Background: Deregulation of Wnt signaling contributes to the development of cystic kidney diseases such as nephronophthisis (NPH).
Results: The NPH protein NPHP4 stabilizes Jade-1, a negative Wnt regulator, and translocates Jade-1 to the nucleus.
Conclusion: NPHP4 and Jade-1 additively decrease canonical Wnt signaling.
Significance: Loss of NPHP4-mediated Wnt repression via Jade-1 may contribute to cystogenesis in NPH.
Keywords: Cilia, Genetic Diseases, Kidney, Signaling, Wnt Signaling, NPHP, Kidney Cyst, Nephronophthisis
Abstract
Nephronophthisis (NPH) is an autosomal-recessive cystic kidney disease and represents the most common genetic cause for end-stage renal disease in children and adolescents. It can be caused by the mutation of genes encoding for the nephrocystin proteins (NPHPs). All NPHPs localize to primary cilia, classifying this disease as a “ciliopathy.” The primary cilium is a critical regulator of several cell signaling pathways. Cystogenesis in the kidney is thought to involve overactivation of canonical Wnt signaling, which is negatively regulated by the primary cilium and several NPH proteins, although the mechanism remains unclear. Jade-1 has recently been identified as a novel ubiquitin ligase targeting the canonical Wnt downstream effector β-catenin for proteasomal degradation. Here, we identify Jade-1 as a novel component of the NPHP protein complex. Jade-1 colocalizes with NPHP1 at the transition zone of primary cilia and interacts with NPHP4. Furthermore, NPHP4 stabilizes protein levels of Jade-1 and promotes the translocation of Jade-1 to the nucleus. Finally, NPHP4 and Jade-1 additively inhibit canonical Wnt signaling, and this genetic interaction is conserved in zebrafish. The stabilization and nuclear translocation of Jade-1 by NPHP4 enhances the ability of Jade-1 to negatively regulate canonical Wnt signaling. Loss of this repressor function in nephronophthisis might be an important factor promoting Wnt activation and contributing to cyst formation.
Introduction
Nephronophthisis (NPH)2 is the most common genetic cause of cystic kidney disease in children, with no causative treatment currently available. The most common juvenile form of the disease manifests with urinary concentration defects in early childhood and leads to end-stage renal failure by an average of 13 years (1, 2). The disease is characterized histologically by a hallmark triad of tubular basement membrane disruption, interstitial fibrosis, and corticomedullary cyst development (1–3). Pathogenesis involves the loss of function of one or more NPHP genes, leading to the disease classification of a “ciliopathy” because all nephrocystin proteins (NPHP) studied to date localize to primary cilia or centrosomes (1, 4).
Primary cilia are assembled from the mother centriole in virtually all postmitotic eukaryotic cell types and have been associated with an increasing number of cell signaling pathways (5). An important example is the inhibition of canonical Wnt signaling by the primary cilium and basal body, which have been shown to promote proteasomal β-catenin turnover (6, 7) and sequester Wnt signaling components (8). Canonical Wnt signaling is required for cellular proliferation and differentiation during kidney development and during self-renewal of several adult tissues (9, 10) and has been implicated in disease processes such as tumor development when deregulated (10). When Wnt signaling is initiated, cytosolic β-catenin escapes degradation and is able to translocate to the nucleus, where it binds T cell factor/lymphoid enhancer factor-1 (TCF/LEF1) transcription factors and initiates Wnt-dependent gene transcription. In the absence of Wnt stimulation, cytosolic β-catenin is constitutively phosphorylated by the GSK3β-Axin2-APC destruction complex, subsequently ubiquitylated, and targeted for proteasomal degradation (10).
Consistent with the role of primary cilia in repressing canonical Wnt signaling, it has been demonstrated that the ciliary von Hippel-Lindau protein (pVHL) stabilizes Jade-1 (PHF17 isoform 1S), which, in turn, can act as an E3-ubiquitin ligase to target cytosolic as well as nuclear β-catenin for proteasomal degradation (11). Thus, Jade-1 might be able to “fine-tune” β-catenin levels. The loss of pVHL, as is the case in VHL disease and in the majority of renal cell carcinomas, therefore, would lead to derepressed canonical Wnt signaling via decreased Jade-1 stabilization. Loss of pVHL also disrupts ciliogenesis and leads to cystic kidneys (12–14). Taking into consideration that mouse mutants with overactive canonical Wnt signaling develop cystic kidneys (15, 16), that the protein products of the PKD genes mutated in autosomal dominant polycystic kidney disease negatively regulate Wnt signaling (17, 18), and that histological samples from patients with NPH show abnormal Wnt up-regulation (19), an emerging model suggests that cystogenesis may in part be due to the overactivation of canonical Wnt signaling, with cilia playing a direct role in Wnt regulation (20, 21).
In line with this model, several of the cilia-associated nephrocystin proteins have also been identified as negative modulators of the canonical Wnt signaling pathway (22–24). However, reports are inconsistent regarding the contribution of NPH proteins to structural integrity of primary cilia (25–27). Furthermore, although some mouse models with a nephronophthisis phenotype present with slight changes of cilia number or length (22, 28), others retain morphologically normal cilia (29, 30). Therefore, it is likely that NPH proteins are involved with the regulation of cilia-mediated cell signaling rather than simply contributing to ciliary structure. Because Jade-1 has been identified as novel regulator of β-catenin levels (11), we questioned in this study whether Jade-1 could be a route through which the nephrocystin proteins influence Wnt signaling.
Nephrocystin-4 (NPHP4) has been shown to localize to the basal body (27, 31) and, furthermore is thought to be involved in docking and organizing ciliary traffic at the transition zone (32, 33). Recently, NPHP4 has been reported to be a negative regulator of canonical Wnt signaling (34), acting in a similar manner to inversin (NPHP2) to regulate Dsh localization. The current study confirms that NPHP4 negatively influences canonical Wnt signaling and points to an additional mechanism of this regulation. We identify Jade-1 as a ciliary protein that interacts with NPHP4, which, in turn, facilitates Jade-1 nuclear translocation, resulting in decreased canonical Wnt signaling. We further demonstrate genetic interaction between NPHP4 and Jade-1 in zebrafish, consistent with a conserved role for NPHP4/Jade-1 in regulating canonical Wnt signaling.
EXPERIMENTAL PROCEDURES
Plasmids, siRNA, and Antibodies
FLAG- or V5-tagged plasmids were generated by PCR from the fetal human kidney cDNA library (Stratagene) and inserted into a modified pcDNA6 vector (Invitrogen) using standard cloning techniques. All plasmids were verified by automated DNA sequencing. M50 Super 8× TOPFlash and M51 Super 8× FOPFlash were generated by the Moon laboratory and received from Addgene (plasmids 12456 and 12457). Renilla luciferase pGL4.74 was purchased from Promega (E6921). Control siRNA and NPHP4 siRNA have been described previously (35). siRNAs purchased from Biomers (Konstanz, Germany) were directed against sequences as follows: NPHP4, 5′-AAGCAACGAGATGGTGCTACA-3′ and scrambled/control, 5′-AAATGTACTGCGCGTGGAGAC-3′. siRNAs purchased from Qiagen were directed again the following sequences: Jade-1 #1, 5′-AAGCGGGAGCAGGATGTCTTA-3′; Jade-1 #5, 5′-AGCAGCGATGCTACGACAATA-3′; and Jade-1 #7, 5′-GAAGACCATCTTAGCAGAGAA-3′. Antibodies were obtained from Sigma-Aldrich (polyclonal rabbit anti-PHF17 (ab1) used for immunofluorescence; monoclonal mouse anti-acetylated-tubulin; polyclonal rabbit anti-γ-tubulin; monoclonal mouse anti-FLAG (M2); polyclonal rabbit anti-FLAG), Proteintech (polyclonal rabbit anti-PHF17 used for Western blot analysis), Genetex (polyclonal rabbit anti-PHF17 used for immunofluorescence); Abcam (polyclonal rabbit fibrillarin), Serotec (monoclonal mouse anti-V5) and the Developmental Studies Hybridoma Bank (mouse monoclonal anti-α-tubulin). Monoclonal mouse anti-NPHP4 was generously provided by Sophie Saunier. Monoclonal mouse anti-NPHP1 was described previously (32).
Cell Culture and Transfections
RPE-1 cells were cultured in DMEM-F12 medium (Sigma) supplemented with 10% FBS, 2.5 mm L-glutamate and 1.2 g/liter sodium bicarbonate. Cilia growth was stimulated by growing cells to 70–80% confluency and then serum-starving them for 24–48 h in the RPE1 medium as described above with no FBS supplement. HEK293T cells were cultured in DMEM (Sigma) supplemented with 10% FBS and seeded in 10-cm or 6-well dishes. For transfection experiments, cells were grown to 60% confluency and transiently transfected with plasmid DNA using the calcium-phosphate method as described previously (36, 37). For siRNA knockdown experiments, cells were grown to 80% confluency and transfected with Lipofectamine 2000 (Invitrogen) according to the instructions of the manufacturer. 24 h after transfection, cells in 6-well dishes were harvested in 1 ml of cold PBS, and the centrifuged pellet was boiled in Laemmli buffer at 95 °C for 10 min to obtain a whole cell lysate. Cells in 10-cm dishes were harvested with 6 ml of cold PBS, and 1 ml was immediately removed from the cell suspension to be used for “whole cell lysate” as described above. The remaining 5 ml were centrifuged, and the cell pellet used for coimmunoprecipitation.
