Abstract
Hypoxia inducible factors (HIFs) regulate a variety of genes to prepare cells to adapt and to survive under a hypoxic environment. Recently, microRNAs (miRNAs) have emerged as a new class of genes regulated by HIF in response to hypoxia, of which miR-210 is the most consistently and predominantly upregulated miRNA. Functional studies have demonstrated that miR-210 is a versatile gene that regulates many aspects of hypoxia pathways, both in physiological and malignant conditions. Here, we summarize recent findings on the mechanism of hypoxia regulation of miR-210 expression and its multifaceted biological functions in normal physiological and malignant conditions and discuss challenges we face in elucidating the biological functions of miR-210 and in exploring its potential use for therapeutics.
Keywords: Hypoxia, HIF, microRNA, miR-210
Introduction of hypoxia and microRNA
MicroRNAs (miRNAs) are a class of single-stranded noncoding RNAs 21–22 nucleotides in length. Genes encoding miRNAs are initially transcribed as part of much longer primary transcripts (pri-miRNAs) [1]. In the nucleus, these primary transcripts are first processed by the type III RNA endonuclease, Drosha, into pre-miRNAs that are 60–70 nucleotides in length and form a stem-loop structure [2]. Subsequent transport of pre-miRNAs into the cytoplasm by exportin-5 results in further processing by another type III RNA endonuclease, Dicer, to generate mature miRNAs 22-nucleotides in length [3]. Mature single-stranded miRNAs form an RNA-protein complex known as RNA-induced silencing complex (RISC) with proteins of the Argonaute family [4]. This complex regulates gene expression through inhibition of RNA translation or degradation of target messenger RNA (mRNA) by base-pairing between their “seed region,” nucleotides 2–8, and their target genes’ 3′ UTR [5] (Figure 1). Our understanding of miRNA biology unfolded slowly: the first miRNA, lin-4, was discovered in Caenorhabditis elegans in 1993 [6, 7], but the second miRNA, let-7, was not discovered until 2000 [8]. The finding that let-7 is well conserved in a wide range of animal species [9] stimulated an explosion in miRNA discovery that is still ongoing. Suddenly, miRNAs have emerged as important regulators in developmental, physiological and pathological settings including cell growth, differentiation, metabolism, viral infection and tumorigenesis [10]. Accumulating evidence shows that miRNAs are frequently dysregulated in human malignancies and can function as oncogenes or tumor suppressor genes [11].
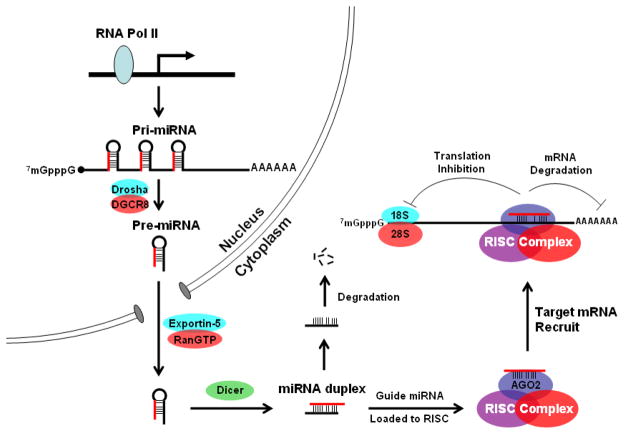
Figure 1.
Schematic view of miRNA biogenesis and functioning pathways. Genes encoding miRNA are transcribed into pri-miRNA by RNA polymerase II (RNA Pol II), the pri-miRNA is first processed by the type III RNA endonuclease, Drosha, into pre-miRNAs that are 60–70 nucleotides in length and have a stem-loop structure. The pre-miRNA is exported out of nucleus by exportin-5 and is further processed by another type III RNA endonuclease, Dicer, to generate a mature miRNA duplex 22-nucleotides in length. The sense strand of the miRNA duplex is then loaded into RISC, while the complementary strand, “star” form (*) of the miRNA duplex is degraded. The RISC complex regulates gene expression through inhibition of RNA translation or degradation of target mRNA by base-pairing the “seed region” of a miRNA to the 3′ UTR of target mRNA.
Hypoxia, the condition of insufficient oxygen supply to tissues, results from physiological or pathological conditions, such as high altitude, anemia, or abnormal and insufficient vasculature. The hypoxia inducible factors (HIFs) control the cellular response to hypoxia by regulating genes that are involved in metabolism, angiogenesis, erythropoiesis, cell proliferation, differentiation and apoptosis. HIF is a heterodimer consisting of an oxygen-sensitive alpha subunit (HIFα) and a constitutively active beta subunit (HIF1β; aryl hydrocarbon receptor nuclear translocator (ARNT)). Although three isoforms of HIFα have been identified, HIF1α and HIF2α appear to modulate the majority of the HIFα response to hypoxia [12]. Under hypoxia, stabilized HIFα binds to HIF1β; the complex regulates expression of downstream genes involved in adaptation to and protection against low oxygen conditions [12].
Given the diverse roles that miRNAs play in numerous aspects of cellular functions, it is not surprising that they also play a role in hypoxia gene regulation. Recently, miRNAs have been implicated in regulating both upstream and downstream signaling of the HIF pathways: miR-199a, miR-17-92 clusters and miR-20b regulate HIF1α under hypoxia [13–15], and miR-23, miR-24, miR-26, miR-107, miR-210 and miR-373 have been shown to be induced by HIFs [16–18]. Following the first report of hypoxia affecting miRNA expression by Ivan and colleagues in 2007 [18], a number of papers have appeared, especially on miR-210, covering almost every aspect of hypoxia biology (Table 1). Although several miRNAs are regulated by hypoxia, miR-210 is the most consistently and robustly induced miRNA under hypoxia [17, 18]. In this review, we summarize the recent findings of miR-210 on the hypoxia regulation of its expression and its biological functions in normal physiological and malignant conditions and discuss challenges we face in comprehensively identifying miR-210 target genes and elucidating miR-210 functions and in exploring its potential use for therapeutics.
Table 1.
