Background: Error-prone aminoacyl-tRNA synthetases clear noncognate aminoacyl-adenylates and misacylated tRNAs within synthetic and editing sites, respectively.
Results: Product release limits the rate of post-transfer editing by leucyl-tRNA synthetase.
Conclusion: Kinetic partitioning of misacylated tRNA determines the relative contribution of cis and trans editing.
Significance: In contrast to DNA polymerases, error correction in class I tRNA synthetases relies on substrate selection by the editing site.
Keywords: Aminoacyl-tRNA Synthetase, Enzyme Mechanisms, Enzymes, Protein Synthesis, Protein-Nucleic Acid Interaction, Editing, Kinetic Partitioning, Leucyl-tRNA Synthetase, Norvaline
Abstract
Comprehensive steady-state and transient kinetic studies of the synthetic and editing activities of Escherichia coli leucyl-tRNA synthetase (LeuRS) demonstrate that the enzyme depends almost entirely on post-transfer editing to endow the cell with specificity against incorporation of norvaline into protein. Among the three class I tRNA synthetases possessing a dedicated post-transfer editing domain (connective peptide 1; CP1 domain), LeuRS resembles valyl-tRNA synthetase in its reliance on post-transfer editing, whereas isoleucyl-tRNA synthetase differs in retaining a distinct tRNA-dependent synthetic site pre-transfer editing activity to clear noncognate amino acids before misacylation. Further characterization of the post-transfer editing activity in LeuRS by single-turnover kinetics demonstrates that the rate-limiting step is dissociation of deacylated tRNA and/or amino acid product and highlights the critical role of a conserved aspartate residue in mediating the first-order hydrolytic steps on the enzyme. Parallel analyses of adenylate and aminoacyl-tRNA formation reactions by wild-type and mutant LeuRS demonstrate that the efficiency of post-transfer editing is controlled by kinetic partitioning between hydrolysis and dissociation of misacylated tRNA and shows that trans editing after rebinding is a competent kinetic pathway. Together with prior analyses of isoleucyl-tRNA synthetase and valyl-tRNA synthetase, these experiments provide the basis for a comprehensive model of editing by class I tRNA synthetases, in which kinetic partitioning plays an essential role at both pre-transfer and post-transfer steps.
Introduction
Aminoacyl-tRNA synthetases (aaRSs)2 covalently couple tRNAs with their cognate amino acids in a two-step aminoacylation reaction that provides the precursors for ribosomal protein synthesis (Fig. 1) (1, 2). Activation of amino acid and transfer of the aminoacyl moiety from an aminoacyl-adenylate (aa-AMP) intermediate to tRNA each occur within the synthetic active site. aaRSs are divided into two classes, I and II (3), with enzymes of each class sharing the architecture of the catalytic domain and some features of the reaction kinetics (4).
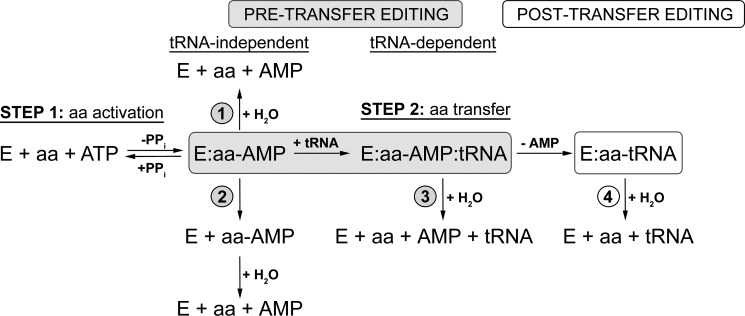
FIGURE 1.
Schematic presentation of editing pathways. Pre-transfer editing can occur through enhanced dissociation of noncognate aminoacyl-adenylate (pathway 2) or its enzymatic hydrolysis (pathways 1 and 3), which may be tRNA-independent (pathway 1) or tRNA-dependent (pathway 3). After transfer, mischarged tRNA can be deacylated through post-transfer editing (pathway 4).
A high level of accuracy in matching the cognate aminoacyl-tRNA (aa-tRNA) pairs is necessary to avoid ambiguity in the genetic code. This is ensured by two mechanisms. First, many of the aaRS discriminate among amino acids and tRNAs with sufficiently high specificity manifested solely in the binding and catalytic steps of the synthetic reaction (5). However, other enzymes are unable to discriminate well among cognate and noncognate amino acids of similar structure. In these cases, noncognate amino acids are cleared by a variety of hydrolytic editing pathways (Fig. 1) (6, 7). Hydrolysis of noncognate aa-AMP, known as pre-transfer editing, can occur by three distinct mechanisms: (i) selective release of aa-AMP followed by nonenzymatic hydrolysis, (ii) enzyme-catalyzed hydrolysis, and (iii) tRNA-stimulated enzyme-catalyzed hydrolysis. If the noncognate amino acid is transferred to tRNA, the misacylated tRNA may be subsequently hydrolyzed via a post-transfer editing pathway (Fig. 1).
Structural (8–10) and biochemical (11, 12) data support the involvement of a distal domain (editing domain) in catalysis of post-transfer editing by both class I and II aaRSs. After misacylation in the synthetic site, the 3′-end of the misacylated tRNA is translocated ∼30 Å to the editing domain, where hydrolysis occurs (10, 13, 14). A trans-editing model has also been proposed for class II aaRSs. In this model, the misacylated tRNA is released from the enzyme but then undergoes hydrolysis after rebinding (15).
Localization of pre-transfer editing within the confines of the synthetic active site was first proposed based on the finding that a modified glutaminyl-tRNA synthetase·tRNAGln complex, which lacks an editing domain, carries out the stereochemically equivalent hydrolysis of glutaminyl-adenylate (16). A growing body of evidence now supports this notion. (i) aaRSs that inherently lack a separate post-transfer editing domain are capable of tRNA-independent pre-transfer editing (17, 18). (ii) aaRS from which the post-transfer domain was excised are capable of tRNA-independent (19, 20) or tRNA-dependent editing (21). (iii) Efficient tRNA-independent (22, 23) and tRNA-dependent (22) editing is catalyzed by enzymes carrying single or double mutations that inactivate the post-transfer editing site. (iv) Substitutions of amino acids located within the synthetic site influence hydrolysis of either cognate or noncognate aa-AMP (24).
The respective contributions of particular pathways to overall editing vary among different aaRSs (25, 26). Using the class I isoleucyl- and valyl-tRNA synthetases (IleRS; ValRS) from Escherichia coli as model editing enzymes, we observed that the extent to which pre-transfer editing is utilized is inversely related to the rate at which the amino acid is transferred to tRNA (22). Fast transfer by ValRS prevented efficient tRNA-dependent pre-transfer editing of threonine; in contrast, the slow transfer by IleRS allowed water to compete for the nucleophilic attack on Val-AMP. These data provided the basis for a synthetic site-based pre-transfer editing model whereby kinetic partitioning of noncognate aa-AMP between transfer and hydrolysis determines the balance between pre- and post-transfer editing (22). In parallel work on class II threonyl-tRNA synthetase (ThrRS), tRNA-dependent pre-transfer editing was induced by slowing of the aminoacyl transfer step by a designed mutation (19). Thus, a close interconnection between the rate of amino acid transfer to tRNA and the utilization of tRNA-dependent pre-transfer editing has been established for aaRSs of both classes.
Leucyl-tRNA synthetase (LeuRS) is a class I enzyme closely related to IleRS and ValRS by the presence of a common large insertion (connective peptide 1; CP1) in the Rossmann fold (27), which forms the post-transfer editing domain (8, 9, 14, 28, 29). E. coli (Ec) LeuRS activates several noncognate amino acids in vitro, including isoleucine, valine, methionine, and norvaline (Nva) (21, 30–33). Among these, norvaline is a nonproteinogenic amino acid that is of particular importance because it can be incorporated into recombinant proteins in place of leucine (34). It was recently demonstrated that the in vivo concentration of norvaline in the cell may rise to 1 mm under anaerobic growth conditions (35). Accumulation of norvaline to such high levels may pose a serious threat to the fidelity of protein biosynthesis under these conditions. Among proteinogenic noncognate amino acids that may be misactivated by EcLeuRS, the best studied is isoleucine (21, 36–39). However, its in vivo concentration is only about 0.1 mm (40), and this level is not elevated by a shift to anaerobic growth conditions (35). Although excesses of either isoleucine or norvaline inhibit the growth of an E. coli strain that relies on an editing-deficient LeuRS, norvaline was recognized as the more potent inhibitor (41). Misincorporation into proteins produced in such strain was also observed (33).
Although the importance of hydrolytic editing of Nva-AMP and Nva-tRNALeu thus appears clear, most mechanistic studies of LeuRS have instead focused on isoleucine as the noncognate amino acid. Several recent studies have included norvaline as an editing substrate but have combined measurements of the pre-transfer and total editing activities toward norvaline, with post-transfer editing of isoleucine (9, 42). Such studies incorporate the untested assumption that the mechanism of editing is identical regardless of the identity of the noncognate amino acid. Reported literature values of the kinetic constants for many aspects of LeuRS synthetic and editing reactions also vary by up to 100-fold or more (21, 30–33), making critical evaluations difficult because some of these data are presumably unreliable.
To address these shortcomings, we have comprehensively measured kinetic parameters for cognate and noncognate synthetic and editing reactions by EcLeuRS, employing a combination of steady-state and pre-steady-state methodologies that we have previously developed in studies of IleRS and ValRS, including new approaches for parallel measurements of AMP and aminoacyl-tRNA formation. In sharp contrast with prior conclusions, these data demonstrate to a high degree of confidence that LeuRS clears noncognate norvaline almost entirely by post-transfer editing and that tRNALeu does not significantly enhance pre-transfer editing. Additional experiments provide significant further mechanistic insight, as follows. (i) The post-transfer editing active site of LeuRS possesses substantial intrinsic substrate preference for norvaline as compared with isoleucine. (ii) The rate-limiting step in editing is product release. Furthermore, by employing a post-transfer editing domain mutant that is partially but not fully disabled in its capacity to clear Nva-tRNALeu, we demonstrate that the CP1-based post-transfer editing active site functions by kinetic partitioning between hydrolysis and dissociation of misacylated tRNA, and that the dissociated misacylated tRNA is capable of rebinding and subsequent hydrolysis. Finally, analyzing synthesis and subsequent hydrolysis of misacylated tRNA by the wild-type (WT) enzyme allows the first reliable estimate of the rate of aminoacyl-tRNA translocation on any editing tRNA synthetase.
EXPERIMENTAL PROCEDURES
Expression and Purification of LeuRS and tRNALeu
The E. coli leuS gene was PCR-amplified from E. coli genomic DNA and cloned as an NheI-EcoRI cassette into expression vector pET28b. The construct was used as a template for site-directed mutagenesis using QuikChange (Stratagene), and the mutations were confirmed by DNA sequencing. Enzyme overexpression and purification procedures were performed as described previously for IleRS (22). We have shown that LeuRS copurifies with Leu-AMP bound in the active site (supplemental Fig. 1), and we designed an additional purification step for its removal (see supplemental Experimental Procedures). The purification procedure is necessary for assays in which enzyme concentration is comparable with or higher than tRNA concentration.
A synthetic gene for E. coli tRNALeuTAA under control of the inducible T7 promoter was inserted as a SalI-BamHI cassette into the pET3a vector upstream of the T7 RNA polymerase terminator site. Overexpression of tRNALeu was achieved as described previously for tRNAVal (22). Plateau aminoacylation assays established that the sample contains 85–90% functional tRNALeu. Prior to use, tRNALeu was denatured by heating for 3 min at 80 °C, followed by addition of MgCl2 to a final concentration of 10 mm. The tRNALeu sample was then renatured by slow-cooling to ambient temperature.