Reagents
Okadaic acid sodium salt was purchased from Calbiochem and resuspended in water. Cells were incubated with 1 μm okadaic acid in DMEM for 45 min. MG132 was purchased from Calbiochem and used at a concentration of 1 μm in DMEM for 4 h. Control cells were incubated with an equal amount of the solvent dimethyl sulfoxide.
Coimmunoprecipitation
Harvested 293T cell pellets were resuspended in 1 ml of lysis buffer (1% Triton X-100, 20 mm Tris-HCl (pH 7.5), 50 mm NaCl, 50 mm NaF, 15 mm Na4P2O7, 2 mm Na3VO4, and protease inhibitors). After 15 min of incubation on ice, cells were centrifuged (20,000 × g, 15 min, 4 °C), and the supernatant was ultracentrifuged (100,000 × g, 30 min, 4 °C). 50 μl of the supernatant was kept as the “IP lysate” and boiled with Laemmli buffer, and the remaining supernatant was incubated at 4 °C with anti-FLAG-agarose beads (M2, Sigma) for 1 h. The beads were washed extensively with lysis buffer, boiled in Laemmli buffer, and resolved by 10% SDS-PAGE.
Subcellular Fractionation
Harvested 293T cells pellets were resuspended in 100 μl of hypotonic buffer containing 10 mm Hepes, 1.5 mm MgCl2, 10 mm KCl, and protease inhibitors. Cells were incubated on ice for 10 min, subsequently disrupted 14 times using a 21-gauge needle, and then centrifuged at 4 °C at 100 × g for 30 min. The supernatant was then ultracentrifuged for 1 h at 100,000 × g to yield the cytosolic fraction, whereas the nuclear fraction was obtained by washing the pellet with 1 ml of cold PBS and centrifuged at 4 °C at 9200 × g for 10 min and boiled immediately or first resuspended in a high-salt buffer, sonicated, and repelleted. Fractions were boiled in Laemmli buffer and resolved by 10% SDS-PAGE.
Luciferase Assay
HEK293T cells were seeded into 96-well plates and transfected at 80% confluency using Lipofectamine 2000 (Invitrogen) according to the instructions of the manufacturer with a total amount of 140 ng of DNA per well (50 ng TOPFlash or FOPFlash firefly luciferase; 25 ng constitutively active Renilla luciferase pGL4.74; 5 ng FLAG.β-catenin; 50 ng V5.Jade1S or empty vector; 10 ng V5.NPHP4 or empty vector). In all cases, experimental plasmids were added to a prepared master mix containing FLAG-tagged β-catenin (F.β-catenin) and the reporter plasmids. Non-stimulated cells received only the reported plasmids balanced with empty vector. For knockdown experiments, 20 nm of siRNA was incubated in Lipofectamine together with plasmids, as indicated in figure legends. After 24 h, cells were lysed with passive lysis 5× buffer (Promega). Firefly and Renilla luciferase activities were analyzed using a dual-luciferase reporter assay system (Promega) in a luminometer (Mithras LB940, Berthold) and normalized for Renilla luciferase activity to control for transfection efficiency. Each transfection was performed in triplicate, and each experiment was repeated at least three times. Data were normalized by setting the “β-catenin plus empty vector” treatment group to 100%, and significance was assessed for all β-catenin stimulated treatment groups using a one-way analysis of variance with Tukey's post-hoc test. Error bars represent S.E. To assess the effect because of Jade-1 siRNA, data were normalized to set β-catenin stimulation to 100% separately for each Jade-1 siRNA condition. The amount that NPHP4 reduced the signal was then analyzed by normalizing so that the reduction by NPHP4 in the negative siRNA condition was set to 100%.
qPCR
HEK 293T cells transiently transfected for 24 h with the plasmids described in the figure legend were washed with PBS, and RNA was extracted using TRIzol (Invitrogen). After DNase treatment (Invitrogen), reverse transcription was performed using a high-capacity cDNA kit (Applied Biosystems). Jade-1 mRNA was assessed by SYBR Green qPCR using the following primers (Jade-1 forward primer, 5′-TGAAGCCCACCCGTAGCGGA-3′ and Jade-1 reverse primer, 5′-CTCCTGTGCAGCCCCCTTGC-3′), using HPRT1 as an endogenous control (HPRT1 forward primer, 5′-TGACACTGGCAAAACAATGCA-3′ and HPRT1 reverse primer, 5′-GGTCCTTTTCACCAGCAAGCT-3′). All qPCR experiments were performed on a real-time PCR system (7900HT, Applied Biosystems). Data were analyzed by Student's t test, and error bars depict S.E.
Immunofluorescence
Cells grown on glass coverslips were fixed with 4% paraformaldehyde (PFA) or ice-cold methanol, as indicated in figure legends, washed with Dulbecco's PBS, permeabilized with 0.05% Triton-X100, and blocked with 5% normal donkey serum. Primary antibodies were incubated sequentially for 1 h each with anti-Jade1 used first, except when combined with endogenous NPHP4 staining, in which case anti-NPHP4 was used first. Secondary antibodies were incubated for 30 min. Coverslips were washed with DPBS between each step and extensively before mounting onto glass slides using Prolong Gold antifade reagent with DAPI (Invitrogen) mounting medium. Cells were visualized using an Axiovert 200 microscope (objective EC Plan Neofluar ×40/1.3 Oil) equipped with an Axiocam MRm and apotome system (Carl Zeiss MicroImaging, Jena, Germany). Images were captured using Axiovision 4.8 (Carl Zeiss MicroImaging).
Zebrafish
Morpholinos were obtained from Gene Tools (Philomath, OR) with sequences as follows: ATG-targeting NPHP4 (5′-GCGCTTCTCCACTCAGACATCAGAG-3′), ATG-targeting Jade-1 (5′-TCGGGACACGGCTTCGCTTCATCCT-3′), and 5′UTR-targeting Jade-1 (5′-CGTGGCCTTCAAGTGATGCGGAAAT-3′) and were diluted in Danieau's buffer (58). Wild-type zebrafish embryos were injected at the 1- to 2-cell stage with 1.5 nl of the morpholino solution at 20 μm (low dose) or 100 μm (high dose). Embryos were kept in E3 buffer (5 mm NaCl, 0.17 mm KCl, 0.33 mm CaCl2, 0.33 mm MgSO4, 0.00001% methylene blue) at 28 °C and after 72 h were analyzed for body curvature. Only embryos displaying a distinct curvature of the tail angled more than 45° from the head-neck axis were scored as positive. Over 150 fish were counted for each data point. Data for low-dose comparisons were collected from three independent experiments per condition, and average phenotype frequencies and standard errors were calculated (Excel software). Significance was assessed on the basis of percent WT fish using a one-way analysis of variance with Tukey's post-hoc test. Error bars represent S.E. Cystic morphants used for sectioning were PFA-fixed and embedded in paraffin. Sections were stained using H&E.