Summary of the cellular processes miR-210 has been implicated in and miR-210 target genes identified to date.
| Cellular Processes miR-210 Implicated in | Target Genes | References |
|---|---|---|
| Apoptosis | N/A | Cheng et al. (2005) |
| N/A | Kulshreshtha et al. (2007) | |
| Angiogenesis | EFNA3 | Fasanaro et al. (2008) |
| Stem Cell Biology | ||
| Stem cell survival | Casp8ap2 | Kim et al. (2009) |
| Osteoblastic differentiation | AcvR1b | Mizuno et al. (2009) |
| Cell cycle regulation | E2F3 | Giannakakis et al. (2007) |
| MNT | Zhang et al. (2009) | |
| DNA damage repair | RAD52 | Crosby et al. (2009) |
| Mitochondrial metabolism | ISCU1/2 | Chan et al. (2009) |
| Tumor growth | HOXA1, FGFRL1 | Huang et al. (2009) |
Regulation of miR-210 expression by hypoxia
Hypoxic regulation of miR-210 was first identified by miRNA microarray in 2007 [18]. The stem-loop of miR-210 is located in an intron of a noncoding RNA, which is transcribed from AK123483 on chromosome 11p15.5. The boundary and length of human pri-miR-210 is predicted with high confidence based on expressed sequence tags, gene expression analysis using the captured 5′ 7-methylguanosine cap of mRNA, the location of transcription start site (TSS), CpG islands and the polyadenylation site [19]. The predicted size of human pri-miR-210 is 2927 bp, which is almost the same as genomic AK123483. Although the mechanism of miR-210 biogenesis has not been rigorously investigated, the AK123483 transcript appears to be the pri-miR-210, which is then processed to pre- and mature miR-210 [20]. Expression of AK123483 is increased in microarray and quantitative PCR assays under hypoxia as compared to normoxia [20, 21]. Interestingly, two genes located within the 33 Kb region downstream of AK123483, RASSF7 and HRAS, also show increased expression under hypoxia. Although RASSF7 is transcribed in the opposite orientation from AK123483 [17, 21], the proximity of these loci suggests that local chromatin structure could play a role in gene expression; this idea is further supported by the recent demonstration that the Jumonji gene family of histone demethylases are regulated by hypoxia [22, 23].
Interestingly, miR-210 is regulated by both HIF1α [16, 17, 21, 24] and HIF2α [20]. HIF1α directly binds to a hypoxia responsive element (HRE) on the proximal miR-210 promoter [17]. The functional HRE element is located approximately 40 bp upstream of the predicted TSS, suggesting that this HRE is responsible for the hypoxic induction of AK123483 and might coordinate expression of the genes downstream of miR-210 under hypoxia. When the miR-210 core promoter is compared across species, this HRE site is highly conserved, indicating the importance of hypoxia in regulating miR-210 expression in these species. Consistent with this observation, induction of mouse miR-210 under hypoxia is dependent on HIF1α [16], although the hypoxic response of this conserved mouse HRE has yet to be experimentally verified. Interestingly, in the close vicinity of miR-210’s HRE site, several other important transcriptional factor binding sites are also highly conserved across species, such as E2F1, Oct4 and PPARγ. Therefore, it is tempting to speculate that miR-210 is involved in cell cycle regulation, stem cell biology and energy metabolism, processes in which hypoxia is well known to play important roles [12].
It is noteworthy that the elevated expression of miR-210 induced by hypoxia remains significantly elevated above its normoxic level even one day after hypoxic exposure [25]. This long-lasting elevated expression of miR-210 after hypoxia could be attributable to its exceptionally long half-life; there is no evidence, however, that miRNA in general possesses slow turnover. Or long-lasting miR-210 expression could serve as a “buffer” that gradually releases the repression of its target genes, easing the transition from hypoxia to normoxia for the cell.
MiR-210 function
Angiogenesis
Virtually all solid tumors contain hypoxic regions, which stimulate angiogenesis to sustain tumor growth. One of the major regulators of tumor angiogenesis is vascular endothelial growth factor (VEGF), a gene regulated by hypoxia [26]. Although increased expression of VEGF promotes angiogenesis, miR-210 might also be involved [25]. MiR-210 targets the receptor tyrosine kinase ligand, Ephrin-A3 (EFNA3) (which is one of the most consistently reported miR-210 target genes [17, 25, 27, 28]), enhancing differentiation of human umbilical vein endothelial cells (HUVEC), a robust cellular model for investigation of physiological and pharmacological effects induced by cytokine stimulation, into capillary-like structures under hypoxia and promoting the migration of HUVEC cells induced by VEGF [25]. Under normoxia, miR-210 expression in HUVEC cells significantly enhances tube formation, indicating that miR-210 alone might be sufficient to augment angiogenesis. Consistent with this observation, miR-210 expression correlates with VEGF expression, hypoxia and angiogenesis in breast cancer patients [29]. However, direct functional evidence is still needed to support a role for miR-210 in enhancing angiogenesis in human cancers. An alternative explanation of these findings could be that the correlation between miR-210 and angiogenesis only reflects the hypoxic status of those tumor samples and miR-210 does not contribute to tumor angiogenesis [30]. However, until an inducible miR-210 knockdown cellular model or miR-210 knockout mouse model is established, comprehensive understanding of the functional role miR-210 plays in tumor angiogenesis will remain elusive although a pro-angiogenic function of miR-210 has been demonstrated in a HUVEC model by targeting Ephrin-A3.
Stem cells and differentiation
Ischemic preconditioning (IP) is a powerful cytoprotective stimulus [31]. HIF1α-mediated gene expression is essential for the protective effect of IP [32]. MiR-210 is a HIF1α target gene that improves the survival of mesenchymal stem cells (MSC) under anoxia by ischemia/reoxygenation (I/R) preconditioning, an in vitro format of IP on cultured cells [24]. More importantly, increased expression of miR-210 also correlates with improved survival of transplanted MSC in a rat model. By downregulating caspase-8-associated protein 2 (Casp8ap2), a pro-apoptotic regulator of Fas-mediated apoptosis [33], miR-210 protects MSC from cell death; this observation provides a mechanistic link between HIF1α-mediated gene expression and IP.
Hypoxia is an important environmental factor that prevents stem cells from differentiating [34]. Hypoxia greatly enhances the generation of induced pluripotent stem cells (iPS) by 2 factors (Oct3/4 and Klf4), 3 factors (Oct3/4, Sox2 and Klf4) or 4 factors (Oct3/4, Sox2, Klf4 and c-myc) in mouse embryonic fibroblasts (MEFs) and by 4 factors in adult human dermal fibroblasts [35]. When gene expression profiles of the transduced MEFs cultured under hypoxia are compared to those under normoxia, the majority of embryonic stem cell-specific genes are upregulated and MEF-specific genes are downregulated, including marked upregulation of the endogenous Oct3/4 and Nanog genes, suggesting that hypoxia creates a permissive microenvironment for more efficient somatic reprogramming. Thus, it is tempting to speculate that miR-210 could enhance somatic cell reprogramming by preventing iPS cell death under hypoxia.
