Abstract
Malignant peripheral nerve sheath tumors (MPNSTs) are highly aggressive soft tissue sarcomas accounting for 3%–10% of all soft tissue sarcomas. Neurofibromatosis type 1 (NF1) is the most important known risk factor. MPNSTs are often diagnosed at an advanced stage when distant metastases have developed. Although surgical resection remains the main treatment for MPNSTs, complete surgical resection is rarely possible. The prognosis for patients with MPNSTs is poor. There is an urgent need for improved therapies. To this end, we investigated whether microRNA (miR), specifically miR-204, might be implicated in MPNSTs because it is located at a cancer-associated genomic region exhibiting high frequency of loss of heterozygosity in tumors. We show that miR-204 expression is downregulated in NF1 and non-NF1 MPNST tumor tissues and in tumor cell lines. Restoring miR-204 expression in MPNST cell lines STS26T (non-NF1), ST88-14 (NF1), and T265p21 (NF1) significantly reduces cellular proliferation, migration, and invasion in vitro. Restoring miR-204 expression in STS26T decreases tumor growth and malignant progression in vivo. We also report that miR-204 inhibits Ras signaling and expression of high mobility group gene A2. These findings support the hypothesis that miR-204 plays critical roles in MPNST development and tumor progression. miR-204 may represent a novel biomarker for diagnosis and a candidate target with which to develop effective therapies for MPNSTs.
Keywords: HMGA2, miR-204, MPNSTs, NF1, RAS
Malignant peripheral nerve sheath tumors (MPNSTs) are highly aggressive soft tissue sarcomas accounting for 3%–10% of all soft tissue sarcomas. MPNSTs arise from peripheral nerve branches or sheaths of peripheral nerve fibers. The tumor typically arises from cells constituting the nerve sheath, such as Schwann cells or perineural cells.1 Neurofibromatosis type 1 (NF1) is the most important known risk factor, because approximately50%–60% of MPNSTs occur in patients with NF1. The incidence of MPNSTs among patients with NF1 is approximately8%–13%.2 The relative risk of MPNSTs in patients with NF1 is 113 times more than that of a control population3; MPNSTs are the leading cause of NF1-related mortality. The second risk factor for MPNSTs is radiation exposure.4 Approximately 10% of MPNSTs are associated with prior radiation.5
Because of its deep anatomic location, MPNSTs are often diagnosed at an advanced stage when primary tumor diameter may exceed 5 cm and metastatic progression has already occurred.6 Metastases are commonly found in the lungs and less frequently in the liver and brain.2,6 Surgical resection remains the main treatment for MPNSTs. However, because of MPNST invasive growth and propensity to metastasize, complete surgical resection is technically rarely possible. Advanced nonresectable and metastatic MPNSTs rarely respond to chemotherapy. Although radiotherapy may provide local control and delay onset of recurrence, it has little effect on long-term survival rate.7 The prognosis for patients with MPNSTs is generally poor,6 with 5-year survival rates of 20%–50%2 and 10-year disease-specific survival of 7.5%.8 There is an urgent need for new therapeutic strategies.
Approximately half of MPNSTs occur in the context of NF1, whereas the other half are sporadic in origin. Compared with individuals without NF1, those with MPNSTs concomitant with NF1 tend to be diagnosed at younger ages and to have a poor prognosis.2,9 These facts suggest that NF1 and deregulation of its signaling pathway likely play important roles in carcinogenesis of MPNSTs. The NF1 protein, neurofibromin, contains a highly conserved guanosine triphosphatase (GTPase)–activating protein-related domain (GRD) that converts active Ras-GTP into inactive Ras-GDP in the Ras signaling pathway.10,11 Loss of NF1 is considered a tumor-promoting event, leading to constitutively increased activation of Ras and its downstream effectors, thus initiating neurofibromas.12 However, loss of neurofibromin alone is not sufficient for malignant transformation, implying that additional genetic alterations are necessary for the development of MPNSTs.13 Mutations of the TP53 locus were reported in up to 75% of MPNSTs.14 Mice that harbor both Nf1 and Tp53 mutations develop MPNSTs.15Associated with development of MPNSTs are homozygous deletions of the CDKN2A gene, which encodes both p16INK4A and p14ARF in alternative reading frames, and inactivation of CDKN1B, a gene that encodes the p27 (Kip1) cell-cycle regulator.16 Also believed to contribute to MPNST pathogenesis are overexpression of epidermal growth factor receptor (EGFR),17 platelet-derived (PD)GFR–α and PDGFR-β,18 c-Kit,19 matrix metalloproteinase 13 (MMP13),20 and genes involved in cell proliferation (MKI67, TOP2A, CCNE2), apoptosis (BIRC5/Survivin, TP73), and the signaling pathways of Ras (RASF2, HMMR/RHAMM) and Hedgehog-Gli (DHH, Ptch2).21 Despite these findings, there is insufficient knowledge of critical pathways initiating and promoting progression of MPNST, limiting the ability to develop effective treatments.
MicroRNAs (miRNAs, miR-) are a new class of small noncoding RNAs of about 19–25 nucleotides (nt) that function as negative posttranscriptional gene regulators.22 miRNAs hybridize to the 3′ untranslated region (UTR) of target mRNAs and repress translation or mediate mRNA cleavage. miRNAs critically regulate tumorigenesis and progression by targeting oncogenes, tumor suppressor genes, or genes related to proliferation, angiogenesis, and apoptosis.23 Different tumor types and tumors at various differentiation stages exhibit unique miRNA profiles. miRNAs show promise as potential biomarkers for cancer diagnostics, progression, and response to treatment.24,25 In considering a role for miRNAs in MPNSTs, Subramanian et al.26 reported that miR-34a was downregulated in MPNSTs, compared with neurofibromas. They concluded that p53 inactivation and subsequent loss of expression of miR-34a may contribute to MPNST development.26 We reported that miR-10b was upregulated in primary Schwann cells isolated from NF1 neurofibromas and in cell lines and tumor tissues from NF1 MPNSTs, but not in cell lines and tumor tissues from non-NF1 MPNSTs.27 Our prior study indicated that miR-10b may be an NF1-specific regulating factor that targets neurofibromin and Ras signaling to regulate NF1 tumorigenesis.27 Except for these 2 miRNAs, pathological roles of other miRNAs in MPNSTs are largely unexplored. One candidate miRNA of interest, miR-204, is located at the cancer-associated genomic region 9q21.1–q22.3 locus and exhibits a high frequency of loss of heterozygosity in tumors.28–30 miR-204 is also significantly downregulated in broad types of tumors such as breast cancer, prostate cancer, and kidney cancer,31 suggesting a role for miR-204 as a tumor suppressor gene. In the present study, our aims were to determine whether miR-204 might play a role in carcinogenesis in MPNSTs.