RESULTS
Endogenous Jade-1 Localizes to the Ciliary Base and Interacts with NPHP4
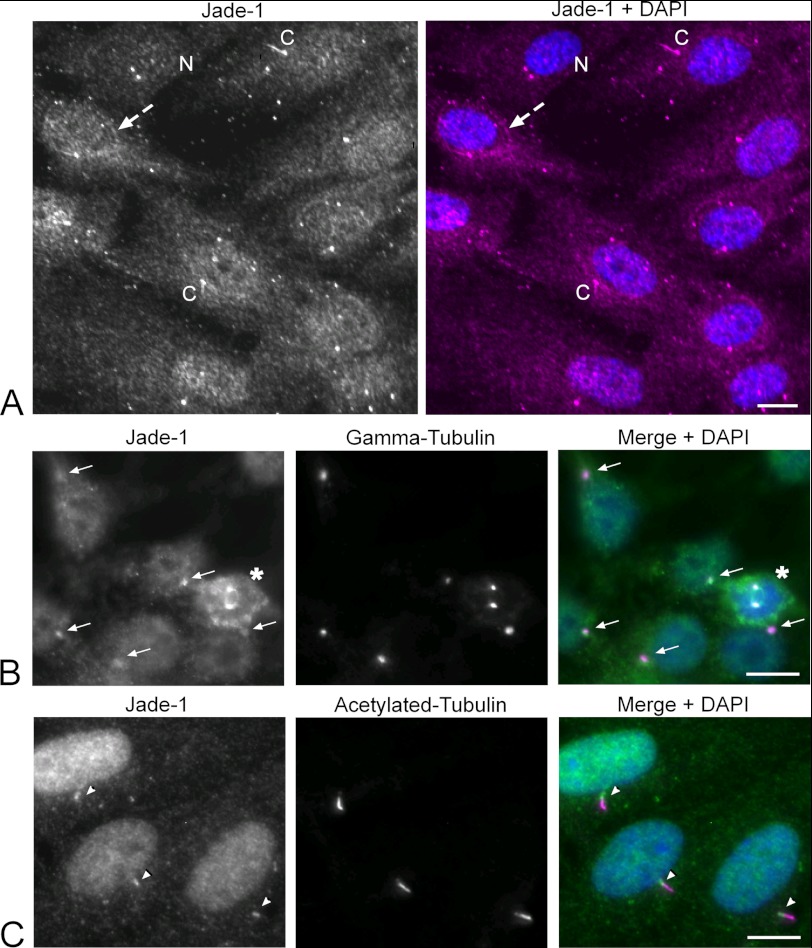
Primary cilia are critical regulators of canonical Wnt signaling (6–8). As Jade-1 is a recently identified Wnt regulator that interacts with the cilia-associated protein pVHL (11), we investigated whether Jade-1 might also localize to cilia or to the ciliary base. In previous studies, Jade-1 was found to be localized both in the cytosol and nucleus, excluding nucleoli (11), which is consistent with our data. In addition, in polarized ciliated retinal pigmented epithelial (RPE-1) cells, we found that Jade-1 primarily localized to the nucleus and perinuclear region, as well as being concentrated at the ciliary base and at times visualized within the cilium proper (Fig. 1A). Some cells did not have a concentration of Jade-1 in the nucleus, which suggests that Jade-1 is a dynamic protein, possibly shuttled in and out of the nucleus under yet to be defined conditions. To confirm the localization of Jade-1 at the centrosome, we costained using appropriate markers and a second Jade-1 antibody in both HEK 293T cells and RPE-1 cells. In both cell types, we again noted a predominantly nuclear staining pattern together with an accumulation at the centrosome, which colabeled with the centrosomal marker γ-tubulin in both dividing and interphase 293T cells (Fig. 1B), and was located at the basal body region at the base of the cilia in RPE-1 cells (Fig. 1C). When the microtubule network was dissociated after 30 min of incubation on ice, Jade-1 could still be identified at the ciliary base (supplemental Fig. 1). This provides evidence that Jade-1 is stably retained at the centrosome, which suggests Jade-1 to be a bona fide centriole-associated protein (38). Further analysis revealed that endogenous Jade-1 localizes both to the basal body and to the transition zone, where it always colocalized with the transition zone marker NPHP1 (supplemental Fig. 2A) and was often also observed in the cilium (supplemental Fig. 2B).
FIGURE 1.
Jade-1 is a dynamic protein that localizes to the basal body region. A, ciliated RPE-1 cells were immunostained for endogenous Jade-1 (magenta) using a polyclonal Jade-1 antibody from Genetex. Jade-1 showed a dynamic expression pattern. In most cells, Jade-1 localized predominantly to the nucleus, but some cells showed an additional concentration of Jade-1 perinuclearly (dashed arrow) or no nuclear concentration of Jade-1 (N). In many cells, Jade-1 labeled structures resembling a primary cilium (C) and in nearly all cells appeared to be centrosomal. B, to confirm centrosomal localization, HEK 293T cells plated on collagen A-coated coverslips were fixed with PFA and immunostained for endogenous Jade-1 (green) using a polyclonal Jade-1 antibody from Sigma-Aldrich. Jade-1 localized to the cytosol and nucleus and colocalized with endogenous γ-tubulin (magenta) at the centrosome (arrows) in interphase as well as in dividing cells (asterisks). C, RPE-1 cells plated on glass coverslips were grown to 60% confluency and then serum-starved for 24 h. Cells were immunostained for endogenous Jade-1 (green, Sigma-Aldrich) and endogenous acetylated-tubulin as a ciliary marker (magenta). Jade-1 was concentrated in the nucleus and at the ciliary base (arrowheads). All stainings were done sequentially with anti-Jade1 used prior to centrosomal/ciliary markers. Specificity of antibody was confirmed by immunofluorescence using secondary antibody only (not shown). Merged images include DAPI. Scale bar = 10 μm.
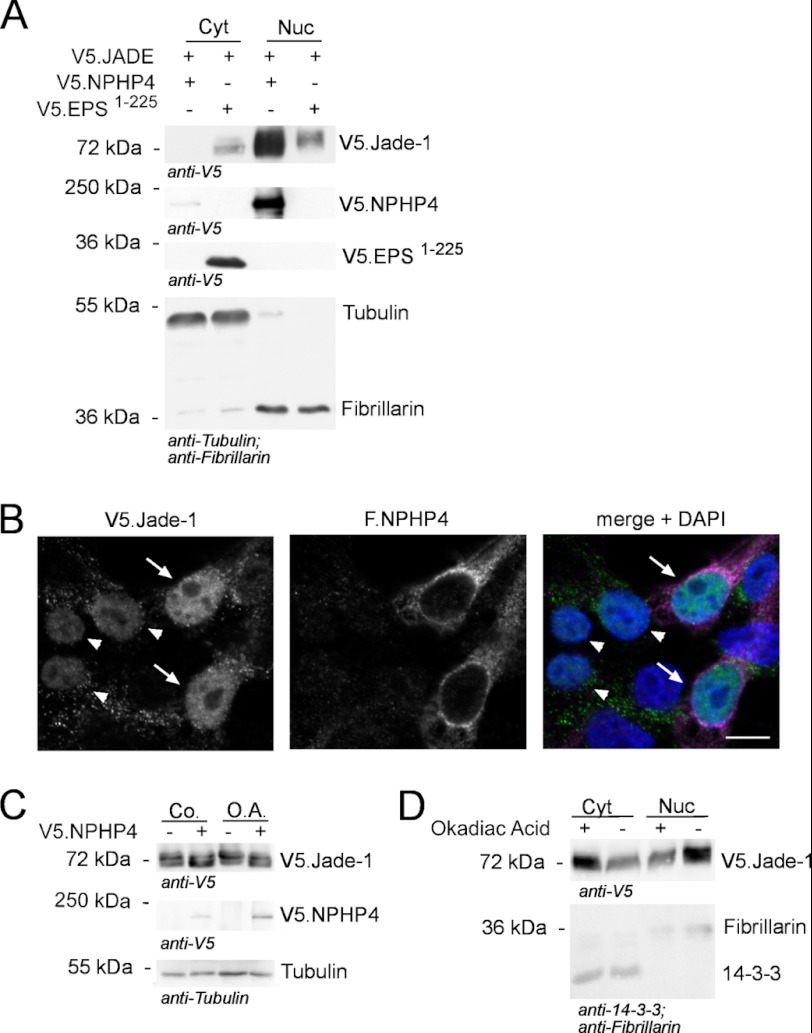
These results demonstrated that the Wnt regulator Jade-1 localizes to the basal body of primary cilia where it is in close proximity to NPHP1, a central component of the NPH protein complex. We therefore investigated whether any of the NPH proteins that have also been described to localize to the ciliary base coprecipitated with Jade-1. Immunoprecipitation (IP) experiments demonstrated that NPHP4 specifically precipitated with Jade-1. V5-tagged Jade-1 (V5.Jade-1) was expressed together with FLAG-tagged NPHP4 (F.NPHP4) or a control protein (F.EPS1–225). V5.Jade-1 coimmunoprecipitated with F.NPHP4 but not with the control protein (Fig. 2A). This finding could be confirmed with the long isoform of endogenous Jade-1 using a specific Jade-1 antibody (Fig. 2C). In the reverse experiment, V5.NPHP4 coimmunoprecipitated with F.Jade-1 but not with F.EPS1–225 (Fig. 2B). These data indicated that Jade-1 and NPHP4 are components of a common protein complex. To assess protein expression independently from localization, a whole cell lysate was taken from the same experiment at the time of cell harvesting. Interestingly, the amount of Jade-1 expressed was increased when coexpressed with NPHP4 in the whole cell lysate but was decreased in the cytosolic IP lysate (Fig. 2A). Because the cytosolic IP lysates lack nuclei, these data suggested that NPHP4 may promote the nuclear localization of Jade-1. Also of interest, immunofluorescence staining of endogenous NPHP4 demonstrated a perinuclear localization in RPE-1 cells (supplemental Fig. 3A, left panel), which is in accordance with previous reports (31). Cells with a perinuclear concentration of endogenous Jade-1 showed colocalization with NPHP4 in this region (supplemental Fig. 3A). NPHP4 is furthermore known to colocalize with NPHP1 in the transition zone of the primary cilium, and it could also be identified in the cilium together with endogenous Jade-1 (supplemental Fig. 3B).
FIGURE 2.
