Background: Na+-NQR is a bacterial respiratory enzyme that catalyzes the oxidation of NADH, the reduction of ubiquinone, and the translocation of Na+ across the membrane.
Results: Mutations at NqrB-G140 and NqrB-G141 impair the reaction of Na+-NQR with ubiquinone.
Conclusion: Residues NqrB-G140 and -G141 are critical for binding and reaction with ubiquinone.
Significance: This work identifies the functional ubiquinone binding site in Na+-NQR.
Keywords: Bioenergetics/Electron Transfer Complex, Electron Transport System (ETS), Energy Metabolism, Enzyme Catalysis, Quinones
Abstract
The Na+-pumping NADH:quinone oxidoreductase (Na+-NQR) is the main entrance for electrons into the respiratory chain of many marine and pathogenic bacteria. The enzyme accepts electrons from NADH and donates them to ubiquinone, and the free energy released by this redox reaction is used to create an electrochemical gradient of sodium across the cell membrane. Here we report the role of glycine 140 and glycine 141 of the NqrB subunit in the functional binding of ubiquinone. Mutations at these residues altered the affinity of the enzyme for ubiquinol. Moreover, mutations in residue NqrB-G140 almost completely abolished the electron transfer to ubiquinone. Thus, NqrB-G140 and -G141 are critical for the binding and reaction of Na+-NQR with its electron acceptor, ubiquinone.
Introduction
The Na+-pumping NADH:quinone oxidoreductase (Na+-NQR)2 is a redox-driven ion pump and the entry point for electrons in the respiratory chain in a number of marine and pathogenic bacteria, such as Vibrio cholerae, Klebsiella pneumoniae, and Yersinia pestis among others (1–3). The enzyme, encoded by the nqr operon, is an integral membrane complex containing six subunits (NqrA–F) that transfers redox equivalents from cytosolically produced NADH to ubiquinone-8. This electron transfer process takes place through a series of five cofactors: an FAD, a 2Fe-2S center, two covalently bound FMN molecules, and one riboflavin (1, 4–10). Several structural motifs have been characterized in the enzyme, including the binding or attachment sites of most of these cofactors. Moreover, the properties of the different redox states of the cofactors and the electron transfer reactions within the enzyme have been intensively studied (4, 6, 11–15). However, very little is known about the location of the ubiquinone binding site.
Hayashi et al. (16) found evidence that glycine 140 in NqrB could form part of the ubiquinone binding site in Na+-NQR from Vibrio alginolyticus. They showed that the enzyme is inhibited by the antibiotic korormicin in the subnanomolar range (Ki = 82 pm) and by 2-n-heptyl-4-hydroxyquinoline N-oxide (HQNO) in the submicromolar range (Ki = 300 nm) (17–19). These authors isolated spontaneous mutants with resistance to korormicin and found that they all had the same single base mutation, which resulted in the replacement of NqrB Gly-140 by valine. Because of the structural similarity of ubiquinone, HQNO, and korormicin, it was suggested that these compounds may bind to the same site, and thus, this residue may be part of the ubiquinone binding site. However, korormicin and HQNO are non-competitive inhibitors with respect to ubiquinone (19), indicating that the inhibitors and ubiquinone do not occupy exactly the same binding site.
To clarify the involvement of NqrB-glycine 141 (V. cholerae numbering) in the binding of ubiquinone, we constructed mutants of two contiguous conserved glycine residues, NqrB-G140 and NqrB-G141, located in transmembrane helix III of NqrB (these residues are homologous to glycine residues 139 and 140 in the V. alginolyticus enzyme). Mutations that alter the size of either of these residues greatly diminish the ability of the enzyme to interact with ubiquinone. Most striking was the result that modifications to NqrB-G140, the glycine residue adjacent to the one altered in the original korormicin-resistant mutants, almost completely abolished binding of this substrate. Analysis of the redox reactions of the enzyme by stopped-flow kinetic methods indicated that the electron transfer pathway of the NqrB-G140A mutant is specifically altered at the step where electrons are transferred to ubiquinone. Redox titrations of the wild-type enzyme and the NqrB-G140A mutant showed that none of the redox cofactor midpoint potentials were altered. Moreover, the mutant had a decreased ability of pumping sodium under steady state conditions, but it was able to build a significant membrane potential (ΔΨ) under partial turnover, which does not depend on the presence of ubiquinone. This clearly indicates a specific impairment of the binding site for ubiquinone in the mutant. IR spectroscopy demonstrated that the overall structure of the enzyme is not disturbed by the mutation and confirmed that the NqrB-G140A and NqrB-G141A mutations specifically affect the ubiquinone binding site of Na+-NQR.
EXPERIMENTAL PROCEDURES
Cell Growth Conditions
V. cholerae cells were grown at 37 °C in New Brunswick BioFlo-5000 fermentors under constant aeration and agitation in Luria Bertani (LB) medium, 50 μg/ml streptomycin, and 100 μg/ml ampicillin. l-Arabinose was added during log phase growth to induce the expression of Na+-NQR.
Protein Purification
Wild-type Na+-NQR and mutant proteins were purified using a nickel-nitrilotriacetic acid resin (Qiagen) as reported before (1). The enzyme was concentrated using Centricon filters (Millipore) with a cutoff of 100 kDa, then frozen in aliquots, and stored at liquid nitrogen temperature until use.
The flavin:protein ratio was determined as described before (13). For all the mutants studied, the flavin:protein ratio was close to 4 as determined in the wild-type enzyme.
Activity Measurements
Measurements of enzymatic activity were made spectrophotometrically in reaction buffer (50 mm Tris-HCl, 1 mm EDTA, 5% glycerol (v/v), and 0.05% n-dodecyl maltoside (w/v), pH 8.0) containing 100 mm NaCl, 250 μm K2-NADH, and 0.5–50 μm ubiquinone-1. Ubiquinone reductase activity was measured at 282 nm as reported before (20).
