Background: Mon1a is known to be involved in membrane trafficking.
Results: Reductions in Mon1a delay ER-to-Golgi trafficking and ERGIC-53-positive vesicle formation. Mon1a interacts with cytoplasmic dynein to efficiently traffic vesicles from ER to Golgi.
Conclusion: Mon1a plays a role in anterograde trafficking in the secretory pathway.
Significance: This work elucidates a role for Mon1a, known to be involved in the endocytic pathway, acting in the secretory pathway.
Keywords: Dynein, Endoplasmic Reticulum (ER), Golgi, Secretion, Transport, Mon1a
Abstract
Mon1a was originally identified as a modifier gene of vesicular traffic, as a mutant Mon1a allele resulted in increased localization of cell surface proteins, whereas reduced levels of Mon1a showed decreased secretory activity. Here we show that Mon1a affects different steps in the secretory pathway including endoplasmic reticulum-to-Golgi traffic. siRNA-dependent reduction of Mon1a levels resulted in a delay in the reformation of the Golgi apparatus after Brefeldin A treatment. Endoglycosidase H treatment of ts045VSVG-GFP confirmed that knockdown of Mon1a delayed endoplasmic reticulum-to-Golgi trafficking. Reductions in Mon1a also resulted in delayed trafficking from Golgi to the plasma membrane. Immunoprecipitation and mass spectrometry analysis showed that Mon1a associates with dynein intermediate chain. Reductions in Mon1a or dynein altered steady state Golgi morphology. Reductions in Mon1a delayed formation of ERGIC-53-positive vesicles, whereas reductions in dynein did not affect vesicle formation. These data provide strong evidence for a role for Mon1a in anterograde trafficking through the secretory apparatus.
Introduction
Membrane trafficking pathways in eukaryotes require highly regulated transfer of vesicles and their contents from a donor to an acceptor compartment. Mon1 was initially identified in Saccharomyces cerevisiae (YMon1)4 as a protein required for vacuolar fusion. YMon1 acts as part of a cis-SNARE complex with Ccz1, and both are required in all pathways where the terminal compartment is the vacuole (1–3). Yeast with a deletion in MON1 or its partner CCZ1 have fragmented vacuoles resulting from a defect in vacuole fusion (1, 4). The Caenorhabditis elegans homologue of Mon1 (SAND-1) was shown to function in the endocytic pathway, although not at lysosomes but rather at early-to-late endosome maturation (5, 6). Mammalian Mon1 (Mon1a) and its homologue Mon1b were shown in both C. elegans and cultured mammalian cells to be involved in ingestion of apoptotic cell corpses (7). Recently, it was reported that Mon1a and Mon1b and the C. elegans homologue SAND-1 are involved in the Rab5-to-Rab7-mediated transition of endosomes (8). Studies in mammals utilized RNAi against both Mon1a and Mon1b to observe changes in the endocytic pathway, but no effects on the secretory pathway were reported.
The first identification of mammalian Mon1a suggested that it plays a role in the movement of molecules through the secretory apparatus (9). In that study, Mon1a was identified in a quantitative trait locus analysis for modifier genes of iron homeostasis. A missense mutation in the Mon1a gene in C57BL/6 mice gives rise to a single amino acid substitution (N374S) at an evolutionarily conserved residue. This substitution results in a “gain-of-function” allele causing an increase in the trafficking of membrane and soluble molecules through the biosynthetic pathway and thus increased localization of proteins at the plasma membrane. Reduction of Mon1a protein by RNAi supports the hypothesis that Mon1a plays a role in the trafficking of newly synthesized proteins through the secretory apparatus. In this study, we show that Mon1a functions in multiple steps in the secretory process including ER-to-Golgi traffic. We show that Mon1a interacts with cytoplasmic dynein and provide evidence that both are required for efficient anterograde trafficking from ER to Golgi.
EXPERIMENTAL PROCEDURES
Mammalian Cells and Constructs
NIH3T3, HeLa, Cos-7, and HeLa cells stably expressing GalNAc-T2-GFP were maintained in DMEM with 10% FBS plus or minus 0.4 g/liter G418 (Sigma-Aldrich). GalNAc-T2-GFP HeLa cells were a generous gift from Dr. Brian Storrie (University of Arkansas). pVSVG-GFP and ts045VSVG-GFP were a generous gift from Dr. Jennifer Lippincott-Schwartz (National Institutes of Health). Mouse Mon1a was cloned into pCMV-Tag2BFLAG (Stratagene, La Jolla, CA) or pEGFPC1 (Clontech). YFP-ER and YFP-Golgi were obtained from Clontech. All constructs were sequence verified prior to use.
Treatment with siRNA Oligonucleotide Pools
Cells were treated with nonspecific, Mon1a-specific, or dynein heavy chain 1-specific oligonucleotides (Dharmacon SiGenome SiRNA SMARTpool) (Dharmacon RNA Technologies, Lafayette, CO) using Oligofectamine reagent (Invitrogen) according to the manufacturer's instructions.
Yeast Strains and Growth Media
Wild type (BY4743), Δmon1, and Δccz1 strains were obtained from the Research Genetics (Invitrogen) homozygous diploid deletion collection. Cells were transformed with pYES2.0 (Invitrogen), pYES2.0mouseMon1a-FLAG, or pYES2.0mouseMon1b-FLAG for GAL1-regulated expression, with pMET3humanc7orf28b-myc for MET3-regulated expression, or with pRS426FLAG-MON1 or pRS416FLAG-MON1. The yeast promoter was defined as 1,000 bp upstream of the MON1 open reading frame. Yeast strains were grown in YPD medium (1% yeast extract, 2% peptone, 2% dextrose) or in complete minimal medium (0.67% yeast nitrogen base without amino acids, 2% dextrose or galactose, and 0.13% amino acid drop-out mix). Complete minimal medium was supplemented with 5 mm ZnCl2, and cells were grown for 2–4 days at 30 °C.
FM4-64 Vacuole Staining
Yeast strains were grown overnight to log phase. 10 optical densities of yeast were pelleted and resuspended in 1.0 ml of growth medium. Cell were given 5.0 μm FM4-64 diluted according to the manufacturer's instructions (Invitrogen), and cells were incubated for 10 min at 30 °C. Cells were washed and placed back in growth media lacking FM4-64 for 45 min at 30 °C. 10 μl of cells were spotted onto concanavalin A-coated coverslips, and vacuole morphology was examined by epifluorescence and Nomarski microscopy. Images were captured using an Olympus BX51 upright microscope with a 100× 1.4NA objective and the Optronics PictureFrame software (Olympus Inc., Melville, NY).
Subcellular Fractionation
NIH3T3 cells were grown on 100-mm tissue culture plates. For crude subcellular fractionation, cells were scraped into phosphate-buffered saline (PBS), pelleted at 1,000 × g for 5 min, and resuspended in 2.0 ml of 0.25 m sucrose, 10 mm Tris-HCl, pH 7.2, 0.5 mm EDTA. Cells were homogenized with a ball-bearing homogenizer and centrifuged at 800 × g for 5 min, and a postnuclear supernatant (PNS) was obtained. The PNS was centrifuged at either 6,552 × g (10,000 rpm) for 20 min or 191,056 × g (54,000 rpm) for 40 min to obtain membrane and cytosolic fractions. For further subcellular fractionation, the PNS was overlaid onto a 23% Percoll (GE Healthcare) gradient and centrifuged at 54,000 rpm for 30 min, and gradients were fractionated bottom to top. Fractions of the gradient were analyzed for organellar markers using SDS-PAGE followed by Western blot. The samples were solubilized in 1.0% Triton X-100, and protein determinations were performed to normalize for protein using the bicinchoninic acid assay (Pierce) and 25 or 50 μg of protein/sample loaded onto SDS-PAGE gels.