Materials and Methods
Human tissues were obtained under human subject protocols approved by the institutional review boards at Maine Institute for Human Genetics and Health and West China Hospital, Sichuan University. Informed consent was obtained from each subject or the subject's guardian. Animal use was in accordance with the guidelines of the Institutional Animal Care and Use Committee at the Jackson Laboratory and West China Hospital, Sichuan University.
Cell Culture
Human NF1-associated MPNST cell lines ST8814 and T265p21 and the non-NF1-associated MPNST cell line STS26T (kindly supplied by Dr. Nancy Ratner) were maintained in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum (FBS). NF1 mutation status for these 3 cell lines has been reported previously by other groups.32,33
Analysis of miRNA Expression by Microarray and qRT-PCR
RNA was extracted from cells or tumor tissues using the RNeasy miRNA kit (Qiagen). miRNA microarrays were performed by LC Sciences. Quantitative (q) real-time and reverse transcriptase (RT) PCR were performed in a 2-step reaction using Taqman miRNA assays (Applied Biosystems), according to the manufacturer's protocols. U6 was used as the internal control. The 2–ΔΔCT method, described by Livak and Schmittgen,34 was used to analyze the data.
Stable Overexpression of miR-204 in MPNST Cell Lines
In order to obtain stable cell lines with high miR-204 expression, miR-204 precursor genes were transfected into MPNST cell lines by a lentiviral expression system. Lentivirus was prepared using the LentiSuite for Lenti-miR-204 miRNA Precursor Expression Construct (System Biosciences) according to the manufacturer's protocol. The pGreenPuro Scramble Hairpin Control Construct (System Biosciences) was used as the control. Lentiviral infection was performed according to the manufacturer's protocol. Stably high-expressing cell lines were selected out by single colony screening of cells cultured in 96-well plates. Expression of miR-204 was assayed by qRT-PCR. Infected cells were analyzed by the various assays described below.
MTT Proliferation Assay
Infected cells at 70%–80% confluence were serum-starved for 24 h, then cultured at a density of 2000/well in 96-well plates at 5% CO2, 37°C. At selected time points, MTT (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium) was added at a final concentration of 0.5 mg/mL. After 4 h incubation, medium was removed, and purple blue sediment was dissolved in 150 µL dimethyl sulfoxide. The relative optical density (OD) for each well was determined using a Wellscan MK3 ELISA kit (Labsystems).
Colony Formation
Infected cells were seeded at a density of 200/well in 6-well plates in 2% FBS-supplemented medium. After culturing for 14 days, cells were fixed with 4% paraformaldehyde for 20 min and stained with 0.1% crystal violet for 30 min. Colonies were counted using bright-field microscopy.
Cell Migration and Invasion
Serum-starved cells were trypsinized, centrifuged, and resuspended in 0.1% FBS-supplemented medium without added growth factor. Cells were plated at a density of 2.5 × 104/well for STS26T cells or 5.0 × 104/well for ST8814 and T265p21 cells in a transwell insert (3 µm pore size; BD Biosciences) for the migration assay or in a matrigel-coated, growth-factor-reduced, invasion chamber (8 µm pore size; BD Biosciences) for the invasion assay. Ten percent FBS-containing medium was added into 24-well plates as a chemoattractant. After 6 h incubation for the migration assay, or after 22 h incubation for the invasion assay, cells were fixed with 4% paraformaldehyde for 1 h. Cells on the apical side of each insert were removed by mechanically scraping. Cells that migrated to the basal side of the membrane were stained with 0.1% crystal violet and visualized under a Zeiss Axiovert 200M microscope. Cell numbers were quantified in an automated mode using the Metamorph analysis software.
Western Blotting
Cells were lysed in radio-immunoprecipitation assay buffer containing Halt Protease Inhibitor Cocktail (Pierce). Protein concentration was determined with the Quick Start Bradford Protein Assay kit (Bio-Rad). The blotting membrane was incubated overnight at 4°C with different primary antibodies: anti–p44/42 MAP kinase (1:1000; Cell Signaling Technology), anti–phospho-p44/42 MAPK (Thr202/Tyr204) (1:1000; Cell Signaling), anti–high mobility group (HMG)A2 (1:500; Sigma), and anti–β-actin (1:1000; Sigma). The blots were incubated for 1 h at room temperature with anti-mouse immunoglobulin (Ig)G (Chemicon) as a horseradish peroxidase–conjugated secondary antibody. Signals were visualized using electrogenerated chemiluminescence (CL) plus CL substrate (Amersham).
HMGA2 3′ UTR Reporter Assay
The 3′ UTR of the HMGA2 gene was amplified from human genomic DNA using primers (forward: 5′-tttactagttgctgatgtatcctttcattcagcta-3′; reverse: and 5′-tttgagctcgaagagatggtgaactcaagccgaa-3′) and subcloned into the 3′ UTR of the pMIR-REPORT miRNA reporter vector (Ambion). The recombined plasmid was confirmed by DNA sequencing. Point mutations within the target sequence for miR-204 in the 3′ UTR (HMGA2 3′ UTRm) were generated by the QuickChange II XL site-directed mutagenesis kit (Stratagene) using primers (forward: 5′-caaaaatcgaagtaagcaaacaaatattgcca-3′; reverse: 5′-ttacttcgatttttgaaaaatgaacataaccttttct-3′). The plasmids containing the HMGA2 3′ UTRm were sequenced to confirm replacement of the targeted residues. The lentivirus mentioned above was used to stably transfect the miR-204 precursor gene or control the sequence into 293T cells. Transfected cells were further cotransfected with pMIR reporter vector with the HMGA2 3′ UTR or the HMGA2 3′ UTRm together with the pMIR-REPORT beta-galactosidase reporter control vector. Cells were collected 24 h after transfection. The ratio of beta-galactosidase to firefly luciferase was measured with the Promega Dual Luciferase Assay kit.