Site-directed Mutagenesis
The QuikChange site-directed mutagenesis kit (Stratagene) was used to construct all mutants in this study. The wild-type nqr operon cloned into the pBAD vector was used as a template for the mutagenesis reactions. The mutations were encoded in the forward primers, which are listed in Table 1. All mutations were confirmed by direct DNA sequencing.
TABLE 1.
Forward primers for site-directed mutagenesis of NqrB-G140 and NqrB-G141
| Mutant | Primer sequencea (5′ →3′) |
|---|---|
| NqrB-G140A | GCTACGGTGTTCATCGTC[b]GCTGGTTTCTGGGAAGTACTGTTC |
| NqrB-G140L | GCTACGGTGTTCATCGTC[b]CTGGGTTTCTGGGAAGTGTTG |
| NqrB-G141A | GCTACGGTGTTCATCGTCGGT[b]GCTTTCTGGGAAGTACTGTTCTG |
| NqrB-G141L | GCTACGGTGTTCATCGTCGGT[b]CTGTTCTGGGAAGTACTGTTCTG |
| NqrB-G141V | GCTACGGTGTTCATCGTCGGT[b]GTTTTCTGGGAAGTACTGTTCTG |
a The mutated codons are bold and underlined.
Fast Kinetics Experiments
The reduction kinetics of NqrB-G140A were studied by stopped flow in buffer containing 250 μm K2-NADH or 0.5 mm ubiquinol-1 in the presence of 100 mm NaCl. The experiments using ubiquinol-1 as a substrate were performed under anaerobic conditions in the presence 5 mm dithiothreitol to avoid the accumulation of ubiquinone-1. Ubiquinol-1 was prepared as reported before (21). Data were averaged and analyzed as reported previously (12).
Redox Titrations
Wild-type Na+-NQR and NqrB-G140A samples were washed with 50 mm Tris-HCl, pH 8.0, 0.05% n-dodecyl maltoside, and 150 mm NaCl or KCl. Typically, 20 μl of the concentrated sample were used to fill the electrochemical thin-layer cell mounted with two gold grids previously modified with a mixture of 2 mm cysteamine and 2 mm mercaptopropionic acid in a 1:1 ratio (22, 23). An Ag/AgCl electrode was used as reference, adding 208 mV for a standard hydrogen electrode. The temperature was maintained at 10 °C throughout the titrations. A mixture of 20 mediators was added to the sample 1 h before starting the titration (24). Spectra were recorded in the UV-visible domain in steps of 20 mV from 0 to −620 mV with an equilibration time of 30- 40 min for each step.
IR Spectroscopy
Mid-IR spectra were recorded in a Bruker 70 spectrometer equipped with a liquid nitrogen-cooled mercury-cadmium-telluride detector. 1 μl of sample was dried under a flux of argon on a silicon attenuated total reflectance crystal. 256 scans were averaged for one spectrum. After performing a base-line correction, band separation of the Amide I signature (1700–1600 cm−1) was done using a multi-Gaussian fit (25).
Membrane Potential Measurements
The NqrB-G140A mutant was reconstituted into liposomes as reported before (13). Formation of ΔΨ was measured spectrophotometrically at 625 minus 587 nm using 3 μm Oxonol VI (13). The reaction buffer contained 100 μm NADH, 100 μm CoQ-1, 100 mm NaCl, 50 mm HEPES, 150 mm KCl, and 1 mm EDTA, pH 7.5. For partial turnover experiments, CoQ-1 was added several seconds after the addition of NADH.
RESULTS
Steady State Kinetics of NqrB-G140 and -G141 Mutants
The binding of ubiquinone to the reduced enzyme cannot be assayed under equilibrium conditions because the enzyme would transfer electrons to this substrate, producing ubiquinol almost immediately. Thus, to determine the role of the NqrB-G140 and NqrB-G141 in the binding of quinone, the Km(app) for this substrate was measured under steady state conditions in the wild-type and in the mutant proteins. Activity was measured at different concentrations of CoQ-1 using fixed and near saturating concentrations of the other two substrates (250 μm NADH and 100 mm NaCl). Under these conditions, the Km(app) for the wild-type Na+-NQR is ∼3 μm (Table 2). For the mutants at position NqrB-G141, the change in the size of this residue increased the Km(app); NqrB-G141L showed a 3-fold increase in Km(app), whereas NqrB-G141A and NqrB-G141V had a 6- and 9-fold increase, respectively. Remarkably, the two mutants at position NqrB-G140, Ala and Leu, showed non-saturating behavior with up to 50 μm ubiquinone, strongly indicating that mutations at these residues greatly decreased the affinity of the binding site.
TABLE 2.
Apparent Km for CoQ and Ki for HQNO for wild-type Na+-NQR and NqrB-G140 and -G141 mutants
| kcat | Km(CoQ) | Ki(HQNO) | |
|---|---|---|---|
| s−1 | μm | μm | |
| WT | 525 | 2.5 ± 0.3 | 0.5 ± 0.1 |
| NqrB-G140A | >200 | >100 | 7.5 ± 1.2 |
| NqrB-G140L | >200 | >100 | NDa |
| NqrB-G141A | 425 | 15 ± 3.6 | ND |
| NqrB-G141L | 463 | 8.8 ± 2.2 | ND |
| NqrB-G141V | 388 | 23.4 ± 5.6 | 1.2 ± 0.3 |
a ND, not determined.
Hayashi et al. (16) showed that a spontaneous mutant at position NqrB-G141 (NqrB-G140V in V. alginolyticus numbering) increased its resistance to korormicin 100,000-fold. Although korormicin and HQNO compete for the same binding site (19), the korormicin-resistant mutant has almost the same sensitivity to HQNO as the wild-type. This indicated that the two inhibitors bind to the same pocket in the enzyme but that they interact with different structures in the protein. Unfortunately, korormicin is not commercially available, so we tested HQNO inhibition on the mutants. The activity the enzyme was titrated with HQNO using nearly saturating concentrations of NADH, ubiquinone, and sodium. Our results confirm the previous observations by Hayashi et al. (16) that two mutants at position NqrB-G141, alanine and valine, have the same sensitivity to the inhibitor as the wild-type Na+-NQR. On the other hand, NqrB-G140A is up to 7 times more resistant to HQNO than the wild-type enzyme. This indicates that NqrB-G140 participates in the binding of both ubiquinone and HQNO.