Brefeldin A Treatment
Cells plated on glass coverslips were incubated with 5 μg/ml brefeldin A (BFA) (Epicenter Biotechnologies, Madison, WI) for 30 min, washed three times, and placed in growth medium for recovery. Wild type VSVG-GFP and ts045VSVG-GFP were expressed in NIH3T3 cells that were maintained at either 32 °C or the restrictive temperature (39 °C) followed by shifting to the permissive temperature (32 °C). VSVG-GFP was considered ER-associated if VSVG-GFP could be visualized within ER compartments (nuclear ring, reticulum), whereas VSVG-GFP was considered Golgi-associated once VSVG-GFP was deposited within Golgi (perinuclear and “stacked”) and plasma membrane once VSVG-GFP could be visualized as a concentrated line at the cell edge as described previously (10). GalNAc-T2-GFP HeLa cells were BFA-treated and recovered at 37 °C. BFA recovery was visualized using an Olympus BX51 upright microscope with a 100× 1.4NA objective and the Optronics PictureFrame software (Olympus).
ts045VSVG-GFP Trafficking and [35S]Methionine Metabolic Labeling
Cells silenced with nonspecific, Mon1a, or DHC1 oligonucleotides were transfected with ts045VSVG-GFP and maintained at the restrictive temperature (39 °C). Cells were moved to the permissive temperature (32 °C), and movement of ts045VSVG-GFP to the plasma membrane over time was assessed by fluorescent microscopy. The data are expressed as the percentage of cells showing VSVG-GFP at the plasma membrane. For metabolic labeling, cells maintained at 39 °C were grown in DMEM-methionine for 2 h. Cells were pulsed with [35S]methionine (PerkinElmer Life Sciences) for 20 min, washed, shifted to 32 °C for various times in the presence of DMEM with excess methionine, lysed in 1.0% Triton X-100, 150 mm NaCl, 0.5 mm EDTA (lysis buffer), and immunoprecipitated with rabbit anti-GFP (Abcam, Cambridge, MA) and protein A/G-agarose (Santa Cruz Biotechnology, Santa Cruz, CA). Following immunoprecipitation, samples were denatured and incubated with or without endoglycosidase H (New England Biolabs, Ipswich, MA) for 2 h, samples were heated to 100 °C for 10 min and separated on SDS-PAGE, and 35S-labeled VSVG-GFP was detected by autoradiography.
Transfections and Western Analysis
HeLa cells, bone marrow-derived macrophages, and NIH3T3 fibroblasts were plated onto tissue culture plates and allowed to grow for 24–48 h to 50–80% confluence. The cells were transfected with various constructs using Amaxa nucleofector technology (Lonza, Walkersville, MD) according to the manufacturer's directions. Protein expression was determined by solubilizing 2–4 × 106 cells in lysis buffer plus 2× protease inhibitor mixture (Roche Applied Science) and 1.0 mm phenylmethylsulfonyl fluoride (Sigma-Aldrich). Samples were analyzed by SDS-PAGE, and Western analysis was performed using mouse anti-FLAG antibody (1:1000; Sigma-Aldrich); rabbit anti-GFP (1:5000, ab6556; Abcam); mouse anti-tubulin (1:1000; GeneTex, San Antonio, TX); mouse anti-p115 (1:500, GeneTex); rabbit anti-ERGIC-53 (1:1000, Sigma-Aldrich); mouse anti-DIC (1:2000, MMS-400R Covance, Princeton, NJ); or DHC1 (1:1000, Abcam) followed by either peroxidase-conjugated goat anti-mouse immunoglobulin IgG (1:5,000; Jackson ImmunoResearch Laboratories, West Grove, PA) or peroxidase-conjugated goat anti-rabbit IgG (1:5,000; Jackson ImmunoResearch Laboratories). Antibodies to Mon1a were generated as described (9). Rabbit anti-Mon1a was used at a concentration of 1:500 followed by peroxidase-conjugated goat anti-rabbit IgG (1:5,000) (Jackson ImmunoResearch Laboratories). The blots were developed using Western Lightning reagent (PerkinElmer Life Sciences). Tubulin was used as a loading control. All experiments were performed a minimum of three times, and error bars represent S.E.
Epifluorescence, Immunofluorescence, and Electron Microscopy
Cells were incubated with 20 μg/ml Texas Red dextran overnight at 37 °C, washed, and chased for 2 h to move all dextran to lysosomes as described previously (11) (12). Cells were incubated with 5 μg/ml Alexa Fluor 488-Tf(Fe)2 for 30 min to load the early endocytic pathway as described previously (13, 14). Live cell images were captured on a confocal microscope or an epifluorescence microscope. Vesicle size was determined manually by measuring the area of the vesicles as described previously using the ImageJ software (15). For immunofluorescence, cells were fixed in 3.7% formaldehyde/PBS, permeabilized in 0.1% saponin/PBS/BSA, and incubated with mouse anti-Sec31 (1:100; BD Biosciences), rabbit anti-Giantin (1:100; Covance), rabbit anti-ERGIC-53 (1:100; Sigma), rabbit anti-GFP (1:500; GeneTex), or mouse anti-p115 (1:500, GeneTex) followed by Alexa Fluor 594-, Alexa Fluor 647-, or Alexa Fluor 488-conjugated goat anti-rabbit or anti-mouse IgG (1:750, Invitrogen). Confocal images were captured on an Olympus FV1000 microscope with a 60× 1.4NA oil immersion objective. Image analysis and quantification were performed using the Volocity software. For electron microscopy (EM), cells were fixed in 2.5% glutaraldehyde, 1% paraformaldehyde, and transmission EM images were captured at the University of Utah EM Core.
Immunoprecipitation
Cells were transfected with pFLAG-Mon1a or GFP-Mon1a, solubilized in lysis buffer, incubated at 0 °C for 30 min, centrifuged at 10,000 × g, 10 min, and immunoprecipitated using mouse anti-FLAG antibody (Sigma) or rabbit anti-GFP (GeneTex) and protein A/G-agarose (Santa Cruz Biotechnology). Proteins in the immunoprecipitate were identified using Western blot analysis as described above, and mass spectrometry was performed at the University of Utah Mass Spectrometry Core as described previously (16).
RESULTS
Overexpression of Mammalian Mon1a or Mon1b in Yeast Does Not Suppress the Phenotypes Associated with Loss of MON1 or CCZ1
Studies of Mon1 in S. cerevisiae have suggested that Mon1 is involved in vacuole fusion (1–3). More recently, Mon1 and its partner Ccz1 have been shown to act as a guanine nucleotide exchange factor of the late endosomal rab7 homolog Ypt7 in S. cerevisiae (17). Mammals have two Mon1 genes, Mon1a and Mon1b. Previously, we determined that reductions in Mon1a did not affect endocytosis or phagocytosis (9). We further determined that reductions in Mon1a did not affect the morphology of lysosomes or early and recycling endosomes (supplemental Fig. 1). To determine whether mammalian Mon1a or Mon1b could complement the loss of Mon1 in S. cerevisiae, we generated yeast plasmids containing epitope-tagged mammalian Mon1a or Mon1b under the regulation of the GAL1 promoter. We determined that Mon1a and Mon1b did not complement the loss of MON1 or CCZ1 in S. cerevisiae based upon vacuole morphology, growth on ZnCl2-containing medium, and carboxypeptidase Y secretion (supplemental Fig. 2). Similarly, expression of the Mon1a or Mon1b along with the putative mammalian homologue of the Mon1 yeast partner protein Ccz1, human c7orf28b, did not complement the loss of MON1 or CCZ1 in S. cerevisiae, although the proteins were expressed (data not shown). We also note that c7orf28b did not complement the loss of CCZ1 (data not shown). These results suggest either that the mammalian homologue cannot interact with the machinery in yeast needed for function or that the mammalian homologues have evolved to perform different functions.
