential management of AA amyloidosis by targeting HS-SAA interaction.
EXPERIMENTAL PROCEDURES
Isolation of HDL Particles by Ultracentrifugation
Plasma SAA levels were experimentally raised in mice by a subcutaneous injection of 0.5 ml of 2% (w/v) AgNO3, inducing an acute inflammatory state. Before blood collection, the syringe was primed with ∼20 μl of EDTA (7%) to prevent blood clotting. HDL particles were isolated from plasma of normal and inflamed mice by sequential density flotation (12). Briefly, the density was adjusted to 1.063 g/ml with NaBr and centrifuged at 175,000 × g for 18 h in a 70.1Ti rotor (Beckman) at 10 °C. After discarding the top layer containing VLDL/LDL, the infranatant was pooled and adjusted to a density of 1.25 g/ml and recentrifuged at 250,000 × g for 24–36 h at 10 °C. The top layer (HDL) was pooled and dialyzed against 20 mm Tris-HCl, 0.15 m NaCl, 0.1% (w/v) EDTA, pH 7.5, for 18 h. The HDL isolated from inflamed mouse plasma is denoted HDL-SAA.
Glycosaminoglycans
Heparin has an average molecular mass of 14 kDa as analyzed by gel chromatography on a Superose 12 column (GE Healthcare). The HS samples were isolated from porcine tissues and characterized by reverse-phase ion-pairing HPLC after enzymatic cleavage with HS/heparin lyases, as described (13). The HS II preparation (isolated from intestine) contained 45% of the trisulfated disaccharide (IdoA2S-GlcNS6S) species, and HS I (isolated from aorta) contained 5% of the species. Based on the content of trisulfated disaccharides, the HS II and HS I preparations were regarded as high sulfated HS and low sulfated HS, respectively. Heparin isolated from pig intestine mucosa was separated into fractions with low and high affinity (HA) for antithrombin (AT), as described (14) (15). The 3H-labled heparin was prepared by N-deacetylation of the heparin followed by re-N-acetylation with N-[3H]acetic anhydride (16). The heparin fragments were prepared by partial degradation with nitrous acid, followed by size exclusion chromatographic separation.
Surface Plasmon Resonance (SPR) Analysis
Interaction of lipoproteins with HS/heparin was evaluated using a Biosensor system (Biacore2000). The lipoproteins were dissolved into a 50 mm NaAc (pH 5.0) (0.5 mg/ml) coupling buffer and immobilized onto a CM5 sensor chip using an amine-coupling kit (GE Healthcare). The kit uses 1-ethyl-3-(3-dimethylaminopropyl) carbodiimide hydrochloride and N-hydroxysuccinimide chemistry to create a covalent link between the surface matrix and free amino groups on the ligand. Injection of lipoproteins (HDL-SAA and HDL) over the CM5 surface for 5 or 15 min generated an immobilization level of ∼10,000 or ∼30,000 RU, respectively. The immobilization procedure was followed by an injection of ethanolamine for 7 min to deactivate the surface and a wash injection of 2 × 2 min of 0.5 m NaCl to remove any unbound material. All experiments and procedures used a flow rate of 20 μl/min. Heparin, HS, or heparin-derived oligosaccharides were injected at a concentration of 7 μm in 50 mm NaAc, 0.1 m NaCl, pH 5.0 or 7.4. After each sample injection, the surface was regenerated by injection of 0.5 m NaCl for 2 min, with exceptions as indicated in respective legends. Generated sensorgrams were evaluated with Biacore evaluation software 2.0. To remove system-related artifacts, the obtained sensorgrams were subtracted by the signal from a reference flow cell and a blank injection. For the immunocapture assay, an anti-SAA-antibody (A183, provided by Prof. G. Westermark, Uppsala University, Sweden) was coupled onto the second flow cell (7,000 RU) using the amino-coupling kit. Lipoproteins (HDL and HDL-SAA; 40 μg/ml) were injected (50 mm NaAc, pH 5.0, 0.1 m NaCl) to confirm the specificity of the antibody for SAA. The surface was regenerated by injecting 0.5 m NaCl and 1% Triton X-100 for 2 min. Thereafter, the fourth flow cell of the sensor chip was prepared for immobilization using the GE amino-coupling kit. The HDL-SAA was injected into the microfluidic system and was initially allowed to pass through the second cell (immobilized with anti-SAA antibody) and then into the fourth cell, where it was immobilized. The N-hydroxysuccinimide/1-ethyl-3-(3-dimethylaminopropyl) carbodiimide hydrochloride reagents were deactivated with ethanolamine. Thereafter, HS II (7 μm) was injected over the SAA-depleted ligand.
AT-Sepharose Affinity Chromatography
The HDL-SAA and HA-heparin co-incubated samples were applied onto an AT-Sepharose column and eluted with equal volume (6 ml) of low- (0.25 m) and high-salt (3 m) solution. Fractions were collected, and the elution profile of 3H-labeled HA-heparin was analyzed with scintillation counting. The SAA concentration was determined using a murine SAA-specific ELISA kit (Invitrogen). The presence of ApoA-I was analyzed using an immune dot-blot assay. Ten microliters of the eluate were applied to a PVDF-Immobilon P membrane (Millipore) and stained with an anti-ApoA antibody (GW20069F, Sigma) for 2 h. The lipoprotein was detected using a horseradish peroxidase-conjugated secondary antibody (anti-chicken IgG; Invitrogen) and quantified using image analysis software (ImageJ). The intensity of each fraction was expressed as the percentage of the total sample intensity acquired.
Turbidity Assay
HDL and HDL-SAA stock solutions were adjusted to the appropriate buffer by gel filtration through a Sepharose G-10 column (8 ml; Sigma) pre-equilibrated in 50 mm sodium acetate and 100 mm NaCl (pH 5.0 or 7) and kept on ice. Heparin and HS were added to the HDL samples and incubated for 30 min at 37 °C followed by absorbance measurements (400 nm).
Homology Modeling
Proteins having sequence similarity to murine SAA1.1 were identified in the Protein Data Bank (PDB) using PSI-BLAST (17). The structures of the proteins that had the highest similarity to SAA1.1 were superimposed and compared with the programs LSQMAN (18) and O (19). Pairwise sequence alignments were used to generate homology models of the central domain of mouse SAA1.1. The domain was generated by part of the sequence of β-xylosidase from Geobacillus stearothermophilus (PDB entry 2EXJ (20); identity 41%). A relevant part was used as templates to build the models with the program SOD (18). The models were adjusted in O, using rotamers that would improve packing in the interior of the protein and accounting for deletions in loop regions. The figure was prepared using MOLSCRIPT (21) and Molray (22). The conserved residues were determined by hidden Markov model-based searches by SAM-T08 server (23) and ConSurf server searches (24).