Tumor Cell Xenograft
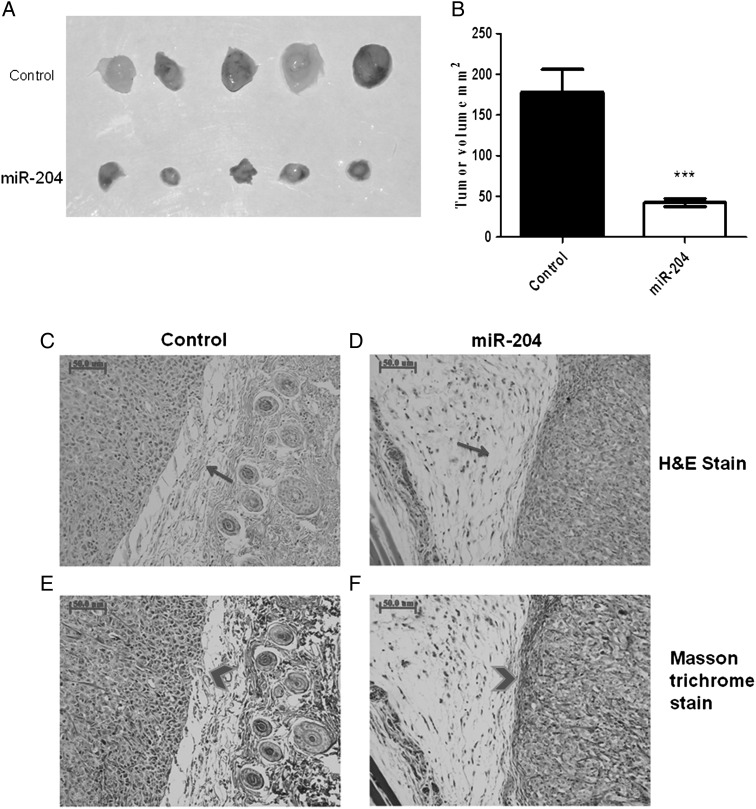
STS26T cells infected with miR-204 virus or control virus were cultured under equivalent conditions. Dissociated cells were collected, rinsed thoroughly, and resuspended in cold 30% matrigel in phosphate buffered saline (BD Biosciences) at a density of 5 × 106/mL. Eight-week old NOD SCID IL-2r gamma −/− (NSG) male mice (10 mice per group) were anesthetized with isoflurane. A cell suspension (1 × 106 cells in 200 μL) was injected subcutaneously into the back of each mouse. Mice were observed daily for the first 3–5 days postoperatively to ensure that the injection site was healthy. Mice were sacrificed on the 14th day after injection. Tumor tissue samples were collected. Tumor volume was calculated by the formula 1/2 ab2, where a is the long axis and b is the short axis. Paraffin-embedded sections of tumor tissues were stained with hematoxylin and eosin (H&E) or Masson's trichrome stains.
Statistical Analyses
Analyses were performed with JMP 8.0 software (SAS). An analysis of variance was used to compare multiple groups, followed by pairwise comparisons if significant differences were detected; a Tukey–Kramer test was used for comparison with a control group, and Dunnett's test was used for comparison of all groups. Unpaired t tests were used to compare 2 groups. Differences were considered statistically significant at P < .05 in a 2-tailed test. Data are expressed as means ± SEM.
Results
Downregulation of miR-204 in Tumor Tissues and Cell Lines from NF1 and non-NF1 MPNSTs
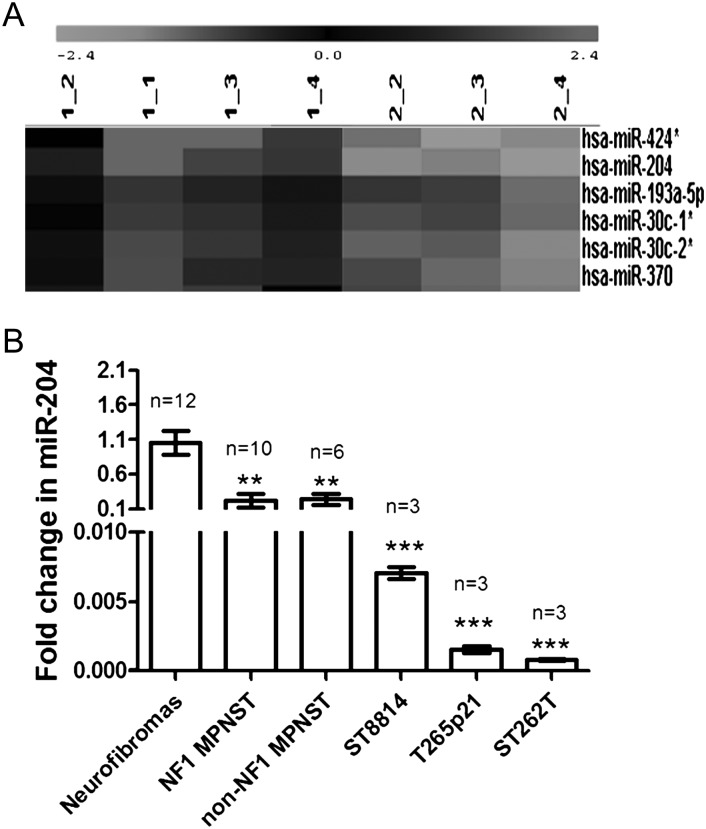
To investigate miRNA expression in tumor tissues, total RNA was isolated from human MPNSTs and benign NF1 neurofibroma tissues; miRNA profiles were studied by miRNA microarray. Twenty-four miRNAs were identified as downregulated, and 6 miRNAs were upregulated in MPNSTs rather than in neurofibromas (Supplementary material, Table S1). miRNAs with a high expression level (density over 300) in either the MPNST group or the neurofibroma group were selected for confirmation by qRT-PCR in tumor tissues and MPNST cell lines. Compared with benign neurofibromas, miR-204 was the only miRNA whose expression was significantly downregulated, while miR-96 was the only miRNA whose expression was significantly upregulated in tumor tissues as well as cell lines from NF1 and non-NF1 MPNSTs (Fig. 1 and Supplementary material, Fig. S1). The consistent deregulation of miR-204 and miR-96 in MPNST tumor tissues and cell lines and in NF1 and non-NF1 genotypes suggests that they may contribute to the carcinogenesis mechanisms in MPNSTs. To further investigate the functional significance of deregulated miRNAs observed in MPNSTs, ST8814 cells were transfected with antisense inhibitors for miR-96, mimic enhancers for miR-204, or with negative controls. Enhancing miR-204 significantly decreased cell proliferation at days 3, 4, and 5 and migration into ST8814 cells, while inhibiting miR-96 expression did not affect proliferation at most time points tested or migration into ST8814 cells (Supplementary material, Fig. S2). These results suggest that miR-204 plays an important role in MPNST tumorigenic mechanisms, while miR-96 may have only a limited role in MPNSTs. Therefore, miR-204 was chosen for further study.