siRNA-mediated Reductions of Mon1a Result in Delayed Secretion
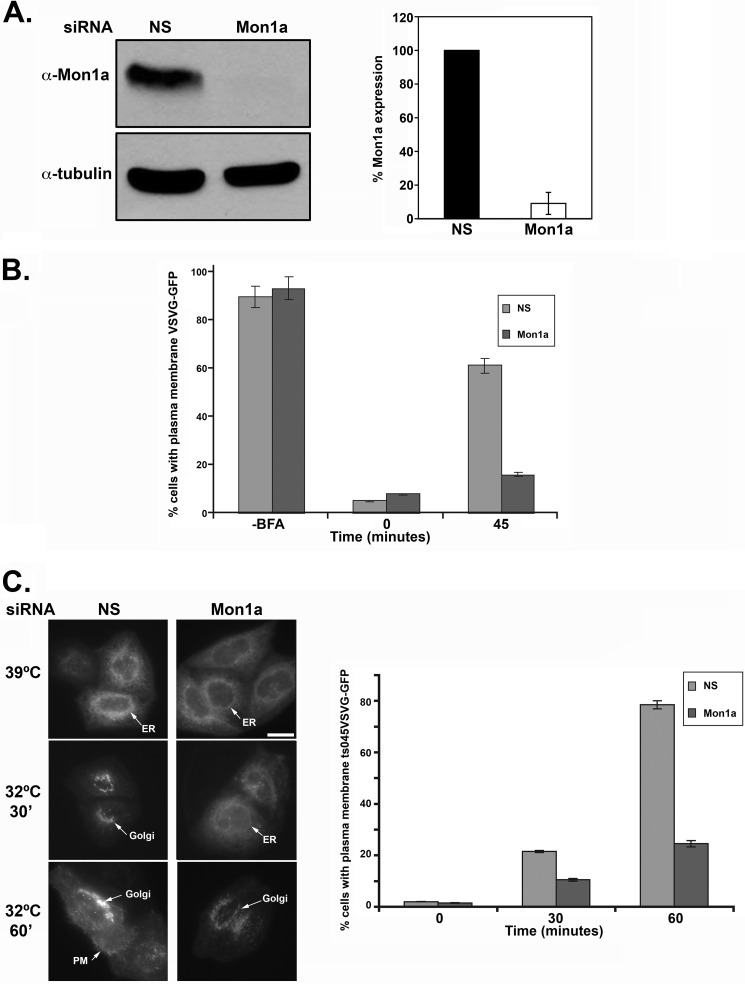
Our previous studies suggest that Mon1a may have a role in the secretory pathway (9). To determine where Mon1a acts in the secretory pathway, we treated NIH3T3 cells with the fungal metabolite BFA, an inhibitor of the GTP exchange factor for ADP-ribosylation factor 1, which ultimately inhibits protein transport from ER to Golgi, redistributing the Golgi apparatus into the ER. Removal of BFA allows recovery of anterograde movement, and proteins will resume trafficking to reassemble the Golgi apparatus. We utilized Mon1a-specific siRNA oligonucleotide pools to reduce the levels of Mon1a in NIH3T3 cells transfected with a plasmid containing vesicular stomatitis virus G protein with a green fluorescent protein tag (VSVG-GFP). We treated cells with BFA and examined the effects of Mon1a reduction on the rate of movement of VSVG-GFP during BFA recovery. Mon1a levels were efficiently reduced in NIH3T3 cells (Fig. 1A). BFA treatment resulted in VSVG-GFP condensing down into a perinuclear ER structure as expected. Reductions in Mon1a did not affect the collapse of the Golgi apparatus in response to BFA. In nonspecifically silenced cells, 60% of cells had VSVG-GFP on the plasma membrane within 45 min of removal of BFA (Fig. 1B). In contrast, at the same time, less than 20% of Mon1a-silenced cells showed VSVG-GFP on the plasma membrane.
FIGURE 1.
Trafficking of VSVG-GFP is delayed in Mon1a-silenced cells. A, NIH3T3 cells expressing VSVG-GFP were silenced with nonspecific (NS) or Mon1a specific oligonucleotide pools for 48–72 h. Cells were lysed, and the levels of Mon1a and tubulin were assessed by Western blot. A representative blot is shown with the graph representing the average knockdown from n = 3 blots. Error bars represent the S.E. B, cells as in A were treated with 5 μg/ml BFA for 30 min at 37 °C, BFA was removed, cells were placed in DMEM at 37 °C, and VSVG-GFP trafficking to the plasma membrane was assessed by epifluorescence microscopy. The data are expressed as the percentage of cells showing VSVG-GFP localization at the plasma membrane −BFA, +BFA (time 0), and 45 minutes, examining localization with 50–100 cells per sample counted. Error bars represent the S.E. C, cells silenced for Mon1a were transfected with ts045VSVG-GFP and maintained at the restrictive temperature (39 °C). Cells were moved to the permissive temperature (32 °C), and movement of ts045VSVG-GFP to the plasma membrane over time was assessed by fluorescent microscopy. Representative images at 39 °C, 32 °C 30 min and 32 °C 60 min are shown. Scale bar = 10 μm. Arrows denote reticular ER, Golgi, or plasma membrane (PM). Images were analyzed and quantified by three separate individuals. The data are expressed as the percentage of cells showing ts045VSVG-GFP at the plasma membrane as described under ‘Experimental Procedures.’ All experiments were performed a minimum of five times. Error bars represent the S.E.
To ensure that the observations made with BFA treatment were not the result of global effects of BFA on all membrane trafficking, we utilized a temperature-sensitive allele of VSVG termed ts045VSVG-GFP that is localized to the ER at the restrictive temperature (39 °C) but moves through the Golgi apparatus to the plasma membrane at the permissive temperature (32 °C) (18). Cells treated with nonspecific or mouse Mon1a-specific siRNA were transfected with ts045VSVG-GFP, grown at the restrictive temperature for 18 h, and then moved to the permissive temperature to assess ts045VSVG-GFP trafficking. Again, reduction of Mon1a levels delayed the movement of ts045VSVG-GFP from ER to plasma membrane (Fig. 1C) when compared with control cells, confirming that the effects seen with BFA treatment were specific to Mon1a knockdown.
Reductions in Mon1a Affect Golgi Apparatus Morphology and Trafficking
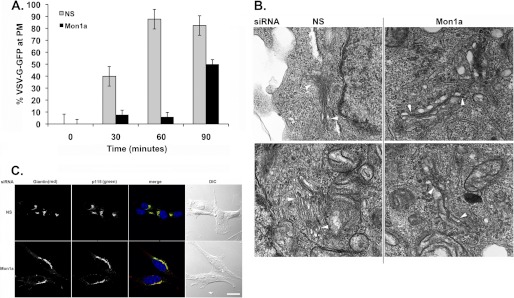
To determine whether reductions in Mon1a affected the trafficking of ts045VSVG-GFP from Golgi to plasma membrane, we placed cells at 15 °C to allow the ts045VSVG-GFP to accumulate in the Golgi and then shifted cells to 24 °C to slow the rate of movement of ts045VSVG-GFP from Golgi to plasma membrane (19). Reductions in Mon1a delayed the movement of ts045VSVG-GFP from the Golgi to the cell surface when compared with control cells (Fig. 2A). Reductions in Mon1a altered the morphology of the Golgi apparatus as assessed by electron microscopy (Fig. 2B) and immunofluorescence (Fig. 2C). Golgi-like compartments were visible in Mon1a-silenced cells; however, the tight organization or stacking of the Golgi apparatus seen in nonspecifically silenced cells was lost. Rather, Mon1a-silenced cells showed more extended, unstacked Golgi morphology.
FIGURE 2.
Reductions in Mon1a alter Golgi morphology. A, cells as in Fig. 1C were shifted from 39 to 15 °C to allow all ts045VSVG-GFP to accumulate in the Golgi (4–8 h). Cells were then shifted to 24 °C to allow movement of ts045VSVG-GFP to the plasma membrane (PM), and images were captured over time. The data are expressed as the percentage at the plasma membrane. All experiments were performed a minimum of three times. Error bars represent the S.E. NS, nonspecific. B, cells silenced as in A were processed for EM as described under ‘Experimental Procedures.’ Magnification = 70,000×. Arrowheads identify Golgi structures in nonspecifically (NS) or Mon1a-silenced cells. C, mouse fibroblasts were silenced with nonspecific or mouse Mon1a-specific oligonucleotides for 96 h, fixed, and processed for immunofluorescence using rabbit anti-Giantin or mouse anti-p115. Images were captured on an Olympus FV1000 confocal microscope. Representative images are shown from n = 3 separate experiments. Scale bar = 10 μm. Arrows denote altered Golgi structures in Mon1a-silenced cells.