Overexpressed and endogenous Jade-1 interacts with NPHP4. A, 293T cells were transiently transfected with V5-tagged Jade-1 (V5.Jade-1) and either FLAG-tagged NPHP4 (F.NPHP4) or a control protein (F.EPS1–225). Immunoprecipitation was performed using an anti-FLAG antibody. V5.Jade-1 was coimmunoprecipitated by F.NPHP4 but not F.EPS1–225. Note that in the cytosolic IP lysates, Jade-1 levels were decreased by NPHP4, whereas in whole cell lysates taken from the same experimental culture dish prior to the IP procedure, Jade-1 levels were increased by NPHP4. Endogenous tubulin staining from whole cell lysates shows equal amounts. IB, immunoblot. B, in the reverse experiment, 293T cells were transiently transfected with V5.NPHP4 and either F.Jade-1 or F.EPS1–225. Immunoprecipitation was performed using an anti-FLAG antibody. F.Jade-1 but not F.EPS1–225 coimmunoprecipitated V5.NPHP4. C, 293T cells were transiently transfected with F.NPHP4 or a control protein. Immunoprecipitation was performed using an anti-FLAG antibody. Endogenous Jade-1 was detected in the precipitates of F.NPHP4 but not in the control lane. The expression of Jade-1 was found to be equal in the lysates. HC, heavy chain. Blots are representative from three experiments.
NPHP4 Stabilizes Jade-1
To confirm that NPHP4 overexpression specifically increased the total protein level of Jade-1, a whole cell lysate using V5.GFP coexpressed with V5.Jade-1 as a transfection control was performed. This experiment demonstrated that F.NPHP4 increased the overall abundance of V5.Jade-1 (Fig. 3A) without affecting V5.GFP expression. F.NPHP4 did not alter the transcription rate of Jade-1, as assessed by qPCR analysis (Fig. 3B). Instead, NPHP4 appears to increase Jade-1 protein levels by blocking proteasomal degradation, as evidenced by a lack of additional Jade-1 protein accumulation in the presence of both NPHP4 and the proteasome inhibitor MG132 (Fig. 3C). Interestingly, NPHP4 predominantly enhanced the abundance of a smaller band of the Jade-1 protein.
FIGURE 3.
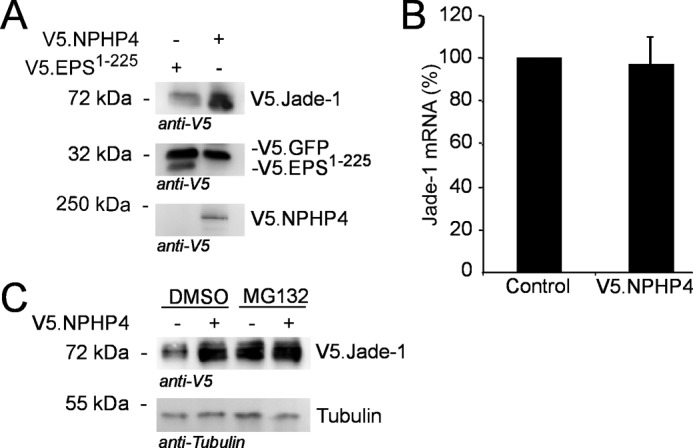
NPHP4 increases protein levels of Jade-1. A, HEK 293T cells were transiently transfected using a plasmid master mix of V5.Jade-1 plus V5.GFP as a transfection control with either V5.NPHP4 or control protein (V5.EPS1–225) added. Cells were harvested after 24 h and processed as a whole cell lysate. V5.Jade-1 expression was up-regulated in the presence of F.NPHP4, but V5.GFP expression remained equal, confirming a specific effect of NPHP4 on Jade-1. B, NPHP4 does not alter endogenous Jade-1 mRNA levels as quantified by qPCR. HEK 293T cells were transiently transfected with V5.NPHP4 or a control protein (V5.EPS1–225) for 24 h (n = 3). C, Jade-1 protein levels are increased by the proteasome inhibitor MG132, and there is no further increase in the presence of V5.NPHP4. HEK 293T cells were transiently transfected for 24 h followed by a 4-h exposure to 1 μm MG132 or dimethyl sulfoxide (DMSO) only before processing as a whole cell lysate. All blots are representative of at least three experiments.
NPHP4 Translocates Jade-1 to the Nucleus
To address whether NPHP4 promotes the nuclear localization of Jade-1, subcellular fractionation was performed following transient transfection of V5.Jade-1 with either V5.NPHP4 or a control protein. These experiments confirmed that expression of V5.NPHP4 reduced the amount of V5.Jade-1 in the cytosol but increased the amount of V5.Jade-1 detected in the nucleus as compared with V5.Jade-1 coexpressed with a control protein (Fig. 4A). The NPHP4-induced nuclear shift of Jade-1 was further supported in immunofluorescence experiments. We expressed low levels of V5.Jade-1 together with F.NPHP4 and observed low levels of nuclear Jade-1 in cells transfected only with V5.Jade-1 but a greatly increased nuclear signal in cells cotransfected with F.NPHP4 (Fig. 4B). It is noteworthy that after cell fraction, overexpressed NPHP4 predominantly associated with the nuclear fraction, in stark contrast to the cytosolic control protein V5.EPS (Fig. 4A), whereas immunofluorescence data reveals that F.NPHP4 does not actually enter the nucleus (B, center panel).
FIGURE 4.
NPHP4 translocates Jade-1 to the nucleus. A, V5.Jade-1 was transiently transfected in HEK293T cells with either a control protein (V5.EPS1–225) or V5.NPHP4, and after 24 h, subcellular fractionation was performed. Cytosolic (Cyt) expression of V5.Jade-1 was decreased in the presence of NPHP4, whereas expression of V5.Jade-1 was increased in the nuclear fraction (Nuc). Tubulin and fibrillarin mark cytoplasmic and nuclear fractions, respectively. The cytosolic fraction was concentrated three times more than the nuclear fraction to visualize cytosolic V5.Jade-1. Note that V5.NPHP4 predominantly associates with the nuclear fraction. Shown is one representative of three experiments. B, HEK 293T cells were transiently transfected with F.NPHP4 and V5.Jade-1. After 24 h, cells were fixed in PFA and immunostained with anti-FLAG antibody (magenta) or anti-V5 antibody (green). Nuclear V5.Jade-1 staining is stronger in cells cotransfected with F.NPHP4 (arrows) than in cells transfected with V5.Jade-1 alone (arrowheads). Note that overexpressed F.NPHP4 does not localize within the nucleus. FLAG and V5 staining in non-transfected cells confirmed that nuclear Jade-1 staining was specific (not shown). Scale bar = 10 μm. C, HEK 293T cells were transiently transfected with V5.NPHP4 or empty vector. After 24 h, cells were incubated with fresh medium containing 1 μm okadaic acid (O.A.) or water (Co.) for 45 min and then processed as a whole cell lysate. Okadaic acid decreased the stabilizing effect of NPHP4 on Jade-1. D, 24 h after HEK 293T cells were transiently transfected with V5.Jade-1, cells were incubated for 45 min with fresh medium containing 1 μm okadaic acid or water and then harvested as a cell fraction. Okadaic acid decreased the amount of Jade-1 in the nucleus. All blots are representative of three experiments.
In our experiments, V5.Jade-1 presents as a double band in whole cell lysates. In the absence of exogenous NPHP4, we generally see the smaller band of Jade-1 in the cytosolic cell fraction, whereas the larger band fractions with the nucleus. As already noted (Fig. 3A), NPHP4 overexpression stabilizes most predominantly the smaller band of V5.Jade-1 and, furthermore, leads to this band fractioning with the nucleus rather than the cytosol (Fig. 4A). We therefore questioned whether the phosphorylation state of V5.Jade-1 could explain the size shift we observed and whether this, in turn, influences the nuclear translocation. Strikingly, the phosphatase inhibitor okadaic acid reduced the ability of NPHP4 to stabilize the smaller band of V5.Jade-1 (Fig. 4C). Furthermore, okadaic acid reduced the amount of Jade-1 in the nucleus compared with the cytosol (Fig. 4D). Taken together, these data suggest that altering the protein level of NPHP4 affects the phosphorylation state and subcellular localization of Jade-1. We then questioned whether this effect would influence the regulatory role of Jade-1 on the cellular response to Wnt signaling.
NPHP4 and Jade-1 Additively Inhibit Canonical Wnt Signaling
Jade-1 has been published previously to be a negative regulator of the canonical Wnt signaling pathway, able to ubiquitylate the major Wnt effector β-catenin in both the cytosol and the nucleus (11). If NPHP4 increases the nuclear abundance of Jade-1, this should enhance the negative effect of Jade-1 on canonical Wnt signaling. Using the TOPFlash TCF/LEF reporter assay, we demonstrated that both NPHP4 and Jade-1 were able to reduce beta-catenin stimulated Wnt reporter activation in a dose-dependent manner. When the transfected amount of V5.NPHP4 and V5.Jade-1 were titrated so that both individually reduced reporter activity to 50%, the combination of the titrated amounts of the two plasmids additively reduced the Wnt reporter activity to near zero (Fig. 5A). In contrast, the co-expression of a control protein (V5. EPS1–225) did not further reduce TCF/LEF reporter activity of either NPHP4 or Jade-1 (Supplemental Fig. 4A). A decrease in canonical Wnt activation by NPHP4 and Jade-1 was corroborated by a decrease in the β-catenin-induced expression of cmyc, a known Wnt target gene (39) (Fig. 5B). It is of interest to note that neither Jade-1 nor NPHP4 were able to reduce the heightened Wnt reporter activity stimulated by a cancer-causing form of β-catenin containing an S33A mutation (Supplemental Fig. 4B). Additionally, Jade-1 consistently led to heightened reporter activity induced by the β-catenin S33A mutant, which mirrors previously published independent results (11).