Fast Kinetics Measurements
Reduction by NADH
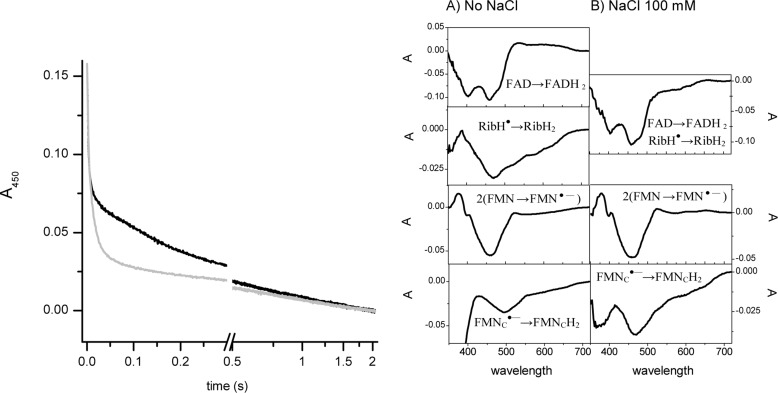
To fully understand the effect of mutating NqrB-G140A, the fast kinetics of reduction with NADH were measured. We have shown previously that the reduction reaction of wild-type Na+-NQR in sodium-free conditions proceeds in four distinguishable kinetic phases (12). Phase I is the two-electron reduction of FAD evidenced by a difference spectrum showing two minima at 390 and 460 nm with a rate constant higher that 250 s−1. Phase II corresponds to the one-electron reduction of the riboflavin neutral radical with a difference spectrum with minima at 575 and 550 and a maximum at 430 nm and a rate of 20–30 s−1. Phase III is the reduction of the two covalently bound FMN molecules to the anionic radical species with a difference spectrum with a minimum at 460 nm and a shoulder at 390 nm and a rate constant of ∼2 s−1. Finally, in phase IV, the reduction of the anionic radical FMNC to the flavohydroquinone can be observed with a minimum at 470–480 nm at a slow rate (Table 3). In the presence of sodium, the reduction rate of the riboflavin neutral radical is increased about 10 times and cannot be individually resolved from the two-electron reduction of FAD. The reductions of the two FMN molecules can be observed in the second and third phases with rates increased 3–8 times.
TABLE 3.
Rate constants of the phases of reduction by NADH of NqrB-G140A mutant in the presence and absence of NaCl
| Redox transitions | Rate constant |
|
|---|---|---|
| No NaCl | 100 mm NaCl | |
| s−1 | ||
| FAD →FADH2 | 330 | 224 |
| RibH• →RibH2 | 28.6 | 224 |
| 2(FMN →FMN⨪) | 2.1 | 34.6 |
| FMNC⨪ →FMNCH2 | 0.2 | 1.4 |
Interestingly, the reduction kinetics for NqrB-G140A show components in the presence or absence of sodium similar to the ones obtained for the wild-type enzyme (Fig. 1 and Table 3). This suggests that the mutation altered the step where electrons are transferred to ubiquinone. To test this hypothesis, the fast reduction reaction using ubiquinol-1 as substrate was measured.
FIGURE 1.
Kinetics of reduction of NqrB-G140A mutant by NADH. The left panel shows the absorbance (Abs) at 450 nm in the absence (black line) and in the presence of 100 mm NaCl (gray line). The right panel shows the differential spectra of the components of the reduction process.
Reduction by Ubiquinol-1
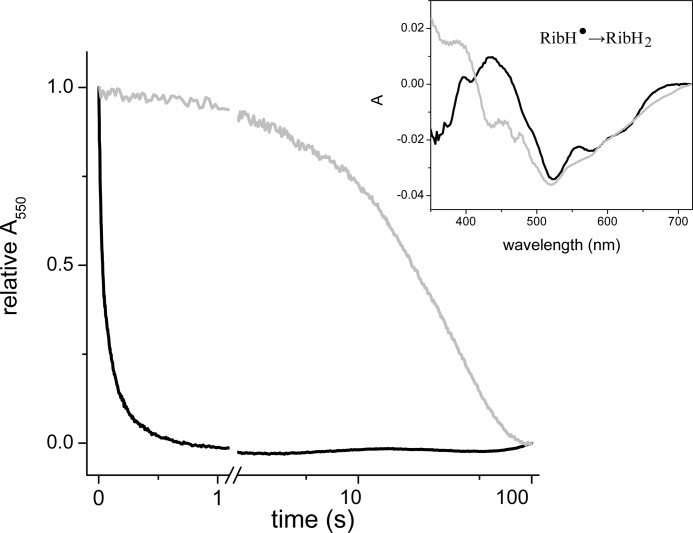
Riboflavin is the last electron carrier in the enzyme, transferring electrons to ubiquinone (14). To elucidate whether NqrB-G140A interferes with the binding of ubiquinone, we studied the kinetics of the reverse flow of electrons using ubiquinol-1 as electron donor. Our previous results demonstrated that the wild-type enzyme can be reduced with this substrate and that the only center that accepts electrons is riboflavin (12, 14). Riboflavin is found as a stable neutral flavosemiquinone (RibH•) in the air-oxidized enzyme, and upon reduction, it is converted into the fully reduced form (RibH2) (14, 26, 27).
The kinetics of reduction of wild type and NqrB-G140A mutant were measured at 550 nm where the reduction of the riboflavin neutral radical has a strong signal. The difference spectrum of the reduction process confirms that riboflavin is the only cofactor involved in the process with a characteristic minimum at 510 nm and shoulders at 575 and 650 nm (Fig. 2). The reduction rate of riboflavin in NqrB-G140A (kobs = 0.02 s−1) decreased by 3 orders of magnitude compared with wild type (kobs = 26 s−1), which conclusively demonstrates that the mutation only affects the interaction of the enzyme with ubiquinone/ubiquinol. In this new set of experiments, the reduction of wild type was considerably faster than in previous reports (14) because experiments were performed under anaerobic conditions and in the presence of dithiothreitol, which chemically reduces ubiquinone, keeping a constant concentration of ubiquinol.