Mon1a Regulates Early and Late Trafficking in the Secretory Pathway
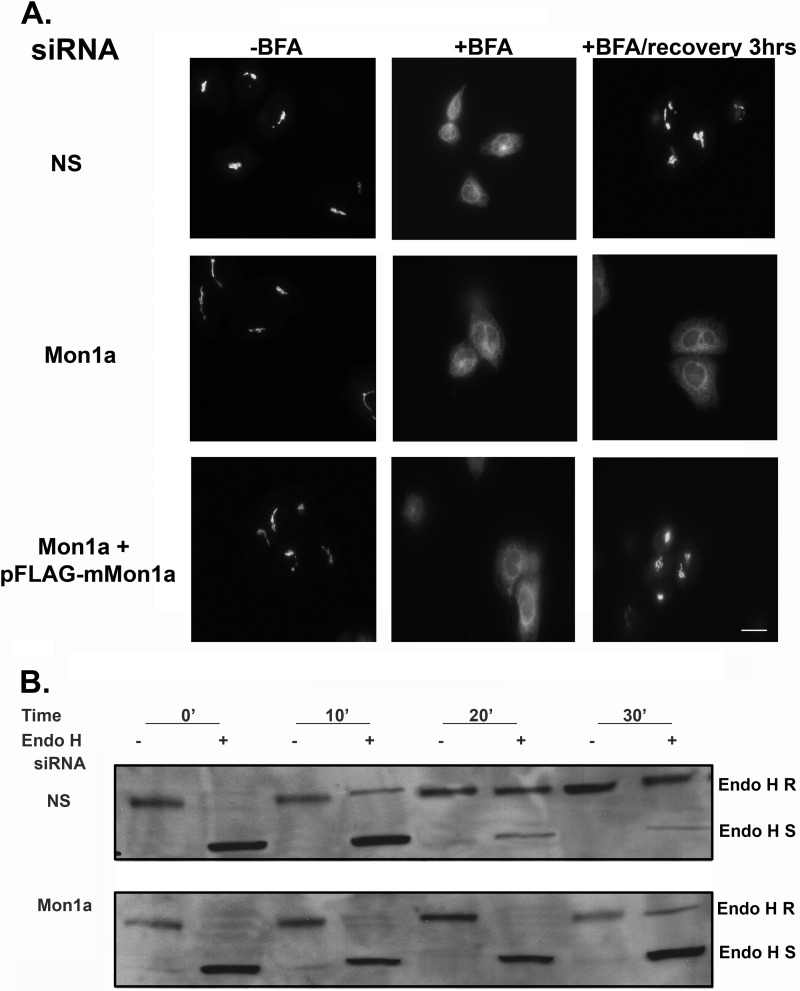
The above experiments show that loss of Mon1a affects trafficking through the secretory pathway and Golgi morphology. We focused our attention on how Mon1a affects ER-to-Golgi trafficking. Mon1a was silenced in a HeLa cell line stably expressing a GFP fusion of the resident Golgi enzyme N-acetylgalactosaminyltransferase-2 (GalNAc-T2). This marker can be used along with BFA treatment to assess the kinetics of Golgi reassembly, which requires functional anterograde transport (20). In the presence of BFA, GalNAc-T2 is redistributed to the ER (Fig. 3A). Complete recovery of the Golgi complex after BFA removal was observed at 2–3 h in control cells, as described previously (20). In contrast, Golgi localization of GalNAc-T2 following BFA treatment was delayed in Mon1a-silenced cells (6–8 h). Expression of an siRNA-resistant FLAG-tagged murine Mon1a allele in HeLa cells silenced for Mon1a restored Golgi recovery, after BFA treatment, back to control. These results demonstrate that the amino terminally FLAG-tagged mouse Mon1a is functional and complemented the loss of human Mon1a and that impaired ER-to-Golgi traffic was a specific effect of Mon1a reduction as opposed to an off-target effect of RNAi.
FIGURE 3.
Reductions in Mon1a affect trafficking from the ER to the Golgi. A, HeLa cells stably expressing GalNAc-T2-GFP were silenced with nonspecific (NS) or human Mon1a specific oligonucleotides for 72 h. Cells were than transfected with empty vector or siRNA-resistant mouse FLAG-Mon1a and incubated for 18 h. Cells were then treated with BFA as in Fig. 1, BFA was washed away, and the movement of GalNAc-T2-GFP to the Golgi apparatus was assessed by epifluorescence microscopy. Representative images are shown from n = 5 experiments. Scale bar = 10 μm. B, NIH3T3 cells nonspecifically or Mon1a-silenced expressing ts045VSVG-GFP were grown in methionine-depleted DMEM for 2 h at 39.5 °C. Cells were then pulsed with [35S]methionine for 30 min at 39.5 °C and shifted to 32 °C for the indicated times. Cells were lysed in 1% Triton X-100, 0.15 m NaCl, 10 mm Tris-HCl, pH 7.2. Lysates were immunoprecipitated for GFP, denatured, and incubated with Endo H for 2 h, and radiolabeled ts045VSVG-GFP was resolved on SDS-PAGE followed by autoradiography. A representative blot (n = 3) is presented. R, Endo H resistant; S, Endo H sensitive.
To examine whether reduction of Mon1a protein levels affects ER-to-Golgi traffic independent of the use of BFA, we utilized a well characterized endoglycosidase H (Endo H) sensitivity assay. Membrane proteins in the ER are N-linked glycosylated, resulting in a high mannose-containing carbohydrate, which is a substrate for endoglycosidase H (for review, see Ref. 21). Once membrane proteins move to the medial-Golgi, the carbohydrate moieties are modified, and N-linked glycosylated proteins become resistant to Endo H. ts045VSVG-GFP is glycosylated in this manner and is sensitive to Endo H when cells are maintained at the restrictive temperature (ER) and becomes resistant when cells are moved to the permissive temperature (Golgi). We took advantage of ts045VSVG-GFP and the Endo H sensitivity of ER-localized glycoproteins to determine whether Mon1a acts in early secretory membrane trafficking. siRNA-treated cells expressing ts045VSVG-GFP were kept at the restrictive temperature. Cells treated with nonspecific or Mon1a-specific oligonucleotide pools showed Endo H-sensitive ts045VSVG-GFP at the nonpermissive temperature (Fig. 3B). Endo H sensitivity of ts045VSVG-GFP was lost in nonspecifically silenced cells as early as 10 min after shifting to the permissive temperature. In contrast, cells silenced for Mon1a showed a significant delay in ts045VSVG-GFP gaining Endo H resistance. These data support the hypothesis that Mon1a functions in ER-to-Golgi trafficking.
Mon1a Is a Cytosolic Protein That Peripherally Associates with the ER
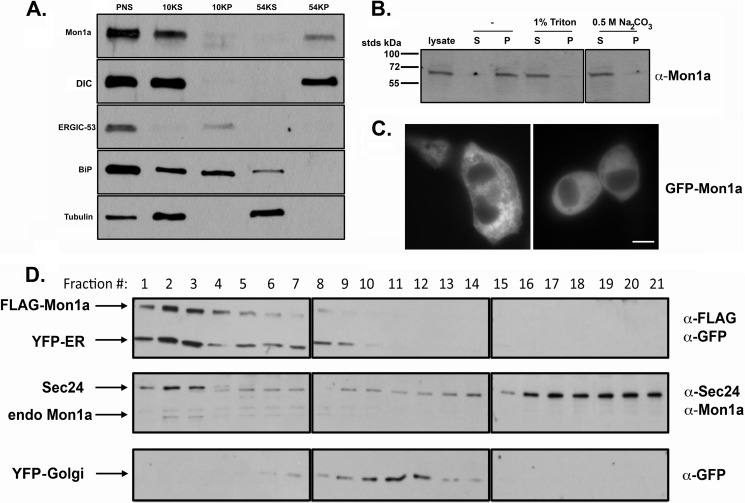
We previously demonstrated that Mon1a is ubiquitously expressed in all tissues of mice and is predicted to be a 68-kDa cytosolic protein (9). To determine the localization of Mon1a, we homogenized mouse NIH3T3 fibroblasts and separated the membrane and cytosolic fractions. A large percentage of endogenous Mon1a was associated with the membrane fraction (Fig. 4A). Treatment of the membrane fraction with Na2CO3, a procedure that disrupts ionic interactions, showed that Mon1a is not an integral membrane protein but rather is a peripheral membrane protein that associates with the membrane in a noncovalent interaction (Fig. 4B). The Mon1a polyclonal antibody used for Western analysis was not effective for immunofluorescence; therefore, we epitope-tagged Mon1a with GFP, confirmed that it was functional similar to FLAG-tagged Mon1a (data not shown), and examined localization by epifluorescence microscopy. GFP-Mon1a was found primarily in the cytosol with some reticular membrane localization (Fig. 4C). One caveat to these results is that GFP-Mon1a is highly overexpressed and may not reflect endogenous localization. We performed subcellular fractionation on cells expressing FLAG-Mon1a and either an ER-localized yellow fluorescent protein (YFP-ER) or a Golgi-localized YFP (YFP-Golgi). Endogenous and FLAG-tagged Mon1a were found primarily coincident with YFP-ER and to a lesser degree with YFP-Golgi (Fig. 4D). These results suggest that Mon1a associates with early secretory pathway membranes.
FIGURE 4.
Mon1a is a cytosolic protein that peripherally associates with the ER. A, NIH3T3 cells were homogenized in 0.25 m sucrose, 10 mm Tris-HCl, 0.5 mm EDTA and centrifuged at 800 × g for 5 min, and PNS was obtained and centrifuged at 6,552 × g (10KP). Supernatant from the 10,000 spin (10KS) was then centrifuged at 191,056 × g for 30 min (54KS and 54KP). Membrane and cytosolic fractions were solubilized in SDS-PAGE sample buffer, and the presence of Mon1a, DIC, ERGIC-53, BiP, and tubulin was determined by Western blot. B, membranes as in A were treated with 1% Triton X-100 or 0.5 m Na2CO3 and repelleted prior to SDS-PAGE, and Western blot analysis for endogenous Mon1a was performed. S, supernatant; P, pellet. C, cells were transfected with pGFP-Mon1a. 24 h after transfection, GFP-Mon1a localization was assessed by epifluorescence microscopy. Two representative images are shown. D, NIH3T3 cells expressing either a YFP-ER or a YFP-Golgi marker and FLAG-Mon1a were homogenized as in A. The postnuclear supernatants (800 × g) from YFP-ER/FLAG-Mon1a or YFP-Golgi/FLAG-Mon1a were top-loaded onto a 23% Percoll gradient, centrifuged at 54,000 × g for 30 min, and fractionated, and the presence of YFP-ER or YFP-Golgi and endogenous Mon1a, Sec24, and FLAG-Mon1a was examined by Western blot analysis. Endogenous Mon1a, Sec24, and FLAG-Mon1a from YFP-ER samples are shown along with YFP-Golgi from a separate gradient to show Golgi localization. All experiments were performed a minimum of three times.