FIGURE 5.
NPHP4 and Jade-1 additively decrease Wnt reporter activity. For all luciferase experiments, experimental plasmids were transiently transfected in HEK 293T cells together with equal amounts of F.β-catenin or empty vector as indicated, plus constitutively active Renilla luciferase and either TCF/LEF-1-dependent luciferase reporter construct TOPFlash (TOP) or non-responsive control reporter FOPFlash (FOP) as indicated. Wnt signaling activity was quantified as relative luciferase units (RLU) as described above. A, V5.NPHP4 and V5.Jade-1 were transiently transfected separately with empty vector or in combination and compared with F.β-catenin activation when transfected with empty vector alone. Independently, both V5.NPHP4 and V5.Jade-1 significantly reduced β-catenin-induced Wnt activation, and the combination yielded a further significant reduction in reporter activity (F(7,21) = 4.836, p = 0.002; Tukey's post hoc test as indicated, ** = p < 0.01, *** = p < 0.001). Error bars indicate mean ± S.E. Experiments were each done in triplicate for TOPFlash. FOPFlash levels consistently showed no activation (one triplicate was done for each treatment group). B, HEK 293T cells were transiently transfected with F.β-catenin and plasmids as indicated. Cells were processed 24 h later as a whole cell lysate, and endogenous levels of the Wnt gene target cMYC were assessed. Expression of V5.NPHP4 and V5.Jade-1 reduced the activation of cMYC by F.β-catenin. C, HEK 293T cells were transiently transfected with F.β-catenin and reporter plasmids as well as either NPHP4 siRNA or control siRNA and either V5.Jade-1 or empty vector. NPHP4 siRNA increased β-catenin-stimulated Wnt reporter activity compared with cells receiving control siRNA. This effect was reversed by the addition of V5.Jade-1, although V5.Jade-1 combined with NPHP4 siRNA did not block Wnt signaling as effectively as V5.Jade-1 combined with scrambled siRNA (F(3,15) = 13.88, p = 0.001; Tukey's post hoc test as indicated, * = p < 0.05, *** = p < 0.001). D, the reduction of F. β-catenin-induced TOPFlash reporter activity by V5.NPHP4 in the presence of control siRNA was normalized to 100% (see “Experimental Procedures”), and this value was compared with the reduction in reporter activity in the presence of three separate Jade siRNAs. Jade siRNA showed a non-significant trend toward reduced NPHP4 efficacy.
We then performed siRNA knockdown experiments to determine the interdependent roles of NPHP4 and Jade-1 in reducing Wnt reporter activity. Knocking down endogenous NPHP4 led to a heightened level of beta-catenin stimulated Wnt reporter activation compared with a scrambled siRNA. This effect was abrogated by expression of V5.Jade-1. However, Jade-1 co-transfected with NPHP4 siRNA was not able to block Wnt activation to the extent that Jade-1 co-transfected with a scrambled siRNA could, indicating that the efficacy of Jade-1 was impaired by NPHP4 siRNA (Fig. 5C). Similarly, knocking down endogenous Jade-1 impaired the ability of NPHP4 to decrease β-catenin-stimulated Wnt reporter activation. The fact that this effect did not reach significance indicates that the dependence of NPHP4 on Jade-1 is less potent than the dependence of Jade-1 on NPHP4 (Fig. 5D). Taken together, these data clearly show that NPHP4 and Jade-1 act in a common pathway to negatively regulate canonical Wnt signaling.
Genetic Interaction of NPHP4 and Jade-1 in Zebrafish
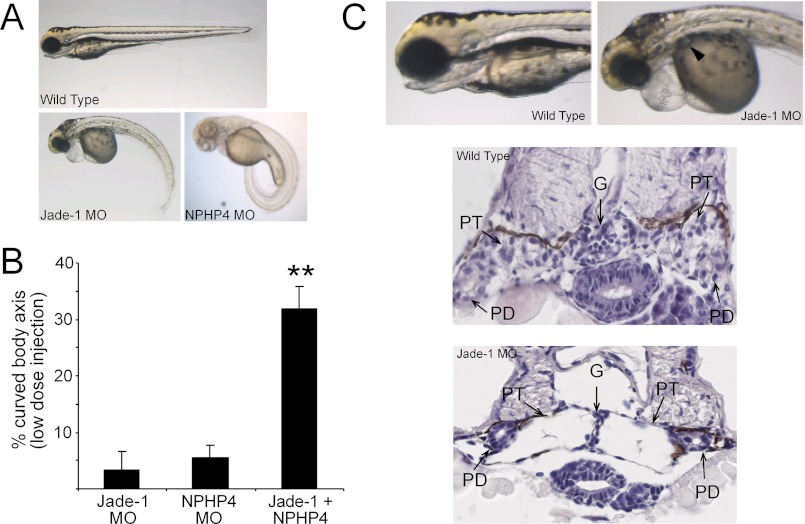
To test whether NPHP4 and Jade-1 act in a common pathway in vivo, we used the developing zebrafish embryo as a model system. The zebrafish embryo is an appropriate model for the functional study of genes related to cystic kidney disease, with zebrafish homologs of human cystoproteins also localizing to cilia and genetic knockdown of these resulting in pronephric cysts in the zebrafish embryo (40, 41). A genetic interaction in vivo was assessed using NPHP4 and Jade-1 morpholinos to knock down gene expression. Injection of NPHP4 ATG-targeting or Jade-1 ATG- or 5′UTR-targeting morpholinos resulted in a body curvature phenotype at 72 h post-fertilization (Fig. 6A). This phenotype has also been reported in zebrafish after knockdown of NPHP2/Inversin, a known cystic kidney disease-causing gene that also regulates canonical Wnt signaling (24), and is associated with ciliary defects and pronephric cysts (42–44). These results also confirm two recent independent reports for NPHP4 knockdown in zebrafish, which further characterized the phenotype to involve ciliary defects (45) and alterations in Wnt signaling (34). A high-dose NPHP4 morpholino injection yielded a body curvature phenotype in 92.0% of embryos, whereas body curvature prevalence was weaker after Jade-1 knockdown (56.5% prevalence) using the same ATG-targeting morpholino dose. However, when the injection doses were individually titrated to yield almost no body curvature phenotype using either morpholino, the combination of NPHP4 and Jade-1 knockdown at this dosage yielded a body curvature phenotype of 31.5%, indicating that NPHP4 and Jade-1 are involved in the same functional pathway in vivo (Fig. 6B). Using the Jade-1 5′UTR-targeting morpholino, we furthermore demonstrated the presence of pronephric cysts (Fig. 6C). These data clearly demonstrated genetic interaction and support a model whereby NPHP4 interacts with Jade-1 to control Jade-1 stability, nuclear translocation, and canonical Wnt signaling.
FIGURE 6.
Jade-1 and NPHP4 additively augment an abnormal body curvature phenotype in vivo. Zebrafish embryos were injected at the 1- to 2-cell stage with either high dose (HD) or low dose (LD) morpholinos against either Jade-1 or NPHP4 and assessed at 72 hours post fertilization for body curvature. Only embryos with a body curvature of the tail angled more than 45° from the head-neck axis were scored as positive. A, representative images of zebrafish embryos injected with high doses of NPHP4 or Jade-1 morpholinos. The abnormal body curvature phenotype has been previously associated with mutations in cystic kidney disease-causing genes, ciliary abnormalities, pronephric cysts, and defective Wnt signaling (see text). B, when NPHP4 and Jade-1 ATG-targeting morpholino injections were individually titrated to a low concentration yielding no obvious phenotype individually, the combination of these two morpholinos at this low concentration resulted in a dramatic increase in the occurrence of an abnormal body curvature (F(2,8) = 25.35; p < 0.001, Tukey's post hoc describes difference between combination and each individual condition, ** = p < 0.01). C, injection of a translation-blocking Jade-1 morpholino resulted in the development of pronephric cysts (PT, pronephric tubule; PD, pronephric duct; G, glomerulum).