Interestingly, miR-210 also promotes osteoblastic differentiation by inhibiting expression of a type 1B receptor of activin A (AcvR1b) [36]. Initially, miR-210 was identified as one of seven miRNAs elevated during osteoblastic differentiation of bone marrow-derived ST2 stromal cells induced by a member of the transforming growth factor-beta (TGFβ) superfamily, bone morphogenetic protein 4 (BMP4). Because hypoxia promotes osteoblastic differentiation of osteocytes [37] and hypoxia also induces BMP4 expression [38], it will be of interest to investigate whether enhanced osteoblastic differentiation found under hypoxic conditions and mediated by inhibition of AcvR1b results from the additive or synergistic induction of miR-210 by both hypoxia and BMP4. MiR-210 appears to be a versatile regulator in stem cell maintenance and differentiation by preventing cell death and promote differentiation, respectively.
Cell cycle regulation
In some cell types, hypoxia can induce cell cycle arrest through HIF1α [39]. MiR-210 is an HIF1α-dependent molecule that regulates cell cycle progression by targeting an E2F family member, E2F3, and an antagonist of Myc-dependent transcriptional activation and cell growth, MNT [20, 40]. Two E2F3 isoforms, a and b, are expressed from alternative promoters [41]. E2F3a expression fluctuates during the cell cycle with maximum expression in G1/S; E2F3b expression is constitutive throughout the cell cycle but peaks in G0 when it is associated with pRb. Based on studies in mouse cells in which E2F3a and E2F3b are individually knocked out, both forms of the gene seem to be involved in cell proliferation, although their contributions are context dependent [42]. Because both isoforms share the same 3′UTR region, miR-210 could regulate the relative balance between these two functionally distinct E2F3 isoforms at different points of cell cycle to control cell cycle progression [40], providing a novel mechanism of cell cycle arrest induced by hypoxia.
Paradoxically, miR-210 also promotes cell cycle progression by downregulating MNT, an antagonist of c-Myc [20]. MNT is a member of the Myc/Max/Mad network with a basic-Helix-Loop-Helix-zipper domain (bHLHzip). MNT represses Myc target genes by binding the E box DNA sequence (CANNTG) after forming heterodimers with Max [43, 44]. Interestingly, E2F3 is a target of Myc [45] and miR-210 [47], suggesting that miR-210 can positively or negatively regulate the Myc pathway at multiple points. Because HIF regulates cell proliferation and metabolism under hypoxia by interacting with c-Myc [46], miR-210 could serve as a rheostat to fine-tune the net output of c-Myc signaling pathway under hypoxic conditions. Interestingly, c-Myc is one of the 4 original transcriptional factors that can reprogram somatic cells to iPS cells [47]. Although later only 3 factors were found to be sufficient for reprogramming, the efficiency was substantially decreased without c-Myc [48], indicating a critical role of c-Myc in facilitating somatic cell reprogramming. Because hypoxia enhances iPS cell generation [35], miR-210 could contribute to this effect by regulating iPS cell cycle through the c-Myc pathway.
DNA damage repair
Hypoxia can increase genetic instability in cancer cells by downregulating DNA damage repair genes, such as MLH1, MSH2 and RAD51 [49]. Repression of these DNA repair genes under hypoxia involves histone deacetylation that alters MLH1 promoter chromatin structure [50], replacement of Myc by HIF1α/SP1 at the MSH2 promoter [51] or increased binding of repressive E2F4/p130 complexes at the RAD51 promoter [52]. MiR-210 represses protein translation but not gene transcription of RAD52, a key gene in homologous recombination (HR) repair, which presents a novel mechanism for downregulating cellular DNA damage repair machinery under hypoxia [16]. Because translation inhibition is a major mechanism by which miRNAs regulate expression of a target gene, it is not surprising that RAD52 was not identified in the original microarray screening for HR genes regulated by hypoxia [53]. Although miR-210 downregulates RAD52, direct functional evidence of miR-210’s involvement in HR and cancer cell genetic instability is lacking.
Regulation of mitochondrial metabolism
When oxygen is abundant, cellular energy can be generated through the tricarboxylic acid (TCA) cycle, in which one mole of glucose generates 38 moles of ATP. Under hypoxic conditions, due to limited oxygen supply, mitochondrial metabolism is suppressed and cellular energy production shifts to the glycolytic pathway where one mole of glucose only generates 2 moles of ATP. Many proteins induced under hypoxia are involved in this metabolic shift including pyruvate dehydrogenase kinase (PDK1), lactate dehydrogenase A (LDHA), cytochrome c oxidase subunit 4-2 (COX4-2) and the mitochondrial protease LON [54]. Recently, miR-210 has emerged as a new player that regulates mitochondrial metabolism under hypoxia by repressing the expression of iron-sulfur (Fe-S) cluster scaffold protein isoforms, ISCU1 and ISCU2 [55]. Fe-S clusters are required for the functions of mitochondrial aconitase, which catalyses the stereospecific isomerization of citrate to isocitrate by cis-aconitate in the TCA cycle [56]. By downregulating ISCU1/2, miR-210 also disrupts the electron transport activity of mitochondria complex I under hypoxia [55], providing mechanistic insight into how hypoxia modulates this metabolic shift by controlling mitochondrial respiration.
More than 80 years ago, the German biochemist Otto Warburg reported that cancer cells consume glucose and produce lactic acid even under aerobic conditions (the Warburg effect) [57]. Numerous hypoxia-regulated genes have been identified that contribute to the Warburg effect, including genes involved in glucose transport, glycolysis and mitochondrial metabolism [58]. Because miR-210 is elevated in many cancer types [20, 21, 28, 29, 59, 60], miR-210 could be a novel molecular link between tumor hypoxia and the Warburg effect.
MiR-210 is truly a multifaceted regulator of many cellular functions. We will not be surprised if many more miR-210 functions, dependent or independent of hypoxia, are discovered, especially if the number of discoveries in the near future reflects what we have learned since its identification as a hypoxia regulated miRNA in 2007.