Fig. 1.
Downregulation of miR-204 expression in MPNSTs. Total RNA was isolated from human MPNST tumor tissues or cell lines. (A) Total RNA was labeled and hybridized on miRNA microarrays. Signals represent median signal values of 3 times. miR-204 along with a few other miRNAs showed low expression levels in NF1 and non-NF1 MPNST tumor tissues (1_1, 1_2, 1_3, 1_4: neurofibromas; 2_2, 2_3: NF1 MPNSTs; 2_4: non-NF1 MPNSTs). (B) The expression of miR-204 was further studied by qRT-PCR. miR-204 expression was significantly lower in NF1 and non-NF1 MPNST tumor tissues, in NF1 MPNST cell lines (ST8814 and T265p21), and the non-NF1 MPNST cell line (STS26T) compared with neurofibromas (**P < .01; ***P < .001; n = 3–12).
Restoring miR-204 Expression Reversed Abnormal Cellular Behaviors in NF1 and Non-NF1 MPNST Cell Lines In Vitro
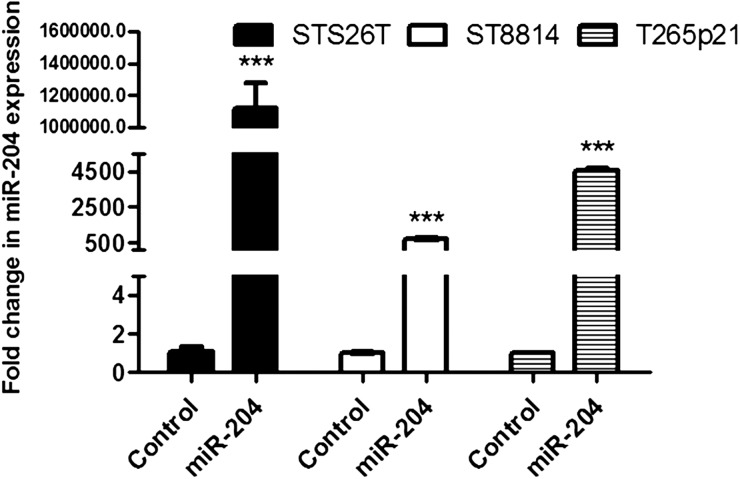
To further investigate the role of miR-204 in MPNST carcinogenesis, the miR-204 gene was stably transfected using a lentiviral system into the NF1 MPNST cell lines, ST8814 and T265p21, and the non-NF1 MPNST cell line, STS26T. This system allows miRNA expression from the constitutive cytomegalovirus promoter. The positive expressing cells can be sorted and monitored as the system contains copepod green fluorescent protein (copGFP) as a reporter. Multiple colonies positive for copGFP in each cell line were selected using fluorescence microscopy. Compared with colonies transfected with the control construct, colonies transfected with miR-204 exhibited significantly higher expression levels of miR-204 (Fig. 2, P < .001).
Fig. 2.
Restoring miR-204 expression in NF1 and non-NF1 MPNST cell lines. miR-204 gene was stably transfected into human NF1-associated MPNST cell lines ST8814 and T265p21 and the non-NF1-associated MPNST cell line STS26T with a lentiviral system. Fluorescence microscopy and qRT-PCR assay were used to select out the colonies with higher expression of miR-204. Assay by qRT-PCR showed that miR-204 expression was significantly higher in ST8814, T265p21, and STS26T cells with exogenous miR-204 gene than in the controls (***compared with controls, P < .001, n = 3).
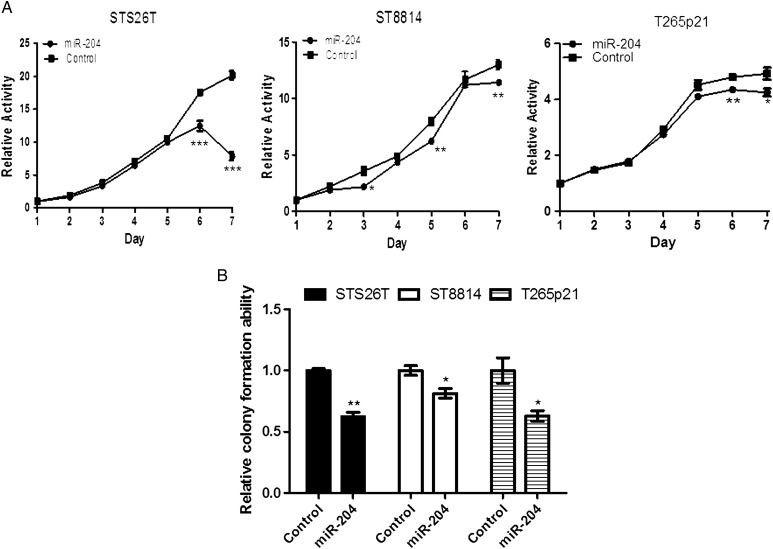
In proliferation assays using MTT activity, restoration of miR-204 expression in STS26T, ST8814, and T265p21 cells significantly reduced cellular proliferation at most time-points during 6–7 days of culture (Fig. 3A). Colonies formed by miR-204 transfected cells were much smaller than those formed by control cells (reduced 38% in STS26T, 37% in T265p21, 27% in ST8814; Fig. 3B, P < .05). The reduction of proliferation was most dramatic in the non-NF1 MPNST cell line STS26T (Fig. 3A and B). These results suggest that miR-204 inhibits cell proliferation in both NF1 and non-NF1 MPNST cell lines.