DISCUSSION
Recent work has demonstrated NPHP4 to be, like many of the nephrocystin proteins, a negative regulator of canonical Wnt signaling (34). This work corroborates this finding and points to a novel mechanism downstream from that suggested previously. Our study identifies Jade-1 as a novel component of the NPH protein complex being regulated by NPHP4. Jade-1, which functions as a single-subunit E3-ubiquitin ligase targeting the downstream canonical Wnt activator β-catenin to the proteasome, modulates both cytosolic and nuclear β-catenin levels and was suggested to fine-tune canonical Wnt stimulation (11). Here, we show that NPHP4 not only stabilizes Jade-1 but shifts the localization of Jade-1 from the cytosol to the nucleus, enhancing the ability of Jade-1 to block β-catenin-induced Wnt gene activation.
Jade-1 appears consistently as a double band in immunoblot analyses of whole cell lysates, and it is the smaller of these that appears to be specifically influenced by NPHP4. We demonstrate that the phosphatase inhibitor okadaic acid reduces the stabilization of this smaller band by NPHP4 and reduces the level of nuclear Jade under non-stimulated conditions. These data suggest that Jade-1 may need to be in a non-phosphorylated form to enter the nucleus, where it presents as a slightly larger band and thus is likely to be rephosphorylated by nuclear kinases. NPHP4 enhances the stability of the smaller size Jade-1 protein form, resulting in increased nuclear translocation. NPHP4 does not alter the transcriptional rate of Jade-1 but rather interacts on a protein level, possibly blocking kinase access to phosphorylation sites. This may also prevent Jade-1 from being directed to the proteasome, as the protein stabilization by NPHP4 mimics that achieved by the proteasome inhibitor MG132.
Jade-1 has been described previously to localize both to the cytosol and the nucleus (11). Our findings corroborate this and further demonstrate that Jade-1 localizes to the ciliary compartment. At the ciliary base, Jade-1 colocalizes with at least two members of the NPH protein complex and was confirmed to interact with NPHP4 by coprecipitation experiments. The factors that influence the ciliary versus centrosomal localization of Jade-1 remain to be determined. That Jade-1 is a highly dynamic protein is further exemplified by its nuclear versus cytosolic localization under basal conditions. Although we most often saw Jade-1 concentrated in the nucleus, we also identified cells within the same experiment where Jade-1 was instead mainly localized in the perinuclear region. In these cells, there was a striking colocalization with endogenous NPHP4, which also localizes perinuclearly. In our biochemical analyses, NPHP4 fractioned predominantly with the nucleus, although it is apparent using immunofluorescence that NPHP4 is not located within the nucleus itself. This is highly suggestive of NPHP4 associating with the nuclear envelope in a prime position to influence nuclear trafficking. These data are of interest in light of the recent parallels drawn between nuclear and ciliary protein transport whereby proteins required for nuclear import and export are also part of the ciliary proteome (46), supporting the possibility of a “ciliary pore” complex in analogy to the nuclear pore complex (33, 47, 48). The presence of NPHP4 at both the cilium and the perinuclear compartment may reflect parallel functions at these regions and warrants further investigation.
Both Jade-1 and NPHP4 have been demonstrated individually to inhibit canonical Wnt signaling (11, 34). Our data demonstrate that these two proteins act in concert. The additive ability of Jade-1 and NPHP4 to further reduce Wnt reporter activity would not have been seen if the two proteins acted completely independently of one another. We further elucidated individual roles in siRNA knockdown experiments. NPHP4 knockdown led to an increase in β-catenin-induced Wnt gene activity, which could be reversed by the overexpression of Jade-1. This reversal did not block Wnt activation to the extent that Jade-1 cotransfected with scrambled siRNA did. This indicates that the function of Jade-1 was reduced by NPHP4 siRNA, likely because of a decreased ability of Jade-1 to translocate to the nucleus under these conditions. Conversely, the degree to which NPHP4 is able to reduce Wnt reporter activity was impaired in the presence of Jade siRNA. However, there was substantial variation in the degree of impairment. Thus, other players are likely involved. Taken together, it is apparent that Jade-1 requires NPHP4 to be fully effective, but NPHP4 might act through several different means. Indeed, the ability of NPHP4 to regulate canonical Wnt signaling by decreasing the protein level of the canonical Wnt activator Dishevelled (Dsh) was demonstrated recently (34). β-Catenin acts downstream from Dsh in the canonical Wnt pathway. Therefore, the effect reported in our study is not likely to involve Dsh. Although the ability of NPHP4 to block Dsh-induced Wnt reporter activity could reflect a downstream effect on β-catenin, it is more likely that NPHP4 influences more than one aspect of the canonical Wnt signaling cascade. This idea is supported by the observation that NPHP4 was much more potent than the downstream regulator Jade-1, both in our Wnt reporter assay as well as in the frequency of our reported zebrafish curvature phenotype. That there are distinct as well as cooperative roles for these proteins is further indicated by observations using a cancer-causing β-catenin mutant. The same transfected dose of this mutant leads to a much stronger activation the Wnt reporter. Neither Jade-1 nor NPHP4 could reduce Wnt reporter activity in this condition, but the presence of Jade-1 consistently led to an increased signal that mirrors previously reported independent data for Jade-1 (11). The regulation of canonical Wnt signaling is complex, with single proteins capable of playing multiple or even opposing roles dependent on cellular conditions (49–51), and it will be intriguing to tease out the roles of Jade-1 in these different conditions.
Clues to reconcile the distinct versus cooperative roles for Jade-1 and NPHP4 may be found in the degree to which the Wnt cascade is stimulated. The ability of Jade-1 to interact with β-catenin is substantially reduced when Wnt signaling is stimulated upstream in the using Wnt3a or the GSK3β inhibitor LiCl (11). In contrast, NPHP4 is active upstream of the destruction complex (34). NPHP4 could thus act as a negative Wnt regulator both by directing cytosolic Dsh to the proteasome and by stabilizing the β-catenin ubiquitin ligase Jade-1, depending on cellular conditions. Bryja et al. (52) demonstrated that under conditions of low-level Wnt3a stimulation, β-catenin is rapidly activated in a Dsh-independent manner but that only prolonged or higher Wnt3a doses lead to the more classically recognized Wnt-mediated phosphorylation of Dsh and inhibition of the β-catenin destruction complex. It is tempting to thus speculate that the NPHP4-mediated stabilization of Jade-1 could contribute to the regulation of Wnt signals of low strength or short intensity, whereas under higher or more frequent Wnt signaling conditions, NPHP4 is able to influence Wnt activation at the more upstream level of Dsh. Such a scheme would allow for a more finely tuned response to various degrees or types of Wnt stimulation.
The cooperative role of Jade-1 and NPHP4 was supported in vivo by demonstrating a genetic interaction of NPHP4 and Jade-1 using morpholino knockdown in zebrafish. The abnormal body curvature phenotype assessed in this study was similar to that reported in a zebrafish knockdown of the known cystic kidney-causing gene NPHP2/Inversin shown previously to be involved in Wnt regulation (24). Of note, the same phenotype was recently also reported by two independent groups after morpholino-based knockdown of NPHP4 (34, 45). Further data provided in these publications demonstrate pronephric cysts and ciliary abnormalities to be associated with the NPHP4 knockdown, which is in accordance with the previously established link between body curvature, pronephric cysts, and ciliary defects (42, 43). Using a translation-blocking morpholino against Jade-1, we also identified pronephric cysts in embryos displaying body axis curvature. The similarity of these phenotypes strongly supports Wnt deregulation and/or ciliary dysfunction in both the NPHP4 and Jade-1 morphants.
It has been proposed that the genetic mutations in ciliary “cystoproteins” can lead to an imbalance between Wnt-PCP and canonical Wnt signaling, contributing to cyst development (6, 7, 20, 34, 53–55). Many components of Wnt signaling pathways localize to primary cilia (53), and phosphorylated β-catenin accumulates around the basal body region (6). Furthermore, the ubiquitin conjugation system is functional in flagella (56). Our data demonstrate that Jade-1, which has been reported to preferentially ubiquitinate phosphorylated β-catenin, also localizes to the ciliary base and is an active member of the nephrocystin protein complex. Recently, Lancaster et al. (8) reported the ciliopathy protein Jbn as the only known β-catenin regulator localizing to cilia. Our results provide evidence of a second ciliary β-catenin regulator and contribute another example to the various means of cilia-mediated Wnt regulation. Functional loss of the ciliary NPH protein complex might lead to an overactivation of the canonical Wnt pathway both by a direct lack of Wnt regulation and by potentially interrupted cross-talk with other signaling pathways (35, 57). This could result in a decreased ability to maintain the balance between canonical and non-canonical Wnt signaling, which may be critical for injury repair in the kidney and facilitate cyst formation when interrupted (20, 29). Elucidating the precise and possibly multiple roles of these proteins will lead to a better understanding of the pathology of this disease as well as the basic mechanisms of Wnt signaling.
Supplementary Material
Acknowledgments
We thank Stefanie Keller, Katrin Walter, Jasmin Manz, Ruth Herzog, and Bettina Maar for excellent technical assistance and members of the laboratories for helpful discussions, with special reference to Martin Höhne and Safiya Khurshid.