ATP-PPi Exchange Assay
The ATP-PPi exchange assay was performed as described previously (16). Briefly, reactions were conducted at 37 °C in 50 mm Hepes (pH 7.5), 20 mm MgCl2, 100 μg/ml BSA, 5 mm DTT, 4 mm ATP, and 1 mm [32P]PPi. WT LeuRS was present at 10 nm, and the concentration of amino acids was varied over the range 0.1–10 times the Km value to determine steady-state parameters.
Aminoacyl-adenylate Synthesis Assay
The reactions (16, 22) were conducted at 37 °C in a buffer containing 100 mm Tris-Cl (pH 7.5), 10 mm MgCl2, 150 mm KCl, 0.3 mm DTT, 100 μg/ml BSA, 0.004 units/μl inorganic pyrophosphatase (IPPase), and 0.5 mm [α-32P]ATP (0.01–0.1 mCi/ml) with (tRNA-dependent) or without (tRNA-independent editing) 15 μm tRNALeu. Enzyme concentrations are provided in Table 4. Steady-state parameters were obtained by varying the concentration of Nva over the range 0.1–10 times the Km value.
TABLE 4.
Steady-state parameters for AMP formation in the presence of norvaline
Data were measured by aminoacyl-adenylate synthesis assay. The values represent the best fit value ± S.E. of three independent experiments.
| EcLeuRS | −tRNA |
+tRNAa |
||||
|---|---|---|---|---|---|---|
| Km(Nva) | kcat | kcat/Km | Km(Nva) | kcat | kcat/Km | |
| mm | s−1 | mm−1 s−1 | mm | s−1 | mm−1 s−1 | |
| WT | 3.6 ± 0.6 | 0.20 ± 0.01 | 0.05b | 4.5 ± 0.5 | 6.2 ± 0.2 | 1.4c |
| D345A | 3.7 ± 0.1 | 0.17 ± 0.01 | 0.05b | 3.1 ± 0.8 | 0.23 ± 0.02 | 0.07b |
| T252R/D345A | 4.3 ± 0.5 | 0.25 ± 0.01 | 0.06b | 2.9 ± 0.4 | 0.23 ± 0.01 | 0.08b |
a tRNALeu was present at 15 μm concentration.
b The enzymes were assayed at 0.5 μm concentration, and norvaline concentration was varied over the range 0.1–10 times the Km value.
c The enzymes were assayed at 25 nm.
Nonenzymatic Hydrolysis of aa-AMP
The aa-AMP stability assay was adapted from previously published procedures (16, 22). Briefly, 4 μm WT LeuRS was incubated with 50 μm [α-32P]ATP (0.01–0.1 mCi/ml) for 5 min at 37 °C in 100 mm Tris-Cl (pH 7.5), 10 mm MgCl2, 150 mm KCl, 1 mm DTT, and 10 μg/ml BSA. Enzymatic synthesis of aa-[32P]AMP was then initiated by addition of 50 mm Nva or 10 mm Leu. Reactions that assessed Leu-AMP stability were supplemented with 0.004 units/μl IPPase to permit accumulation of Leu-[32P]AMP. Synthesis of Nva- or Leu-[32P]AMP was allowed to proceed for 15 or 30 min, respectively, after which unlabeled Mg2+-ATP was added in 2000-fold molar excess, and aliquots were taken in subsequent time courses.
Preparation and Analysis of [32P]tRNA for Use in Kinetic Assays
[32P]tRNALeu was prepared using tRNA nucleotidyltransferase as described previously (43, 44). Based on comparisons with reactions conducted with unlabeled tRNA in which formation of [14C]Leu-tRNALeu was followed, a decrease in tRNALeu plateau aminoacylation from 90 to 60% was detected after the radiolabeling reaction with tRNA nucleotidyltransferase. Similar decreases in aminoacylation efficiencies were previously observed for tRNALys (45) and tRNASer (46). Steady-state rate constants for aa-[32P]tRNALeu formation were therefore adjusted by a factor (0.6) that takes into account the proportion of [32P]tRNALeu that is functional. After correction, the aminoacylation rate constants obtained with [32P]tRNALeu were identical to those obtained with 14C-labeled amino acid, indicating that the nonfunctional tRNA does not function as an inhibitor.
In all assays where radiolabeled tRNA was followed, the reactions were quenched, and tRNA was hydrolyzed using P1 nuclease and TLC analysis performed as described previously (22).
Determination of AMP/aa-tRNA Ratios
[32P]AMP and aa-[32P]tRNALeu formations were followed in parallel steady-state assays performed in a buffer containing 100 mm Hepes-KOH (pH 7.5), 10 mm MgCl2, 150 mm KCl, 1 mm DTT, 100 μg/ml BSA, 0.004 units/μl IPPase, and 15 μm tRNALeu, with the further addition of either 5 mm Leu or 30 mm Nva (saturating concentrations). Two hundred μm ATP supplemented with trace quantities of [α-32P]ATP was also added to reactions when AMP formation was followed. When aa-tRNA was followed, [32P]tRNALeu was present at 20–50 nm. WT and mutant LeuRS enzymes were used at 5 or 10 nm concentrations in reactions containing Leu or Nva, respectively. In these reactions, use of a lower concentration of ATP as compared with that employed in the aminoacyl-adenylate synthesis assay ensures a better signal to noise ratio, enabling reliable quantitation. Reaction time points were quenched in either formic acid (1 m final concentration) or in a mixture of NaOAc (pH 4.5, 0.5 m final concentration) and SDS (final 0.1%) when AMP or aa-tRNALeu formation was followed, respectively.
Steady-state Deacylation Assay
32P-Labeled aa-tRNAs were prepared by incubation of 20 μm D345A EcLeuRS (from which the Leu-AMP copurifying intermediate was removed; see supplemental Fig. 1 and supplemental Experimental Procedures) with 20 μm tRNALeu and ∼50 nm [32P]tRNALeu for 40 min at 37 °C, in a buffer containing 100 mm Hepes-KOH (pH 7.5), 150 mm KCl, 10 μg/ml BSA, 10 mm MgCl2, 0.008 units/μl IPPase, 1 mm DTT, and 2 mm ATP. Nva, Ile, or Leu were present at 10, 0.25, or 5 mm, respectively. aa-tRNA was recovered by phenol extraction and ethanol precipitation. The tRNA pellet was dissolved in 50 mm NaOAc (pH 4.5), and the sample then purified by gel filtration on a Micro Bio-Spin P30 column (Bio-Rad), followed by dialysis against 15 mm NaOAc (pH 5.0). The amount of tRNA was estimated by comparison of radioactivity present in the aminoacylation mixture prior to phenol extraction, with radioactivity present in the sample after the dialysis step. Loss of radioactivity then corresponds to loss of tRNA during the purification procedure. The percentage of aa-tRNA was established through P1 nuclease digestion of the tRNA sample followed by TLC analysis. On average, aa-tRNA included 50–60% of total tRNA. The aa-tRNA was stored at −20 °C and renatured prior to use, as described above.
The steady-state deacylation assay was performed in 100 mm Hepes-KOH (pH 7.5), 10 mm MgCl2, 150 mm KCl, 1 mm DTT, and 100 μg/ml BSA. aa-tRNA was generally present at 6–8 μm concentrations; higher aa-tRNALeu concentrations (up to 15 μm) were employed in some experiments. The LeuRS concentrations used in each reaction are provided in Table 2. Reactions were initiated by addition of aa-tRNALeu and stopped by adding 2-μl aliquots of reaction mixture to 4 μl of a quench solution containing 0.75 m NaOAc (pH 4.5) and 0.15% SDS. Reaction rates were corrected for nonenzymatic hydrolysis of aa-tRNAs measured under the same reaction conditions, with enzyme omitted from the reaction mixture (for further details see supplemental Fig. 2).
TABLE 2.
Steady-state deacylation of aminoacyl-tRNAs
The values represent the mean value ± S.E. of at least two independent experiments. Aminoacyl-tRNA was present at 6–8 μm concentration.
| EcLeuRS |
kobsa |
||
|---|---|---|---|
| Nva-tRNALeu | Ile-tRNALeu | Leu-tRNALeu | |
| s−1 | |||
| WT | 5.8 ± 0.5b | 2.07 ± 0.05c,d | (10 ± 3) × 10−3e |
| D345A | (1.4 ± 0.1) × 10−3f | NDg | ND |
| T252R | 0.38 ± 0.02h | (3.3 ± 0.3) × 10−3f | (3.8 ± 0.1) × 10−3f |
| T252R/D345A | (0.6 ± 0.1) × 10−3f | ND | ND |
a kobs values were corrected for nonenzymatic hydrolysis of aminoacyl-tRNA. Because saturation was established for aminoacyl-tRNA, these values are equivalent to kcat.
b The enzymes were assayed at 2.5 nm concentration.
c The enzymes were assayed at 3 nm concentration.
d A similar value was obtained with a separate preparation of [14C]Ile-tRNALeu.
e The enzymes were assayed at 100 nm concentration.
f The enzymes were assayed at 500 nm concentration.
g ND means not determined because activity was too low for reliable detection.
h The enzymes were assayed at 20 nm concentration.
Single-turnover Deacylation Assay
aa-tRNAs were prepared similarly as described for the steady-state assay. Briefly, both D345A LeuRS and tRNALeu were present at 25 μm, and [32P]tRNALeu was present at roughly a 300 nm concentration. tRNA was recovered by phenol extraction and subjected to two subsequent gel filtration steps (Micro Bio-Spin P30 columns), followed by dialysis against 15 mm NaOAc (pH 5.0).
Single-turnover deacylations were performed with a KinTek RQF-3 instrument or by hand sampling for longer reactions. Equal volumes of 20 μm LeuRS (in a solution consisting of 200 mm Hepes-KOH (pH 7.5), 150 mm KCl, 10 μg/ml BSA, 10 mm MgCl2 and 1 mm DTT) and 200–500 nm aa-[32P]tRNA were combined to initiate the reactions. The reactions were quenched with a buffer containing 0.8 m NaOAc (pH 4.5) (final concentration); the collection tubes also contained SDS (final w/v 0.1%). Data were fit to the single exponential equation y = Y0 + A × e−kdeacyl × t, where Y0 is the y intercept; A is the amplitude; kdeacyl is the apparent deacylation rate constant, and t is time.
Single-turnover Transfer Step
The transfer step in the aminoacylation reaction was measured by mixing LeuRS·aa-AMP preformed in situ with [32P]tRNALeu, by a procedure similar to that described for IleRS and ValRS (22). The LeuRS·aa-AMP complexes were preformed by incubation of 20 μm LeuRS with 5 mm Leu or 30 mm Nva in 100 mm Tris-Cl (pH 7.5), 10 mm MgCl2, 150 mm KCl, 1 mm DTT, 10 μg/ml BSA, 0.008 units/μl IPPase, and 8 mm ATP for 5 min at 37 °C. The single-turnover transfer reactions were then performed with the KinTek RQF-3 instrument by mixing equal volumes of 20 μm LeuRS·aa-AMP incubated in one syringe with 2 μm 32P-labeled tRNA incubated in the second syringe. Reactions were quenched with NaOAc (pH 4.5, final concentration 0.8 m), and the collection tubes contained SDS (final w/v 0.1%).