MiR-210 in cancer
MiR-210 is frequently elevated in cancer, including glioblastoma [59], melanoma [20, 60], clear cell renal cell carcinoma [61], lung [62–66], pancreatic [28] and breast cancer [21, 29, 65], consistent with miR-210 promoting cell cycle progression [20]. However, because miR-210 is the most robustly induced miRNA under hypoxia [17], it is possible that elevated miR-210 expression only reflects the in vivo status of tumor hypoxia. Even under nonmalignant physiological conditions in which hypoxia plays a causal role, for example in the placenta from preeclampsia patients, miR-210 expression is increased [67, 68], confirming hypoxic regulation of miR-210 expression in vivo. Given the well established roles for tumor hypoxia in promoting metastasis and in predicting poor prognosis of cancer patients [69], it is not unexpected that increased expression of miR-210 correlates with poor prognosis in breast and pancreatic cancer patients [21, 28, 70]. MiR-210 expression correlated significantly with the expression of classical hypoxia inducible genes in primary tumor samples from head and neck cancer patients, suggesting miR-210 might be a surrogate marker for tumor hypoxia [17]. Therefore, it remains elusive whether miR-210 is merely a consequence of tumor hypoxia or whether it functionally contributes to tumor aggression [26]. Co-localization studies of well-established hypoxia targets such as carbonic anhydrase IX (CA9), major glucose transporter GLUT1 or HIF1α with miR-210 on primary tumor sections should address this question.
Interestingly, decreased expression of miR-210 and genomic deletion of miR-210 are frequently observed in ovarian cancer [40, 71], suggesting a potential tumor suppressor function. Indeed, miR-210 represses tumor cell growth initiation when ectopically expressed in human pancreatic or head and neck cancer cell lines [17]. Inhibition of miR-210 also elevates caspase activity [18, 72], although no obvious cell growth inhibition is observed in one of the reports [72]. Based on these studies, we propose that miR-210 acts as a homeostasis rheostat and that disturbing the balance of cellular miR-210 levels is detrimental to cell survival. Clearly, miR-210 is a gene with multifaceted roles in tumorigenesis and future careful dissection of miR-210’s function in different genetic backgrounds and in different human tumor types is warranted.
Circulating miR-210 as a novel cancer biomarker
Because of the close correlation between miR-210 expression and tumor hypoxia, circulating miR-210 might be a prognostic marker in cancer patients. MiR-210 levels are higher in serum from patients with diffuse large B-cell lymphoma compared to those of healthy controls [73], providing an exciting proof-of-principle that circulating miR-210 could be used as a cancer biomarker. More recently, increased levels of miR-210 have also been detected in the plasma of pancreatic cancer patients compared to healthy controls [74, 75]. Because pancreatic tumors are highly hypoxic [76], these reports suggest that circulating miR-210 could be used as a novel biomarker for the non-invasive detection of tumor hypoxia.
Future challenges
Although much progress has been made since the first report of hypoxic regulation of miRNA three years ago [18], tremendous challenges still lie ahead (Box 1). One challenge common to the entire miRNA research field is the accurate identification of miRNA targets, because ultimately miRNA exerts its biological function through regulating cellular protein levels. Computational prediction of miRNA targets is based on the complementarity of target gene 3′ UTR sequence to the “seed region” sequence of a miRNA [5]. Many online databases are readily available at no cost. However, since the miRNA “seed region” only consists of 6–7 nucleotides, false positive prediction is a major limit of this approach. Argonaute protein immunoprecipitation (miRNP-IP) is an efficient way to identify miR-210 targets [17, 27]. With the lowering cost of next-generation sequencing (NGS), the combination of miRNP-IP and NGS will provide more complete and accurate identification of miR-210 targets in the near future. Because miRNA-IP captures miRNA exerting its function in action and it is highly feasible as a routine laboratory operation, miRNA-IP could become the main path to experimentally identify miRNA targets. Proteomic profiling has also been utilized to identify miRNA targets [77], including miR-210 [27]. However, the requirement for sophisticated and expensive equipment limits a broad application of this promising technology. Microarray profiling technology is mature and widely used in identifying miRNA targets [5]. However, because miRNA frequently regulates its target gene by inhibiting protein translation but not by changing its mRNA expression, relying exclusively on microarray profiling might miss some authentic miRNA targets. Ideally, when proteomics, miRNP-IP and NGS, and computational prediction approaches are utilized in different cell types and experimental conditions, we will have a comprehensive picture of miR-210 targets, which will facilitate our efforts to fully understand the functional role of miR-210 in hypoxia or other pathways, and provide potential targets for cancer detection and treatment (Figure 2).
Box 1. Outstanding questions.
How is miR-210 transcribed and processed?
What are the other transcriptional factors regulating miR-210 expression, especially in the context of stem cell and cellular metabolism?
How does miR-210 repress tumor growth initiation, the mechanism?
What is the role miR-210 plays in maintenance of an established tumor?
What is the role miR-210 plays in other diseases where hypoxia plays important role (i.e. preeclampsia)?
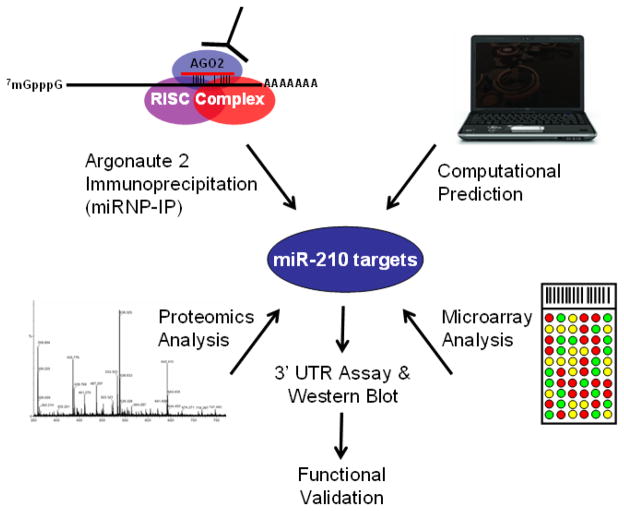
Figure 2.