Mon1a Interacts with the Microtubule-based Molecular Motor Dynein
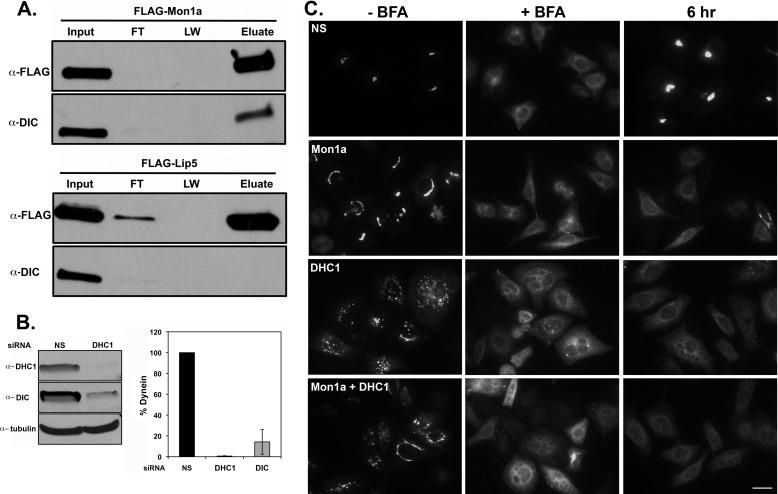
To determine how Mon1a functions in the early secretory pathway, we utilized our epitope-tagged Mon1a (FLAG-Mon1a) to immunoprecipitate Mon1a and examined the immunoprecipitate for possible partners by mass spectrometry. Immunoprecipitated Mon1a (Fig. 5A) was subsequently analyzed by LC/MS/MS, and dynein intermediate chain (DIC) was found associated with Mon1a. We confirmed that DIC coimmunoprecipitated with FLAG-Mon1a using Western blot analysis. This interaction was specific as another FLAG-tagged protein, Lip5, which is involved in multivesicular body formation (16), did not coimmunoprecipitate DIC. Dynein is a multisubunit motor protein involved in many vesicle trafficking events, and its function in the secretory pathway is well documented (22–24). Dynein attaches to membranes often through the intermediate chain and to microtubules through the heavy chain to move vesicles on microtubule tracks, and a role for dynein in vesicle formation from the ER has been suggested (23, 24). We silenced DHC1 and noted that loss of DHC1 resulted in a concomitant loss of DIC (Fig. 5B). Cells silenced for DHC1 showed a fragmented Golgi morphology (Fig. 5C, −BFA and supplemental Fig. 3A). Reductions in both Mon1a and DHC1 showed a reduced fragmentation of the Golgi when compared with DHC1 alone, suggesting that Mon1a is needed for the Golgi fragmentation seen in DHC1-silenced cells.
FIGURE 5.
FLAG-Mon1a interacts with dynein, and reductions in DHC1 delay trafficking from ER to Golgi. A, NIH3T3 cells were transfected with pFLAG-Mon1a or pFLAG-Lip5 (as a negative control). 24 h after transfection, cells were harvested, incubated in lysis buffer at 0 °C for 30 min, and centrifuged for 30 min at 22,000 × g, and the supernatant was incubated with mouse anti-FLAG antibody and protein A/G beads for 18 h at 4 °C. Beads were washed and eluted in 2× sample buffer at 100 °C for 10 min, proteins in the lysate (Input), flow-through (FT), and last wash of three (LW) and eluate were examined for FLAG-Mon1a by Western blot, and mass spectrometry was performed on these samples (data not shown). FLAG-Mon1a immunoprecipitates were probed for DIC. All experiments were performed a minimum of three times. B, NIH3T3 cells were silenced with nonspecific (NS) or DHC1 specific oligonucleotide pools for 72–96 h. Cells were lysed, and the levels of DHC1, DIC, and tubulin were assessed by Western blot. A representative blot is shown. Blots were quantified using tubulin as a loading control n = 3. C, HeLa cells stably expressing GalNAc-T2-GFP were silenced with nonspecific, Mon1a, and/or human DHC1 specific oligonucleotides for 72 h. Cells were then treated with BFA as in Fig. 1, BFA was washed away, and the movement of GalNAc-T2-GFP to the Golgi apparatus was assessed by epifluorescence microscopy. Representative images for −BFA and +BFA and at 6 h of recovery are shown from n = 5 experiments. Scale bar = 10 μm.
Previous studies have shown that the dynein-dynactin complexes are important in coordinating microtubule-mediated exit from the ER (23). Those studies showed that ER exit is mediated through dynactin directly interacting with Sec23, a COPII coat protein necessary for vesicle budding off the ER (25). BFA treatment of DHC1-silenced cells showed delayed reformation of the Golgi apparatus similar to reductions in Mon1a (Fig. 5C). Reductions in DHC1 also resulted in a delay in ts045VSVG-GFP gaining Endo H resistance (supplemental Fig. 3B), confirming that dynein is important in anterograde trafficking between the ER and the Golgi.
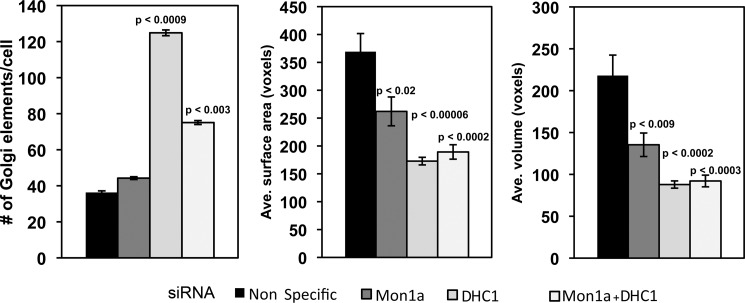
Confocal microscopic analysis of the Golgi after Mon1a and DHC1 silencing was performed using the GalNAc-T2-GFP HeLa cells. The Volocity software was used to quantify the alterations in Golgi morphology observed in Mon1a and DHC1-silenced cells. There were significant changes in the average surface area and the average volume of the Golgi apparatus in Mon1a- or DHC1-silenced cells, demonstrating a role for both proteins in maintaining Golgi morphology (Fig. 6). DHC1-silenced cells also showed an increased number of Golgi elements indicative of fragmentation. Silencing of Mon1a in DHC1-silenced cells reduced the amount of fragmentation seen with loss of DHC1 alone; however, the number of elements still remained higher than in cells that were treated with nonspecific oligonucleotides. The average surface area and volume of the Golgi elements remained similar to silencing DHC1. This quantification suggests that the high level of Golgi fragmentation seen in DHC1-silenced cells is affected by the levels of Mon1a.
FIGURE 6.
Volocity analysis and quantification of changes in Golgi morphology in Mon1a, dynein heavy chain, or Mon1a + dynein heavy chain silenced cells. Cells silenced as in Fig. 5 were imaged using confocal microscopy, and images were quantified using the Volocity software. The Golgi-labeled channel was quantified by setting the lower size cutoff limit at 10 cubic voxels, which is just above the minimum optically resolvable volume to filter out digital noise. Cells were cropped out of the image and measured individually to get cell-by-cell data. Signal was thresholded using the same intensity values for all silenced groups. The data are expressed as the number of Golgi elements/cell, and the average (Ave.) Golgi surface area and average Golgi volume are expressed as voxels (cubic pixel area). All experiments were performed a minimum of three times. Error bars represent the S.E. p values were determined using a two-tailed Student's t test statistical analysis comparing them with the nonspecifically silenced cells.
Reductions in Mon1a Delay the Formation of ER-derived Vesicles
ER exit sites (ERES) and vesicle formation require the recruitment of COPII onto ER exit sites (for review, see Zanetti et al. (31)). To determine whether COPII recruitment was affected by reductions in Mon1a or DHC1, we examined the localization of the COPII protein Sec31 in Mon1a- or DHC1-silenced cells during BFA recovery. Sec31, a component of the outer COPII coat, was recruited to ER exit sites in the presence or absence of Mon1a or DHC1 (supplemental Fig. 4). We note that Sec31 showed a high degree of coincidence with the Golgi marker GalNAc-T2-GFP as reported previously (26); however, the localization was not different in the presence or absence of Mon1a or DHC1. DHC1 silencing frequently resulted in an increased number of nuclei per cell. We also note that there appeared to be increased numbers of Sec31-positive sites present in Mon1a- or DHC1-silenced cells, possibly as a consequence of delayed reformation of the Golgi apparatus. These data demonstrate that Mon1a and DHC1 are not necessary for COPII recruitment and that they function after coat assembly.