Transient Formation of Nva-tRNALeu
Accumulation and subsequent deacylation of Nva-[32P]tRNALeu by T252R LeuRS was followed using the KinTek RQF-3 instrument under single-turnover conditions at 37 °C. As use of T252R allows the tRNA to undergo multiple rounds of transfer and deacylation, observation of a single cycle of aminoacylation and deacylation was ensured by using limiting the amounts of ATP. First, 30 μm T252R LeuRS was incubated with 200 μm Nva for 10 min at 37 °C in 100 mm Hepes (pH 7.5), 150 mm KCl, 10 μg/ml BSA, 10 mm MgCl2, and 1 mm DTT. The mixture was placed on ice and cooled prior to addition of ATP and IPPase (final concentrations of 10 μm and 0.008 units/μl, respectively). The low temperature was maintained at this stage to limit tRNA-independent hydrolysis of Nva-AMP. Furthermore, the syringe used for loading the mixture was cooled in ice prior to use, and a construction was made that enables ice cooling of the syringe mounted on the sample port throughout data collection. Nva-AMP formation was allowed only during a 2-min preincubation in the RQF-3 instrument at 37 °C. In the subsequent reaction, the preincubated T252R enzyme in one syringe was reacted with an equal volume of 2 μm [32P]tRNALeu added from the second syringe. Reactions were quenched with NaOAc (pH 4.5, final concentration 0.8 m), and the collection tubes contained SDS (final w/v 0.1%). As T252R LeuRS was not saturated with Nva-AMP preformed in situ, the rate of Nva-[32P]tRNALeu formation (kform) in this assay does not represent the rate of the isolated transfer step. Data were fit to a minimal kinetic scheme that describes Nva-tRNALeu formation with a rate kform and its subsequent hydrolysis with a rate kdeacyl as two consecutive irreversible reactions as follows: y = A0 × kform/(kdeacyl − kform) × (e−kform × t − e−kdeacyl × t), where A0 represents the initial concentration of tRNALeu, and t is time (47). The parameters obtained from this fit were corroborated by the use of kinetic modeling and the curve fit function in Berkeley Madonna (Version 8.3.14; Macey and Oster, University of California, Berkeley, data not shown). We have attempted isolation of the T252R LeuRS·Nva-AMP complex preformed with millimolar levels of ATP and Nva by gel filtration on Sephadex G-25. However, because of tRNA-independent hydrolysis of Nva-AMP, the isolated complex was not stable at 37 °C in the RQF-3 instrument during the course of data collection (data not shown).
RESULTS
Amino Acid Discrimination in Activation via Ground State Binding
Norvaline is a nonproteinogenic amino acid that at least partially evades proofreading mechanisms and is thus incorporated into proteins under some conditions in vivo (34, 35). It is efficiently activated by LeuRS; turnover numbers for leucine and norvaline activations are comparable, although the Km for the noncognate substrate is increased 100-fold (Table 1). This results in a low discrimination factor (116) and implicates the need for hydrolytic editing to prevent mistranslation (42). Similar observations were made in the case of valine and threonine activation by IleRS and ValRS, respectively (22, 25, 48, 49). For all three enzymes, specificity at the activation step is manifested through a significantly increased Km parameter for the noncognate amino acid, suggesting that the synthetic site (in the absence of tRNA) predominantly discriminates against noncognate amino acids based on ground state binding. The need for editing then arises because there is little to no discrimination at the level of kcat.
TABLE 1.
Steady-state parameters for amino acid activation by wild-type EcLeuRS
Data were measured by the ATP-PPi exchange assay.
| Amino acids | Km (amino acids) | kcat | kcat/Km | (kcat/Km)rel | Discrimination factora |
|---|---|---|---|---|---|
| mm | s−1 | mm−1s−1 | |||
| Leu | 0.05 ± 0.01 | 66 ± 2 | 1320 | 1 | 1 |
| Nva | 4.9 ± 0.4 | 56 ± 1 | 11.4 | 86 × 10−4 | 116 |
a Discrimination factor is defined as 1/(kcat/Km)rel. The values represent the best fit value ± S.E. of at least two independent experiments. WT LeuRS was present at 10 nm, and concentrations of Leu and Nva were varied over the range 0.1–10 times the Km value.
Specificity of the CP1 Domain in Post-transfer Editing
Post-transfer editing by EcLeuRS can be independently studied by a deacylation assay in which preformed aminoacylated tRNA (aa-tRNA) is used as substrate. Because radiolabeled norvaline is not commercially available, to permit comparative studies we employed [32P]tRNALeu prepared using tRNA nucleotidyltransferase (43, 44) in reactions monitoring deacylation of either Nva-tRNALeu or Ile-tRNALeu. Use of [32P]tRNA results in much higher sensitivity and broader applicability of the assay, as compared with labeled amino acid. Despite these advantages, only a few prior studies report use of [32P]tRNA to measure deacylation, and this approach has not been employed for any LeuRS (16, 50–53). Nva-[32P]tRNALeu and Ile-[32P]tRNALeu (each at 60% misacylation level) were produced using D345A LeuRS, which is inactive in post-transfer editing (9). D345A LeuRS purified from cells contains bound Leu-AMP necessitating additional controls and purification steps to ensure that the misacylated tRNA is not contaminated with cognate Leu-tRNALeu (see supplemental Experimental Procedures and supplemental Fig. 1). These are the first measurements of Nva-tRNALeu hydrolysis catalyzed by the CP1 domain of any LeuRS.
The specificity of the CP1-editing site was first examined in steady-state deacylation reactions using WT LeuRS and saturating concentrations of misacylated tRNAs (8 μm Nva-[32P]tRNALeu or Ile-[32P]tRNALeu). Steady-state rates were corrected by subtracting the rate of nonenzymatic deacylation obtained at the same concentration of aa-[32P]tRNALeu and in the same time window (supplemental Fig. 2). Inspection of the corrected rate constants (Table 2) establishes that LeuRS hydrolyzes Ile-[32P]tRNALeu (2.07 s−1) about 3-fold more slowly than Nva-[32P]tRNALeu (5.8 s−1).
To investigate if discrimination resides in the first-order hydrolytic step on the enzyme, the hydrolysis of Nva-tRNALeu and Ile-tRNALeu was studied under single-turnover conditions. Twenty μm LeuRS was mixed with a limiting amount of either Nva-[32P]tRNALeu or Ile-[32P]tRNALeu using a rapid chemical quench instrument. Decay of Nva-[32P]tRNALeu fit well to a single exponential equation with a rate constant of 310 s−1 (Fig. 2A and Table 3). Thus, hydrolysis of Nva-tRNALeu within the CP1-editing site is a very fast process. Interestingly, hydrolysis of Ile-tRNALeu by LeuRS proceeds an order of magnitude more slowly (Fig. 2A and Table 3). This demonstrates that the efficiency of the CP1-editing site varies significantly between different editing substrates. There were no changes in the single-turnover hydrolytic rates when measured under conditions of doubled enzyme concentration, indicating that aa-tRNA binding was not rate-limiting in these experiments.
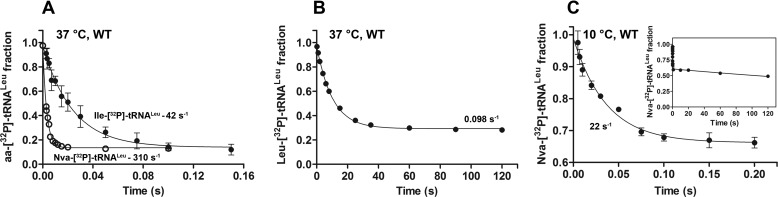
FIGURE 2.
Single-turnover deacylation of Ile-, Nva-, or Leu-tRNALeu by WT EcLeuRS. A, single-turnover deacylation of Ile-[32P]tRNALeu (●) or Nva-[32P]tRNALeu (○) by WT LeuRS at 37 °C (pH 7.5). B, single-turnover deacylation of Leu-[32P]tRNALeu by WT LeuRS at 37 °C (pH 7.5). Note the much longer time scale on the abscissa as compared with A. C, single-turnover deacylation of Nva-[32P]tRNALeu by WT LeuRS at 10 °C (pH 7.0). The inset shows the data over an extended time course and thus depicts the biphasic nature of Nva-tRNALeu deacylation. The rate constant of the slow phase (1.8 × 10−3 s−1) is distinguished from the nonenzymatic hydrolysis that proceeds with a rate constant of 3.3 × 10−4 s−1 (supplemental Fig. 2).
TABLE 3.
Single-turnover deacylation of aminoacyl-tRNAs at 37 °C
The values represent the best fit value ± S.E. of at least three independent experiments. Aminoacyl-tRNAs were present at 100–500 nm and enzymes at 10–20 μm concentration. The rate constant for Nva-tRNALeu deacylation by WT EcLeuRS is 22 ± 4 s−1 at 10 °C.
| EcLeuRS |
kdeacyl |
||
|---|---|---|---|
| Nva-tRNALeu | Ile-tRNALeu | Leu-tRNALeu | |
| s−1 | |||
| WT | (31 ± 2) × 10 | 42 ± 7 | 0.098 ± 0.003 |
| D345A | (6.2 ± 0.7) × 10−3 | (0.30 ± 0.02) × 10−3 | (2.1 ± 0.1) × 10−3 |
| T252R | 2.5 ± 0.2 | (13.2 ± 0.6) × 10−3 | (17 ± 1) × 10−3 |
| T252R/D345A | (1.5 ± 0.2) × 10−3 | (0.63 ± 0.03) × 10−3 | (0.4 ± 0.2) × 10−3 |
Because of fast Nva-tRNALeu hydrolysis at 37 °C (310 s−1), more than half of the reaction took place within the dead time of the instrument (t½ is 2.2 ms;, Fig. 2A). Therefore, to observe the missing part of the curve, the same experiment was performed at 10 °C. Data were again well fit to a single exponential equation; the extracted rate constant was 22 s−1 (Fig. 2C and Table 3). The low amplitude obtained at 10 °C prompted us to monitor the process on a longer time scale, revealing a second phase with a significantly lower rate constant (1.8 × 10−3 s−1; Fig. 2C, inset). The biphasic nature of deacylation was previously observed for Val-tRNAIle hydrolysis by IleRS (48, 54), although the significance of the slower phase remains unclear.
Comparison of the single-turnover and steady-state rate constants shows that the former are 53- and 20-fold greater for Nva-tRNALeu and Ile-tRNALeu hydrolysis, respectively. Thus, either product release or a conformational change preceding product release is rate-limiting for CP1-based post-transfer editing of either substrate by EcLeuRS. Prior conclusions regarding the mechanism of post-transfer editing by LeuRS, which were based entirely on steady-state deacylation (36–38), should be reconsidered in light of these findings. Our data demonstrate that the catalytic mechanism of post-transfer editing by the CP1 domain can only be definitively established by single-turnover kinetics.
We confirmed the crucial role of Asp-345 in the EcLeuRS CP1 domain by incubating 20 μm D345A LeuRS with a limiting amount of aa-[32P]tRNALeu followed by manual sampling. D345A LeuRS displayed 105-fold decreases in single-turnover deacylation rates for both Nva-[32P]tRNALeu and Ile-[32P]tRNALeu (Table 3). This provides the first direct proof of the prominent role of Asp-345 in the hydrolytic chemistry of post-transfer editing. Crystallographic models suggest that Asp-345 participates in a salt bridge with the α-amino group of norvaline (9). The 105-fold decrease in the rate of the hydrolytic step by D345A suggests that this interaction plays a crucial role in positioning the carbonyl carbon atom of the ester linkage in Nva-tRNALeu with respect to a bound nucleophilic water molecule.
tRNA-independent Hydrolytic Editing within the Synthetic Site
Because D345A LeuRS is disabled in post-transfer editing, this enzyme allows analysis of pre-transfer hydrolytic reactions using approaches we have described for IleRS and ValRS, which permit separate measurement of aa-[32P]AMP (if it accumulates) and [32P]AMP formation rates (16, 22). In tRNA-independent reactions, LeuRS produces more [32P]AMP in the presence of norvaline as compared with cognate leucine, implying the existence of editing correction as observed previously (Fig. 3A) (42). Because a detailed steady-state kinetic analysis was not previously reported, we further determined Km and kcat for this reaction (Table 4).