RESULTS
HS/Heparin Promotes SAA Fibrillization
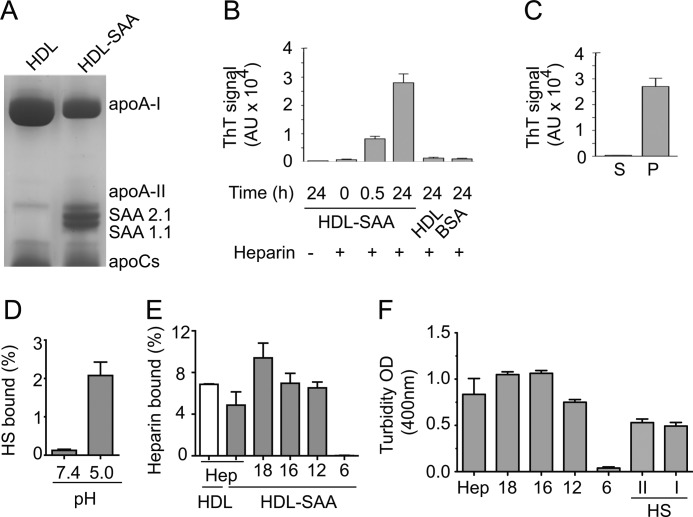
HDL particles were isolated from normal and inflamed mouse plasma by sequential density flotation. SDS-urea-PAGE analysis revealed high SAA contents in HDL isolated from inflamed mouse plasma (HDL-SAA) (Fig. 1A). Heparin efficiently promoted the formation of amyloid-like fibrils when incubated with HDL-SAA in 50 mm NaAc buffer, pH 5.0 (Fig. 1B), as analyzed by thioflavin-T fluorescence (25, 26). In comparison, the addition of heparin to HDL isolated from normal mouse plasma did not result in fibril formation. Fibrils isolated from AA amyloid-laden mouse spleen were used as positive control for the thioflavin-T fluorescence assay (supplemental Fig. S1). The effect of heparin on HDL-SAA fibrillization was time-dependent as prolonged incubation resulted in a considerably higher degree of fibril formation (Fig. 1B). The effective recovery of fibrils in the pellet upon centrifugation (10 min at 8,100 × g) further demonstrates the aggregated property of the HDL-SAA co-incubated with heparin (Fig. 1C). To examine the binding of HS/heparin to the lipoproteins, we incubated HDL and HDL-SAA with 3H-labeled polysaccharide. The amounts of 3H-labeled HS/heparin bound to the particles were analyzed using a nitrocellulose filter trapping method (27). In line with earlier findings (10), HS binding to HDL-SAA was pH-dependent, with substantial binding at pH 5.0, but essentially no binding at neutral condition (Fig. 1D). The binding was affected by the total negative charge in the polysaccharide as heparin (which contains more sulfate groups when compared with HS) bound considerably stronger than HS (Fig. 1, D and E). Notably, heparin also bound to HDL isolated from normal mouse plasma (Fig. 1E), in agreement with a previous study (28). Particularly, the interaction was size-dependent as short heparin fragments (6-mers) failed to bind with HDL-SAA (Fig. 1E). This size-dependent effect was further illustrated by examination of HDL-SAA aggregation; heparin efficiently promoted HDL-SAA aggregation, as measured by the degree of turbidity (optical density at 400 nm) after 30 min of incubation in 50 mm NaAc buffer at pH 5.0, whereas the heparin 6-mers failed to induce aggregation (Fig. 1F). The effect of HS on HDL-SAA aggregation was also evaluated using two HS preparations with different degrees of sulfation (HS I with low sulfation and HS II with high sulfation, as described under “Experimental Procedures”). A reduction relative to full-length heparin was observed, whereas no difference in effect between the two preparations was seen.
FIGURE 1.
pH- and size-dependent interaction between HDL-SAA and HS/heparin. A, HDL (from normal mouse plasma) and HDL-SAA (from inflamed mouse plasma) were analyzed by SDS-urea-PAGE (12.5%). B, HDL and HDL-SAA (0.6 mg/ml) were dissolved in 50 mm NaAc buffer, pH 5.0, and incubated with (+) (0.2 mg/ml) and without (−) heparin at the indicated time periods. The samples were neutralized in PBS-containing thioflavin-T (ThT). The fibrillization was measured with thioflavin-T fluorescence. C, HDL-SAA and heparin were co-incubated for 24 h, and the sample was centrifuged (10 min at 8,100 × g). Thioflavin-T fluorescence (20 μm) was used to detect fibrillar aggregates in the supernatant (S) and pellet (P), respectively. D, HDL-SAA binding to 3H-labeled HS (10,000 cpm) at neutral and mild acidic condition. E, binding of 3H-labeled heparin (Hep) and heparin-derived oligosaccharides (10,000 cpm) to HDL (white bar) and HDL-SAA (gray bars) in mild acidic condition. The binding was analyzed using a nitrocellulose filter trapping method. F, HDL-SAA aggregation induced by heparin of different chain lengths and HS (HS II, high sulfation degree; HS I, low sulfation degree, see “Experimental Procedures”) were assessed by the degree of absorbance at 400 nm. OD, optical density. Error bars in B, C, E, and F correspond to the S.E. values of three independent experiments.
HS/Heparin-HDL-SAA Interaction Leads to Dissociation of SAA
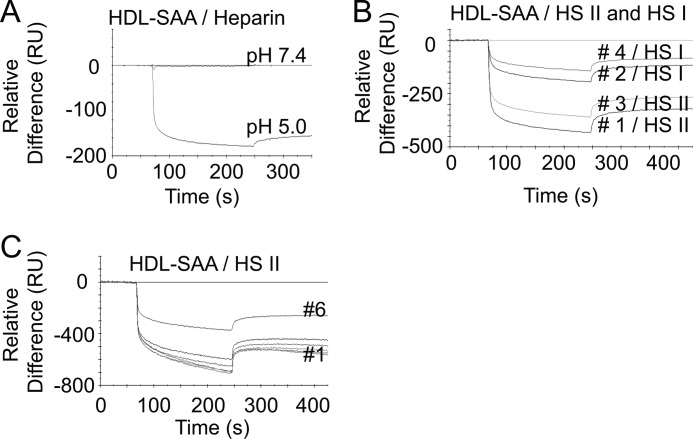
The molecular mechanisms of HS/heparin and HDL-SAA interaction were examined with the SPR spectroscopy. HDL-SAA was immobilized onto a CM5 Biacore sensor chip, and heparin was allowed to interact (20 μl/min) with the immobilized HDL-SAA in 50 mm NaAc buffer, 150 mm NaCl, at different pH conditions. As observed in the free solution, heparin did not interact with the HDL-SAA at pH 7.4, but interacted at pH 5.0 (Fig. 2A). Unexpectedly, the interaction resulted in a negative curve. The generation of negative curves has previously been linked to a ligand-induced structural change; for example, the immobilized component may undergo a decrease of its hydrodynamic radius (29, 30). It is plausible that the negative curve obtained here is due to a heparin-induced conformational change of the lipoprotein.
FIGURE 2.
HS/heparin interaction with HDL-SAA causes mass dissociation. A–C, SPR analysis of the binding of HS/heparin (7 μm) to HDLs. A, HDL-SAA was immobilized to a Biacore CM5 sensor chip (10,000 RU), and heparin was injected over the surface at neutral (pH 7.4) and mild acidic condition (pH 5.0). B, alternate injections of HS II (high sulfation degree) and HS I (low sulfation degree) over immobilized HDL-SAA (15,000 RU) at mild acidic condition (pH 5.0) in the indicated order. C, overlay graph of repeated injections of HS II over HDL-SAA (34,000 RU) at pH 5.0. Experiments presented in A–C were repeated at least twice, and representative sensorgrams are shown.