Fig. 3.
Restoring miR-204 expression inhibited proliferation in NF1 and non-NF1 MPNST cell lines. miR-204 gene was stably transfected into MPNST cells with a lentiviral system. (A) MTT assay showed that restoring miR-204 in MPNST cells inhibited cell proliferation at most time points during days 6–7 (n = 5 at each time point). (B) Overexpressing miR-204 significantly reduced colony formation in NF1 MPNST cell lines T265p21 and ST8814 cells, and in the non-NF1 MPNST cell line STS26T (n = 3) compared with controls, *P < .05; **P < .01; ***P < .001).
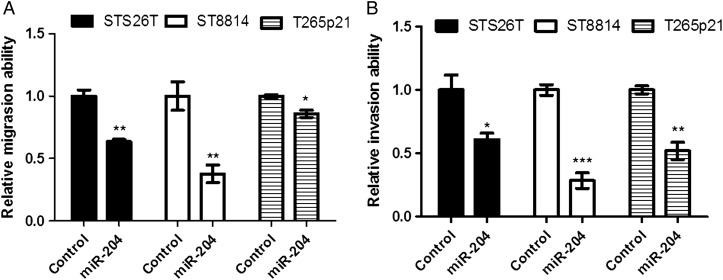
Restoring miR-204 reduced the cellular migration by 26% in STS26T, 62% in ST8814, and 35% in T265p21 (Fig. 4A, P < .05). Restoring miR-204 also reduced cellular invasion by 53% in STS26T, 72% in ST8814, and 48% in T265p21 (Fig. 4B, P < .05). The maximal inhibition of migration and invasion occurred in the NF1 MPNST cell line ST8814. These results support the hypothesis that miR-204 may play an important role in regulation of both NF1 and non-NF1 MPNST development and progression.
Fig. 4.
Restoring miR-204 expression inhibited migration and invasion in NF1 and non-NF1 MPNST cell lines. The miR-204 precursor gene was stably transfected into MPNST cells with a lentiviral system. Cellular migration and invasion were studied with transwell inserts or invasion chambers. (A) Overexpressing miR-204 significantly reduced cellular migration in NF1 MPNST cell lines T265p21 and ST8814 cells and in the non-NF1 MPNST cell line STS26T. (B) Overexpressing miR-204 significantly reduced cellular invasion in NF1 MPNST cell lines T265p21 and ST8814 cells, and in the non-NF1 MPNST cell line STS26T. (Compared with controls, *P < .05; **P < .01; ***P < .001. n = 3).
Restoring miR-204 Expression Inhibited MPNST Tumor Formation In Vivo
Tumor cell xenografts in immunodeficient mice represent standard strategies to model human tumor cell growth in vivo. Prior research reported that ST8814 and T265p21 cell xenografts in immunodeficient mice either failed to form tumors35,36 or took up to 3 months to form tumors.37 Therefore, we selected STS26T cells for xenografts. All mice exhibited tumor growth within 14 days after STS26T cells were implanted. Average tumor volume of the miR-204 group was about a fourth (42.3 mm3 vs 178 mm3, P < .001, Fig. 5A andB) of the control group. Tumor xenografts in control mice were irregular in shape and hard to dissect from surrounding normal tissues. In contrast, most tumors in the miR-204–expressing cell xenografts were close to spherical in shape and easy to dissect. H&E staining demonstrated less cell mitosis and invasion in the miR-204 group compared with controls (Fig. 5C and D). Masson trichrome staining showed that collagen around tumor xenograft tissues in controls was thin and noncontiguous. In contrast, collagen in the miR-204–expressing cell xenografts was thick, integrated, and continuous (Fig. 5E and F). These in vivo data indicate that increasing miR-204 expression reduced tumor size and malignancy (tissue invasion), supporting the hypothesis that miR-204 plays a key role in regulating malignant transformation and progression in MPNSTs.
Fig. 5.
Restoring miR-204 expression reduced MPNST tumor formation in vivo. STS26T cells containing the exogenous miR-204 or the control construct were injected subcutaneously into 8-week old NSG mice. The mice were euthanized 14 days after injection. The tumor volume was calculated by using formula: 1/2 ab2 (a: long axis, b: short axis). Compared with controls, restoring miR-204 reduced tumor size (A) and volume (B) after xenograft (miR-204 group versus control group: ***P < .001, n = 10). (C–F) Compared with tumors in the control group, the tumor in the miR-204 group showed loose connection with surrounding tissue (arrow), collagen around tumor tissue in the miR-204 group was thick, integrated, and continuous (arrow head) (C, D: H&E stain; E, F: Masson's trichrome stain).