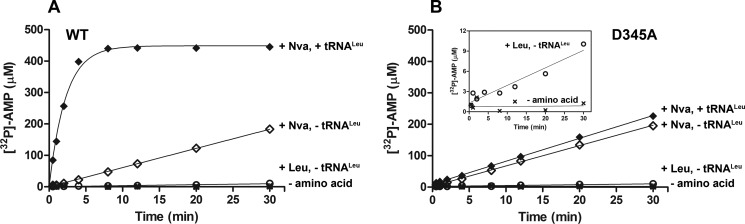
FIGURE 3.
Editing by WT and D345A EcLeuRS. A, [32P]AMP formation by 500 nm WT LeuRS in the absence of amino acid (×), in the presence of 15 mm Leu and absence of tRNALeu (○), and in the presence of 30 mm Nva and presence (♦) or absence (◊) of 15 μm tRNALeu. B, [32P]AMP formation by 500 nm D345A LeuRS in the absence of amino acid (×), in the presence of 15 mm Leu and absence of tRNALeu (○), and in the presence of 30 mm Nva and presence (♦) or absence (◊) of 15 μm tRNALeu. The inset shows [32P]AMP formation in the absence (×) or presence of 15 mm Leu (○) on a plot with a narrower y axis scale to more clearly depict the existence of tRNA-independent editing of cognate Leu.
The AMP produced may originate from enzymatic aa-AMP hydrolysis (tRNA-independent editing) or from nonenzymatic hydrolysis of aa-AMP after its dissociation from the enzyme (Fig. 1, pathways 1 and 2, respectively). To distinguish these possibilities, the nonenzymatic hydrolysis of Nva-AMP was followed by the cold chase assay (16). LeuRS was incubated with Nva and [α-32P]ATP to accumulate Nva-[32P]AMP in solution. A large molar excess of unlabeled ATP was then added, and reaction time points were taken. The rate of Nva-[32P]AMP nonenzymatic hydrolysis was found to be 140-fold lower than the rate of [32P]AMP production in the presence of Nva (14 × 10−4 s−1 versus 0.2 s−1, see supplemental Table 1 and Table 4, respectively). Thus, nearly all of the AMP observed in tRNA-independent pre-transfer editing originates from LeuRS-catalyzed Nva-AMP hydrolysis. It appears that tRNA-independent hydrolytic editing is a universal feature of LeuRS, because analogous findings were made in the case of Aquifex aeolicus (23) and human LeuRS enzymes (55). Very similar kcat/Km parameters were found to characterize tRNA-independent editing of valine by IleRS and of threonine by ValRS (22). Thus, all three class I editing enzymes are similarly efficient at this level of proofreading.
In addition to [32P]AMP, Nva-[32P]AMP accumulation was also observed and independently quantified. Steady-state parameters were extracted (supplemental Table 2) establishing similar Km and 4-fold lower kcat values than in [32P]AMP formation. Because Nva-AMP is hydrolyzed slowly in solution, the lower accumulation of Nva-AMP with respect to AMP supports the notion that nonenzymatic hydrolysis is a minor LeuRS-editing pathway.
To address the location of tRNA-independent editing in EcLeuRS, the AMP formation activity was also studied in the D345A mutant. Steady-state parameters in aa-AMP synthesis assay revealed no difference between the WT LeuRS and D345A variant (0.2 s−1 versus 0.17 s−1, respectively, see Table 4). Thus, inactivation of the CP1-editing site by replacement of the essential Asp-345 did not influence tRNA-independent editing. This strongly suggests that tRNA-independent hydrolysis of Nva-AMP does not occur within the CP1-editing site, indicating instead that this reaction occurs within the synthetic editing site.
Post-transfer Editing Dominates Error Correction by EcLeuRS
Addition of tRNALeu stimulates norvaline-dependent AMP production by LeuRS (Fig. 3A). At the level of kcat/Km, AMP formation is 25-fold more efficient in the presence than in the absence of tRNA, indicating predominance of the tRNA-dependent pathway in LeuRS editing (Table 4). Similarly, 12- and 30-fold stimulation of threonine and valine editing by ValRS and IleRS, respectively, was observed in the presence of cognate tRNA (22). In the presence of tRNA, both aminoacyl transfer and deacylation of aa-tRNA may also occur. Thus, the observed stimulation can reflect tRNA-dependent pre-transfer editing, post-transfer editing, or a combination of both (Fig. 1, pathways 3 and 4, respectively). The two pathways are not distinguishable with this assay because they both accumulate the same AMP product (Fig. 1). To isolate the tRNA-dependent pre-transfer editing step, the D345A variant inactivated in post-transfer editing (Tables 2 and 3) was used in the aa-AMP synthesis assay in the presence of tRNALeu.
Steady-state data show that tRNA fails to stimulate AMP formation by D345A LeuRS (Fig. 3B); kcat/Km values in tRNA-independent and tRNA-dependent editing revealed no significant difference (0.05 mm−1 s−1 versus 0.07 mm−1 s−1, respectively, see Table 4). Thus, disabling the post-transfer pathway seems to eliminate tRNA-dependent editing, indicating its predominance as a mode of error correction. Under the conditions of relatively high enzyme concentration (500 nm) required for substantial [32P]AMP accumulation in these experiments, nearly all of the tRNA is rapidly misacylated (in Fig. 4A, the y-intercept of the time course corresponds to the concentration of aminoacylable tRNA), and remains misacylated during the course of the assay. The same kcat value obtained in tRNA-dependent and tRNA-independent editing (Table 4) demonstrates that misacylated tRNA does not stimulate pre-transfer editing by LeuRS. To further examine whether any pre-transfer editing occurs in the presence of uncharged tRNA, formation of AMP and Nva-tRNALeu by D345A LeuRS was monitored using either [α-32P]ATP or [32P]tRNA, respectively, in otherwise identical parallel reactions. Significantly lower enzyme concentrations (10 nm) were used to extract initial rates on a time scale that allows manual sampling. These measurements revealed that highly similar catalytic constants characterize [32P]AMP and Nva-[32P]tRNALeu formation (Fig. 5C), producing an AMP/Nva-tRNALeu ratio close to 1. Therefore, Nva-AMP hydrolysis does not take place to any considerable extent within the confines of the D345A LeuRS·tRNA complex, and the Nva-AMP formed in the first step yields Nva-tRNALeu quantitatively. These data further support the notion that tRNA-dependent pre-transfer editing does not contribute significantly to error correction by EcLeuRS.
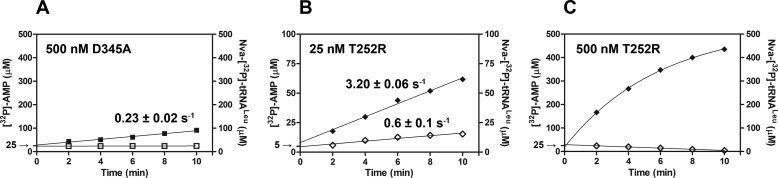
FIGURE 4.
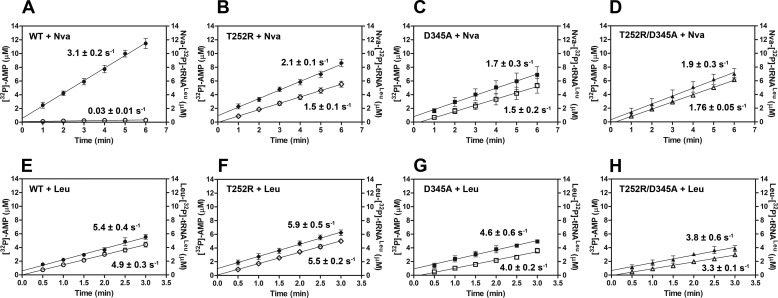
[32P]AMP and Nva-[32P]tRNALeu production in parallel reaction assays. Time courses following [32P]AMP production are illustrated with filled symbols, and time courses following Nva-[32P]tRNALeu production are illustrated with open symbols. Product formation was followed under conditions optimized for the aa-AMP synthesis assay, except that tRNALeu is present at higher concentration (31 μm instead of 15 μm) to facilitate visualization of both products on the same y axis scale. Nva-[32P]tRNALeu and [32P]AMP production by 500 nm D345A EcLeuRS (A). B, 25 nm T252R EcLeuRS; C, 500 nm T252R EcLeuRS. All rate constants were obtained from three independent experiments, and the values represent the mean value ± S.E.
FIGURE 5.
Stoichiometry of AMP and aa-tRNALeu production in the presence of Leu or Nva in parallel steady-state reactions by WT and mutant EcLeuRS enzymes. Time courses following [32P]AMP production are illustrated by filled symbols, and time courses following aa-[32P]tRNALeu production are illustrated by open symbols. A–D represent parallel production of [32P]AMP and Nva-[32P]tRNALeu by 10 nm WT, T252R, D345A, or T252R/D345A LeuRS, respectively. Because of the low Nva-tRNALeu accumulation by WT LeuRS, the formation rate of 0.03 s−1 (A) should be taken as an approximate value. E–H represent parallel production of [32P]AMP and Leu-[32P]tRNALeu by 5 nm WT, T252R, D345A, or T252R/D345A LeuRS, respectively. The rate constants represent the best fit value ± S.E. of three independent experiments.
Previously, robust tRNA-dependent pre-transfer editing of norvaline by EcLeuRS was proposed based on kinetic analysis of a different CP1 site mutant T252R (42). However, in this study post-transfer editing of Nva-tRNALeu was not followed directly; instead, conclusions about this activity were inferred from deacylation of Ile-tRNALeu. Because we have shown that the CP1 domain of EcLeuRS exhibits specificity among aminoacyl-tRNA substrates (Fig. 2A), we inferred that the conclusions of the prior study might be unreliable because the measurements on which they depended were not consistently made using the same noncognate amino acid. We therefore produced the T252R EcLeuRS mutant and tested its activity in the deacylation assay. The architecture of the amino acid pocket in the CP1 domain of EcLeuRS appears optimized for linear hydrophobic side chains, because the WT enzyme preferentially deacylates Nva-tRNALeu as compared with Ile-tRNALeu. Interestingly, increasing steric hindrance in the pocket by introduction of T252R further skews the substrate preference in favor of Nva. The single-turnover rate constant toward Ile-[32P]tRNALeu was more than 103-fold lower compared with WT LeuRS (0.013 s−1 versus 42 s−1, respectively; Table 3), whereas deacylation of Nva-[32P]tRNALeu was also impaired but not to the same extent (2.5 s−1 versus 310 s−1; Table 3). Similar results were obtained in steady-state deacylation assays (Table 2). Thus, T252R is able to catalyze post-transfer editing of Nva-[32P]tRNALeu with substantial efficiency, rendering this mutant inappropriate for isolating pre-transfer editing because it permits AMP accumulation from both pre- and post-transfer reactions. We infer that the activity previously assigned as tRNA-dependent pre-transfer hydrolysis, in the T252R variant, in fact arises from post-transfer editing (42).
To abolish the post-transfer pathway within the T252R CP1-editing site, the T252R/D345A double mutant was also produced. As expected, the T252R/D345A mutant abolished steady-state and single-turnover deacylations of Nva-[32P]tRNALeu (0.6 × 10−3 and 1.5 × 10−3 s−1, Tables 2 and 3, respectively). When the double mutant was tested in the aa-AMP synthesis assay, no tRNA-dependent stimulation of [32P]AMP formation was found (0.06 mm−1 s−1 versus 0.08 mm−1 s−1; Table 4). Thus, inactivation of the post-transfer pathway via D345A eliminates tRNA-dependent editing whether T252R is present or not. The initial rates of [32P]AMP and Nva-[32P]tRNALeu formation were shown to be highly similar (1.9 and 1.76 s−1; Fig. 5D), again confirming absence of tRNA-dependent pre-transfer editing. The T252R/D345A variant exhibits completely analogous editing capabilities as D345A LeuRS (Tables 2–4), supporting the notion that the lack of tRNA-dependent pre-transfer editing is an inherent feature of EcLeuRS.