FIGURE 2.
Kinetics of reduction of wild-type Na+-NQR and NqrB-G140A mutant by ubiquinol-1. The inset shows the difference spectra of the different components of the reduction process. Abs, absorbance.
Membrane Potential Formation
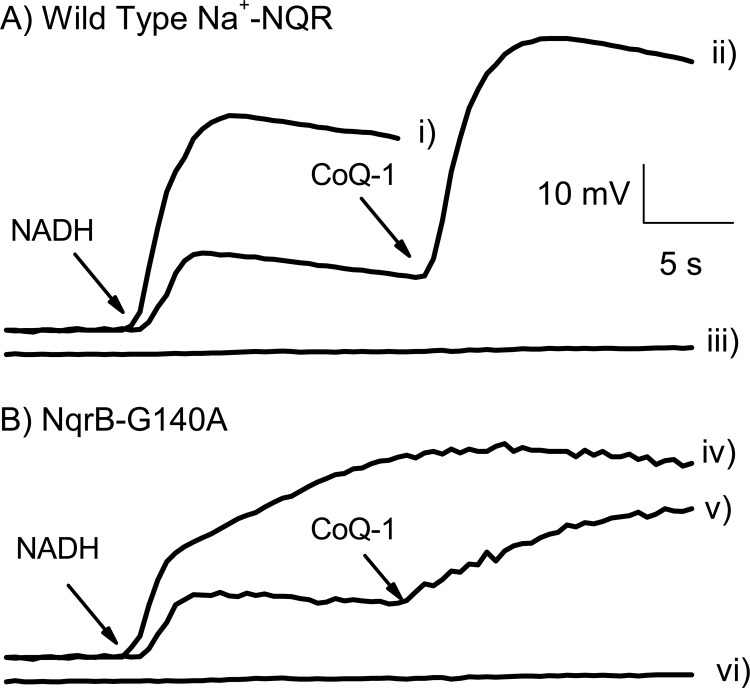
The sodium-pumping activity of NqrB-G140A mutant was measured spectrophotometrically in proteoliposomes using the ΔΨ-sensitive dye Oxonol VI (13). Fig. 3 shows the pumping activities of wild-type Na+-NQR and the NqrB-G140A mutant under partial turnover and under steady state conditions. We have reported previously that Na+-NQR can pump a small amount of sodium in the presence of NADH and in the absence of ubiquinone, which corresponds to a partial turnover of the enzyme (11). Top middle (wild type) and bottom middle (NqrB-G140A) traces show that the sodium-pumping activity of the mutant is not affected under partial turnover conditions. Under steady state conditions (in the presence of NADH, CoQ-1, and Na+), the mutant exhibits a biphasic behavior (bottom left trace). A fast phase probably corresponds to the partial turnover, and a slower phase corresponds to true steady state sodium pumping, which is impaired in the mutant. This result corroborates that the NqrB-G140A mutation is specifically altering the ability of the enzyme to interact with ubiquinone.
FIGURE 3.
Formation of ΔΨ by reconstituted wild-type Na+-NQR and NqrB-G140A mutant. ΔΨ was measured using Oxonol VI as described before (13). For steady state conditions, 100 μm NADH, 100 μm CoQ-1, and 100 mm NaCl were used. For partial turnover conditions, CoQ-1 was added after NADH. Traces i and iv: steady turnover for wild-type and NqrB-G140A mutant, respectively. Traces ii and v: partial turnover for wild-type and NqrB-G140A mutant, respectively. Traces iii and vi show the effect of Na+ ionophore ETH-157. The signal was calibrated against membrane potential produced with 1 μg/ml valinomycin at different potassium concentrations according to the Nernst equation. Abs, absorbance.
Redox Titrations
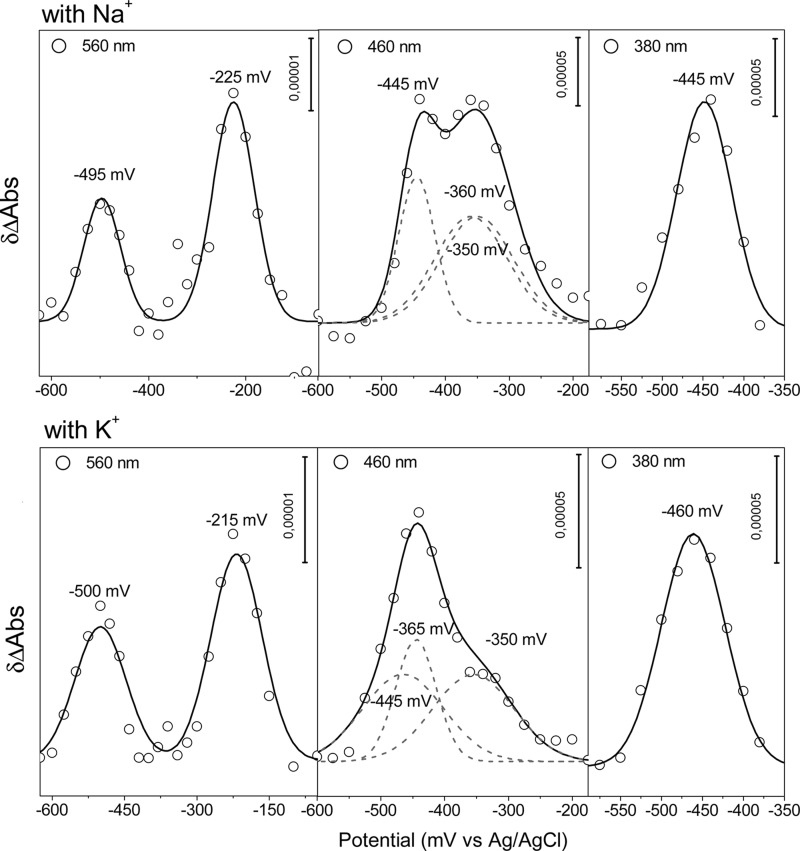
To further corroborate that NqrB-G140A is affecting exclusively the binding of ubiquinone, the redox properties of the mutant were studied. A spectropotentiometric redox titration from 0 to −620 mV with steps of 20 mV was performed, and the redox-dependent development of the UV-visible spectra was recorded.