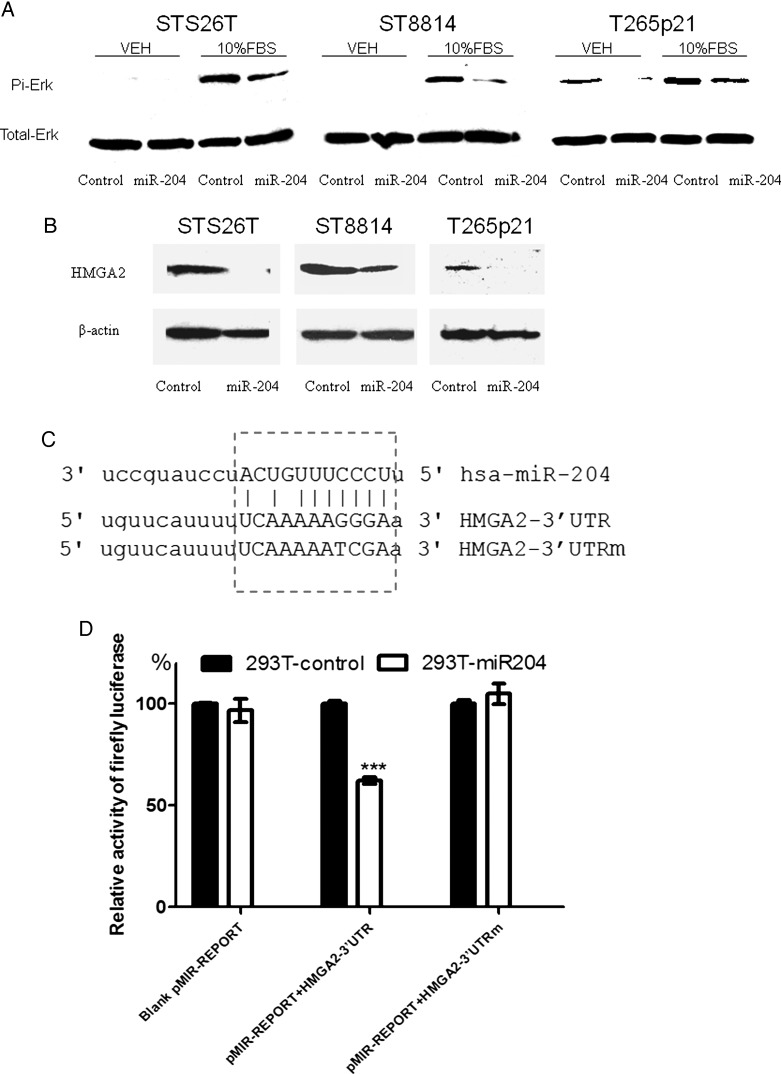
miR-204 Modulated RAS Signaling and Inhibited HMGA2 Expression to Regulate Carcinogenesis Progression in MPNSTs
Using Web-based informatics software (miRecords, http://mirecords.biolead.org/index.php), we identified certain genes in the Ras signaling pathways as candidate targets for miR-204 (Table 1). Because most NF1-related tumors display abnormal Ras signaling pathways and because genes involved in the Ras pathways, such as RASF2 and HMMR/RHAMM, were deregulated in MPNSTs,21 we wondered if miR-204 targets Ras signaling pathways in MPNST cell lines. Compared with controls, both NF1 (ST8814 and T265p21) and non-NF1 (STS26T) MPNST cells transfected with miR-204 exhibited less phosphorylated extracellular-signal-regulated kinase (ERK) after serum stimulation (Fig. 6A). This result indicated that miR-204 may target molecules within Ras signaling pathways.
Table 1.
Predicted targets for miR-204
| Symbol | Description | Miranda | Mirtarget2 | Pictar | Pita | Rnahybrid | Targetscan |
|---|---|---|---|---|---|---|---|
| CCND2 | Cyclin D2 | 1 | 1 | 0 | 1 | 1 | 0 |
| CCNI2 | Cyclin I family, member 2 | 1 | 1 | 0 | 1 | 1 | 0 |
| CCNJ | Cyclin J | 0 | 1 | 1 | 1 | 1 | 1 |
| CCNT2 | Cyclin T2 | 1 | 1 | 1 | 1 | 1 | 1 |
| CCNY | Cyclin Y | 1 | 1 | 0 | 1 | 1 | 0 |
| CNNM4 | Cyclin M4 | 0 | 0 | 1 | 1 | 1 | 1 |
| CNTD1 | Cyclin N-terminal domain containing 1 | 1 | 1 | 0 | 1 | 1 | 0 |
| HDAC9 | Histone deacetylase 9 | 1 | 1 | 1 | 1 | 1 | 0 |
| HMGA2 | High mobility group AT-hook 2 | 1 | 0 | 1 | 1 | 1 | 1 |
| RAB10 | RAB10, member Ras oncogene family | 1 | 1 | 1 | 1 | 1 | 1 |
| RAB13 | RAB13, member Ras oncogene family | 1 | 1 | 0 | 1 | 1 | 0 |
| RAB14 | RAB14, member Ras oncogene family | 1 | 0 | 1 | 1 | 1 | 1 |
| RAB1A | RAB1A, member Ras oncogene family | 1 | 0 | 1 | 1 | 1 | 1 |
| RAB22A | RAB22A, member Ras oncogene family | 1 | 1 | 1 | 1 | 1 | 1 |
| RAB27B | RAB27B, member Ras oncogene family | 1 | 1 | 0 | 1 | 1 | 0 |
| RAB40B | RAB40B, member Ras oncogene family | 1 | 1 | 0 | 1 | 1 | 0 |
| RAB5C | RAB5C, member Ras oncogene family | 1 | 0 | 1 | 1 | 1 | 0 |
| RAB6IP1 | RAB6 interacting protein 1 | 0 | 1 | 1 | 1 | 1 | 1 |
| RAP2C | RAP2C, member of Ras oncogene family | 1 | 1 | 1 | 1 | 1 | 1 |
| RASL10A | Ras-like, family 10, member A | 1 | 1 | 0 | 1 | 1 | 0 |
| RASSF2 | Ras association (RalGDS/AF-6) domain family member 2 | 1 | 1 | 0 | 1 | 1 | 0 |
| RASSF8 | Ras association (RalGDS/AF-6) domain family member 8 | 1 | 1 | 0 | 1 | 1 | 0 |
| RHOT1 | Ras homolog gene family, member T1 | 0 | 1 | 0 | 1 | 1 | 1 |
| TP53INP1 | Tumor protein p53 inducible nuclear protein 1 | 1 | 0 | 1 | 1 | 1 | 1 |
| TP63 | Tumor protein p63 | 1 | 1 | 0 | 1 | 1 | 0 |
| TPRG1L | Tumor protein p63 regulated 1–like | 1 | 1 | 0 | 1 | 1 | 0 |
| TRIAP1 | TP53 regulated inhibitor of apoptosis 1 | 0 | 0 | 1 | 1 | 1 | 1 |
| COL14A1 | Collagen, type XIV, alpha 1 | 0 | 0 | 1 | 1 | 0 | 0 |
| COL5A1 | Collagen, type V, alpha 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| DNA2 | DNA replication helicase 2 homolog (yeast) | 0 | 0 | 1 | 1 | 0 | 0 |
| MMP16 | Matrix metallopeptidase 16 (membrane-inserted) | 0 | 0 | 1 | 1 | 0 | 0 |
Abbreviations: 1, interaction predicted, 0 interaction not predicted.