Strategies for identifying miR-210 targets. Schematic view of four major strategies utilized to identify miRNA targets. By immunoprecipitating a major component of the RISC complex, Argonaute 2 protein (AGO2), miRNP-IP pulls down the mRNA targets associated with miRNA within RISC. The mRNA targets are identified by next-generation sequencing or microarray. Because miRNA regulates target gene expression through the inhibition of RNA translation or degradation of target mRNA, two strategies can be used to identify miRNA targets, proteomics analysis or mRNA microarray profiling. When miRNA targets identified through experimental approaches are combined with computational prediction, which is mainly based on the homology of the short miRNA “seed region” to the 3′ UTR sequence of target gene, a list of candidate miRNA target genes are generated. 3′ UTR sequences of the putative targets are cloned into a reporter construct and a luciferase reporter assay is then performed to measure inhibition of protein production. Western blotting is also performed to verify decreased protein levels. The verified miRNA target gene is then subject to appropriate functional assays for validation.
Loss-of-function by RNA interference is a powerful approach to study gene function, which also applies to elucidating miR-210 function Transient inhibition of miRNA expression has been achieved using locked nucleic acids (LNA) [78]; stable suppression has been achieved with a miRNA sponge [79]. However, due to the lack of standards for designing the sponge sequences and the difficulty in maintaining homogeneously high expression levels of the sponge, this method has not been widely adopted by the miRNA research community when compared to short hairpin RNA (shRNA) technology. The relative number of published reports utilizing each of these approaches reflects this preference. To dissect miR-210’s function in tumor cells, which are usually under a hypoxic microenvironment in vivo, inducible stable suppression of miR-210 is desirable. After establishment of tumor mass in a xenograft model, knockdown of miR-210 in this setting could specifically address what role miR-210 plays in adaptation and survival of tumor cells under hypoxia, mechanistically addressing whether elevated miR-210 expression contributes to poor survival in cancer patients. Furthermore, a knockout mouse model will be also be valuable in answering questions concerning miR-210’s function during embryonic development, given the critical role hypoxia is known to play [80]. If this mouse model is crossed with other mouse models for cancer or other diseases, several fundamental questions, such as miR-210’s role in tumorigenesis and progression can be addressed in relatively physiological conditions.
In summary, miR-210 plays a critical role in the cellular response to hypoxia (Figure 3). Studies have thus far focused on understanding miR-210 in specific pathways. A systems biology approach will help clarify the net cellular effect of miR-210 expression. By combining the transcriptome, proteome, metabolome and phenotypic changes that result from the manipulation of miR-210 expression levels in cell or mouse models, we will then have a comprehensive understanding of miR-210 function. Furthermore, miR-210 has been implicated in tumor initiation and expansion, so it could be a useful biomarker for tumor hypoxia. However, manipulating miR-210 expression alters cellular homeostasis, and can result in either tumor growth inhibition or tumor cell apoptosis, depending on the cellular context. Therefore, a challenge for the future is to better define the contextual effects of miR-210 inhibition in normal tissues and tumors so that the potential of therapeutically targeting miR-210 can be explored.
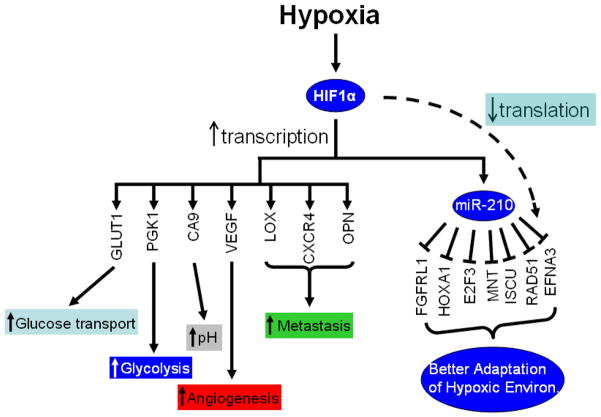
Figure 3.
Genes regulated by HIF1α. HIF1α regulates a broad spectrum of genes under hypoxia, including genes involved in glucose transport (glucose transport 1 (GLUT1)), glycolysis (phosphoglycerate kinase 1 (PGK1)), angiogenesis (VEGF), tumor metastasis (lysyl oxidase (LOX), chemokine (C-X-C motif) receptor 4 (CXCR4), osteopontin (OPN)), and pH control (CA9). Recently, miR-210 was identified as a HIF1α target under hypoxia. MiR-210 suppresses the expression of a number of genes involved in a variety of cellular functions. By up-regulating miR-210 expression under hypoxia, HIF1α can indirectly inhibit the expression of a subset of genes so cells can adjust and adapt to the hypoxic environment.
Acknowledgments
This work is supported by NIH grants P01-CA67166 (QTL, AJG), R01-CA118582 (QTL) and funding from Magee-Womens Research Institute (XH).
Glossary
- Locked nucleic acid (LNA)
LNA is a modified nucleotide with an extra bridge connecting the 2′ oxygen and 4′ carbon on the ribose moiety. The locked ribose confirmation significantly increases the thermal stability of oligonucleotide where LNA is incorporated. Because of the short target sequence (~22 nt) of mature miRNA, LNA nucleotide is usually incorporated into the antisense RNA oligonucleotide for efficient miRNA knockdown
- MiRNA sponge
a competitive miRNA inhibitor composed of multiple, tandem miRNA binding sites transcribed from a strong promoter. By competing with endogenous miRNA targets binding to miRNA, it is designed to saturate cellular miRNA and therefore, relieve endogenous miRNA targets from repression
- Ischemic preconditioning (IP)
an experimental practice of repeatedly exposing myocardial cells to short-term ischemia, usually achieved by occluding coronary artery in an animal, followed by reperfusion in order to protect the cells from later prolonged ischemia. Tissues become resistant to the loss of blood supply and, thus oxygen, after IP treatment