Fast Transfer of Norvaline to tRNALeu by EcLeuRS
We have recently proposed a general kinetic partitioning model in which tRNA-dependent pre-transfer editing occurs in the synthetic active site when transfer of amino acid to tRNA is relatively slow, allowing a water molecule to compete with the tRNA A76 ribose for nucleophilic attack on aa-AMP (22). According to this model, LeuRS should therefore exhibit fast transfer of the aminoacyl moiety to tRNA. To isolate the transfer step, a molar excess of D345A or T252R/D345A EcLeuRS (20 μm) was incubated with amino acid and ATP (allowing formation of aa-AMP in situ) and then mixed with a limiting amount of [32P]tRNALeu (2 μm) using a rapid chemical quench instrument. For both leucine and norvaline, the transfer step was shown to be fast (Table 5) and exhibited high amplitude (supplemental Fig. 3A). No significant difference in leucyl transfer was observed between D345A and WT enzymes, confirming that D345A has no influence on the transfer step (Table 5). Furthermore, transfers of cognate Leu and noncognate Nva proceed with almost identical rate constants (Table 5), showing that EcLeuRS does not control the specificity of aminoacylation through the transfer step. This was previously observed as well for several other class I and class II aaRSs (19, 22) indicating it as a general feature of these enzymes. These findings are consistent with the chemical partitioning model, substantiating the notion that EcLeuRS does not exhibit tRNA-dependent pre-transfer editing because the fast transfer of norvaline to tRNA does not allow a hydrolytic water molecule to effectively compete for attack on Nva-AMP.
TABLE 5.
Single-turnover transfers of amino acids to tRNAs at 37 °C
The values represent the best fit value ± S.E. of at least three independent experiments. tRNALeu was present at 1 μm and enzymes at 10 μm concentration. Transfer of leucine by WT EcLeuRS was 59 s−1.
| EcLeuRS |
ktrans |
|
|---|---|---|
| Leu | Nva | |
| s−1 | ||
| D345A | 58 ± 5 | 64 ± 8 |
| T252R/D345A | 48 ± 3 | 75 ± 6 |
When formation of Nva-tRNALeu is followed under rapid chemical quench conditions by WT LeuRS in the presence of saturating ATP and Nva but a limiting concentration of tRNA, high amplitude in Nva-tRNALeu formation (∼40–45%) is observed (supplemental Fig. 3B). Steady-state accumulation of Nva-tRNALeu relative to total tRNALeu is determined by the ratio ktrans/(ktrans+ khydr) (56), where ktrans represents the first-order rate constant for Nva-tRNALeu formation (transfer step), and khydr represents the first-order rate constant for Nva-tRNALeu decay (hydrolysis). Based on the separately determined transfer and deacylation rate constants (Tables 3 and 5), accumulation of Nva-tRNALeu to a plateau level of up to 17% of total tRNA was expected. The observed higher amplitude indicates that the Nva-tRNALeu formed in situ is deacylated with a rate constant of 80 s−1 (the value represents the lower limit). This presumably reflects the translocation rate, because the “chemistry” of deacylation, measured by single-turnover kinetics, is about 4-fold faster than this (Fig. 2A and Table 3). These measurements provide the first reliable insight into the kinetics of the tRNA translocation step by any editing aaRS. Although earlier studies attempted to measure translocation by monitoring rebinding of a fluorescent ATP analog to the synthetic active site, these experiments only observed events occurring directly in that site and could not distinguish the identity of the translocating species (57).
The high amplitude in Nva-tRNALeu formation also suggests that robust tRNA-dependent pre-transfer editing by EcLeuRS is highly unlikely. This supports the notion that the inability of tRNA to stimulate norvaline editing by D345A LeuRS is a consequence of its inactivated post-transfer pathway. EcLeuRS thus almost exclusively relies on post-transfer editing to eliminate activated norvaline.
Partitioning of aa-tRNA between Dissociation and Hydrolysis Determines the Balance between Aminoacylation and Editing
Because the synthetic pathway is not highly discriminative toward noncognate amino acids in either LeuRS (supplemental Fig. 3A and Tables 1 and 5), IleRS, or ValRS (22), it was of interest to inquire into the extent to which cognate and noncognate amino acids might be discriminated by the editing pathways. Previous steady-state data showed that Leu-tRNALeu can be hydrolyzed by EcLeuRS (38), but no data addressing this question are available for the pre-transfer editing pathways.
Both Leu-[32P]AMP and [32P]AMP were observed in tRNA-independent pre-transfer editing assays in which Leu was employed as the amino acid substrate. [32P]AMP formation was pronounced (kobs at 15 mm Leu was (11.7 ± 0.3) × 10−3 s−1; Fig. 3B, inset), while Leu-[32P]AMP accumulated only slightly above the enzyme concentration. Furthermore, the cold chase assay returned a rate constant ∼10-fold lower, indicating that the majority of the AMP originates from enzyme-catalyzed pre-transfer editing (supplemental Table 1). Hydrolytic activity toward the cognate Leu-AMP was also not altered by the D345A substitution (kobs for WT LeuRS and for D345A LeuRS are 11.7 × 10−3 and 10.4 × 10−3 s−1, respectively).
Post-transfer editing of cognate Leu-[32P]tRNALeu by WT LeuRS was then tested under both multiple-turnover and single-turnover conditions. In both cases, weak hydrolytic activity was observed (Tables 2 and 3 and Fig. 2B). Single-turnover hydrolysis proceeded faster than the steady-state reaction (measured rate constants of 0.098 s−1 versus 0.010 s−1, respectively), indicating that a step after hydrolysis limits the steady-state deacylation rate. As observed for post-transfer editing of misacylated tRNAs, Leu-[32P]tRNALeu hydrolysis was also impaired by the D345A substitution within the CP1-editing site (Tables 2 and 3). Taken together, the data demonstrate that EcLeuRS is capable of Leu-AMP and Leu-tRNALeu hydrolysis within the synthetic and CP1-editing sites, respectively. However, the extent to which hydrolysis occurs significantly differs; hydrolysis of the cognate intermediate is attenuated by less than 20-fold as compared with hydrolysis of Nva-AMP, while hydrolysis of Leu-tRNALeu is much more significantly (∼103-fold) decreased compared with Nva-tRNALeu hydrolysis. This implies that the CP1-editing site manifests higher specificity in rejection of the cognate substrates than does the synthetic editing site.
The capacity of LeuRS to weakly hydrolyze Leu-AMP and Leu-tRNALeu does not necessarily indicate that leucine is indeed proofread during aminoacylation. Instead, under aminoacylation conditions, kinetic partitioning between synthesis and editing reactions is expected. To better address the extent of cognate substrate editing during aminoacyl-tRNA formation, we therefore monitored initial rates of [32P]AMP and Leu-[32P]tRNALeu formation using differently labeled substrates ([α-32P]ATP or [32P]tRNALeu, respectively) in otherwise parallel assays, as described above for D345A and T252R/D345A LeuRS. This allows estimation of the number of ATP consumed per aa-tRNA formed in the course of aminoacylation (Fig. 5). In these assays both ATP and enzyme concentrations were kept low (200 μm and 5 or 10 nm, respectively) to ensure high sensitivity and initial rate conditions.
Measurements of ATP consumption during cognate aminoacylation by WT LeuRS revealed that the rate constants for [32P]AMP and Leu-[32P]tRNALeu formation were highly similar (5.4 and 4.9 s−1, respectively; Fig. 5E), leading to an AMP/Leu-tRNALeu ratio close to 1. This demonstrates that leucine is not edited during aminoacylation, presumably because both leucine transfer to tRNA and dissociation of Leu-tRNALeu are fast with respect to the hydrolytic reactions within the synthetic and CP1-editing sites, respectively. Initial rates of [32P]AMP and Leu-[32P]tRNALeu formation were also followed for all LeuRS variants (Fig. 5, F–G). In all cases the AMP/Leu-tRNA ratio was the same as in the reaction of WT LeuRS. A similar absence of observed hydrolytic reactions for the cognate substrate was also found for class II histidyl-tRNA synthetase and ThrRS (19, 58).
In contrast, in WT LeuRS norvaline-dependent [32P]AMP formation (3.1 s−1) was significantly faster than Nva-[32P]tRNALeu formation (∼0.03 s−1), as predicted by the efficient post-transfer editing displayed by WT LeuRS toward norvaline (Fig. 5A). Nva-[32P]tRNALeu did not substantially accumulate in this reaction, confirming that fast hydrolysis within the CP1-editing site efficiently competes with Nva-[32P]tRNALeu dissociation. However, a significant disparity in the rates of [32P]AMP and Nva-[32P]tRNALeu formation is not observed for the T252R mutant (the measured rate constants are 2.1 and 1.5 s−1, respectively; Fig. 5B), implying that under these conditions tRNA-dependent editing is insignificant (AMP/Nva-tRNALeu ratio is 1.4). This is analogous to the results obtained with D345A and T252R/D345A LeuRS (Fig. 5, C and D), which are entirely defective in post-transfer editing. Because T252R has an attenuated yet still active post-transfer editing pathway, it appears that the majority of the Nva-[32P]tRNALeu may escape hydrolytic correction due to a dissociation step that efficiently competes with hydrolytic clearance. The accumulation of Nva-[32P]tRNALeu to a similar extent as observed for the mutants unable to hydrolyze misacylated tRNA supports this inference (Fig. 5, B–D). Thus, the fundamental notion suggested by these observations is that the flux through the post-transfer editing pathway is determined by partitioning of misacylated tRNA between hydrolysis and dissociation from the CP1-editing site. Most of the Nva-tRNALeu formed by T252R LeuRS evades editing in cis because the slowed hydrolytic step does not efficiently compete with dissociation.
Is Editing in Trans a Mopping Up Activity?
To test the hypothesis that misacylated tRNA is partitioned between hydrolysis and dissociation from the CP1 domain, we measured parallel formation of [32P]AMP and Nva-[32P]tRNALeu using a higher T252R concentration (25 and 500 nm) and doubled concentration of tRNA (Fig. 4, B and C, respectively). In both cases we observe significantly higher accumulation of [32P]AMP relative to Nva-[32P]tRNALeu. This finding is consistent with kinetic partitioning, because the higher concentrations used will have the effect of promoting the second-order rate constant for reassociation of Nva-tRNALeu with the CP1 domain of LeuRS.
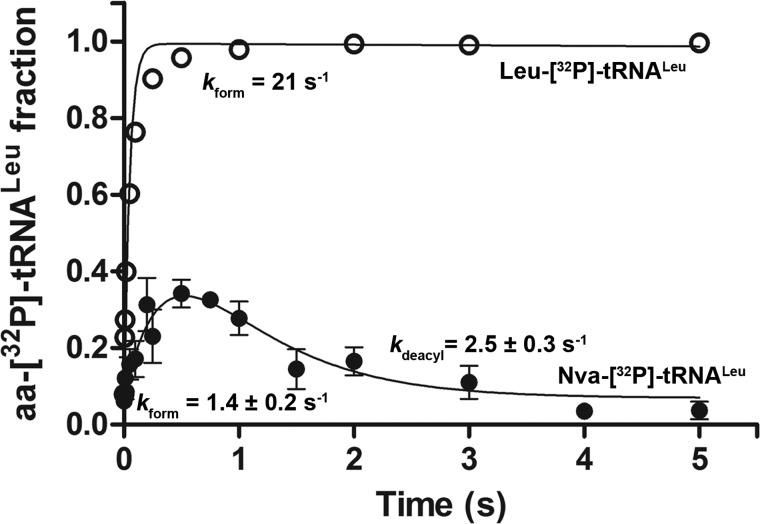
To demonstrate more clearly that tRNA-dependent editing by T252R LeuRS proceeds through a misacylation-deacylation pathway, we followed formation and decay of Nva-[32P]tRNALeu under single-turnover conditions with limiting amounts of ATP and tRNALeu. Thirty μm T252R, 200 μm Nva, and 10 μm ATP were mixed with 1 μm [32P]tRNALeu in a rapid chemical quench instrument. The data clearly show transient accumulation of Nva-[32P]tRNALeu reaching a maximum of 40% (Fig. 6). Estimated rate constants that account for such a progress curve are 1.4 s−1 for Nva-[32P]tRNALeu formation and 2.5 s−1 for its subsequent hydrolysis (see under “Experimental Procedures”). Cognate aminoacylation reactions were performed as a control (Fig. 6). Rate constants for Nva-[32P]tRNALeu and Leu-[32P]tRNALeu formation were significantly lower than measured in the isolated transfer steps. This suggests that under conditions of limiting ATP concentration, aa-AMP formation is slow and limits the rate of aa-tRNA formation. In contrast, the rate constant for Nva-tRNALeu hydrolysis is the same as the constant obtained in the single-turnover deacylation assay (Table 3).