Direct fitting of the ΔA against the applied potential to the Nernst equation is not possible because the different spectral transitions of the flavins overlap each other. However, at selected wavelengths, the specific transitions of the different flavins and their reaction intermediates are distinguishable (28). At 560 nm, for example, major contributions from the one-electron reduction of the 2Fe-2S center (2Fe-2S+ ↔ 2Fe-2S2+) and the riboflavin neutral radical (FlH− ↔ FlH•) transitions can be depicted. At this wavelength, the FlH− ↔ Fl⨪ transition from FMNC has a large contribution, but it is practically canceled by the FlH− ↔ Fl transition from FAD because these two transitions appear concurrently, and the contribution of FAD is negative. At 460 nm, the major contributions arise from the two-electron reduction of FAD (FlH− ↔ Fl) and the one-electron reduction of the two covalently bound FMN molecules (Fl⨪ ↔ Fl). At 380 nm, the signals of the FlH− ↔ Fl transition from FAD and the FlH− ↔ Fl• transition of FMNC are the predominant signals.
Data obtained at 560, 380 and 460 nm, respectively, were fitted to a two or three component model based on the Nernst equation (Fig. 4). The midpoint potentials for the cofactors in the NqrB-G140A mutant are practically the same as the ones obtained for the wild-type enzyme (28) (Table 4).
FIGURE 4.
Plot of δΔA (Abs) at 560, 460, and 480 nm against the applied potential of the NqrB-G140A mutant in the presence of Na+ (top) and K+ (bottom).
TABLE 4.
Redox potentials obtained for the wild-type Na+-NQR and NqrB-G140A mutant versus Ag/AgCl (add 208 mV for standard hydrogen electrode)
The calculated error is not more than ±15 mV.
| Cofactor | Redox transition | Number of electrons |
Em |
|||
|---|---|---|---|---|---|---|
| Wild-type |
NqrB-G140A |
|||||
| Na+ | K+ | Na+ | K+ | |||
| mV vs. Ag/AgCl | ||||||
| FAD | FlH− ↔ Fl | 2 | −440 | −450 | −445 | −445 |
| FMNC | FlH− ↔ Fl⨪ | 1 | −440 | −465 | −440 | −460 |
| FMNC | Fl⨪ ↔ Fl | 1 | −345 | −450 | −350 | −460 |
| FMNB | Fl⨪ ↔ Fl | 1 | −360 | −360 | −360 | −360 |
| Riboflavin | FlH− ↔ FlH• | 1 | −215 | −220 | −220 | −215 |
| 2Fe-2S | [2Fe-2S]+ ↔ [2Fe-2S]2+ | 1 | −500 | −500 | −490 | −500 |
Infrared Spectroscopy
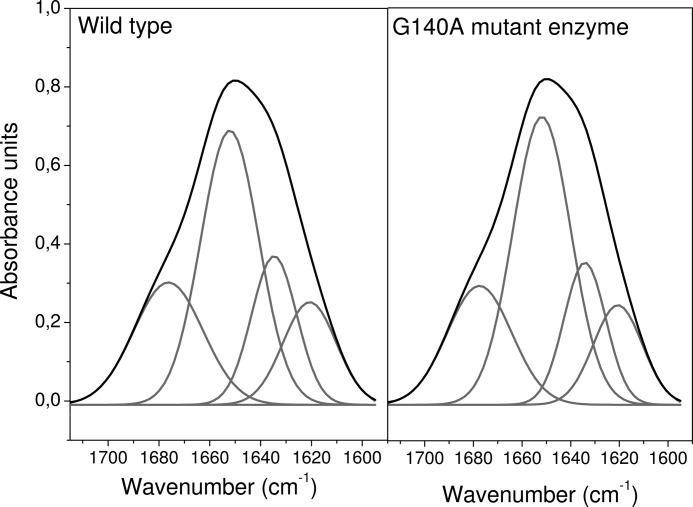
To further verify that the NqrB-G140A mutation does not perturb the secondary structure of the enzyme, we compared the mid-IR spectrum of the mutant with that of wild-type Na+-NQR. The mid-IR spectral region of proteins includes vibrations that arise from the amide backbone. These amide bands provide a direct window into the secondary structure of proteins and have been assigned as follows (29): Amide I (1700–1600 cm−1), C=O stretching mode; Amide II (1575–1480 cm−1), coupled N–H bending/C–N stretching mode; Amide III (1320–1220 cm−1), N–H bending, C–N bending, C=O stretching, O=C–N bending mode. The Amide I signature has been used to determine the secondary structure of proteins. By means of curve fitting, it is possible to determine the proportions of different classes of secondary structure, including α-helix, β-turn, and β-sheet. Comparing the IR absorbance spectra of a wild-type protein and a mutant can thus show whether the mutation caused significant changes in secondary structure.
We recorded the IR absorbance spectra of wild-type Na+-NQR and the NqrB-G140A mutant using an attenuated total reflectance method. The spectra of the wild-type enzyme and the NqrB-G140A mutant are clearly similar in Amide I, II, and III regions, suggesting that the secondary structure is not perturbed in the mutant (data not shown.)
The best fit for band separation of the Amide I signature is obtained with four components (Fig. 5). The area of each component with respect to the total area of the Amide I signal gives the relative contribution of each class of secondary structure to the overall folding of the protein (25). The results for the wild type and the NqrB-G140A mutant differ less than 1%, which is well within the maximum expected error of 5% (Table 5). Therefore, we conclude that the mutation does not have a significant effect on the overall structure of the enzyme.
FIGURE 5.