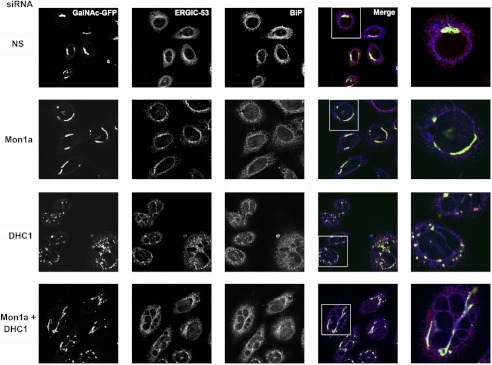
Once COPII proteins are recruited to ERES, then the newly formed vesicles can acquire the mannose-specific membrane lectin ERGIC-53, which acts as a receptor for glycoprotein trafficking to the ERGIC or vesicular-tubular compartment between the ER and Golgi (27). It is possible that reductions in Mon1a or DHC1 affect the ability to recruit ERGIC-53, and subsequently, GalNAc-T2-GFP would not be trafficked in the reformation of the Golgi apparatus. We examined the localization of ERGIC-53 in Mon1a- and DHC1-silenced GalNAc-T2-GFP cells using confocal microscopy. Mon1a-silenced cells showed an increased perinuclear localization of ERGIC-53 surrounding the nucleus when compared with nonspecifically silenced cells (Fig. 7). DHC1-silenced cells also showed increased perinuclear ERGIC-53 localization similar to Mon1a-silenced cells. We note that the resident ER protein BiP appeared unchanged, suggesting that reductions in Mon1a or DHC1 do not significantly affect ER morphology.
FIGURE 7.
Reductions in Mon1a or DHC1 affect steady state Golgi and ERGIC-53 morphology. HeLa cells expressing GalNAc-T2-GFP (green) incubated with nonspecific (NS), Mon1a, or DHC1 oligonucleotides. 72 h after siRNA, cells were fixed, permeabilized, and incubated with mouse anti-ERGIC-53 (red) and rabbit anti-BiP (pseudocolored blue) followed by Alexa Fluor 594-conjugated goat anti-mouse and Alexa Fluor 647-conjugated goat anti-rabbit and imaged using a 60× oil immersion objective and an Olympus FV1000 confocal microscopy. Z stacks were captured every 0.5 μm, and a single plane projection showing coincidence is shown (n = 3). Images are representative fields with the far right panel a zoom of the boxed area. Scale bar = 10 μm.
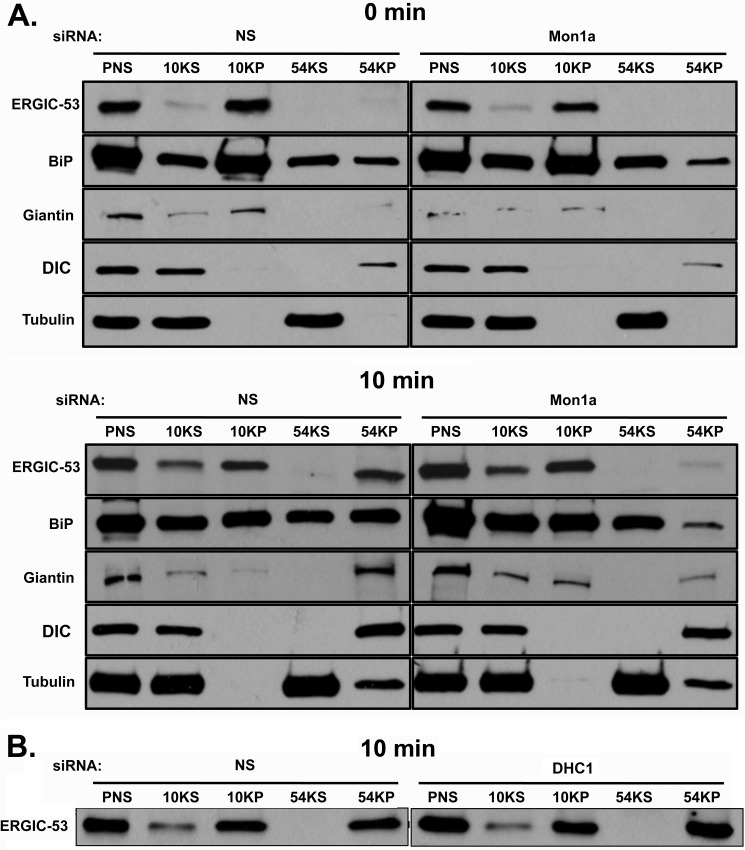
The loss of Mon1a or dynein could affect the budding of vesicles from the ER or the fusion of budded vesicles with the ERGIC or Golgi. To determine whether the loss of Mon1a or DHC1 and concomitantly DIC affects the formation of ER-derived vesicles, we followed the movement of ERGIC-53 into ER-derived vesicles. Microsomal membranes can be pelleted at low speeds, whereas ER-derived vesicles require higher speed centrifugation (28). Nonspecifically silenced, Mon1a-silenced, or DHC1-silenced cells were treated with BFA for 30 min to collapse the Golgi and move all ERGIC-53 into the ER. BFA was removed, and the movement of ERGIC-53 into vesicles was assessed by Western blot. 10 minutes after removal of BFA, most ERGIC-53 was found in the high-speed pellet in nonspecifically silenced cells. In contrast, little ERGIC-53 was found in the high-speed pellet in Mon1a-silenced cells (Fig. 8A). Similarly, the Golgi marker Giantin was found in the high-speed pellet in nonspecifically silenced cells, but a reduced amount was present in the high-speed pellet from Mon1a-silenced cells. These data support a role for Mon1a in the formation of vesicles at ERES. We note that BiP was found in all samples, but was most predominant in the 10,000 pellet, as would be predicted for an ER marker. We also note a slight tubulin contamination in the high-speed pellet at 10 min; however, this tubulin was present in all silenced samples. DIC, which associates with many types of vesicles, remained associated with the vesicle fraction in Mon1a-silenced cells, as expected. Silencing of DHC1 did not affect the ability of ERGIC-53 to be incorporated into high-speed pelleted vesicles (Fig. 8B). This suggests that dynein is required for efficient anterograde trafficking, but is dispensable in vesicle formation at ERES.
FIGURE 8.
Silencing of Mon1a delays formation of ERGIC-53-positive vesicles off the ER. A, NIH3T3 cells nonspecifically (NS) or Mon1a-silenced were treated with BFA as in Fig. 1. BFA was removed, cells were incubated for 10 min at 37 °C and homogenized, homogenates were centrifuged at 800 × g for 5 min (PNS), supernatant was centrifuged at 6,552 × g, 30 min (10KS and 10KP) to obtain an ER membrane fraction, and the remaining supernatant was centrifuged at 191,065 × g for 30 min (54KS and 54KP) to obtain ER-derived vesicles. Supernatants and membranes were solubilized in lysis buffer and resolved on SDS-PAGE, and ERGIC-53, BiP, Giantin, DIC, and tubulin localization was determined by Western blot analysis. A representative blot is shown (n = 3). B, cells as in A were nonspecifically or DHC1-silenced and were treated as in A, and ERGIC-53 movement into high-speed vesicles pellets was assessed. A representative blot of the 10 min recovery is shown (n = 3).
DISCUSSION
Mon1a was identified initially through a genetic screen as a gene that affected the localization and trafficking of cell surface and secreted proteins (9). A naturally occurring mutant allele of Mon1a resulted in increased movement of the mammalian iron transporter ferroportin through the secretory pathway. In this current study, we determined that reductions in Mon1a delay the reformation of the Golgi apparatus after BFA disruption and that Mon1a functions in efficient movement of molecules from the ER to the Golgi and from the Golgi to the plasma membrane. We further determined that Mon1a interacts with DIC in the dynein-dynactin complex to efficiently generate ER-derived ERGIC-53-positive vesicles.