FIGURE 6.
Transient formation of Nva-[32P]tRNALeu by T252R EcLeuRS. Formation and hydrolysis of Nva-[32P]tRNALeu or Leu-[32P]tRNALeu by T252R LeuRS were followed in single-turnover reactions at 37 °C. Equal volumes of 30 μm T252R LeuRS with 200 μm Nva or Leu and 10 μm ATP were mixed with 2 μm [32P]tRNALeu. The estimated rate constant (kform) for Nva-[32P]tRNALeu is 1.4 s−1 and that for deacylation is 2.5 s−1, whereas the rate constant for Leu-[32P]tRNALeu formation in the same reaction conditions is 21 s−1, with no hydrolysis evident within the observed time scale. The rate constants for Nva-[32P]tRNALeu formation and deacylation were estimated from four independent experiments.
Promotion of editing at high enzyme concentrations suggests that T252R LeuRS is capable of editing Nva-tRNALeu in trans. Under these conditions, Nva-tRNALeu is rapidly formed and accumulated to high levels in solution (see intercepts at y axes on Fig. 4, B and C). This strongly encourages reformation of the Nva-tRNALeu·T252R LeuRS complex and editing in trans. Although this pathway is naturally suppressed by fast deacylation of misacylated tRNA within the CP1-editing site in WT LeuRS, it may nonetheless represent a mopping up activity for any misacylated tRNAs that escape cis editing. A dissociation-reassociation pathway by which misacylated tRNAs are released and rebound by the aaRS editing domains has been proposed as a significant editing pathway for class II aaRSs (15). Explicit measurements of association and dissociation rates for cognate and noncognate aa-tRNAs for class I editing enzymes will offer more quantitative insight into this phenomenon.
DISCUSSION
LeuRS Resembles ValRS in Utilizing Post-transfer Editing as a Dominant Error Correction Mode
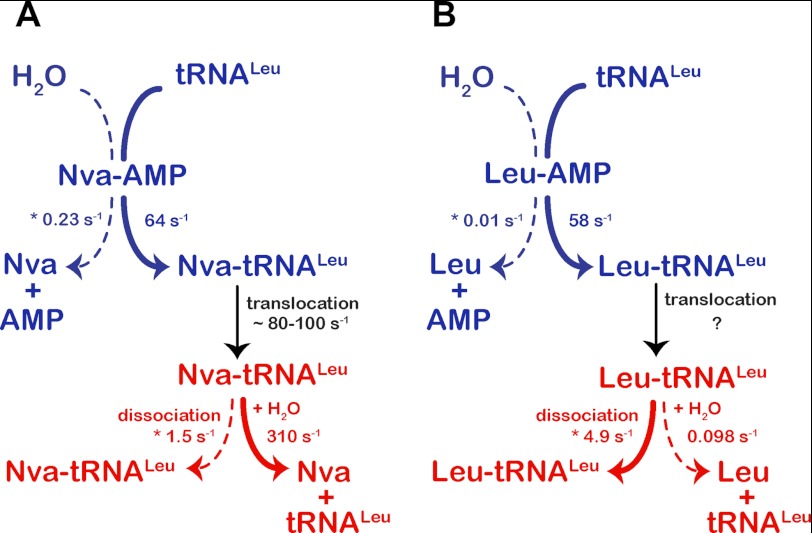
The detailed mechanistic studies of E. coli LeuRS presented here further validate and extend a model in which kinetic competition between water and tRNA for nucleophilic attack on aa-AMP determines partitioning between the pre- and post-transfer editing pathways. In LeuRS and ValRS, the fast transfer of the noncognate amino acid to tRNA prevents pre-transfer editing (Tables 4 and 5) (22). Hence, misincorporation of the noncognate amino acid into cellular proteins is primarily prevented by post-transfer editing after translocation of the tRNA 3′-end to the CP1 domain. This is further demonstrated by the following. (i) Measurements on both enzymes show that inactivation of post-transfer editing concomitantly inactivates all tRNA-dependent editing. (ii) Our finding that the rate constants for formation and hydrolysis of Nva-tRNALeu are very rapid ensures that this species is a competent kinetic intermediate (47). Previously, it was shown that both Thr-tRNAVal and α-aminobutyryl-tRNAVal are rapidly hydrolyzed by ValRS (49, 59); therefore, it appears that LeuRS and ValRS behave similarly in this respect.
In contrast, for IleRS, the rate of aminoacyl-tRNA formation on the enzyme is about 100-fold slower than in ValRS or LeuRS. This allows a water nucleophile to attack the misacylated adenylate prior to transfer. In IleRS, CP1 domain mutants that are wholly inactive in post-transfer editing retain active tRNA-dependent editing catalyzed in the synthetic site (22). Because misacylated Val-tRNAIle is still synthesized at a significant rate by these mutants, both pre-transfer and post-transfer editing significantly contribute to overall editing. The slow tRNA transfer step by IleRS is unusual not only within the subgroup of three class I editing synthetases possessing the CP1 domain but also more generally among tRNA synthetases of either class, where fast tRNA transfer is commonly observed (4, 60, 61). The unique structural determinants of the IleRS·tRNAIle complex responsible for this behavior are unknown.
Common features among all three canonical class I editing aaRS possessing CP1 domains are now also clearly seen. All three enzymes catalyze a synthetic site-based tRNA-independent pre-transfer editing activity of unknown physiological significance that corresponds to about 3% of the tRNA-dependent editing. This activity is in all cases insensitive to CP1 domain mutants that disable post-transfer editing (Table 4 and Fig. 3) (22). Furthermore, our experiments also demonstrate that nonenzymatic hydrolysis of noncognate aminoacyl-adenylate after dissociation from the enzyme (kinetic proofreading) does not contribute significantly in any of the three enzymes. Finally, IleRS, ValRS, and LeuRS each transfer at least some noncognate amino acids onto tRNA at rates comparable with that of cognate aminoacyl-tRNA formation on the enzymes, demonstrating that there is little specificity against misincorporation at this step.
Previous work on E. coli LeuRS concluded that robust tRNA-dependent pre-transfer editing of norvaline occurs, but we have shown here that this conclusion is in error because it was based on the incorrect assertion that the T252R substitution fully inactivates post-transfer editing of Nva-tRNALeu (42). It was also recently suggested that isoleucine may be edited via tRNA-dependent pre-transfer editing by ΔCP1 LeuRS, a variant deprived of the entire CP1 post-transfer editing domain (21). It was hypothesized that this activity is dormant in WT LeuRS but is activated by deletion of the CP1 domain. In accordance with the model we advance here, we suggest that such activation of the pre-transfer pathway in ΔCP1 LeuRS may arise from a decrease in the rate of the transfer step. This would parallel the effect observed in a ThrRS variant possessing a decreased rate of transfer (19).
Another experiment provides additional evidence against the notion that LeuRS catalyzes robust tRNA-dependent pre-transfer editing. Because tRNA-dependent pre-transfer editing was investigated under conditions in which Nva-tRNALeu was rapidly formed (Fig. 4A and Table 4), we further excluded the possibility that uncharged tRNA stimulates this activity by following initial rates of [32P]AMP and aa-[32P]tRNA formation in parallel assays where only a small fraction of the input tRNA is misacylated (Fig. 5C). These experiments require careful correction for the proportion of tRNA that is aminoacylable after 32P labeling by tRNA nucleotidyltransferase. Our approach is similar to that used in studies of class II ThrRS editing where aa-AMP and aa-tRNA formations were followed with radiolabel in ATP and the amino acid, respectively (19, 58).
Product Release Likely Limits Overall Editing by LeuRS
Overall editing expressed through its steady-state rate (6.2 s−1; Table 4) is significantly slower than the first-order steps of norvaline activation, norvaline transfer, and Nva-tRNALeu deacylation (56, 64, and 310 s−1, respectively). This implies that the rate-limiting step is either translocation of the 3′-end of the misacylated tRNA from the synthetic to the editing site or product release (including conformational change associated with release). When transient formation of misacylated tRNALeu by WT LeuRS was monitored using the complete reaction mixture in a rapid quench instrument, a higher steady-state Nva-tRNALeu plateau (up to 40–45%) was observed than expected (17%). This allows estimation of a rate constant for deacylation of Nva-tRNALeu formed in situ (80–100 s−1, see supplemental Fig. 3B) that presumably reflects the kinetics of the translocation step. Additional experiments are in progress to isolate and measure the translocation step explicitly. Because translocation appears to be rapid, product release represents the rate-limiting step. Indeed, there is good agreement between the steady-state rate of overall editing (6.2 s−1; Table 4) and the steady-state rate of the isolated post-transfer step (5.8 s−1; Table 2).
In single-turnover experiments in which deacylation of aminoacylated tRNALeu was monitored, the time course of product formation is well fit to a single exponential function (Fig. 2). By contrast, in editing T7 and human mitochondrial DNA polymerases, biphasic curves were observed in the analogous single-turnover hydrolytic reactions, suggesting that DNA is distributed between the synthetic and exonuclease active sites (62, 63). The faster phase represented hydrolysis in hydrolytic site, whereas the slower phase represented translocation of DNA from polymerase to the exonuclease site. We have estimated that the translocation rate in EcLeuRS is ∼4-fold lower than the rate of deacylation in the CP1 domain. If this estimate is correct, then a distribution of aminoacylated tRNA 3′-end binding between synthetic and editing active sites would be expected to produce a similar biphasic curve, because the two first-order rate constants are similar. The observation that a single exponential function fits the product formation curves thus implies that the 3′-end of aminoacylated tRNA binds directly to the editing site. This may be an important feature of trans editing in circumstances where the deacylation kinetics are slowed sufficiently to render this a significant pathway (see below).
This suggestion that the 3′-end of aminoacylated tRNA may bind directly to the editing site is consistent with inhibition experiments in which it was shown that the benzoxaborole AN2690 compound covalently traps uncharged tRNA in the CP1-editing site (64). Thus, the favorable binding mode of tRNALeu with its 3′-end positioned in the CP1 domain may be obtained whether the amino acid is attached or not. Displacement of the 3′-end of the tRNA from the CP1-editing site by a post-transfer editing substrate analog suggests that misacylated tRNALeu may possess higher affinity for the CP1 site than does uncharged tRNA (65).
Late Steps in Post-transfer Editing by LeuRS
The kinetic data presented here show that the fate of aminoacylated tRNA on LeuRS is determined by kinetic partitioning between deacylation and dissociation (Fig. 7). This is demonstrated by the behavior of the T252R mutant in the CP1 domain: the enzyme retains activity in post-transfer editing when provided with misacylated Nva-tRNALeu as substrate (Tables 2 and 3), but comparison of the initial steady-state rates of [32P]AMP and Nva-[32P]tRNALeu formation in the overall reaction suggests that no editing occurs (excess AMP does not significantly accumulate; Fig. 5B). Thus, we infer that a portion of the misacylated tRNA dissociates from the enzyme, either before or after translocation of the 3′-end to the CP1 domain. The observed increase in the ratio of [32P]AMP to Nva-[32P]tRNALeu formation when the concentrations of enzyme and tRNA are increased supports the notion that editing proceeds through dissociation and rebinding, because such higher concentrations would promote the second-order rebinding step.