To verify whether the negative signal is a property of the interaction between heparin and HDL-SAA, we also tested the two HS preparations (HS I with low sulfation and HS II with high sulfation). Alternate injections of HS I and HS II produced negative signals, similar in shape to that seen with heparin. The magnitude of the negative deflection correlated with the degree of sulfation in that HS I (injection 2) appeared less effective than HS II (injection 1) (Fig. 2B). This phenomenon reoccurred with consecutive injections of the HS II (injection 3) and HS I (injection 4) samples. However, the negativity of the signal was reduced. The decreasing negative signals were further confirmed by six sequential injections of HS II alone (Fig. 2C). Notably, the signal was substantially reduced after injection 6.
The progressive decrease in negative deflection could be due to an incomplete surface regeneration following each run or partial inactivation of the ligand. Examination of the biosensor surface revealed a reduction in the base-line mass after each HS injection (supplemental Fig. S2), indicating a mass loss from the sensor surface following each HS interaction. Because SAA is a major component of HDL-SAA and because it is known to bind HS/heparin, it was our primary candidate for the glycosaminoglycan-mediated mass removal.
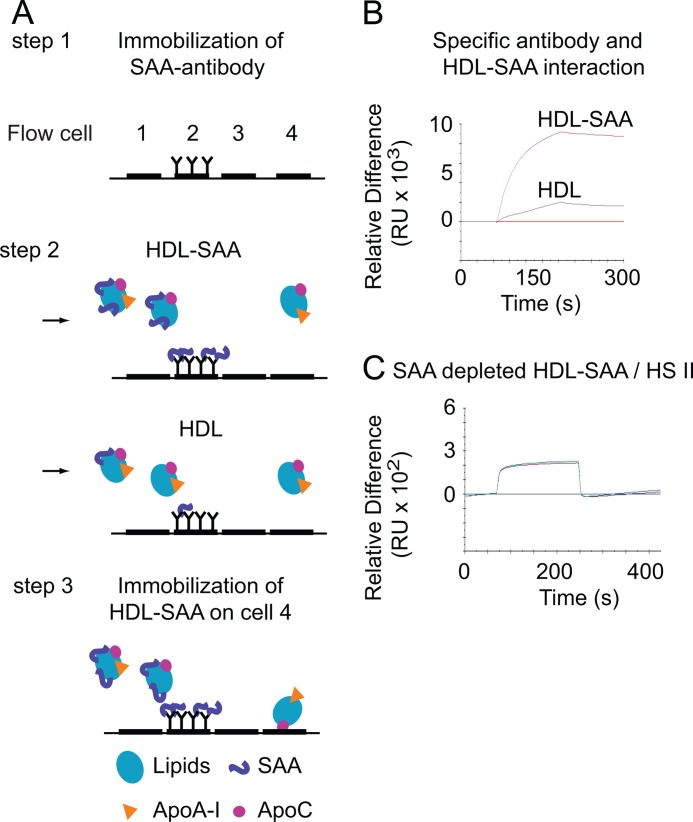
To demonstrate that SAA is the primary component that is removed from HDL-SAA upon interaction with HS, we designed an immunocapture assay to selectively remove SAA from the HDL-SAA complex. An anti-SAA antibody was immobilized onto the second flow cell of a CM5 Biacore sensor chip, as illustrated in Fig. 3A, step 1. Injection of HDL-SAA through the cell resulted in substantial binding (Fig. 3A, step 2; Fig. 3B); in contrast, injection of HDL onto the cell produced negligible signals, confirming the specific binding of the anti-SAA antibody with SAA. Then, HDL-SAA was injected into the flow system. After passing through the second flow cell (where the SAA was selectively removed by the immobilized anti-SAA antibody), the lipoprotein was thereafter immobilized on the fourth flow cell (as described under “Experimental Procedures”) (Fig. 3A, step 3). Then, injection of HS II over the fourth cell, immobilized with SAA-depleted HDL-SAA, resulted in positive binding signals (Fig. 3C). The results demonstrate that the HS/heparin-induced negative deflection is related to the SAA content of the lipoprotein, resulting in dissociation of SAA from the complex.
FIGURE 3.
HS/heparin-induced mass dissociation is dependent on SAA content of HDL. A, steps 1–3, an illustration of the experimental design and the flow cells arrangement in a Biacore2000 instrument. A, step 1, the sensor chip has four flow cells over which a reagent can be directed. Antibody against SAA was immobilized (7,000 RU) onto the second flow cell of the sensor chip. To confirm the specificity of the antibody for SAA, lipoproteins (40 μg/ml) of HDL-SAA (isolated form inflamed plasma) or HDL (from noninflamed plasma) were injected (B). A, steps 2 and 3, HDL-SAA was thereafter immobilized on flow cell 4 after passing through the second flow cell (the anti-SAA surface) (step 2). After immobilization, duplicate injections of HS II were made over the SAA-depleted lipoprotein (flow cell 4) (C) (step 3).
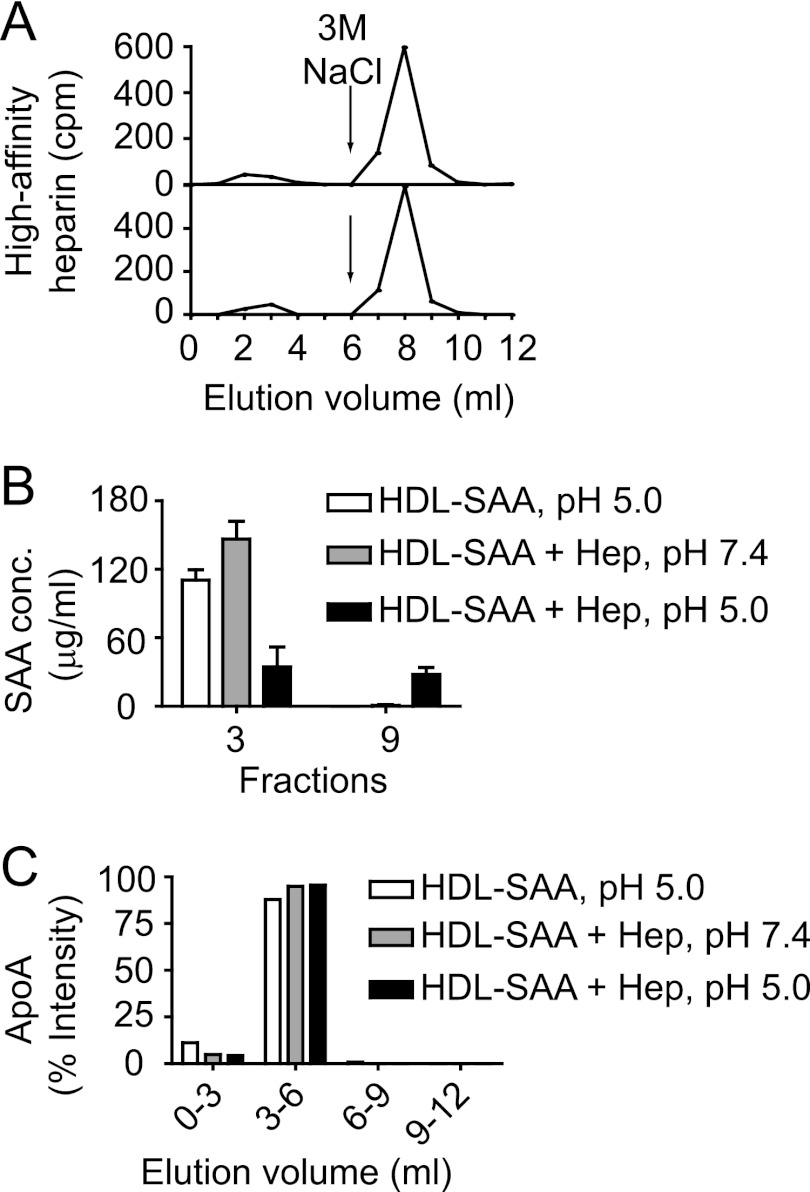
To confirm that HS/heparin indeed selectively dissociated SAA from the HDL-SAA complex, the lipoprotein was incubated with a mixture of 3H-labeled and unlabeled heparin having HA for AT as described under “Experimental Procedures.“ The mixture of HDL-SAA and the HA-heparin sample was incubated for 30 min at 37 °C in 50 mm NaAc buffer, 150 mm NaCl, at either pH 5.0 or 7.4. The samples were then applied onto an AT-Sepharose column that was eluted with an equal volume (6 ml) of low (0.25 m) and high (3 m) concentrations of NaCl. Fractions were collected and analyzed by scintillation counting. As expected, the majority of 3H-labeled heparin was detected in the high-salt fractions (Fig. 4A, upper panel), displaying an identical pattern to that when HA heparin alone was applied to the same column (Fig. 4B, lower panel). This indicates that the HDL-SAA and HA-heparin interaction did not affect the binding of heparin to antithrombin. Furthermore, selected fractions from low-salt (fraction 3) and high-salt (fraction 9) eluate were analyzed with an SAA-specific ELISA. SAA was only detected in the low-salt fraction when HDL-SAA was incubated alone or with heparin at pH 7.4. In contrast, SAA was substantially recovered in the high-salt fraction, in addition to the low-salt fraction, when HDL-SAA was co-incubated with heparin at pH 5.0 (Fig. 4B). To investigate whether HS/heparin also dissociated other apolipoproteins from the HDL-SAA complex, the collected fractions were analyzed for ApoA-I, using an immuno-dot-blot assay. ApoA-I was detected exclusively in the low-salt fractions, independent of the incubation conditions (Fig. 4C and supplemental Fig. S3), demonstrating that no ApoA-I was co-eluted with the HA-heparin fraction.