Band separation of the Amide I signature of the wild-type Na+-NQR and the NqrB-G140A mutant.
TABLE 5.
Secondary structure content of wild type Na+-NQR and NqrB-G140A mutant measured by attenuated total reflectance
The contribution to the secondary structure was measured by the relative area of each component with respect of the total area of the Amide I signal.
| Wave number | Secondary structure | Percentage in the WT | Percentage in the NqrB-G140A |
|---|---|---|---|
| cm−1 | % | % | |
| 1676 | β-Turn | 23 | 22 |
| 1652 | α-Helix | 44 | 45 |
| 1634 | β-Sheet | 18 | 17 |
| 1620 | β-Sheet | 15 | 16 |
DISCUSSION
Several structural motifs have been identified in Na+-NQR: the binding sites for NADH, 2Fe-2S center, and FAD as well as the attachment sites of the two covalently bound FMN cofactors (4, 6, 8, 9, 30). Moreover, recent studies have also identified several negatively charged residues that could form part of the sodium binding sites (11, 13). However, the ubiquinone binding site has remained largely uncharacterized. Hayashi et al. (16) identified that the mutation of the glycine residue 141 to valine of NqrB subunit confers resistance to the antibiotic korormicin. Because korormicin is a ubiquinone analog, it was suggested that this site should also be involved in ubiquinone binding. However, it was also demonstrated that korormicin is a non-competitive inhibitor against ubiquinone, complicating this interpretation. The inhibition patterns of HQNO and korormicin produced double reciprocal plots with intersecting lines consistent with a non-competitive inhibition with respect to ubiquinone (19). Nakayama et al. (19) also determined the Ki constants for the free enzyme and for the substrate-enzyme form, obtaining identical values, indicating that the binding of ubiquinone does not interfere with the binding of the inhibitor, and in consequence, they might not compete for the same site. An alternative explanation for this behavior is that the inhibitor may compete with ubiquinone for its binding site but is also able to interact with another form of the enzyme, which could produce inhibition with competitive and uncompetitive components. Until now, it has not been clear whether HQNO and ubiquinone bind to the same site on the enzyme. The results found here demonstrate that mutations at residue NqrB-G140 decrease the ability of the enzyme to interact with HQNO as well with ubiquinone, suggesting that this residue is part of the binding site of both molecules.
Recently, Casutt et al. (31) characterized the binding of ubiquinone to Na+-NQR using a photoreactive biotinylated quinone. Their results suggest that the NqrA subunit is involved in ubiquinone binding: the photoaffinity-labeled quinone binds to a band on their SDS gel of a molecular weight similar to that of NqrA, and the isolated NqrA subunit is able to bind ubiquinone. However, the binding to the isolated NqrA may not be catalytically relevant especially because the saturation curve shows a strong negative cooperativity, a feature not found in the titrations of the activity reported here and in previous studies (32). Moreover, the authors showed that the ubiquinone analogs occupy the site of a “tightly bound” ubiquinone. The role of this tightly bound ubiquinone is not clear because it is found in substoichiometric amounts (1, 10). Also, independent studies have shown that this ubiquinone does not participate in the redox reactions and does not affect the stability or activity of the enzyme (31). Furthermore, Hayashi et al. (32) have shown that NADH and ubiquinone react with the enzyme following a Ping-Pong mechanism. According to this mechanism, the ubiquinone binding site would not be available to the inhibitor or to ubiquinone until the enzyme is reduced and the NAD is released. Thus, the reaction of the ubiquinone analog with the oxidized form (or free form) of the enzyme might not label the catalytically relevant binding site.
In the present study, we show that the conserved residues NqrB-G140 and NqrB-G141 are part of a functional ubiquinone binding site. Enzymes with mutations in these residues show non-saturating kinetics versus ubiquinone. When the reduction of NqrB-G140A was followed in the reverse direction using ubiquinol, it was clear that the reduction rate of the reaction was severely perturbed, strongly indicating that the ubiquinone binding site is severely perturbed. Further characterization of NqrB-G140A indicates that all other redox properties of this enzyme remained unchanged, including the reduction kinetics by NADH in the forward direction and the midpoint potential of the cofactors.
Hayashi et al. (16) identified NqrB-G141 as part of the korormicin binding site by selecting mutants that were resistant to this antibiotic. It is possible that mutations at NqrB-G140 were not identified by these authors because such mutations would be deleterious, greatly decreasing the activity of the enzyme.
Ubiquinone might not interact directly with NqrB-G140 because glycine residues lack functional groups that can form strong electrostatic interactions with this substrate. However, in hydrophobic environments, the methylene group of glycine can form hydrogen bonds with nearby oxygen atoms (33). Thus, the role of NqrB-G140 in binding ubiquinone could be consistent with two possibilities. 1) The mutation disturbs the packing of helices, which affects the immediate environment of the quinone binding site. In contrast with soluble globular proteins in which glycine residues are common helix breakers, these residues have an important role in the stability and folding of polytopic membrane proteins by allowing strong electrostatic interactions between the backbones of contiguous helices and by creating space for interdigitation (34). Examples of these types of interactions can be found in glycine zippers (35). 2) NqrB-G140 creates a cavity that is part of the ubiquinone binding site, so mutations altering the size of this residue would directly sterically interfere with ubiquinone, decreasing its binding. Indeed, glycine 122 of rhodopsin forms a pocket for the binding of retinol. Substitution of this residue by a larger group decreases the ability of the enzyme to interact with its cofactor (36). In the case of Na+-NQR, the first possibility seems unlikely because a disruption of the packing should have a larger effect on the enzyme and not a localized effect. For instance, the kinetics of electron transfer in the forward and reverse directions should have been disturbed because the alteration of structure would modify the distance between cofactors, which is one of the main factors controlling the rate of electron transfer (37). Also, a global change in the structure of the protein could expose the cofactors to different environments, which could modify their redox potentials. However, functional studies and IR spectroscopy indicate that the structure of the enzyme is not perturbed by the mutation. Thus, data here support a localized effect of the mutations in which NqrB-G140 and NqrB-G141 in transmembrane helix III are directly involved in the functional binding of ubiquinone to Na+-NQR.