FIGURE 7.
Kinetic partitioning of aminoacyl-adenylate and aminoacylated tRNA within the synthetic (blue) and editing (red) sites, respectively. First-order rate constants for aminoacyl transfer and aa-tRNA deacylation were determined by single-turnover kinetics. Aminoacyl-adenylate hydrolysis and aa-tRNA dissociation are represented by steady-state rate constants (marked with an asterisk) that impose lower limits on the true values of the first-order microscopic rate constants for these steps. Dynamics of the cognate aa-tRNA translocation has not yet been investigated.
Recent work on class II phenylalanyl-tRNA synthetase (PheRS) demonstrated editing in trans, because PheRS efficiently competes with the EF-Tu·GTP complex for binding to Tyr-tRNAPhe under conditions close to physiological (15). The rapid dissociation of Tyr-tRNAPhe from PheRS further supported the notion of trans editing in this case. trans editing is more easily rationalized for class II aaRS, because for these enzymes aa-tRNA dissociation is generally faster than the overall aminoacylation rate (4, 6). In contrast, product dissociation is generally rate-limiting in class I aaRSs, suggesting that trans editing should be less likely in these enzymes (4, 15). Indeed, trans editing in T252R LeuRS is very likely observed only because deacylation is slowed by 100-fold; in WT LeuRS, rapid deacylation in general precludes dissociation and rebinding. Consistent with this notion, a preference for the misacylation-deacylation pathway, without resampling of Thr-tRNAVal, was shown for WT ValRS (49). Rapid deacylation is a general feature of CP1-editing sites (48, 49, 59),3 where it competes efficiently with dissociation of aa-tRNA.
Comparisons with Editing by DNA Polymerases
The experiments that we have reported here and elsewhere (22) also allow clear contrasts to be drawn between the strategies for generation of specificity in editing tRNA synthetases versus editing DNA polymerases (66). In T7 DNA polymerase, translocation (transfer) of DNA from the polymerase to the exonuclease site seems to be substantially slower than the “chemical” hydrolytic step in the exonuclease site (62). Moreover, discrimination between matched and mismatched DNA preferentially occurs in translocation and not at the hydrolytic step (67). Finally, a large drop in the rate of incorporation following a mismatched base contributes most to selectivity by allowing time to transfer mismatched DNA to the hydrolytic site (68, 69).
In contrast, we have shown that the CP1 post-transfer editing site in LeuRS displays high selectivity against the cognate Leu-tRNALeu substrate, because the decrease in kchem is as high as 103 for cognate versus misacylated tRNAs. The confidence that indeed hydrolysis (not slow translocation from the synthetic site if Leu-tRNALeu preferentially binds there) is measured during the single-turnover arises because the D345A substitution decreases the rate by 50-fold. Comparison of editing DNA polymerases and aaRSs then unveils two independently developed proofreading models. Interestingly, however, both models rely on kinetic partitioning to achieve high specificity and rapid product formation. In DNA polymerases, the synthetic site is much more specific, as noncognate mismatched bases are excluded based on a slower chemical step of incorporation and an enhanced dissociation rate (70–72). In these enzymes, the hydrolytic editing site is less selective (63). Because of this, the hydrolytic correction of the cognate incoming nucleotide is maintained low by partitioning that favors fast polymerization over slow transfer to hydrolytic site (67). In contrast, aaRSs activate and transfer cognate and noncognate substrates with highly similar rates at saturating conditions within the synthetic site but distinguish these in a highly selective hydrolytic site (Fig. 7).
Supplementary Material
Acknowledgments
We are indebted to Ivana Weygand-Durasevic (University of Zagreb, Croatia) for access to research facilities. We thank Andrea Hoffmeier and Mario Mörl (University of Leipzig, Germany) for a generous gift of tRNA nucleotidyltransferase overexpression plasmid.
This work was supported, in whole or in part, by National Institutes of Health Grants 1R03TW008024 (to J. J. P. and I. G. S.) and GM63713 (to J. J. P.) This work was also supported by Ministry of Science, Education, and Sports of the Republic of Croatia Grant 119-0982913-1358 (to I. Weygand-Durasevic).

This article contains supplemental Figs. 1–3, Tables 1 and 2, Experimental Procedures, and additional references.
M. Dulic and I. Gruic-Sovulj, unpublished data.
- aaRS
- aminoacyl-tRNA synthetase
- aa-AMP
- aminoacyl-adenylate
- aa-tRNA
- aminoacyl-tRNA
- CP1
- connective peptide 1
- IleRS
- isoleucyl-tRNA synthetase
- IPPase
- inorganic pyrophosphatase
- LeuRS
- leucyl-tRNA synthetase
- Nva
- norvaline
- ThrRS
- threonyl-tRNA synthetase
- PheRS
- phenylalanyl-tRNA synthetase
- ValRS
- valyl-tRNA synthetase.
REFERENCES
- 1. Ibba M., Soll D. (2000) Aminoacyl-tRNA synthesis. Annu. Rev. Biochem. 69, 617–650 [DOI] [PubMed] [Google Scholar]
- 2. Francklyn C., Perona J. J., Puetz J., Hou Y. M. (2002) Aminoacyl-tRNA synthetases. Versatile players in the changing theater of translation. RNA 8, 1363–1372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Eriani G., Delarue M., Poch O., Gangloff J., Moras D. (1990) Partition of tRNA synthetases into two classes based on mutually exclusive sets of sequence motifs. Nature 347, 203–206 [DOI] [PubMed] [Google Scholar]
- 4. Zhang C. M., Perona J. J., Ryu K., Francklyn C., Hou Y. M. (2006) Distinct kinetic mechanisms of the two classes of aminoacyl-tRNA synthetases. J. Mol. Biol. 361, 300–311 [DOI] [PubMed] [Google Scholar]
- 5. Ibba M., Sever S., Praetorius-Ibba M., Söll D. (1999) Transfer RNA identity contributes to transition state stabilization during aminoacyl-tRNA synthesis. Nucleic Acids Res. 27, 3631–3637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Yadavalli S. S., Ibba M. (2012) Quality control in aminoacyl-tRNA synthesis, Its role in translational fidelity. Adv. Protein Chem. Struct. Biol. 86, 1–43 [DOI] [PubMed] [Google Scholar]
- 7. Mascarenhas A. P., An S., Rosen A. E., Martinis S. A., Musier-Forsyth K. (2008) in Protein Engineering, Nucleic Acids and Molecular Biology (Köhrer C., RajBhandary U. L., eds) pp. 155–203, Springer-Verlag, Berlin/Heidelberg [Google Scholar]
- 8. Nureki O., Vassylyev D. G., Tateno M., Shimada A., Nakama T., Fukai S., Konno M., Hendrickson T. L., Schimmel P., Yokoyama S. (1998) Enzyme structure with two catalytic sites for double-sieve selection of substrate. Science 280, 578–582 [DOI] [PubMed] [Google Scholar]
- 9. Lincecum T. L., Jr., Tukalo M., Yaremchuk A., Mursinna R. S., Williams A. M., Sproat B. S., Van Den Eynde W., Link A., Van Calenbergh S., Grøtli M., Martinis S. A., Cusack S. (2003) Structural and mechanistic basis of pre- and post-transfer editing by leucyl-tRNA synthetase. Mol. Cell 11, 951–963 [DOI] [PubMed] [Google Scholar]
- 10. Dock-Bregeon A. C., Rees B., Torres-Larios A., Bey G., Caillet J., Moras D. (2004) Achieving error-free translation; the mechanism of proofreading of threonyl-tRNA synthetase at atomic resolution. Mol. Cell 16, 375–386 [DOI] [PubMed] [Google Scholar]
- 11. Lin L., Hale S. P., Schimmel P. (1996) Aminoacylation error correction. Nature 384, 33–34 [DOI] [PubMed] [Google Scholar]
- 12. Betha A. K., Williams A. M., Martinis S. A. (2007) Isolated CP1 domain of Escherichia coli leucyl-tRNA synthetase is dependent on flanking hinge motifs for amino acid editing activity. Biochemistry 46, 6258–6267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Silvian L. F., Wang J., Steitz T. A. (1999) Insights into editing from an Ile-tRNA synthetase structure with tRNAIle and mupirocin. Science 285, 1074–1077 [PubMed] [Google Scholar]
- 14. Fukai S., Nureki O., Sekine S., Shimada A., Tao J., Vassylyev D. G., Yokoyama S. (2000) Structural basis for double-sieve discrimination of l-valine from l-isoleucine and l-threonine by the complex of tRNAVal and valyl-tRNA synthetase. Cell 103, 793–803 [DOI] [PubMed] [Google Scholar]
- 15. Ling J., So B. R., Yadavalli S. S., Roy H., Shoji S., Fredrick K., Musier-Forsyth K., Ibba M. (2009) Resampling and editing of mischarged tRNA prior to translation elongation. Mol. Cell 33, 654–660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Gruic-Sovulj I., Uter N., Bullock T., Perona J. J. (2005) tRNA-dependent aminoacyl-adenylate hydrolysis by a nonediting class I aminoacyl-tRNA synthetase. J. Biol. Chem. 280, 23978–23986 [DOI] [PubMed] [Google Scholar]
- 17. Gruic-Sovulj I., Rokov-Plavec J., Weygand-Durasevic I. (2007) Hydrolysis of noncognate aminoacyl-adenylates by a class II aminoacyl-tRNA synthetase lacking an editing domain. FEBS Lett. 581, 5110–5114 [DOI] [PubMed] [Google Scholar]
- 18. Splan K. E., Ignatov M. E., Musier-Forsyth K. (2008) Transfer RNA modulates the editing mechanism used by class II prolyl-tRNA synthetase. J. Biol. Chem. 283, 7128–7134 [DOI] [PubMed] [Google Scholar]
- 19. Minajigi A., Francklyn C. S. (2010) Aminoacyl transfer rate dictates choice of editing pathway in threonyl-tRNA synthetase. J. Biol. Chem. 285, 23810–23817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hati S., Ziervogel B., Sternjohn J., Wong F. C., Nagan M. C., Rosen A. E., Siliciano P. G., Chihade J. W., Musier-Forsyth K. (2006) Pre-transfer editing by class II prolyl-tRNA synthetase. Role of aminoacylation active site in “selective release” of noncognate amino acids. J. Biol. Chem. 281, 27862–27872 [DOI] [PubMed] [Google Scholar]
- 21. Boniecki M. T., Vu M. T., Betha A. K., Martinis S. A. (2008) CP1-dependent partitioning of pretransfer and post-transfer editing in leucyl-tRNA synthetase. Proc. Natl. Acad. Sci. U.S.A. 105, 19223–19228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Dulic M., Cvetesic N., Perona J. J., Gruic-Sovulj I. (2010) Partitioning of tRNA-dependent editing between pre- and post-transfer pathways in class I aminoacyl-tRNA synthetases. J. Biol. Chem. 285, 23799–23809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Zhu B., Yao P., Tan M., Eriani G., Wang E. D. (2009) tRNA-independent pretransfer editing by class I leucyl-tRNA synthetase. J. Biol. Chem. 284, 3418–3424 [DOI] [PubMed] [Google Scholar]
- 24. Gruic-Sovulj I., Dulic M., Weygand-Durasevic I. (2011) Pre-transfer editing of serine hydroxamate within the active site of methanogenic-type seryl-tRNA synthetase. Croat. Chem. Acta 84, 179–184 [Google Scholar]
- 25. Jakubowski H., Fersht A. R. (1981) Alternative pathways for editing noncognate amino acids by aminoacyl-tRNA synthetases. Nucleic Acids Res. 9, 3105–3117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Jakubowski H. (2012) Quality control in tRNA charging. Wiley Interdiscip. Rev. RNA 3, 295–310 [DOI] [PubMed] [Google Scholar]