Initial studies in S. cerevisiae and C. elegans suggested that YMon1 and SAND-1, respectively, are involved in the endocytic pathway; however, the precise step in the endocytic pathway is different (1, 3, 4, 29). Based upon those studies and the sequence homology with YMon1, we expected that Mon1a would also function in the endocytic pathway. Surprisingly, reductions in Mon1a levels did not affect endocytosis, phagocytosis, or trafficking through the endocytic pathway. Additionally, the steady state size of organelles in the endocytic pathway was not affected by reduced levels of Mon1a. A recent study proposed that SAND-1 in C. elegans acts in ingestion of apoptotic cell corpses, a specific phagocytic event, and that mammalian Mon1a and Mon1b might have a similar function acting in complex with a mammalian homologue of Ccz1 (c7orf28b) (7). The S. cerevisiae homologues Mon1 and Ccz1 have been shown to be involved in autophagy (29). We previously reported that loss of Mon1a in macrophages did not affect erythrophagocytosis (9), consistent with the report by Kinchen and Ravichandran (7) that silencing of mammalian Mon1a or its homologue Mon1b did not affect phagocytosis. In the C. elegans study, silencing of both Mon1a and Mon1b, however, did affect acidification of the apoptotic cell-containing phagosome. A second role for Mon1a in the endocytic pathway was suggested by Poteryaev et al. (5). These authors proposed that SAND-1/Mon1a/b functions in endosome maturation by acting as a switch from Rab5-to-Rab7-positive endosomes (8). The authors noted that effects on the endocytic pathway were best seen when both Mon1a and Mon1b protein levels were reduced. Confirmatory results were reported in S. cerevisiae (17), supporting the role of Mon1 proteins in the endocytic pathway. Importantly, none of these studies reported whether loss of Mon1a, Mon1b, or SAND-1 affected vesicle trafficking in the secretory pathway.
We provide evidence that reductions in Mon1a affect transfer of vesicular contents from ER to Golgi and from Golgi to plasma membrane. Further, reduced levels of Mon1a lead to delayed assembly of the Golgi apparatus following BFA treatment. If Mon1a is required for ER-to-Golgi trafficking, one might expect that Golgi markers and the apparatus would be found more localized with the ER in Mon1a-silenced cells, as is seen in point mutants in Sec24 or Sar1 proteins (20, 30). This is not the case; reductions in Mon1a give rise to subtle but consistent changes in the morphology of the Golgi apparatus with longer extensions and less tight stacking of the Golgi. These results can be explained either by the fact that siRNA is not complete or by the fact that Mon1a also acts in retrograde trafficking from Golgi to ER. Studies are currently underway to address the role of Mon1a in retrograde trafficking to the ER.
Loss of Mon1a affects the generation of ER-derived ERGIC-53-positive vesicles. We show that Mon1a interacts with cytoplasmic DIC, which has been previously shown to be involved in trafficking at ER exit sites (23, 24). We observed that reductions in dynein result in Golgi fragmentation, but we did not see any diminution in ERGIC-53-positive vesicle formation. That dynein is involved in trafficking at ERES is not surprising. What is novel is the identification of Mon1a as a dynein-interacting protein that is important in efficient trafficking out of the ER. Reductions in Mon1a or DHC1 did not affect the ability to recruit Sec31 to ER exit sites, suggesting that Mon1a and DHC1 are recruited after COPII proteins. The dynein-dynactin complex then interacts with microtubules to efficiently guide vesicles and move them to the ERGIC and subsequently to the Golgi. A recent review by Zanetti et al. (31) suggests that vesicle formation at ERES remains to be further characterized. We propose a model where Mon1a is functioning at ER exit sites to assist in vesicle formation, that the dynein-dynactin complex binds to microtubules, Sec23, and Mon1a at ER exit sites, and that DIC binds Mon1a to efficiently traffic vesicles away from the ER (Fig. 9). Microtubules and motor proteins, including the dynein-dynactin complex, are clearly involved in trafficking. Perhaps Mon1a is an accessory protein that may act to stabilize the formation of vesicles exiting the ERES.
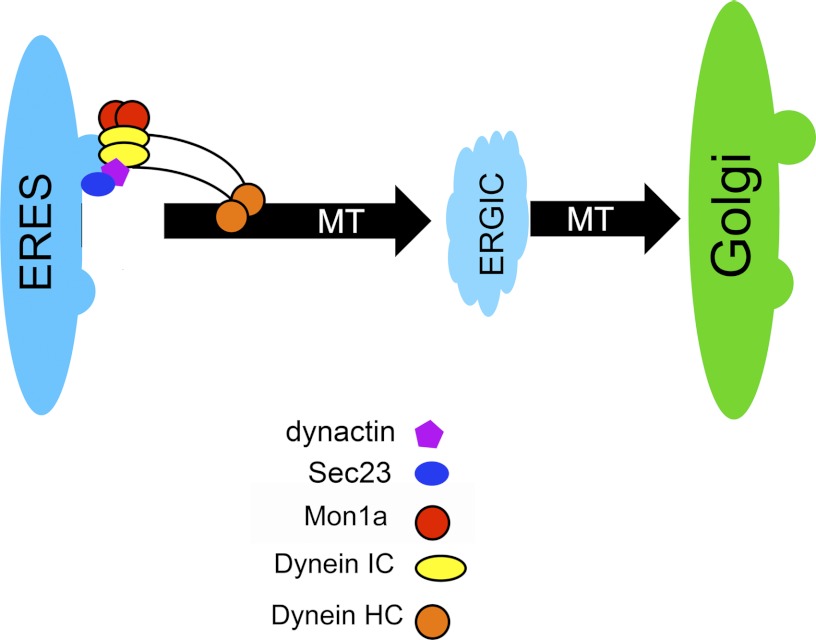
FIGURE 9.
Model of Mon1a and dynein acting in vesicle formation off the ER. A model showing dynein-dynactin complex interacts with vesicles through the DIC and with microtubules (MT) through the DHC1. Dynein-dynactin interacts with Sec23 on the ERES (25). Mon1a interacts with dynein to efficiently traffic vesicles from ER to Golgi. IC, intermediate chain; HC, heavy chain.
A finding that loss of Mon1a affects ER-to-Golgi membrane traffic does not preclude a role for Mon1a in other forms of vesicular traffic; in fact, we have shown that reductions in Mon1a can affect Golgi-to-plasma membrane sorting as well. As suggested by others, the SAND-1 family of proteins may be involved in many trafficking events (1, 3, 4, 7, 8, 17, 29). Indeed, many vesicle trafficking events require dynein and dynein-interacting proteins, possibly Mon1a. The finding that reduced levels of Mon1a in mammalian cells affects trafficking through the secretory apparatus may also be reconciled by studies that show that loss of SAND-1 affects autophagy in C. elegans (7). Studies in S. cerevisiae have shown that components of the secretory pathway including Sec1, Sec12, Sec16, and Sec23 are required for autophagy (32, 33). These studies suggest that the ER, and not the specific COPII vesicle components, is necessary for autophagosome formation by providing membrane (33, 34). It is possible that the effects on the ingestion of apoptotic cell corpses seen in C. elegans SAND-1 mutants or in mammalian cells silenced for Mon1a and its homologue Mon1b may reflect a common defect at the level of the secretory pathway. Why vertebrates have two Mon1 proteins remains to be determined. Outside of the SAND domain of mammalian Mon1a and Mon1b, the homology of the two mammalian Mon1 proteins is less conserved. It is possible that these two proteins function in completely different membrane trafficking pathways. Studies are underway to further characterize how Mon1a interacts with dynein and whether there are other molecules that play a role in efficient intracellular vesicle trafficking.
Supplementary Material
Acknowledgments
We express our appreciation to members of the Kaplan/Ward laboratory for editing this manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grant HL26922 (to D. M. W.).

This article contains supplemental Figs. 1–4.
- YMon1
- yeast Mon1
- BFA
- brefeldin A
- DHC1
- dynein-heavy chain
- Endo H
- endoglycosidase H
- ER
- endoplasmic reticulum
- ERES
- ER exit sites
- ERGIC
- ER-Golgi intermediate compartment
- GalNAc-T2-GFP
- Golgi protein N-acetylgalactosaminyltransferase-2-green fluorescent protein
- PNS
- postnuclear supernatant
- VSVG-GFP
- vesicular stomatitis virus G protein-GFP
- DIC
- dynein intermediate chain
- COPII
- coat complex II.