Molecules in the Ras signaling pathways are highlighted.
Fig. 6.
miR-204 inhibited RAS signaling and HMGA2 expression. (A) NF1 and non-NF1 MPNST cells with the exogenous miR-204 or the control construct were serum-starved for 24 h and then stimulated with serum-free medium (vehicle, VEH) or medium supplemented with 10% FBS for 10 min. Phosphorylated ERK and total ERK were studied by Western blotting. Compared with the VEH controls, treatment with 10% FBS increased phosphorylated ERK in all 3 MPNST cell lines. Cells with the exogenous miR-204 showed less phosphorylated ERK than cells with the control construct. (B) MPNST cells with the exogenous miR-204 or the control construct were serum-starved for 24 h. The expression of HMGA2 was studied by Western blotting. Compared with the controls, HMGA2 expression was lower in MPNST cells with the exogenous miR-204. (C) The 3′ UTR of the HMGA2 gene was analyzed by TargetScan, and a target sequence for miR-204 was identified. The strategy to make point mutations within the target sequence was listed as HMGA2–3′ UTRm. (D) The miR-204 precursor gene or the control was stably transfected into 293T cells. The transfected cells were further cotransfected with pMIR reporter vector with HMGA2 3′ UTR or HMGA2 3′UTRm. The Dual Luciferase Assay kit was used to perform the HMGA2 3′ UTR reporter assay. Overexpression of miR-204 reduced the levels of luciferase activity by about 40% in the cells containing the HMGA2 3′ UTR (HMGA2 3′ UTR) but not in the cells with tmutated seed sequence (HMGA2 3′ UTRm). These results indicated that HMGA2 3′ UTR was directly targeted by miR-204 (***P < .001 vs control; n = 3).
Our bioinformatics analysis showed that miR-204 may also target HMGA2, suggesting that additional mechanisms could contribute to carcinogenesis under miR-204 control (Table 1). Interestingly, HMGA2 expression is significantly higher in invasive than in noninvasive tumors38 and is strongly upregulated in all MPNSTs.32 Compared with controls, NF1 (ST8814 and T265p21) and non-NF1 (STS26T) MPNST cells transfected with miR-204 exhibited a lower level of HMGA2 (Fig. 6B). To further investigate whether miR-204 targets HMGA2 directly, we cloned the HMGA2 3′ UTR and placed it in the 3′ UTR of a luciferase reporter expression cassette. Cotransfection in 293T cells with either the miR-204 expression vector or a control vector showed that overexpression of miR-204 significantly reduced luciferase activity from the construct containing the HMGA2 3′ UTR (Fig. 6D; P < .001). miR-204 failed to reduce luciferase activity when its target sequence in the HMGA2 3′ UTR was mutated (Fig. 6D). These results indicate that miR-204 likely directly targets the 3′ UTR of HMGA2 mRNA, representing an alternative pathway that could regulate malignant transformation and progression in MPNSTs.
Discussion
miRNAs are implicated in the carcinogenesis of several tumors and are under investigation as candidate oncology therapeutic targets.24,39 miR-204 is located within the sixth intron of the host gene transient receptor potential melastatin 3 cation channel (TRPM3, NM_020952) and is transcribed in the same direction as TRPM3.40 miR-204 appears to be an important regulator in cell differentiation, in apoptosis, in stress response and inflammation,41,42 in lens and retinal development,43 and in the maintenance of axonal structure and function.44 Interestingly, miR-204 is located within the cancer-associated genomic region 9q21.1–q22.3 locus that exhibits high frequency of loss of heterozygosity in some types of tumors, such as squamous cell carcinoma of the head and neck.28,29 Compared with normal tissues, miR-204 expression is significantly lower, by 0.07% to 5%, in tumors in 5 of the 9 tissue types (brain, kidney, ovary, hematological cells, and colon).31 Expression of miR-204 is lower in the National Cancer Institute–60 tumor cell line panel compared with its expression in 13 normal tissues.31 Significant downregulation of miR-204 was recently reported in a subtype of acute myeloid leukemia-bearing cytoplasmic mutated nucleophosmin and in 3 Burkitt B-cell lymphoma cell lines.45 miR-204 is also downregulated in gastric cancer cells and gastric carcinomas.46 Taken as a whole, these reports strongly suggest that miR-204 functions as a tumor suppressor.
In the present study, we found that miR-204 was downregulated in NF1 and non-NF1 MPNST tumor tissues, in NF1 MPNST tumor cell lines ST8814 and T265p21, and in the non-NF1 MPNST tumor cell line STS26T, supporting the hypothesis that downregulation of miR-204 occurs in MPNSTs. Restoring miR-204 expression in NF1 and non-NF1 MPNST tumor cell lines significantly reduced cellular proliferation, migration, and invasion in vitro (Figs 3 and 4). Importantly, restoring miR-204 expression in non-NF1 MPNST tumor cell lines significantly reduced tumorigenesis in vivo (Fig. 5). Our data support the concept that miR-204 functions as a tumor suppressor and may critically regulate tumor formation and malignant progression in MPNSTs.