This work was supported by Deutsche Forschungsgemeinschaft Grants SCHE1562 and SFB832 (to B. S.), BE2212 and SFB829 (to T. B.), and SFB 572 (to T. B. and M. H.) and by the Center for Molecular Medicine Cologne (to T. B. and B. S.).

This article contains supplemental Figs. S1–S4.
- NPH
- nephronophthisis
- NPHP
- nephrocystin protein
- TCF/LEF1
- T-cell factor/lymphoid enhancer factor 1
- pVHL
- von Hippel-Lindau protein
- qPCR
- quantitative PCR
- RPE-1
- retinal pigmented epithelial-1
- IP
- immunoprecipitation
- Dsh
- Dishevelled
- PFA
- paraformaldehyde.
REFERENCES
- 1. Hildebrandt F., Attanasio M., Otto E. (2009) Nephronophthisis. Disease mechanisms of a ciliopathy. J. Am. Soc. Nephrol. 20, 23–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Salomon R., Saunier S., Niaudet P. (2009) Nephronophthisis. Pediatr. Nephrol. 24, 2333–2344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Krishnan R., Eley L., Sayer J. A. (2008) Urinary concentration defects and mechanisms underlying nephronophthisis. Kidney Blood Press. Res. 31, 152–162 [DOI] [PubMed] [Google Scholar]
- 4. van Reeuwijk J., Arts H. H., Roepman R. (2011) Scrutinizing ciliopathies by unraveling ciliary interaction networks. Hum. Mol. Genet. 20, R149–157 [DOI] [PubMed] [Google Scholar]
- 5. Gerdes J. M., Davis E. E., Katsanis N. (2009) The vertebrate primary cilium in development, homeostasis, and disease. Cell 137, 32–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Corbit K. C., Shyer A. E., Dowdle W. E., Gaulden J., Singla V., Chen M. H., Chuang P. T., Reiter J. F. (2008) Kif3a constrains β-catenin-dependent Wnt signalling through dual ciliary and non-ciliary mechanisms. Nat. Cell Biol. 10, 70–76 [DOI] [PubMed] [Google Scholar]
- 7. Gerdes J. M., Liu Y., Zaghloul N. A., Leitch C. C., Lawson S. S., Kato M., Beachy P. A., Beales P. L., DeMartino G. N., Fisher S., Badano J. L., Katsanis N. (2007) Disruption of the basal body compromises proteasomal function and perturbs intracellular Wnt response. Nat. Genet. 39, 1350–1360 [DOI] [PubMed] [Google Scholar]
- 8. Lancaster M. A., Schroth J., Gleeson J. G. (2011) Subcellular spatial regulation of canonical Wnt signalling at the primary cilium. Nat. Cell Biol. 13, 700–707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Barker N. (2008) The canonical Wnt/beta-catenin signalling pathway. Methods Mol. Biol. 468, 5–15 [DOI] [PubMed] [Google Scholar]
- 10. Clevers H. (2006) Wnt/beta-catenin signaling in development and disease. Cell 127, 469–480 [DOI] [PubMed] [Google Scholar]
- 11. Chitalia V. C., Foy R. L., Bachschmid M. M., Zeng L., Panchenko M. V., Zhou M. I., Bharti A., Seldin D. C., Lecker S. H., Dominguez I., Cohen H. T. (2008) Jade-1 inhibits Wnt signalling by ubiquitylating β-catenin and mediates Wnt pathway inhibition by pVHL. Nat. Cell Biol. 10, 1208–1216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Esteban M. A., Harten S. K., Tran M. G., Maxwell P. H. (2006) Formation of primary cilia in the renal epithelium is regulated by the von Hippel-Lindau tumor suppressor protein. J. Am. Soc. Nephrol. 17, 1801–1806 [DOI] [PubMed] [Google Scholar]
- 13. Schermer B., Ghenoiu C., Bartram M., Müller R. U., Kotsis F., Höhne M., Kühn W., Rapka M., Nitschke R., Zentgraf H., Fliegauf M., Omran H., Walz G., Benzing T. (2006) The von Hippel-Lindau tumor suppressor protein controls ciliogenesis by orienting microtubule growth. J. Cell Biol. 175, 547–554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Thoma C. R., Frew I. J., Hoerner C. R., Montani M., Moch H., Krek W. (2007) pVHL and GSK3β are components of a primary cilium-maintenance signalling network. Nat. Cell Biol. 9, 588–595 [DOI] [PubMed] [Google Scholar]
- 15. Qian C. N., Knol J., Igarashi P., Lin F., Zylstra U., Teh B. T., Williams B. O. (2005) Cystic renal neoplasia following conditional inactivation of apc in mouse renal tubular epithelium. J. Biol. Chem. 280, 3938–3945 [DOI] [PubMed] [Google Scholar]
- 16. Saadi-Kheddouci S., Berrebi D., Romagnolo B., Cluzeaud F., Peuchmaur M., Kahn A., Vandewalle A., Perret C. (2001) Early development of polycystic kidney disease in transgenic mice expressing an activated mutant of the β-catenin gene. Oncogene 20, 5972–5981 [DOI] [PubMed] [Google Scholar]
- 17. Chapin H. C., Caplan M. J. (2010) The cell biology of polycystic kidney disease. J. Cell Biol. 191, 701–710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lal M., Song X., Pluznick J. L., Di Giovanni V., Merrick D. M., Rosenblum N. D., Chauvet V., Gottardi C. J., Pei Y., Caplan M. J. (2008) Polycystin-1 C-terminal tail associates with β-catenin and inhibits canonical Wnt signaling. Hum. Mol. Genet 17, 3105–3117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Bellavia S., Dahan K., Terryn S., Cosyns J. P., Devuyst O., Pirson Y. (2010) A homozygous mutation in INVS causing juvenile nephronophthisis with abnormal reactivity of the Wnt/β-catenin pathway. Nephrol. Dial. Transplant. 25, 4097–4102 [DOI] [PubMed] [Google Scholar]
- 20. Lancaster M. A., Gleeson J. G. (2010) Cystic kidney disease. The role of Wnt signaling. Trends Mol. Med. 16, 349–360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lienkamp S., Ganner A., Walz G. (2012) Inversin, Wnt signaling and primary cilia. Differentiation 83, S49–55 [DOI] [PubMed] [Google Scholar]
- 22. Bergmann C., Fliegauf M., Brüchle N. O., Frank V., Olbrich H., Kirschner J., Schermer B., Schmedding I., Kispert A., Kränzlin B., Nürnberg G., Becker C., Grimm T., Girschick G., Lynch S. A., Kelehan P., Senderek J., Neuhaus T. J., Stallmach T., Zentgraf H., Nürnberg P., Gretz N., Lo C., Lienkamp S., Schäfer T., Walz G., Benzing T., Zerres K., Omran H. (2008) Loss of nephrocystin-3 function can cause embryonic lethality, Meckel-Gruber-like syndrome, situs inversus, and renal-hepatic-pancreatic dysplasia. Am. J. Hum. Genet. 82, 959–970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kim Y. S., Kang H. S., Jetten A. M. (2007) The Krüppel-like zinc finger protein Glis2 functions as a negative modulator of the Wnt/β-catenin signaling pathway. FEBS Lett. 581, 858–864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Simons M., Gloy J., Ganner A., Bullerkotte A., Bashkurov M., Krönig C., Schermer B., Benzing T., Cabello O. A., Jenny A., Mlodzik M., Polok B., Driever W., Obara T., Walz G. (2005) Inversin, the gene product mutated in nephronophthisis type II, functions as a molecular switch between Wnt signaling pathways. Nat. Genet. 37, 537–543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Delous M., Hellman N. E., Gaudé H. M., Silbermann F., Le Bivic A., Salomon R., Antignac C., Saunier S. (2009) Nephrocystin-1 and nephrocystin-4 are required for epithelial morphogenesis and associate with PALS1/PATJ and Par6. Hum. Mol. Genet. 18, 4711–4723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Fliegauf M., Benzing T., Omran H. (2007) When cilia go bad. Cilia defects and ciliopathies. Nat. Rev. Mol. Cell Biol. 8, 880–893 [DOI] [PubMed] [Google Scholar]
- 27. Mollet G., Silbermann F., Delous M., Salomon R., Antignac C., Saunier S. (2005) Characterization of the nephrocystin/nephrocystin-4 complex and subcellular localization of nephrocystin-4 to primary cilia and centrosomes. Hum. Mol. Genet. 14, 645–656 [DOI] [PubMed] [Google Scholar]