Acknowledgments
We thank Dr. Joel Morgan for critical reading of the manuscript and the Microbiology and Analytical Biochemistry Core Facilities at the Center for Biotechnology and Interdisciplinary Studies for providing essential infrastructure.
This work was supported in part by National Science Foundation Division of Molecular and Cellular Biosciences Grant 1052234.
- Na+-NQR
- Na+-pumping NADH:quinone oxidoreductase
- HQNO
- 2-n-heptyl-4-hydroxyquinoline N-oxide
- ΔΨ
- membrane potential.
REFERENCES
- 1. Barquera B., Hellwig P., Zhou W., Morgan J. E., Häse C. C., Gosink K. K., Nilges M., Bruesehoff P. J., Roth A., Lancaster C. R., Gennis R. B. (2002) Purification and characterization of the recombinant Na+-translocating NADH:quinone oxidoreductase from Vibrio cholerae. Biochemistry 41, 3781–3789 [DOI] [PubMed] [Google Scholar]
- 2. Bertsova Y. V., Bogachev A. V. (2004) The origin of the sodium-dependent NADH oxidation by the respiratory chain of Klebsiella pneumoniae. FEBS Lett. 563, 207–212 [DOI] [PubMed] [Google Scholar]
- 3. Häse C. C., Fedorova N. D., Galperin M. Y., Dibrov P. A. (2001) Sodium ion cycle in bacterial pathogens: evidence from cross-genome comparisons. Microbiol. Mol. Biol. Rev. 65, 353–370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Barquera B., Nilges M. J., Morgan J. E., Ramirez-Silva L., Zhou W., Gennis R. B. (2004) Mutagenesis study of the 2Fe-2S center and the FAD binding site of the Na+-translocating NADH:ubiquinone oxidoreductase from Vibrio cholerae. Biochemistry 43, 12322–12330 [DOI] [PubMed] [Google Scholar]
- 5. Barquera B., Zhou W., Morgan J. E., Gennis R. B. (2002) Riboflavin is a component of the Na+-pumping NADH-quinone oxidoreductase from Vibrio cholerae. Proc. Natl. Acad. Sci. 99, 10322–10324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Barquera B., Häse C. C., Gennis R. B. (2001) Expression and mutagenesis of the NqrC subunit of the NQR respiratory Na+ pump from Vibrio cholerae with covalently attached FMN. FEBS Lett. 492, 45–49 [DOI] [PubMed] [Google Scholar]
- 7. Hayashi M., Nakayama Y., Unemoto T. (2001) Recent progress in the Na+-translocating NADH-quinone reductase from the marine Vibrio alginolyticus. Biochim. Biophys. Acta 1505, 37–44 [DOI] [PubMed] [Google Scholar]
- 8. Hayashi M., Nakayama Y., Yasui M., Maeda M., Furuishi K., Unemoto T. (2001) FMN is covalently attached to a threonine residue in the NqrB and NqrC subunits of Na+-translocating NADH-quinone reductase from Vibrio alginolyticus. FEBS Lett. 488, 5–8 [DOI] [PubMed] [Google Scholar]
- 9. Nakayama Y., Yasui M., Sugahara K., Hayashi M., Unemoto T. (2000) Covalently bound flavin in the NqrB and NqrC subunits of Na+-translocating NADH-quinone reductase from Vibrio alginolyticus. FEBS Lett. 474, 165–168 [DOI] [PubMed] [Google Scholar]
- 10. Pfenninger-Li X. D., Albracht S. P., van Belzen R., Dimroth P. (1996) NADH:ubiquinone oxidoreductase of Vibrio alginolyticus: purification, properties, and reconstitution of the Na+ pump. Biochemistry 35, 6233–6242 [DOI] [PubMed] [Google Scholar]
- 11. Juárez O., Morgan J. E., Nilges M. J., Barquera B. (2010) Energy transducing redox steps of the Na+-pumping NADH:quinone oxidoreductase from Vibrio cholerae. Proc. Natl. Acad. Sci. U.S.A. 107, 12505–12510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Juárez O., Morgan J. E., Barquera B. (2009) The electron transfer pathway of the Na+-pumping NADH:quinone oxidoreductase from Vibrio cholerae. J. Biol. Chem. 284, 8963–8972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Juárez O., Athearn K., Gillespie P., Barquera B. (2009) Acid residues in the transmembrane helices of the Na+-pumping NADH:quinone oxidoreductase from Vibrio cholerae involved in sodium translocation. Biochemistry 48, 9516–9524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Juárez O., Nilges M. J., Gillespie P., Cotton J., Barquera B. (2008) Riboflavin is an active redox cofactor in the Na+-pumping NADH:quinone oxidoreductase (Na+-NQR) from Vibrio cholerae. J. Biol. Chem. 283, 33162–33167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Casutt M. S., Huber T., Brunisholz R., Tao M., Fritz G., Steuber J. (2010) Localization and function of the membrane-bound riboflavin in the Na+-translocating NADH:quinone oxidoreductase (Na+-NQR) from Vibrio cholerae. J. Biol. Chem. 285, 27088–27099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hayashi M., Shibata N., Nakayama Y., Yoshikawa K., Unemoto T. (2002) Korormicin insensitivity in Vibrio alginolyticus is correlated with a single point mutation of Gly-140 in the NqrB subunit of the Na+-translocating NADH-quinone reductase. Arch. Biochem. Biophys. 401, 173–177 [DOI] [PubMed] [Google Scholar]
- 17. Yoshikawa K., Takadera T., Adachi K., Nishijima M., Sano H. (1997) Korormicin, a novel antibiotic specifically active against marine Gram-negative bacteria, produced by a marine bacterium. J. Antibiot. 50, 949–953 [DOI] [PubMed] [Google Scholar]