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Lee Y, et al. MicroRNA maturation: stepwise processing and subcellular localization. EMBO J. 2002;21(17):4663–4670. doi: 10.1093/emboj/cdf476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lee Y, et al. The nuclear RNase III Drosha initiates microRNA processing. Nature. 2003;425(6956):415–419. doi: 10.1038/nature01957. [DOI] [PubMed] [Google Scholar]
- 3.Kim VN. MicroRNA biogenesis: coordinated cropping and dicing. Nat Rev Mol Cell Biol. 2005;6(5):376–385. doi: 10.1038/nrm1644. [DOI] [PubMed] [Google Scholar]
- 4.Gregory RI, et al. Human RISC Couples MicroRNA Biogenesis and Posttranscriptional Gene Silencing. Cell. 2005;123(4):631–640. doi: 10.1016/j.cell.2005.10.022. [DOI] [PubMed] [Google Scholar]
- 5.Bartel DP. MicroRNAs: Target Recognition and Regulatory Functions. Cell. 2009;136(2):215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75(5):843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- 7.Wightman B, Ha I, Ruvkun G. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell. 1993;75(5):855–862. doi: 10.1016/0092-8674(93)90530-4. [DOI] [PubMed] [Google Scholar]
- 8.Reinhart BJ, et al. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature. 2000;403(6772):901–906. doi: 10.1038/35002607. [DOI] [PubMed] [Google Scholar]
- 9.Pasquinelli AE, et al. Conservation of the sequence and temporal expression of let-7 heterochronic regulatory RNA. Nature. 2000;408(6808):86–89. doi: 10.1038/35040556. [DOI] [PubMed] [Google Scholar]
- 10.Bushati N, Cohen SM. microRNA Functions. Annual Review of Cell and Developmental Biology. 2007;23(1):175–205. doi: 10.1146/annurev.cellbio.23.090506.123406. [DOI] [PubMed] [Google Scholar]
- 11.Ventura A, Jacks T. MicroRNAs and Cancer: Short RNAs Go a Long Way. Cell. 2009;136(4):586–591. doi: 10.1016/j.cell.2009.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kaelin WG, Ratcliffe PJ. Oxygen sensing by metazoans: The central role of the HIF hydroxylase pathway. Molecular Cell. 2008;30(4):393–402. doi: 10.1016/j.molcel.2008.04.009. [DOI] [PubMed] [Google Scholar]
- 13.Taguchi A, et al. Identification of Hypoxia-Inducible Factor-1a as a Novel Target for miR-17–92 MicroRNA Cluster. Cancer Res. 2008;68(14):5540–5545. doi: 10.1158/0008-5472.CAN-07-6460. [DOI] [PubMed] [Google Scholar]
- 14.Rane S, et al. Downregulation of MiR-199a Derepresses Hypoxia-Inducible Factor-1a and Sirtuin 1 and Recapitulates Hypoxia Preconditioning in Cardiac Myocytes. Circ Res. 2009;104(7):879–886. doi: 10.1161/CIRCRESAHA.108.193102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lei Z, et al. Regulation of HIF-1a and VEGF by miR-20b Tunes Tumor Cells to Adapt to the Alteration of Oxygen Concentration. PLoS ONE. 2009;4(10):e7629. doi: 10.1371/journal.pone.0007629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Crosby ME, et al. MicroRNA Regulation of DNA Repair Gene Expression in Hypoxic Stress. Cancer Res. 2009;69(3):1221–1229. doi: 10.1158/0008-5472.CAN-08-2516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huang X, et al. Hypoxia-Inducible mir-210 Regulates Normoxic Gene Expression Involved in Tumor Initiation. Molecular Cell. 2009;35(6):856–867. doi: 10.1016/j.molcel.2009.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kulshreshtha R, et al. A MicroRNA Signature of Hypoxia. Mol Cell Biol. 2007;27(5):1859–1867. doi: 10.1128/MCB.01395-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Saini HK, Griffiths-Jones S, Enright AJ. Genomic analysis of human microRNA transcripts. Proceedings of the National Academy of Sciences. 2007;104(45):17719–17724. doi: 10.1073/pnas.0703890104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang Z, et al. MicroRNA miR-210 modulates cellular response to hypoxia through the MYC antagonist MNT. Cell Cycle. 2009;8(17):2756–2768. doi: 10.4161/cc.8.17.9387. [DOI] [PubMed] [Google Scholar]
- 21.Camps C, et al. hsa-miR-210 is induced by hypoxia and is an independent prognostic factor in breast cancer. Clinical Cancer Research. 2008;14(5):1340–1348. doi: 10.1158/1078-0432.CCR-07-1755. [DOI] [PubMed] [Google Scholar]
- 22.Krieg AJ, et al. Regulation of the Histone Demethylase JMJD1A by Hypoxia-Inducible Factor 1{alpha} Enhances Hypoxic Gene Expression and Tumor Growth. Mol Cell Biol. 2010;30(1):344–353. doi: 10.1128/MCB.00444-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yang J, et al. Role of Hypoxia-Inducible Factors in Epigenetic Regulation via Histone Demethylases. Annals of the New York Academy of Sciences. 2009;1177:185–197. doi: 10.1111/j.1749-6632.2009.05027.x. (Hypoxia and Consequences From Molecule to Malady) [DOI] [PubMed] [Google Scholar]
- 24.Kim HW, et al. Ischemic Preconditioning Augments Survival of Stem Cells via miR-210 Expression by Targeting Caspase-8-associated Protein 2. Journal of Biological Chemistry. 2009;284(48):33161–33168. doi: 10.1074/jbc.M109.020925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fasanaro P, et al. MicroRNA-210 Modulates Endothelial Cell Response to Hypoxia and Inhibits the Receptor Tyrosine Kinase Ligand Ephrin-A3. Journal of Biological Chemistry. 2008;283(23):15878–15883. doi: 10.1074/jbc.M800731200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu Y, et al. Hypoxia Regulates Vascular Endothelial Growth Factor Gene Expression in Endothelial Cells : Identification of a 5′ Enhancer. Circ Res. 1995;77(3):638–643. doi: 10.1161/01.res.77.3.638. [DOI] [PubMed] [Google Scholar]
- 27.Fasanaro P, et al. An Integrated Approach for Experimental Target Identification of Hypoxia-induced miR-210. Journal of Biological Chemistry. 2009;284(50):35134–35143. doi: 10.1074/jbc.M109.052779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Greither T, et al. Elevated expression of microRNAs 155, 203, 210 and 222 in pancreatic tumors is associated with poorer survival. International Journal of Cancer. 2009;126(1):73–80. doi: 10.1002/ijc.24687. [DOI] [PubMed] [Google Scholar]