- 27. Starzyk R. M., Webster T. A., Schimmel P. (1987) Evidence for dispensable sequences inserted into a nucleotide fold. Science 237, 1614–1618 [DOI] [PubMed] [Google Scholar]
- 28. Cusack S., Yaremchuk A., Tukalo M. (2000) The 2-Å crystal structure of leucyl-tRNA synthetase and its complex with a leucyl-adenylate analogue. EMBO J. 19, 2351–2361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Fukunaga R., Yokoyama S. (2005) Aminoacylation complex structures of leucyl-tRNA synthetase and tRNALeu reveal two modes of discriminator-base recognition. Nat. Struct. Mol. Biol. 12, 915–922 [DOI] [PubMed] [Google Scholar]
- 30. Lue S. W., Kelley S. O. (2005) An aminoacyl-tRNA synthetase with a defunct editing site. Biochemistry 44, 3010–3016 [DOI] [PubMed] [Google Scholar]
- 31. Lue S. W., Kelley S. O. (2007) A single residue in leucyl-tRNA synthetase affecting amino acid specificity and tRNA aminoacylation. Biochemistry 46, 4466–4472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Chen J. F., Guo N. N., Li T., Wang E. D., Wang Y. L. (2000) CP1 domain in Escherichia coli leucyl-tRNA synthetase is crucial for its editing function. Biochemistry 39, 6726–6731 [DOI] [PubMed] [Google Scholar]
- 33. Tang Y., Tirrell D. A. (2002) Attenuation of the editing activity of the Escherichia coli leucyl-tRNA synthetase allows incorporation of novel amino acids into proteins in vivo. Biochemistry 41, 10635–10645 [DOI] [PubMed] [Google Scholar]
- 34. Apostol I., Levine J., Lippincott J., Leach J., Hess E., Glascock C. B., Weickert M. J., Blackmore R. (1997) Incorporation of norvaline at leucine positions in recombinant human hemoglobin expressed in Escherichia coli. J. Biol. Chem. 272, 28980–28988 [DOI] [PubMed] [Google Scholar]
- 35. Soini J., Falschlehner C., Liedert C., Bernhardt J., Vuoristo J., Neubauer P. (2008) Norvaline is accumulated after a down-shift of oxygen in Escherichia coli W3110. Microb. Cell Fact. 7, 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Mursinna R. S., Lincecum T. L., Jr., Martinis S. A. (2001) A conserved threonine within Escherichia coli leucyl-tRNA synthetase prevents hydrolytic editing of leucyl-tRNALeu. Biochemistry 40, 5376–5381 [DOI] [PubMed] [Google Scholar]
- 37. Mursinna R. S., Martinis S. A. (2002) Rational design to block amino acid editing of a tRNA synthetase. J. Am. Chem. Soc. 124, 7286–7287 [DOI] [PubMed] [Google Scholar]
- 38. Mursinna R. S., Lee K. W., Briggs J. M., Martinis S. A. (2004) Molecular dissection of a critical specificity determinant within the amino acid editing domain of leucyl-tRNA synthetase. Biochemistry 43, 155–165 [DOI] [PubMed] [Google Scholar]
- 39. Xu M. G., Li J., Du X., Wang E. D. (2004) Groups on the side chain of T252 in Escherichia coli leucyl-tRNA synthetase are important for discrimination of amino acids and cell viability. Biochem. Biophys. Res. Commun. 318, 11–16 [DOI] [PubMed] [Google Scholar]
- 40. Bennett B. D., Kimball E. H., Gao M., Osterhout R., Van Dien S. J., Rabinowitz J. D. (2009) Absolute metabolite concentrations and implied enzyme active site occupancy in Escherichia coli. Nat. Chem. Biol. 5, 593–599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Karkhanis V. A., Boniecki M. T., Poruri K., Martinis S. A. (2006) A viable amino acid editing activity in the leucyl-tRNA synthetase CP1-splicing domain is not required in the yeast mitochondria. J. Biol. Chem. 281, 33217–33225 [DOI] [PubMed] [Google Scholar]
- 42. Tan M., Zhu B., Zhou X. L., He R., Chen X., Eriani G., Wang E. D. (2010) tRNA-dependent pre-transfer editing by prokaryotic leucyl-tRNA synthetase. J. Biol. Chem. 285, 3235–3244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Wolfson A. D., Uhlenbeck O. C. (2002) Modulation of tRNAAla identity by inorganic pyrophosphatase. Proc. Natl. Acad. Sci. U.S.A. 99, 5965–5970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Ledoux S., Uhlenbeck O. C. (2008) 3-32P-Labeling tRNA with nucleotidyltransferase for assaying aminoacylation and peptide bond formation. Methods 44, 74–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Levengood J., Ataide S. F., Roy H., Ibba M. (2004) Divergence in noncognate amino acid recognition between class I and class II lysyl-tRNA synthetases. J. Biol. Chem. 279, 17707–17714 [DOI] [PubMed] [Google Scholar]
- 46. Dulic M., Pozar J., Bilokapic S., Weygand-Durasevic I., Gruic-Sovulj I. (2011) An idiosyncratic serine ordering loop in methanogen seryl-tRNA synthetases guides substrates through seryl-tRNASer formation. Biochimie 93, 1761–1769 [DOI] [PubMed] [Google Scholar]
- 47. Fersht A. (1999) Structure and Mechanism in Protein Science: A Guide to Enzyme Catalysis and Protein Folding, pp. 132–242, W. H. Freeman, New York [Google Scholar]
- 48. Fersht A. R. (1977) Editing mechanisms in protein synthesis. Rejection of valine by the isoleucyl-tRNA synthetase. Biochemistry 16, 1025–1030 [DOI] [PubMed] [Google Scholar]
- 49. Fersht A. R., Kaethner M. M. (1976) Enzyme hyperspecificity. Rejection of threonine by the valyl-tRNA synthetase by misacylation and hydrolytic editing. Biochemistry 15, 3342–3346 [DOI] [PubMed] [Google Scholar]
- 50. Minajigi A., Deng B., Francklyn C. S. (2011) Fidelity escape by the unnatural amino acid β-hydroxynorvaline. An efficient substrate for Escherichia coli threonyl-tRNA synthetase with toxic effects on growth. Biochemistry 50, 1101–1109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Pasman Z., Robey-Bond S., Mirando A. C., Smith G. J., Lague A., Francklyn C. S. (2011) Substrate specificity and catalysis by the editing active site of alanyl-tRNA synthetase from Escherichia coli. Biochemistry 50, 1474–1482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. McLaughlin H. M., Sakaguchi R., Giblin W., NISC Comparative Sequencing Program, Wilson T. E., Biesecker L., Lupski J. R., Talbot K., Vance J. M., Züchner S., Lee Y. C., Kennerson M., Hou Y. M., Nicholson G., Antonellis A. (2012) A recurrent loss-of-function alanyl-tRNA synthetase (AARS) mutation in patients with Charcot-Marie-Tooth disease type 2N (CMT2N). Hum. Mutat. 33, 244–253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Kumar S., Das M., Hadad C. M., Musier-Forsyth K. (2012) Substrate specificity of bacterial prolyl-tRNA synthetase editing domain is controlled by a tunable hydrophobic pocket. J. Biol. Chem. 287, 3175–3184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Eldred E. W., Schimmel P. R. (1972) Rapid deacylation by isoleucyl transfer ribonucleic acid synthetase of isoleucine-specific transfer ribonucleic acid aminoacylated with valine. J. Biol. Chem. 247, 2961–2964 [PubMed] [Google Scholar]
- 55. Chen X., Ma J. J., Tan M., Yao P., Hu Q. H., Eriani G., Wang E. D. (2011) Modular pathways for editing noncognate amino acids by human cytoplasmic leucyl-tRNA synthetase. Nucleic Acids Res. 39, 235–247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Gutfreund H. (1995) Kinetics for the Life Sciences: Receptors, Transmitters, and Catalysts, pp. 103–137, Cambridge University Press, Cambridge, UK [Google Scholar]
- 57. Nomanbhoy T. K., Hendrickson T. L., Schimmel P. (1999) Transfer RNA-dependent translocation of misactivated amino acids to prevent errors in protein synthesis. Mol. Cell 4, 519–528 [DOI] [PubMed] [Google Scholar]
- 58. Guth E. C., Francklyn C. S. (2007) Kinetic discrimination of tRNA identity by the conserved motif 2 loop of a class II aminoacyl-tRNA synthetase. Mol. Cell 25, 531–542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Fersht A. R., Dingwall C. (1979) Establishing the misacylation/deacylation of the tRNA pathway for the editing mechanism of prokaryotic and eukaryotic valyl-tRNA synthetases. Biochemistry 18, 1238–1245 [DOI] [PubMed] [Google Scholar]
- 60. Uter N. T., Perona J. J. (2004) Long range intramolecular signaling in a tRNA synthetase complex revealed by pre-steady-state kinetics. Proc. Natl. Acad. Sci. U.S.A. 101, 14396–14401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Minajigi A., Francklyn C. S. (2008) RNA-assisted catalysis in a protein enzyme. The 2′-hydroxyl of tRNAThr A76 promotes aminoacylation by threonyl-tRNA synthetase. Proc. Natl. Acad. Sci. U.S.A. 105, 17748–17753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Donlin M. J., Patel S. S., Johnson K. A. (1991) Kinetic partitioning between the exonuclease and polymerase sites in DNA error correction. Biochemistry 30, 538–546 [DOI] [PubMed] [Google Scholar]
- 63. Johnson A. A., Johnson K. A. (2001) Exonuclease proofreading by human mitochondrial DNA polymerase. J. Biol. Chem. 276, 38097–38107 [DOI] [PubMed] [Google Scholar]
- 64. Rock F. L., Mao W., Yaremchuk A., Tukalo M., Crépin T., Zhou H., Zhang Y. K., Hernandez V., Akama T., Baker S. J., Plattner J. J., Shapiro L., Martinis S. A., Benkovic S. J., Cusack S., Alley M. R. (2007) An antifungal agent inhibits an aminoacyl-tRNA synthetase by trapping tRNA in the editing site. Science 316, 1759–1761 [DOI] [PubMed] [Google Scholar]
- 65. Tukalo M., Yaremchuk A., Fukunaga R., Yokoyama S., Cusack S. (2005) The crystal structure of leucyl-tRNA synthetase complexed with tRNALeu in the post-transfer editing conformation. Nat. Struct. Mol. Biol. 12, 923–930 [DOI] [PubMed] [Google Scholar]
- 66. Francklyn C. S. (2008) DNA polymerases and aminoacyl-tRNA synthetases: shared mechanisms for ensuring the fidelity of gene expression. Biochemistry 47, 11695–11703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Johnson K. A. (1993) Conformational coupling in DNA polymerase fidelity. Annu. Rev. Biochem. 62, 685–713 [DOI] [PubMed] [Google Scholar]
- 68. Patel S. S., Wong I., Johnson K. A. (1991) Pre-steady-state kinetic analysis of processive DNA replication, including complete characterization of an exonuclease-deficient mutant. Biochemistry 30, 511–525 [DOI] [PubMed] [Google Scholar]
- 69. Wong I., Patel S. S., Johnson K. A. (1991) An induced-fit kinetic mechanism for DNA replication fidelity. Direct measurement by single-turnover kinetics. Biochemistry 30, 526–537 [DOI] [PubMed] [Google Scholar]
- 70. Tsai Y. C., Johnson K. A. (2006) A new paradigm for DNA polymerase specificity. Biochemistry 45, 9675–9687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Johnson K. A. (2008) Role of induced fit in enzyme specificity. A molecular forward/reverse switch. J. Biol. Chem. 283, 26297–26301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Johnson K. A. (2010) The kinetic and chemical mechanism of high fidelity DNA polymerases. Biochim. Biophys. Acta 1804, 1041–1048 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.