FIGURE 4.
SAA is selectively removed from HDL-SAA by heparin. HDL-SAA dissolved in sodium acetate buffer (pH 5.0 or 7.4) was incubated with heparin fractions (mixture of 3H-labeled and unlabeled) of high affinity for antithrombin at 37 °C for 30 min. Samples were applied onto an antithrombin-Sepharose column and eluted with equal volume (6 ml) of low- (0.25 m) and high- (3.0 m) NaCl solution. A, elution profile of 3H-labeled heparin (1,000 cpm) after incubation with (above) or without HDL-SAA (below). B, SAA concentrations (SAA conc.) were measured in a low-salt eluted fraction (fraction 3) and in a high-salt eluted fraction (fraction 9) using an SAA-specific ELISA. Hep, heparin. C, 10 μl of the pools of low-salt fractions (0–3 ml; 3–6 ml) and the high-salt fractions (6–9 ml; 9–12 ml) were applied onto a PVDF membrane and stained with antibodies against ApoA-I (supplemental Fig. 3). Error bars correspond to the S.E. values of three independent experiments. Densitometry analysis of the ApoA-I dot-blot is presented in C.
To investigate whether the negative curve relates to a conformational change of HDL-SAA, apart from the dissociation of the SAA polypeptide, we treated the immobilized HDL-SAA surface with 0.1% sodium dodecyl sulfate (SDS) for 5 min to alter the lipoprotein structure as illustrated in supplemental Fig. S4A. Injection of HS over the delipidated HDL-SAA complex produced a positive binding curve (supplemental Fig. S4B), implying that the structural integrity of the HDL-SAA lipoparticle is important for the generation of negative signals. Further, the positive binding response is likely to be accounted for by the residual apolipoproteins that are still covalently attached to the sensor surface, such as SAA and to some extent also ApoA-I, ApoE, and ApoC (i.e. those that originally served to anchor the lipoprotein).
Chain Length Is Critical for the Binding Activity of HS/Heparin
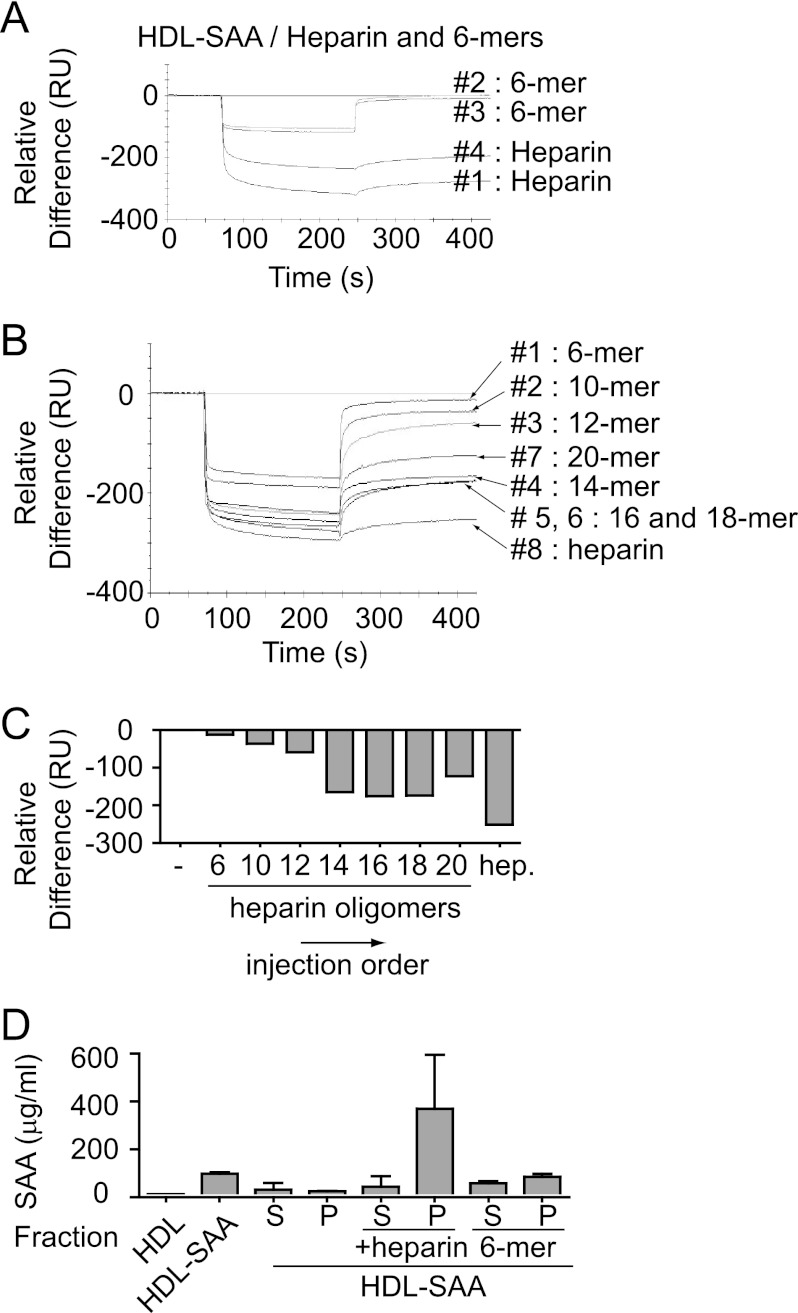
Heparin-derived fragments with less than 12-sugar residues were unable to bind HDL-SAA and consequently failed to induce HDL-SAA aggregation (Fig. 1, E and F). To determine the molecular basis for this finding, we examined the interaction of short heparin fragments (e.g. 6-mers) with HDL-SAA by SPR. Fig. 5A shows the result of sequential injections of heparin and the 6-mers in 50-mm NaAc, 150 mm NaCl, pH 5.0, over the Biacore CM5 sensor chip immobilized with HDL-SAA. Similar to the HS injections (Fig. 2B), the first injection of heparin (injection 1) produced a substantial negative deflection curve, and the second injection of heparin (injection 4) resulted in slightly reduced binding activity. In contrast, injections of the 6-mers (injections 2 and 3) between the two heparin injections generated limited negative signals with apparent rapid on- and off-rates (Fig. 5A). The identical binding curves of the two consecutive injections and their rapid return to base line indicate negligible mass loss from the sensor surface by injection of the short heparin fragment. The different effects of heparin and heparin fragments on the reduction of biosensor surface mass is further illustrated in supplemental Fig. S5.