- 29.Foekens JA, et al. Four miRNAs associated with aggressiveness of lymph node-negative, estrogen receptor-positive human breast cancer. Proceedings of the National Academy of Sciences. 2008;105(35):13021–13026. doi: 10.1073/pnas.0803304105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Carmeliet P, Jain RK. Angiogenesis in cancer and other diseases. Nature. 2000;407(6801):249–257. doi: 10.1038/35025220. [DOI] [PubMed] [Google Scholar]
- 31.Kloner RA, Jennings RB. Consequences of Brief Ischemia: Stunning, Preconditioning, and Their Clinical Implications: Part 1. Circulation. 2001;104(24):2981–2989. doi: 10.1161/hc4801.100038. [DOI] [PubMed] [Google Scholar]
- 32.Cai Z, et al. Complete loss of ischaemic preconditioning-induced cardioprotection in mice with partial deficiency of HIF-1a. Cardiovasc Res. 2008;77(3):463–470. doi: 10.1093/cvr/cvm035. [DOI] [PubMed] [Google Scholar]
- 33.Imai Y, et al. The CED-4-homologous protein FLASH is involved in Fas-mediated activation of caspase-8 during apoptosis. Nature. 1999;398(6730):777–785. doi: 10.1038/19709. [DOI] [PubMed] [Google Scholar]
- 34.Keith B, Simon MC. Hypoxia-Inducible Factors. Stem Cells, and Cancer. Cell. 2007;129(3):465–472. doi: 10.1016/j.cell.2007.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yoshida Y, et al. Hypoxia Enhances the Generation of Induced Pluripotent Stem Cells. Cell Stem Cell. 2009;5(3):237–241. doi: 10.1016/j.stem.2009.08.001. [DOI] [PubMed] [Google Scholar]
- 36.Mizuno Y, et al. miR-210 promotes osteoblastic differentiation through inhibition of AcvR1b. FEBS Letters. 2009;583(13):2263–2268. doi: 10.1016/j.febslet.2009.06.006. [DOI] [PubMed] [Google Scholar]
- 37.Hirao M, et al. Oxygen tension is an important mediator of the transformation of osteoblasts to osteocytes. Journal of Bone and Mineral Metabolism. 2007;25(5):266–276. doi: 10.1007/s00774-007-0765-9. [DOI] [PubMed] [Google Scholar]
- 38.Maegdefrau U, et al. Bone morphogenetic protein 4 is induced in hepatocellular carcinoma by hypoxia and promotes tumour progression. The Journal of Pathology. 2009;218(4):520–529. doi: 10.1002/path.2563. [DOI] [PubMed] [Google Scholar]
- 39.Goda N, et al. Hypoxia-Inducible Factor 1a Is Essential for Cell Cycle Arrest during Hypoxia. Mol Cell Biol. 2003;23(1):359–369. doi: 10.1128/MCB.23.1.359-369.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Giannakakis A, et al. miR-210 links hypoxia with cell cycle regulation and is deleted in human epithelial ovarian cancer. Cancer Biol Ther. 2007;7(2):255–264. doi: 10.4161/cbt.7.2.5297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.He Y, et al. Identification of E2F-3B, an alternative form of E2F-3 lacking a conserved N-terminal region. Oncogene. 2000;19(30):3422–3433. doi: 10.1038/sj.onc.1203682. [DOI] [PubMed] [Google Scholar]
- 42.Chong JL, et al. E2f3a and E2f3b Contribute to the Control of Cell Proliferation and Mouse Development. Mol Cell Biol. 2009;29(2):414–424. doi: 10.1128/MCB.01161-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hurlin PJ, Queva C, Eisenman RN. Mnt, a novel Max-interacting protein is coexpressed with Myc in proliferating cells and mediates repression at Myc binding sites. Genes & Development. 1997;11(1):44–58. doi: 10.1101/gad.11.1.44. [DOI] [PubMed] [Google Scholar]
- 44.Meroni G, et al. Rox, a novel bHLHZip protein expressed in quiescent cells that heterodimerizes with Max, binds a non-canonical E box and acts as a transcriptional repressor. EMBO J. 1997;16(10):2892–2906. doi: 10.1093/emboj/16.10.2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Leone G, et al. Myc Requires Distinct E2F Activities to Induce S Phase and Apoptosis. Molecular Cell. 2001;8(1):105–113. doi: 10.1016/s1097-2765(01)00275-1. [DOI] [PubMed] [Google Scholar]
- 46.Gordan JD, Thompson CB, Simon MC. HIF and c-Myc: Sibling Rivals for Control of Cancer Cell Metabolism and Proliferation. Cancer Cell. 2007;12(2):108–113. doi: 10.1016/j.ccr.2007.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Takahashi K, Yamanaka S. Induction of Pluripotent Stem Cells from Mouse Embryonic and Adult Fibroblast Cultures by Defined Factors. Cell. 2006;126(4):663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 48.Nakagawa M, et al. Generation of induced pluripotent stem cells without Myc from mouse and human fibroblasts. Nat Biotech. 2008;26(1):101–106. doi: 10.1038/nbt1374. [DOI] [PubMed] [Google Scholar]
- 49.Bindra R, Crosby M, Glazer P. Regulation of DNA repair in hypoxic cancer cells. Cancer and Metastasis Reviews. 2007;26(2):249–260. doi: 10.1007/s10555-007-9061-3. [DOI] [PubMed] [Google Scholar]
- 50.Mihaylova VT, et al. Decreased Expression of the DNA Mismatch Repair Gene Mlh1 under Hypoxic Stress in Mammalian Cells. Mol Cell Biol. 2003;23(9):3265–3273. doi: 10.1128/MCB.23.9.3265-3273.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Koshiji M, et al. HIF-1a Induces Genetic Instability by Transcriptionally Downregulating MutSa Expression. Molecular Cell. 2005;17(6):793–803. doi: 10.1016/j.molcel.2005.02.015. [DOI] [PubMed] [Google Scholar]
- 52.Bindra RS, Glazer PM. Repression of RAD51 gene expression by E2F4//p130 complexes in hypoxia. Oncogene. 2006;26(14):2048–2057. doi: 10.1038/sj.onc.1210001. [DOI] [PubMed] [Google Scholar]
- 53.Bindra RS, et al. Down-Regulation of Rad51 and Decreased Homologous Recombination in Hypoxic Cancer Cells. Mol Cell Biol. 2004;24(19):8504–8518. doi: 10.1128/MCB.24.19.8504-8518.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Semenza GL. Oxygen-dependent regulation of mitochondrial respiration by hypoxia-inducible factor 1. Biochem J. 2007;405(1):1–9. doi: 10.1042/BJ20070389. [DOI] [PubMed] [Google Scholar]