- 18. Yoshikawa K., Nakayama Y., Hayashi M., Unemoto T., Mochida K. (1999) Korormicin, an antibiotic specific for Gram-negative marine bacteria, strongly inhibits the respiratory chain-linked Na+-translocating NADH: quinone reductase from the marine Vibrio alginolyticus. J. Antibiot. 52, 182–185 [DOI] [PubMed] [Google Scholar]
- 19. Nakayama Y., Hayashi M., Yoshikawa K., Mochida K., Unemoto T. (1999) Inhibitor studies of a new antibiotic, korormicin, 2-n-heptyl-4-hydroxyquinoline N-oxide and Ag+ toward the Na+-translocating NADH-quinone reductase from the marine Vibrio alginolyticus. Biol. Pharm. Bull. 22, 1064–1067 [DOI] [PubMed] [Google Scholar]
- 20. Juárez O., Shea M. E., Makhatadze G. I., Barquera B. (2011) The role and specificity of the catalytic and regulatory cation-binding sites of the Na+-pumping NADH:quinone oxidoreductase from Vibrio cholerae. J. Biol. Chem. 286, 26383–26390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Trumpower B. L., Edwards C. A. (1979) Purification of a reconstitutively active iron-sulfur protein (oxidation factor) from succinate·cytochrome c reductase complex of bovine heart mitochondria. J. Biol. Chem. 254, 8697–8706 [PubMed] [Google Scholar]
- 22. Moss D., Nabedryk E., Breton J., Mäntele W. (1990) Redox-linked conformational changes in proteins detected by a combination of infrared spectroscopy and protein electrochemistry. Evaluation of the technique with cytochrome c. Eur. J. Biochem. 187, 565–572 [DOI] [PubMed] [Google Scholar]
- 23. Hellwig P., Scheide D., Bungert S., Mäntele W., Friedrich T. (2000) FT-IR spectroscopic characterization of NADH:ubiquinone oxidoreductase (complex I) from Escherichia coli: oxidation of FeS cluster N2 is coupled with the protonation of an aspartate or glutamate side chain. Biochemistry 39, 10884–10891 [DOI] [PubMed] [Google Scholar]
- 24. Behr J., Hellwig P., Mäntele W., Michel H. (1998) Redox dependent changes at the heme propionates in cytochrome c oxidase from Paracoccus denitrificans: direct evidence from FTIR difference spectroscopy in combination with heme propionate 13C labeling. Biochemistry 37, 7400–7406 [DOI] [PubMed] [Google Scholar]
- 25. Goormaghtigh E., Cabiaux V., Ruysschaert J. M. (1994) Determination of soluble and membrane protein structure by Fourier transform infrared spectroscopy. III. Secondary structures. Subcell. Biochem. 23, 405–450 [DOI] [PubMed] [Google Scholar]
- 26. Barquera B., Ramirez-Silva L., Morgan J. E., Nilges M. J. (2006) A new flavin radical signal in the Na+-pumping NADH:quinone oxidoreductase from Vibrio cholerae. An EPR/electron nuclear double resonance investigation of the role of the covalently bound flavins in subunits B and C. J. Biol. Chem. 281, 36482–36491 [DOI] [PubMed] [Google Scholar]
- 27. Barquera B., Morgan J. E., Lukoyanov D., Scholes C. P., Gennis R. B., Nilges M. J. (2003) X- and W-band EPR and Q-band ENDOR studies of the flavin radical in the Na+-translocating NADH:quinone oxidoreductase from Vibrio cholerae. J. Am. Chem. Soc. 125, 265–275 [DOI] [PubMed] [Google Scholar]
- 28. Neehaul Y., Juárez O., Barquera B., Hellwig P. (2012) Thermodynamic contribution to the regulation of electron transfer in the Na+-pumping NADH:quinone oxidoreductase from Vibrio cholerae. Biochemistry 51, 4072–4077 [DOI] [PubMed] [Google Scholar]
- 29. Barth A., Zscherp C. (2002) What vibrations tell us about proteins. Q. Rev. Biophys. 35, 369–430 [DOI] [PubMed] [Google Scholar]
- 30. Rich P. R., Meunier B., Ward F. B. (1995) Predicted structure and possible ionmotive mechanism of the sodium-linked NADH-ubiquinone oxidoreductase of Vibrio alginolyticus. FEBS Lett. 375, 5–10 [DOI] [PubMed] [Google Scholar]
- 31. Casutt M. S., Nedielkov R., Wendelspiess S., Vossler S., Gerken U., Murai M., Miyoshi H., Möller H. M., Steuber J. (2011) Localization of ubiquinone-8 in the Na+-pumping NADH:quinone oxidoreductase from Vibrio cholerae. J. Biol. Chem. 286, 40075–44082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Hayashi M., Miyoshi T., Sato M., Unemoto T. (1992) Properties of respiratory chain-linked Na+-independent NADH-quinone reductase in a marine Vibrio alginolyticus. Biochim. Biophys. Acta 1099, 145–151 [PubMed] [Google Scholar]
- 33. Derewenda Z. S., Lee L., Derewenda U. (1995) The occurrence of C–H … O hydrogen bonds in proteins. J. Mol. Biol. 252, 248–262 [DOI] [PubMed] [Google Scholar]
- 34. Javadpour M. M., Eilers M., Groesbeek M., Smith S. O. (1999) Helix packing in polytopic membrane proteins: role of glycine in transmembrane helix association. Biophys. J. 77, 1609–1618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Dalbey R. E., Wang P., Kuhn A. (2011) Assembly of bacterial inner membrane proteins. Annu. Rev. Biochem. 80, 161–187 [DOI] [PubMed] [Google Scholar]
- 36. Han M., Lin S. W., Smith S. O., Sakmar T. P. (1996) The effects of amino acid replacements of glycine 121 on transmembrane helix 3 of rhodopsin. J. Biol. Chem. 271, 32330–32336 [DOI] [PubMed] [Google Scholar]
- 37. Page C. C., Moser C. C., Chen X., Dutton P. L. (1999) Natural engineering principles of electron tunnelling in biological oxidation-reduction. Nature 402, 47–52 [DOI] [PubMed] [Google Scholar]