REFERENCES
- 1. Wang C. W., Stromhaug P. E., Kauffman E. J., Weisman L. S., Klionsky D. J. (2003) Yeast homotypic vacuole fusion requires the Ccz1-Mon1 complex during the tethering/docking stage. J. Cell Biol. 163, 973–985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Hoffman-Sommer M., Kucharczyk R., Piekarska I., Kozlowska E., Rytka J. (2009) Mutations in the Saccharomyces cerevisiae vacuolar fusion proteins Ccz1, Mon1, and Ypt7 cause defects in cell cycle progression in a num1Δ background. Eur. J. Cell Biol. 88, 639–652 [DOI] [PubMed] [Google Scholar]
- 3. Kucharczyk R., Hoffman-Sommer M., Piekarska I., von Mollard G. F., Rytka J. (2009) The Saccharomyces cerevisiae protein Ccz1p interacts with components of the endosomal fusion machinery. FEMS Yeast Res. 9, 565–573 [DOI] [PubMed] [Google Scholar]
- 4. Hoffman-Sommer M., Migdalski A., Rytka J., Kucharczyk R. (2005) Multiple functions of the vacuolar sorting protein Ccz1p in Saccharomyces cerevisiae. Biochem. Biophys. Res. Commun. 329, 197–204 [DOI] [PubMed] [Google Scholar]
- 5. Poteryaev D., Spang A. (2005) A role of SAND family proteins in endocytosis. Biochem. Soc. Trans. 33, 606–608 [DOI] [PubMed] [Google Scholar]
- 6. Poteryaev D., Fares H., Bowerman B., Spang A. (2007) Caenorhabditis elegans SAND-1 is essential for RAB-7 function in endosomal traffic. EMBO J. 26, 301–312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kinchen J. M., Ravichandran K. S. (2010) Identification of two evolutionarily conserved genes regulating processing of engulfed apoptotic cells. Nature 464, 778–782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Poteryaev D., Datta S., Ackema K., Zerial M., Spang A. (2010) Identification of the switch in early-to-late endosome transition. Cell 141, 497–508 [DOI] [PubMed] [Google Scholar]
- 9. Wang F., Paradkar P. N., Custodio A. O., McVey Ward D., Fleming M. D., Campagna D., Roberts K. A., Boyartchuk V., Dietrich W. F., Kaplan J., Andrews N. C. (2007) Genetic variation in Mon1a affects protein trafficking and modifies macrophage iron loading in mice. Nat. Genet. 39, 1025–1032 [DOI] [PubMed] [Google Scholar]
- 10. Ryan S. D., Bhanot K., Ferrier A., De Repentigny Y., Chu A., Blais A., Kothary R. (2012) Microtubule stability, Golgi organization, and transport flux require dystonin-a2-MAP1B interaction. J. Cell Biol. 196, 727–742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Perou C. M., Kaplan J. (1993) Chediak-Higashi syndrome is not due to a defect in microtubule-based lysosomal mobility. J. Cell Sci. 106, 99–107 [DOI] [PubMed] [Google Scholar]
- 12. Zhang Y., Zolov S. N., Chow C. Y., Slutsky S. G., Richardson S. C., Piper R. C., Yang B., Nau J. J., Westrick R. J., Morrison S. J., Meisler M. H., Weisman L. S. (2007) Loss of Vac14, a regulator of the signaling lipid phosphatidylinositol 3,5-bisphosphate, results in neurodegeneration in mice. Proc. Natl. Acad. Sci. U.S.A. 104, 17518–17523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Raiborg C., Bremnes B., Mehlum A., Gillooly D. J., D'Arrigo A., Stang E., Stenmark H. (2001) FYVE and coiled-coil domains determine the specific localization of Hrs to early endosomes. J. Cell Sci. 114, 2255–2263 [DOI] [PubMed] [Google Scholar]
- 14. de Renzis S., Sönnichsen B., Zerial M. (2002) Divalent Rab effectors regulate the subcompartmental organization and sorting of early endosomes. Nat. Cell Biol. 4, 124–133 [DOI] [PubMed] [Google Scholar]
- 15. Durchfort N., Verhoef S., Vaughn M. B., Shrestha R., Adam D., Kaplan J., Ward D. M. (2012) The enlarged lysosomes in beigej cells result from decreased lysosome fission and not increased lysosome fusion. Traffic 13, 108–119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ward D. M., Vaughn M. B., Shiflett S. L., White P. L., Pollock A. L., Hill J., Schnegelberger R., Sundquist W. I., Kaplan J. (2005) The role of LIP5 and CHMP5 in multivesicular body formation and HIV-1 budding in mammalian cells. J. Biol. Chem. 280, 10548–10555 [DOI] [PubMed] [Google Scholar]
- 17. Nordmann M., Cabrera M., Perz A., Bröcker C., Ostrowicz C., Engelbrecht-Vandré S., Ungermann C. (2010) The Mon1-Ccz1 complex is the GEF of the late endosomal Rab7 homolog Ypt7. Curr. Biol. 20, 1654–1659 [DOI] [PubMed] [Google Scholar]
- 18. Altan-Bonnet N., Sougrat R., Liu W., Snapp E. L., Ward T., Lippincott-Schwartz J. (2006) Golgi inheritance in mammalian cells is mediated through endoplasmic reticulum export activities. Mol. Biol. Cell 17, 990–1005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Saraste J., Palade G. E., Farquhar M. G. (1986) Temperature-sensitive steps in the transport of secretory proteins through the Golgi complex in exocrine pancreatic cells. Proc. Natl. Acad. Sci. U.S.A. 83, 6425–6429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Storrie B., White J., Röttger S., Stelzer E. H., Suganuma T., Nilsson T. (1998) Recycling of Golgi-resident glycosyltransferases through the ER reveals a novel pathway and provides an explanation for nocodazole-induced Golgi scattering. J. Cell Biol. 143, 1505–1521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Helenius A., Aebi M. (2004) Roles of N-linked glycans in the endoplasmic reticulum. Annu. Rev. Biochem. 73, 1019–1049 [DOI] [PubMed] [Google Scholar]
- 22. Presley J. F., Cole N. B., Schroer T. A., Hirschberg K., Zaal K. J., Lippincott-Schwartz J. (1997) ER-to-Golgi transport visualized in living cells. Nature 389, 81–85 [DOI] [PubMed] [Google Scholar]
- 23. Watson P., Forster R., Palmer K. J., Pepperkok R., Stephens D. J. (2005) Coupling of ER exit to microtubules through direct interaction of COPII with dynactin. Nat. Cell Biol. 7, 48–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Iyadurai S. J., Robinson J. T., Ma L., He Y., Mische S., Li M. G., Brown W., Guichard A., Bier E., Hays T. S. (2008) Dynein and Star interact in EGFR signaling and ligand trafficking. J. Cell Sci. 121, 2643–2651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Fromme J. C., Orci L., Schekman R. (2008) Coordination of COPII vesicle trafficking by Sec23. Trends Cell Biol. 18, 330–336 [DOI] [PubMed] [Google Scholar]
- 26. Townley A. K., Feng Y., Schmidt K., Carter D. A., Porter R., Verkade P., Stephens D. J. (2008) Efficient coupling of Sec23-Sec24 to Sec13-Sec31 drives COPII-dependent collagen secretion and is essential for normal craniofacial development. J. Cell Sci. 121, 3025–3034 [DOI] [PubMed] [Google Scholar]
- 27. Hauri H. P., Kappeler F., Andersson H., Appenzeller C. (2000) ERGIC-53 and traffic in the secretory pathway. J. Cell Sci. 113, 587–596 [DOI] [PubMed] [Google Scholar]
- 28. Rowe T., Aridor M., McCaffery J. M., Plutner H., Nuoffer C., Balch W. E. (1996) COPII vesicles derived from mammalian endoplasmic reticulum microsomes recruit COPI. J. Cell Biol. 135, 895–911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Wang C. W., Stromhaug P. E., Shima J., Klionsky D. J. (2002) The Ccz1-Mon1 protein complex is required for the late step of multiple vacuole delivery pathways. J. Biol. Chem. 277, 47917–47927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Fromme J. C., Ravazzola M., Hamamoto S., Al-Balwi M., Eyaid W., Boyadjiev S. A., Cosson P., Schekman R., Orci L. (2007) The genetic basis of a craniofacial disease provides insight into COPII coat assembly. Dev. Cell 13, 623–634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Zanetti G., Pahuja K. B., Studer S., Shim S., Schekman R. (2012) COPII and the regulation of protein sorting in mammals. Nat. Cell Biol. 14, 20–28 [DOI] [PubMed] [Google Scholar]
- 32. Ishihara N., Hamasaki M., Yokota S., Suzuki K., Kamada Y., Kihara A., Yoshimori T., Noda T., Ohsumi Y. (2001) Autophagosome requires specific early Sec proteins for its formation and NSF/SNARE for vacuolar fusion. Mol. Biol. Cell 12, 3690–3702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Hamasaki M., Noda T., Ohsumi Y. (2003) The early secretory pathway contributes to autophagy in yeast. Cell Struct. Funct. 28, 49–54 [DOI] [PubMed] [Google Scholar]
- 34. Di Bartolomeo S., Corazzari M., Nazio F., Oliverio S., Lisi G., Antonioli M., Pagliarini V., Matteoni S., Fuoco C., Giunta L., D'Amelio M., Nardacci R., Romagnoli A., Piacentini M., Cecconi F., Fimia G. M. (2010) The dynamic interaction of AMBRA1 with the dynein motor complex regulates mammalian autophagy. J. Cell Biol. 191, 155–168 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.