- 55.Chan SY, et al. MicroRNA-210 Controls Mitochondrial Metabolism during Hypoxia by Repressing the Iron-Sulfur Cluster Assembly Proteins ISCU1/2. Cell Metabolism. 2009;10(4):273–284. doi: 10.1016/j.cmet.2009.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tong WH, Rouault TA. Functions of mitochondrial ISCU and cytosolic ISCU in mammalian iron-sulfur cluster biogenesis and iron homeostasis. Cell Metabolism. 2006;3(3):199–210. doi: 10.1016/j.cmet.2006.02.003. [DOI] [PubMed] [Google Scholar]
- 57.Warburg O. On the Origin of Cancer Cells. Science. 1956;123(3191):309–314. doi: 10.1126/science.123.3191.309. [DOI] [PubMed] [Google Scholar]
- 58.Kim J-w, Gao P, Dang C. Effects of hypoxia on tumor metabolism. Cancer and Metastasis Reviews. 2007;26(2):291–298. doi: 10.1007/s10555-007-9060-4. [DOI] [PubMed] [Google Scholar]
- 59.Malzkorn B, et al. Identification and Functional Characterization of microRNAs Involved in the Malignant Progression of Gliomas. Brain Pathology. 2009;9999(9999) doi: 10.1111/j.1750-3639.2009.00328.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Satzger I, et al. MicroRNA-15b represents an independent prognostic parameter and is correlated with tumor cell proliferation and apoptosis in malignant melanoma. International Journal of Cancer. 2009;126(11):2553–2562. doi: 10.1002/ijc.24960. [DOI] [PubMed] [Google Scholar]
- 61.Juan D, et al. Identification of a MicroRNA Panel for Clear-cell Kidney Cancer. Urology. 2010 doi: 10.1016/j.urology.2009.10.033. [DOI] [PubMed] [Google Scholar]
- 62.Cho WCS, Chow ASC, Au JSK. Restoration of tumour suppressor hsa-miR-145 inhibits cancer cell growth in lung adenocarcinoma patients with epidermal growth factor receptor mutation. European Journal of Cancer. 2009;45(12):2197–2206. doi: 10.1016/j.ejca.2009.04.039. [DOI] [PubMed] [Google Scholar]
- 63.Miko E, et al. Differentially expressed microRNAs in small cell lung cancer. Exp Lung Res. 2009;35(8):646–664. doi: 10.3109/01902140902822312. [DOI] [PubMed] [Google Scholar]
- 64.Raponi M, et al. MicroRNA Classifiers for Predicting Prognosis of Squamous Cell Lung Cancer. Cancer Res. 2009;69(14):5776–5783. doi: 10.1158/0008-5472.CAN-09-0587. [DOI] [PubMed] [Google Scholar]
- 65.Volinia S, et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proceedings of the National Academy of Sciences of the United States of America. 2006;103(7):2257–2261. doi: 10.1073/pnas.0510565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yanaihara N, et al. Unique microRNA molecular profiles in lung cancer diagnosis and prognosis. Cancer Cell. 2006;9(3):189–198. doi: 10.1016/j.ccr.2006.01.025. [DOI] [PubMed] [Google Scholar]
- 67.Zhu X-m, et al. Differential expression profile of microRNAs in human placentas from preeclamptic pregnancies vs normal pregnancies. American Journal of Obstetrics and Gynecology. 2009;200(6):661.e1–661.e7. doi: 10.1016/j.ajog.2008.12.045. [DOI] [PubMed] [Google Scholar]
- 68.Pineles BL, et al. Distinct subsets of microRNAs are expressed differentially in the human placentas of patients with preeclampsia. American Journal of Obstetrics and Gynecology. 2007;196(3):261.e1–261.e6. doi: 10.1016/j.ajog.2007.01.008. [DOI] [PubMed] [Google Scholar]
- 69.Sullivan R, Graham C. Hypoxia-driven selection of the metastatic phenotype. Cancer and Metastasis Reviews. 2007;26(2):319–331. doi: 10.1007/s10555-007-9062-2. [DOI] [PubMed] [Google Scholar]
- 70.Gee HE, et al. MicroRNA-10b and breast cancer metastasis. Nature. 2008;455(7216):E8–E9. doi: 10.1038/nature07362. [DOI] [PubMed] [Google Scholar]
- 71.Wyman SK, et al. Repertoire of microRNAs in Epithelial Ovarian Cancer as Determined by Next Generation Sequencing of Small RNA cDNA Libraries. PLoS ONE. 2009;4(4):e5311. doi: 10.1371/journal.pone.0005311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Cheng AM, et al. Antisense inhibition of human miRNAs and indications for an involvement of miRNA in cell growth and apoptosis. Nucl Acids Res. 2005;33(4):1290–1297. doi: 10.1093/nar/gki200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lawrie CH, et al. Detection of elevated levels of tumour-associated microRNAs in serum of patients with diffuse large B-cell lymphoma. British Journal of Haematology. 2008;141(5):672–675. doi: 10.1111/j.1365-2141.2008.07077.x. [DOI] [PubMed] [Google Scholar]
- 74.Wang J, et al. MicroRNAs in Plasma of Pancreatic Ductal Adenocarcinoma Patients as Novel Blood-Based Biomarkers of Disease. Cancer Prev Res. 2009;2(9):807–813. doi: 10.1158/1940-6207.CAPR-09-0094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ho AS, et al. Circulating miR-210 is a novel hypoxia marker in pancreatic cancer. Translational Oncology. 2010;3(2):109–113. doi: 10.1593/tlo.09256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Koong AC, et al. Pancreatic tumors show high levels of hypoxia. Int J Radiation Oncology Biol Phys. 2000;48(4):919–922. doi: 10.1016/s0360-3016(00)00803-8. [DOI] [PubMed] [Google Scholar]
- 77.Selbach M, et al. Widespread changes in protein synthesis induced by microRNAs. Nature. 2008;455(7209):58–63. doi: 10.1038/nature07228. [DOI] [PubMed] [Google Scholar]
- 78.Vester B, Wengel J. LNA (Locked Nucleic Acid): High-Affinity Targeting of Complementary RNA and DNA. Biochemistry. 2004;43(42):13233–13241. doi: 10.1021/bi0485732. [DOI] [PubMed] [Google Scholar]
- 79.Ebert MS, Neilson JR, Sharp PA. MicroRNA sponges: competitive inhibitors of small RNAs in mammalian cells. Nat Meth. 2007;4(9):721–726. doi: 10.1038/nmeth1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Dunwoodie SL. The Role of Hypoxia in Development of the Mammalian Embryo. Developmental Cell. 2009;17(6):755–773. doi: 10.1016/j.devcel.2009.11.008. [DOI] [PubMed] [Google Scholar]