FIGURE 5.
SAA dissociation requires a minimal heparin chain length. A–C, binding of heparin and heparin-derived oligomers to HDL-SAA was assessed by SPR spectroscopy. A, sequential injection of heparin and 6-mers (7 μm) over immobilized HDL-SAA (15,000 RU) at pH 5.0, in the indicated order. B, sequential injection of oligosaccharides with different sizes (18-mer, 16-mer, etc.) over immobilized HDL-SAA (15,000 RU) at mild acidic condition (pH 5.0), in the indicated order. The response level (RU) at interaction end-point (425 s) for each sample is shown in C. D, HDL-SAA was dissolved in mild acidic buffer (50 mm NaAc, pH 5.0) and incubated with heparin and heparin 6-mers for 24 h. After incubation, the samples was centrifuged, and the SAA levels were analyzed in the supernatant (S) and pellet (P), respectively. Error bars correspond to the S.E. values of three independent experiments.
Sequential injection of oligosaccharides with increasing chain length over the immobilized HDL-SAA resulted in a progressive increase in negativity of the binding signal, appearing as two distinct populations (Fig. 5, B and C). The saccharides of 10- and 12-mers produced similar patterns to the 6-mers, with rapid on- and off-rates. In contrast, the oligosaccharides of 14-mers or larger produced patterns similar to heparin. The results suggest that an oligosaccharide composed of minimum 14 sugar residues is needed to induce substantial SAA dissociation. Unexpectedly, the 20-mer generated a lower degree of negative deflection when compared with the 14–18-mer preparations. We have yet to fully understand this effect, but the result supports the idea that optimal chain lengths exist for SAA displacement. The critical effect of chain length of heparin fragments is also confirmed for SAA aggregation as no aggregate was detected in the pellet of HDL-SAA after co-incubation with 6-mers; in comparison, substantial SAA was found in the pellets upon co-incubation of HDL-SAA with heparin (Fig. 5D).
HS/Heparin-induced Dissociation Involves More than One Binding Site on HDL-SAA
It appears that the heparin injection is able to saturate all available binding sites on HDL-SAA as the binding curves reach a plateau state almost immediately. To verify this observation, heparin was injected repeatedly over the HDL-SAA particle without the standard surface regeneration procedure (wash with 500 mm NaCl for 2 min, 20 μl/min) after each injection. The first injection generated, as observed previously (Figs. 2, A and B, and 5A), a substantial negative binding curve. However, the subsequent heparin injections (injections 2–4) failed to generate any measurable signals (supplemental Fig. S6, A and B), indicating that the ligands were blocked by the first injection. Because the binding activity of the ligand can only be retrieved by the surface regeneration procedure, this indicates a saturable reaction. This seems inconsistent with the observation that repeated injection of HS/heparin resulted in multiple dissociation events (Figs. 2 and 5).
To find out the mechanisms behind this apparent discrepancy, we assessed the binding of heparin to other components of the HDL complex. HDL isolated from non-inflamed plasma was immobilized onto a CM5 Biacore sensor chip, and heparin was allowed to interact with the lipoprotein in 50 mm NaAc buffer, 150 mm NaCl at different pH conditions. As observed for the HDL-SAA and heparin interaction, heparin was found to bind the normal HDL at mild acidic conditions (pH 5.0), suggesting a pH-dependent reaction (supplemental Fig. S7A). As ApoA-I is the major component of HDL (Fig. 1A), the apolipoprotein most likely accounts for the HDL interaction with heparin. Analysis of the murine ApoA-I sequence demonstrates that the apolipoprotein holds a basic amino acid residue cluster that may interact with HS/heparin (31) (supplemental Fig. S7B).
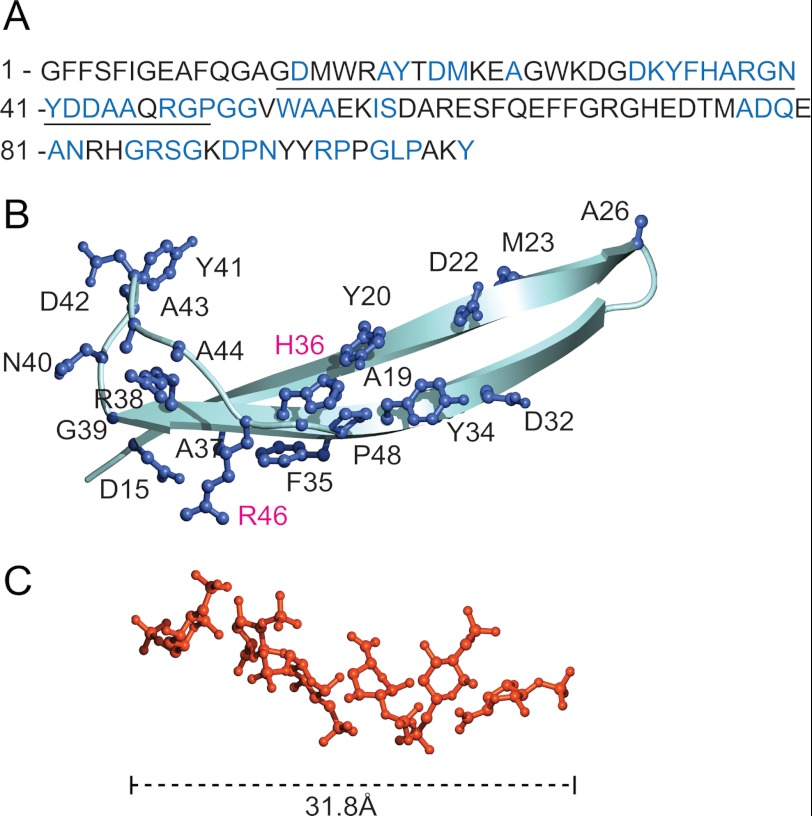
If SAA dissociation induced by HS/heparin involves binding to both SAA and ApoA-I, only heparin structures exceeding a certain minimal length would be expected to simultaneously bind both polypeptides. A pH-dependent HS/heparin binding site was previously identified on the central domain of SAA (10). A model generated using homology-modeling software shows that this binding site has a length of 25 Å (Fig. 6B), a similar dimension to that of a heparin 6-mer (32 Å) (Fig. 6C) (PDB ID 1HPN (32)). Further, the length of the proposed HS/heparin binding site on ApoA-I (30 Å) was also found to correspond to that of a 6-mer (supplemental Fig. S8). Taken together, these findings concur with the observation that a minimum of 12–14-sugar sequence is required for efficient displacement of SAA from HDL-SAA.