- 28. Hossain Z., Ali S. M., Ko H. L., Xu J., Ng C. P., Guo K., Qi Z., Ponniah S., Hong W., Hunziker W. (2007) Glomerulocystic kidney disease in mice with a targeted inactivation of Wwtr1. Proc. Natl. Acad. Sci. U.S.A. 104, 1631–1636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lancaster M. A., Louie C. M., Silhavy J. L., Sintasath L., Decambre M., Nigam S. K., Willert K., Gleeson J. G. (2009) Impaired Wnt-β-catenin signaling disrupts adult renal homeostasis and leads to cystic kidney ciliopathy. Nat. Med. 15, 1046–1054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Olsen O., Funke L., Long J. F., Fukata M., Kazuta T., Trinidad J. C., Moore K. A., Misawa H., Welling P. A., Burlingame A. L., Zhang M., Bredt D. S. (2007) Renal defects associated with improper polarization of the CRB and DLG polarity complexes in MALS-3 knockout mice. J. Cell Biol. 179, 151–164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Sang L., Miller J. J., Corbit K. C., Giles R. H., Brauer M. J., Otto E. A., Baye L. M., Wen X., Scales S. J., Kwong M., Huntzicker E. G., Sfakianos M. K., Sandoval W., Bazan J. F., Kulkarni P., Garcia-Gonzalo F. R., Seol A. D., O'Toole J. F., Held S., Reutter H. M., Lane W. S., Rafiq M. A., Noor A., Ansar M., Devi A. R., Sheffield V. C., Slusarski D. C., Vincent J. B., Doherty D. A., Hildebrandt F., Reiter J. F., Jackson P. K. (2011) Mapping the NPHP-JBTS-MKS protein network reveals ciliopathy disease genes and pathways. Cell 145, 513–528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Liebau M. C., Höpker K., Müller R. U., Schmedding I., Zank S., Schairer B., Fabretti F., Höhne M., Bartram M. P., Dafinger C., Hackl M., Burst V., Habbig S., Zentgraf H., Blaukat A., Walz G., Benzing T., Schermer B. (2011) Nephrocystin-4 regulates Pyk2-induced tyrosine phosphorylation of nephrocystin-1 to control targeting to monocilia. J. Biol. Chem. 286, 14237–14245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Williams C. L., Li C., Kida K., Inglis P. N., Mohan S., Semenec L., Bialas N. J., Stupay R. M., Chen N., Blacque O. E., Yoder B. K., Leroux M. R. (2011) MKS and NPHP modules cooperate to establish basal body/transition zone membrane associations and ciliary gate function during ciliogenesis. J. Cell Biol. 192, 1023–1041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Burcklé C., Gaudé H. M., Vesque C., Silbermann F., Salomon R., Jeanpierre C., Antignac C., Saunier S., Schneider-Maunoury S. (2011) Control of the Wnt pathways by nephrocystin-4 is required for morphogenesis of the zebrafish pronephros. Hum. Mol. Genet. 20, 2611–2627 [DOI] [PubMed] [Google Scholar]
- 35. Habbig S., Bartram M. P., Müller R. U., Schwarz R., Andriopoulos N., Chen S., Sägmüller J. G., Hoehne M., Burst V., Liebau M. C., Reinhardt H. C., Benzing T., Schermer B. (2011) NPHP4, a cilia-associated protein, negatively regulates the Hippo pathway. J. Cell Biol. 193, 633–642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Benzing T., Gerke P., Höpker K., Hildebrandt F., Kim E., Walz G. (2001) Nephrocystin interacts with Pyk2, p130(Cas), and tensin and triggers phosphorylation of Pyk2. Proc. Natl. Acad. Sci. U.S.A. 98, 9784–9789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Schermer B., Höpker K., Omran H., Ghenoiu C., Fliegauf M., Fekete A., Horvath J., Köttgen M., Hackl M., Zschiedrich S., Huber T. B., Kramer-Zucker A., Zentgraf H., Blaukat A., Walz G., Benzing T. (2005) Phosphorylation by casein kinase 2 induces PACS-1 binding of nephrocystin and targeting to cilia. EMBO J. 24, 4415–4424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Wiens C. J., Tong Y., Esmail M. A., Oh E., Gerdes J. M., Wang J., Tempel W., Rattner J. B., Katsanis N., Park H. W., Leroux M. R. (2010) Bardet-Biedl syndrome-associated small GTPase ARL6 (BBS3) functions at or near the ciliary gate and modulates Wnt signaling. J. Biol. Chem. 285, 16218–16230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. He T. C., Sparks A. B., Rago C., Hermeking H., Zawel L., da Costa L. T., Morin P. J., Vogelstein B., Kinzler K. W. (1998) Identification of c-MYC as a target of the APC pathway. Science 281, 1509–1512 [DOI] [PubMed] [Google Scholar]
- 40. Drummond I. A. (2005) Kidney development and disease in the zebrafish. J. Am. Soc. Nephrol. 16, 299–304 [DOI] [PubMed] [Google Scholar]
- 41. Drummond I. A., Davidson A. J. (2010) Zebrafish kidney development. Methods Cell Biol. 100, 233–260 [DOI] [PubMed] [Google Scholar]
- 42. DiBella L. M., Park A., Sun Z. (2009) Zebrafish Tsc1 reveals functional interactions between the cilium and the TOR pathway. Hum. Mol. Genet. 18, 595–606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Sun Z., Amsterdam A., Pazour G. J., Cole D. G., Miller M. S., Hopkins N. (2004) A genetic screen in zebrafish identifies cilia genes as a principal cause of cystic kidney. Development 131, 4085–4093 [DOI] [PubMed] [Google Scholar]
- 44. Drummond I. A., Majumdar A., Hentschel H., Elger M., Solnica-Krezel L., Schier A. F., Neuhauss S. C., Stemple D. L., Zwartkruis F., Rangini Z., Driever W., Fishman M. C. (1998) Early development of the zebrafish pronephros and analysis of mutations affecting pronephric function. Development 125, 4655–4667 [DOI] [PubMed] [Google Scholar]
- 45. Slanchev K., Pütz M., Schmitt A., Kramer-Zucker A., Walz G. (2011) Nephrocystin-4 is required for pronephric duct-dependent cloaca formation in zebrafish. Hum. Mol. Genet. 20, 3119–3128 [DOI] [PubMed] [Google Scholar]
- 46. Ishikawa H., Thompson J., Yates J. R., 3rd, Marshall W. F. (2012) Proteomic analysis of mammalian primary cilia. Curr. Biol. 22, 414–419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Dishinger J. F., Kee H. L., Jenkins P. M., Fan S., Hurd T. W., Hammond J. W., Truong Y. N., Margolis B., Martens J. R., Verhey K. J. (2010) Ciliary entry of the kinesin-2 motor KIF17 is regulated by importin-β2 and RanGTP. Nat. Cell Biol. 12, 703–710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Kee H. L., Dishinger J. F., Blasius T. L., Liu C. J., Margolis B., Verhey K. J. (2012) A size-exclusion permeability barrier and nucleoporins characterize a ciliary pore complex that regulates transport into cilia. Nat. Cell Biol. 14, 431–437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Dale T. (2006) Kinase cogs go forward and reverse in the Wnt signaling machine. Nat. Struct. Mol. Biol. 13, 9–11 [DOI] [PubMed] [Google Scholar]
- 50. Davidson G., Wu W., Shen J., Bilic J., Fenger U., Stannek P., Glinka A., Niehrs C. (2005) Casein kinase 1 γ couples Wnt receptor activation to cytoplasmic signal transduction. Nature 438, 867–872 [DOI] [PubMed] [Google Scholar]
- 51. Zeng X., Tamai K., Doble B., Li S., Huang H., Habas R., Okamura H., Woodgett J., He X. (2005) A dual-kinase mechanism for Wnt co-receptor phosphorylation and activation. Nature 438, 873–877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Bryja V., Schulte G., Arenas E. (2007) Wnt-3a utilizes a novel low dose and rapid pathway that does not require casein kinase 1-mediated phosphorylation of Dvl to activate β-catenin. Cell Signal 19, 610–616 [DOI] [PubMed] [Google Scholar]
- 53. Gerdes J. M., Katsanis N. (2008) Ciliary function and Wnt signal modulation. Curr. Top. Dev. Biol. 85, 175–195 [DOI] [PubMed] [Google Scholar]
- 54. Germino G. G. (2005) Linking cilia to Wnts. Nat. Genet. 37, 455–457 [DOI] [PubMed] [Google Scholar]
- 55. Kishimoto N., Cao Y., Park A., Sun Z. (2008) Cystic kidney gene seahorse regulates cilia-mediated processes and Wnt pathways. Dev. Cell 14, 954–961 [DOI] [PubMed] [Google Scholar]
- 56. Huang K., Diener D. R., Rosenbaum J. L. (2009) The ubiquitin conjugation system is involved in the disassembly of cilia and flagella. J. Cell Biol. 186, 601–613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Varelas X., Miller B. W., Sopko R., Song S., Gregorieff A., Fellouse F. A., Sakuma R., Pawson T., Hunziker W., McNeill H., Wrana J. L., Attisano L. (2010) The Hippo pathway regulates Wnt/β-catenin signaling. Dev. Cell 18, 579–591 [DOI] [PubMed] [Google Scholar]
- 58. Nasevicius A., Ekker S. C. (2000) Effective targeted gene ‘knockdown’ in zebrafish. Nat. Genet. 26, 216–220 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.