FIGURE 6.
Modeling of the HS/heparin binding sites on SAA. A, the amino acid sequence of mouse SAA1.1, with conserved residues marked in blue. The central domain, which comprises the pH-dependent HS/heparin binding site, is underlined. B, a model of the domain was generated by homology software (see “Experimental Procedures”). The domain was found to have a length of 25 Å. Conserved residues are displayed with ball-and-stick models in blue. The basic amino acids that are aligned in the same direction in space, and therefore expected to be critical for HS/heparin binding, are marked in magenta. The histidine 36 residue, which accounts for the pH-dependent HS/heparin and HDL-SAA interaction (10), is located in the center of the domain. C, structure of a heparin 6-mer is shown (32 Å) (PDB ID: 1HPN (32)).
DISCUSSION
The central issue in the pathogenesis of AA amyloidosis is the conversion of circulating SAA into amyloid fibrils. Several lines of evidence have demonstrated that the disease progression is dependent on HS. We have previously found that transgenic mice overexpressing human heparanase, an HS-degrading enzyme, were resistant to experimental induction of AA amyloidosis (11), demonstrating a decisive role for HS in the pathogenic process. The present study aimed to get further insight on the molecular mechanisms of the HS and SAA interaction.
Incubation of heparin, a commonly used analog of HS, with HDL-SAA isolated from inflamed mouse plasma induced fibrillization of the polypeptide at mild acidic condition (pH 5.0) (Fig. 1). The pH-dependent effect may reflect the in vivo situation, where mild acidic condition is expected in proximity to membrane-anchored HS proteoglycans (33, 34). This effect of HS/heparin is also size-dependent as shorter fragments, e.g. a 6-mer, failed to bind to HDL-SAA in free solution (Fig. 1E) and accordingly lost their capacity to induce SAA aggregation (Fig. 1F). This is consistent with previous in vivo data where the heparanase-overexpressing mice did not develop amyloidosis upon induction (11) due to the shortening of the HS chains in the mice.
To further assess the HS/heparin and HDL-SAA interaction, we applied SPR spectroscopy. In contrast to the positive binding curves that are typical for SPR, injection of HS/heparin over HDL-SAA generated negative signals (Fig. 2, A–C). SPR is commonly employed to study molecular interactions because a mass change at the sensor surface (e.g. due to a analyte-ligand binding) causes a concomitant change in refractive index. Negative signals can be observed when the immobilized ligand undergoes a conformational change, often related to an analyte-induced decrease of its hydrodynamic radius. This change in conformation results in a decrease of the refractive index (29, 30). Thus, the negative signals observed for HS/heparin interaction with HDL-SAA likely correspond to a conformational change of the lipoprotein. This hypothesis is confirmed by the positive binding curves obtained when HDL-SAA were disassembled by SDS treatment (supplemental Fig. S4, A and B), prior to HS binding analysis.
Repeated injections of HS or heparin caused a mass reduction of the HDL-SAA complex, demonstrated by the decrease of base-line mass after each injection (supplemental Fig. S2). Affinity chromatographic analysis demonstrated that HS/heparin binding to HDL-SAA caused dissociation of SAA at mild acidic pH (Fig. 4). This dissociation required a minimal chain length as short heparin fragments did not cause any base-line change (Fig. 5A and supplemental Fig. S2). The size-dependent effect suggests that multiple HS-binding sites on HDL-SAA are involved in SAA dissociation.
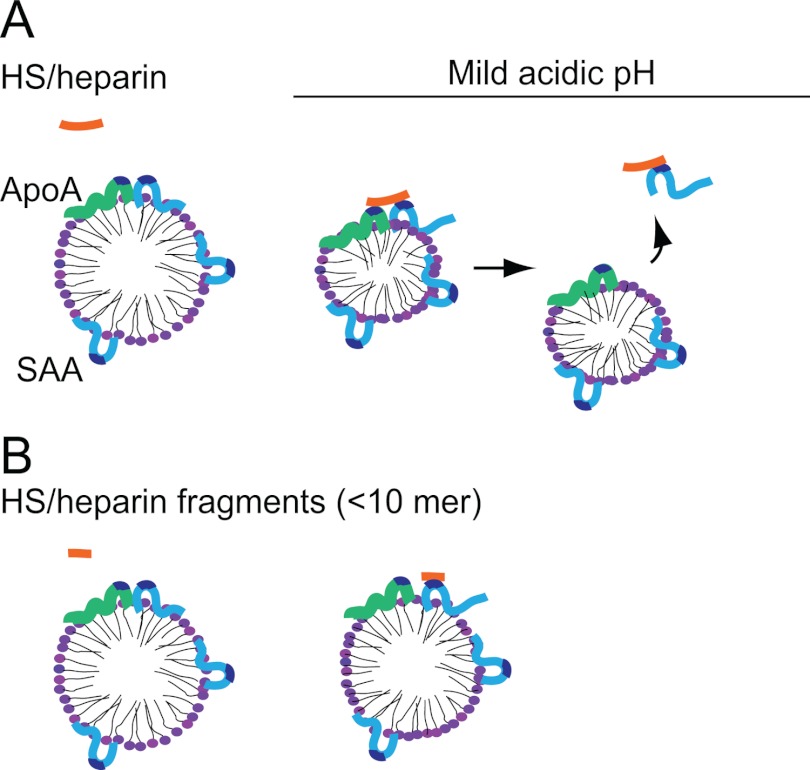
Further, the SPR data demonstrated that repeated HS/heparin injections over HDL-SAA generated stepwise dissociation of SAA. Despite this gradual removal, the curves indicated that each consecutive interaction of (excess) heparin with HDL-SAA resulted in complete saturation of all binding sites of HDL-SAA (Fig. 5A and supplemental Fig. S6, A and B). These findings suggest that HS/heparin binds to an additional HDL-associated component and that the availability of this site limits the dissociation process. Because ApoA-I is the other major protein constituent of HDL-SAA (Fig. 1A), we considered whether simultaneous binding of HS/heparin to ApoA-I and SAA might be required for SAA-dissociation. Accordingly, HS was found to bind normal HDL, abundant in ApoA-I (supplemental Fig. S7A). This interaction was only observed at mild acidic pH, consistent with the condition at which HS/heparin-mediated SAA displacement occurred. Analysis of ApoA-I sequence identified a basic amino acid motif that fits the consensus sequence for HS/heparin binding (31). This proposed binding site includes a histidine residue, likely contributing to the pH dependence seen in the ApoA-I and heparin interaction. The molecular dimensions of the HS/heparin binding sites on ApoA-I and SAA each accommodate an HS/heparin 6-mer (31.8Å). Thus, a heparin fragment of 12–14-mer can simultaneously bind to two such sites, whereas a fragment of 6-mer can only bind to one site. These data propose that simultaneous binding of HS/heparin to SAA and ApoA-I is a prerequisite for the displacement of SAA (Fig. 7). Notably, our study does not exclude the possibility that the HS/heparin-mediated displacement of SAA occurs through co-binding of HS/heparin to SAA and other HDL-associated apolipoproteins (e.g. ApoE, ApoC). Such an interpretation warrants consideration because HS/heparin binding sites have been identified on ApoE (35). However, it should be noted that the proportion of ApoE on HDL is relatively small (<5%) (36).
FIGURE 7.
A proposed model of HS/heparin-mediated SAA dissociation from acute-phase HDL. Mild acidic condition activates the pH-dependent binding motifs on SAA (light blue) (10) and ApoA-I (light green), resulting in HS/heparin (orange) binding to the apolipoproteins. A and B, oligosaccharides of sufficient length (>12 monosaccharide units) bind the two apolipoproteins simultaneously, resulting in the dissociation of SAA from HDL-SAA.
The importance of heparin chain length has previously been demonstrated for the antithrombin inhibition of thrombin. Simultaneous binding of heparin to both antithrombin and thrombin accelerates the interaction between the molecules (37). A minimal sequence of 18 sugar units is required for this reaction (38), a length similar to that observed for the proposed binding of heparin to ApoA-I and SAA. Further, studies on the interaction between HS and CXCL12, an inflammation-related cytokine, demonstrated that HS is capable of simultaneously binding to two sites on the protein (39).
SAA is a well conserved protein throughout evolution, and many different physiological functions have been suggested (40). Several studies implicate SAA in cholesterol metabolism as SAA-derived peptides have been demonstrated to inhibit acyl coenzyme A cholesterol acyltransferase and enhance cholesterol esterase activities in cholesterol-laden macrophages (41, 42). Therefore, remodeling of HDL-SAA by HS may reflect a physiological setting that relates to the function of SAA in cholesterol metabolism, which is worthy of further investigation.
Earlier studies have demonstrated a temporal and structural relationship between AA amyloid and HS, suggesting a role for HS in the pathogenic process. Accumulated evidence shows that the molecular size of heparin is critical for aggregation of different types of amyloid proteins (43, 44). It is believed that long HS/heparin structures provide a scaffold for the assembly of amyloid peptides, thereby promoting their polymerization into fibrillar structures.
Our results point to a novel role for HS/heparin in AA amyloidosis in which a critical length is required for separation of SAA from HDL. These results also offer an explanation for our early in vivo finding (11) that molecular size of HS is critical for SAA deposition as shorter fragments of the sugar chains are insufficient to cause dissociation of SAA from HDL-SAA complex. This finding may have potential value for preventing AA amyloidosis by specifically targeting HS-SAA interaction.
Supplementary Material
Acknowledgments
We thank Prof. Ulf Lindahl and Paul O'Callaghan (Uppsala University) for critical reading of the manuscript.
The study was supported by grants from the European Commission (EURAMY European Rare Amyloidosis Network), the Swedish Research Council (K2009-67X-21128-01-3), the Mizutani Foundation for Glycoscience, the Swedish Cancerfonden (09 0717), and the Polysackaridforskning Foundation (Uppsala, Sweden).

This article contains supplemental Figs. S1–S8.
- SAA
- serum amyloid A
- AA
- amyloid A
- HS
- heparan sulfate
- ApoA-I
- apolipoprotein A-I
- AT
- antithrombin
- SPR
- surface plasmon resonance
- RU
- resonance units
- HA
- high affinity.
REFERENCES
- 1. Morrow J. F., Stearman R. S., Peltzman C. G., Potter D. A. (1981) Induction of hepatic synthesis of serum amyloid A protein and actin. Proc. Natl. Acad. Sci. U.S.A. 78, 4718–4722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Gabay C., Kushner I. (1999) Acute-phase proteins and other systemic responses to inflammation. N. Engl. J. Med. 340, 448–454 [DOI] [PubMed] [Google Scholar]
- 3. Westermark G. T., Westermark P. (2009) Serum amyloid A and protein AA: molecular mechanisms of a transmissible amyloidosis. FEBS Lett. 583, 2685–2690 [DOI] [PubMed] [Google Scholar]
- 4. Wall J. S., Richey T., Stuckey A., Donnell R., Macy S., Martin E. B., Williams A., Higuchi K., Kennel S. J. (2011) In vivo molecular imaging of peripheral amyloidosis using heparin-binding peptides. Proc. Natl. Acad. Sci. U.S.A. 108, E586–594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Gillmore J. D., Lovat L. B., Persey M. R., Pepys M. B., Hawkins P. N. (2001) Amyloid load and clinical outcome in AA amyloidosis in relation to circulating concentration of serum amyloid A protein. Lancet 358, 24–29 [DOI] [PubMed] [Google Scholar]
- 6. Lundmark K., Westermark G. T., Olsén A., Westermark P. (2005) Protein fibrils in nature can enhance amyloid protein A amyloidosis in mice: cross-seeding as a disease mechanism. Proc. Natl. Acad. Sci. U.S.A. 102, 6098–6102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Axelrad M. A., Kisilevsky R., Willmer J., Chen S. J., Skinner M. (1982) Further characterization of amyloid-enhancing factor. Lab. Invest. 47, 139–146 [PubMed] [Google Scholar]
- 8. Snow A. D., Willmer J., Kisilevsky R. (1987) A close ultrastructural relationship between sulfated proteoglycans and AA amyloid fibrils. Lab. Invest. 57, 687–698 [PubMed] [Google Scholar]
- 9. Elimova E., Kisilevsky R., Szarek W. A., Ancsin J. B. (2004) Amyloidogenesis recapitulated in cell culture: a peptide inhibitor provides direct evidence for the role of heparan sulfate and suggests a new treatment strategy. FASEB J. 18, 1749–1751 [DOI] [PubMed] [Google Scholar]
- 10. Elimova E., Kisilevsky R., Ancsin J. B. (2009) Heparan sulfate promotes the aggregation of HDL-associated serum amyloid A: evidence for a proamyloidogenic histidine molecular switch. FASEB J. 23, 3436–3448 [DOI] [PubMed] [Google Scholar]
- 11. Li J. P., Galvis M. L., Gong F., Zhang X., Zcharia E., Metzger S., Vlodavsky I., Kisilevsky R., Lindahl U. (2005) In vivo fragmentation of heparan sulfate by heparanase overexpression renders mice resistant to amyloid protein A amyloidosis. Proc. Natl. Acad. Sci. U.S.A. 102, 6473–6477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Havel R. J., Eder H. A., Bragdon J. H. (1955) The distribution and chemical composition of ultracentrifugally separated lipoproteins in human serum. J. Clin. Invest. 34, 1345–1353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Staatz W. D., Toyoda H., Kinoshita-Toyoda A., Chhor K., Selleck S. B. (2001) Analysis of proteoglycans and glycosaminoglycans from Drosophila. Methods Mol. Biol. 171, 41–52 [DOI] [PubMed] [Google Scholar]
- 14. Lindahl U., Bäckström G., Thunberg L., Leder I. G. (1980) Evidence for a 3-O-sulfated d-glucosamine residue in the antithrombin-binding sequence of heparin. Proc. Natl. Acad. Sci. U.S.A. 77, 6551–6555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Höök M., Björk I., Hopwood J., Lindahl U. (1976) Anticoagulant activity of heparin: separation of high-activity and low-activity heparin species by affinity chromatography on immobilized antithrombin. FEBS Lett. 66, 90–93 [DOI] [PubMed] [Google Scholar]
- 16. Hook G. E., Gilmore L. B. (1982) Hydrolases of pulmonary lysosomes and lamellar bodies. J. Biol. Chem. 257, 9211–9220 [PubMed] [Google Scholar]
- 17. Altschul S. F., Madden T. L., Schäffer A. A., Zhang J., Zhang Z., Miller W., Lipman D. J. (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kleywegt G. J., Zou J.Y., Kjeldgaard M., Jones T.A. (2001) Around O, in International Tables for Crystallography (Rossmann M. G., Arnold E., eds) pp. 353–356, Kluwer Academic, Dordrecht, The Netherlands [Google Scholar]
- 19. Jones T. A., Zou J. Y., Cowan S. W., Kjeldgaard M. (1991) Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A 47, 110–119 [DOI] [PubMed] [Google Scholar]
- 20. Brüx C., Ben-David A., Shallom-Shezifi D., Leon M., Niefind K., Shoham G., Shoham Y., Schomburg D. (2006) The structure of an inverting GH43 β-xylosidase from Geobacillus stearothermophilus with its substrate reveals the role of the three catalytic residues. J. Mol. Biol. 359, 97–109 [DOI] [PubMed] [Google Scholar]
- 21. Kraulis P. J. (1991) MOLSCRIPT: a program to produce both detailed and schematic plots of protein structures. J. Appl. Crystallogr. 24, 946–950 [Google Scholar]
- 22. Harris M., Jones T. A. (2001) Molray–a web interface between O and the POV-Ray ray tracer. Acta Crystallogr. D Biol. Crystallogr. 57, 1201–1203 [DOI] [PubMed] [Google Scholar]
- 23. Karplus K., Karchin R., Barrett C., Tu S., Cline M., Diekhans M., Grate L., Casper J., Hughey R. (2001) What is the value added by human intervention in protein structure prediction? Proteins Suppl. 5, 86–91 [DOI] [PubMed] [Google Scholar]
- 24. Berezin C., Glaser F., Rosenberg J., Paz I., Pupko T., Fariselli P., Casadio R., Ben-Tal N. (2004) ConSeq: the identification of functionally and structurally important residues in protein sequences. Bioinformatics 20, 1322–1324 [DOI] [PubMed] [Google Scholar]
- 25. Naiki H., Higuchi K., Hosokawa M., Takeda T. (1989) Fluorometric determination of amyloid fibrils in vitro using the fluorescent dye, thioflavin T1. Anal. Biochem. 177, 244–249 [DOI] [PubMed] [Google Scholar]
- 26. Lee J., Culyba E. K., Powers E. T., Kelly J. W. (2011) Amyloid-β forms fibrils by nucleated conformational conversion of oligomers. Nat. Chem. Biol. 7, 602–609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kreuger J., Lindahl U., Jemth P. (2003) Nitrocellulose filter binding to assess binding of glycosaminoglycans to proteins. Methods Enzymol. 363, 327–339 [DOI] [PubMed] [Google Scholar]
- 28. Ancsin J. B., Kisilevsky R. (1999) The heparin/heparan sulfate-binding site on apo-serum amyloid A: implications for the therapeutic intervention of amyloidosis. J. Biol. Chem. 274, 7172–7181 [DOI] [PubMed] [Google Scholar]
- 29. Salamon Z., Cowell S., Varga E., Yamamura H. I., Hruby V. J., Tollin G. (2000) Plasmon resonance studies of agonist/antagonist binding to the human δ-opioid receptor: new structural insights into receptor-ligand interactions. Biophys. J. 79, 2463–2474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Gestwicki J. E., Hsieh H. V., Pitner J. B. (2001) Using receptor conformational change to detect low molecular weight analytes by surface plasmon resonance. Anal. Chem. 73, 5732–5737 [DOI] [PubMed] [Google Scholar]
- 31. Cardin A. D., Weintraub H. J. (1989) Molecular modeling of protein-glycosaminoglycan interactions. Arteriosclerosis 9, 21–32 [DOI] [PubMed] [Google Scholar]
- 32. Mulloy B., Forster M. J., Jones C., Davies D. B. (1993) NMR and molecular modeling studies of the solution conformation of heparin. Biochem. J. 293, 849–858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Cevc G. (1990) Membrane electrostatics. Biochim. Biophys. Acta 1031, 311–382 [DOI] [PubMed] [Google Scholar]
- 34. Mulgrew-Nesbitt A., Diraviyam K., Wang J., Singh S., Murray P., Li Z., Rogers L., Mirkovic N., Murray D. (2006) The role of electrostatics in protein-membrane interactions. Biochim. Biophys. Acta 1761, 812–826 [DOI] [PubMed] [Google Scholar]
- 35. Saito H., Dhanasekaran P., Nguyen D., Baldwin F., Weisgraber K. H., Wehrli S., Phillips M. C., Lund-Katz S. (2003) Characterization of the heparin binding sites in human apolipoprotein E. J. Biol. Chem. 278, 14782–14787 [DOI] [PubMed] [Google Scholar]
- 36. Krimbou L., Tremblay M., Davignon J., Cohn J. S. (1997) Characterization of human plasma apolipoprotein E-containing lipoproteins in the high-density lipoprotein size range: focus on pre-β1-LpE, pre-β2-LpE, and α-LpE. J. Lipid Res. 38, 35–48 [PubMed] [Google Scholar]
- 37. Lindahl U., Li J. P. (2009) Interactions between heparan sulfate and proteins-design and functional implications. Int. Rev. Cell Mol. Biol. 276, 105–159 [DOI] [PubMed] [Google Scholar]
- 38. Danielsson A., Raub E., Lindahl U., Björk I. (1986) Role of ternary complexes, in which heparin binds both antithrombin and proteinase, in the acceleration of the reactions between antithrombin and thrombin or factor Xa. J. Biol. Chem. 261, 15467–15473 [PubMed] [Google Scholar]
- 39. Lortat-Jacob H. (2009) The molecular basis and functional implications of chemokine interactions with heparan sulfate. Curr. Opin. Struct. Biol. 19, 543–548 [DOI] [PubMed] [Google Scholar]
- 40. Tam S. P., Kisilevsky R., Ancsin J. B. (2008) Acute-phase HDL remodeling by heparan sulfate generates a novel lipoprotein with exceptional cholesterol efflux activity from macrophages. PLoS One 3, e3867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Tam S. P., Flexman A., Hulme J., Kisilevsky R. (2002) Promoting export of macrophage cholesterol: the physiological role of a major acute-phase protein, serum amyloid A 2.1. J. Lipid Res. 43, 1410–1420 [DOI] [PubMed] [Google Scholar]
- 42. Tam S. P., Ancsin J. B., Tan R., Kisilevsky R. (2005) Peptides derived from serum amyloid A prevent, and reverse, aortic lipid lesions in apoE−/− mice. J. Lipid Res. 46, 2091–2101 [DOI] [PubMed] [Google Scholar]
- 43. Jha S., Patil S. M., Gibson J., Nelson C. E., Alder N. N., Alexandrescu A. T. (2011) Mechanism of amylin fibrillization enhancement by heparin. J. Biol. Chem. 286, 22894–22904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Noborn F., O'Callaghan P., Hermansson E., Zhang X., Ancsin J. B., Damas A. M., Dacklin I., Presto J., Johansson J., Saraiva M. J., Lundgren E., Kisilevsky R., Westermark P., Li J. P. (2011) Heparan sulfate/heparin promotes transthyretin fibrillization through selective binding to a basic motif in the protein. Proc. Natl. Acad. Sci. U.S.A. 108, 5584–5589 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.