Multiple lines of evidence support the importance of the Raf–mitogen-activated protein kinase–ERK pathway, downstream of Ras in regulating proliferation of MPNSTs in general, and of NF1-related MPNSTs in particular.47,48 Phosphorylated ERK (pERK) is a surrogate marker for activated Ras. The tumorigenic effect of pERK is supported by its overexpression in more than 90% of MPNST tissues compared with its expression in 21% of benign neurofibromas.8 In the present study, we report that both NF1 and non-NF1 MPNST cell lines with exogenous miR-204 exhibited a lower level of pERK after serum stimulation (Fig. 6A), indicating that miR-204 targets molecules within Ras signaling pathways. It has been reported that miR-204 is downregulated in gastric cancer cells and gastric carcinomas.46 miR-204 inhibited ezrin, which activates Ras by remodeling the cortical actin cytoskeleton46 and has been implicated in metastatic progression of certain cancers. By contributing to ezrin upregulation, miR-204 downregulation represents a novel mechanism for aberrant Ras activation in gastric carcinogenesis.46 It seems plausible that miR-204 targets molecules such as ezrin, which may deregulate Ras signaling in a way that promotes malignant transformation and progression in MPNSTs.
Our bioinformatics analysis suggested that miR-204 might target HMGA2, part of the high mobility group HMGA protein family, which are architectural transcription factors that both positively and negatively regulate the transcription of a variety of genes. Overexpression of HMGA2 was associated with malignant transformation, because HMGA2 expression was significantly higher in invasive than in noninvasive tumors.38 Supporting this concept, inactivation of the HMGA2 gene prevented cell transformation.49 Compared with its expression in normal human Schwann cells, HMGA2 was upregulated in all MPNSTs,32 implicating HMGA2 as a factor in the promotion of MPNST carcinogenesis. Indeed, HMGA2 expression was reduced by about 70% when miR-204 function was ectopically enhanced in the JSQ3 squamous cell carcinoma of the head and neck cell line.50 Our data showed that NF1 (ST8814 and T265p21) and non-NF1 (STS26T) MPNST cells transfected with miR-204 exhibited less HMGA2 than their controls (Fig. 6B). Further study confirmed that miR-204 targets HMGA2 directly (Fig. 6D). We speculate that miR-204 may contribute to deregulation mechanisms promoting progression of MPNSTs via HMGA2 pathways.
Our data suggest that miR-204 may critically regulate malignant transformation and progression in MPNSTs. It is still not clear whether miR-204 alone is sufficient to promote carcinogenesis in MPNSTs. Subramanian et al.26 reported that p53 inactivation and subsequent loss of expression of miR-34a may contribute to MPNST development. Our earlier research reported that miR-10b targets neurofibromin and Ras signaling to regulate NF1 tumorigenesis.27 In the present study, we found that miR-204 is downregulated in NF1 and non-NF1 MPNST tumor tissues, as well as in tumor cell lines. Restoring miR-204 expression in MPNST cell lines significantly reduced cellular proliferation, migration, and invasion in vitro and tumor growth and malignant transformation in vivo. We speculate that miR-204 may cooperate with other miRNAs, such as miR-10b and miR-34a, to promote MPNST development and progression. These miRNAs may represent novel selective and specific candidate targets for more effective therapies of MPNSTs.
Supplementary Material
Funding
National Natural Science Foundation of China (No. 30872632, No. 81072190 to X. Yu; No. 81101920 to J. Ma); US Army Medical Research Command research contract (W81XWH-07 -02-0116 to J. Hock).
Supplementary Material
Acknowledgments
Some work for this study was conducted by Dr. M. Gong in Dr. Yu's Maine Institute of Human Genetics and Health laboratory at the Jackson Laboratory, Bar Harbor, ME.
Conflict of interest statement. None declared.
References
- 1.Doorn PF, Molenaar WM, Buter J, Hoekstra HJ. Malignant peripheral nerve sheath tumors in patients with and without neurofibromatosis. Eur J Surg Oncol. 1995;21:78–82. doi: 10.1016/s0748-7983(05)80073-3. [DOI] [PubMed] [Google Scholar]
- 2.Ducatman BS, Scheithauer BW, Piepgras DG, Reiman HM, Ilstrup DM. Malignant peripheral nerve sheath tumors. A clinicopathologic study of 120 cases. Cancer. 1986;57:2006–2021. doi: 10.1002/1097-0142(19860515)57:10<2006::aid-cncr2820571022>3.0.co;2-6. [DOI] [PubMed] [Google Scholar]
- 3.King AA, Debaun MR, Riccardi VM, Gutmann DH. Malignant peripheral nerve sheath tumors in neurofibromatosis 1. Am J Med Genet. 2000;93:388–392. [PubMed] [Google Scholar]
- 4.Ducatman BS, Scheithauer BW. Postirradiation neurofibrosarcoma. Cancer. 1983;51:1028–1033. doi: 10.1002/1097-0142(19830315)51:6<1028::aid-cncr2820510610>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- 5.Foley KM, Woodruff JM, Ellis FT, Posner JB. Radiation-induced malignant and atypical peripheral nerve sheath tumors. Ann Neurol. 1980;7:311–318. doi: 10.1002/ana.410070405. [DOI] [PubMed] [Google Scholar]
- 6.Anghileri M, Miceli R, Fiore M, et al. Malignant peripheral nerve sheath tumors: prognostic factors and survival in a series of patients treated at a single institution. Cancer. 2006;107:1065–1074. doi: 10.1002/cncr.22098. [DOI] [PubMed] [Google Scholar]
- 7.Gupta G, Mammis A, Maniker A. Malignant peripheral nerve sheath tumors. Neurosurgery Clinics of North America. 2008;19:533–543. doi: 10.1016/j.nec.2008.07.004. v. [DOI] [PubMed] [Google Scholar]
- 8.Zou C, Smith KD, Liu J, et al. Clinical, pathological, and molecular variables predictive of malignant peripheral nerve sheath tumor outcome. Annals of Surgery. 2009;249:1014–1022. doi: 10.1097/SLA.0b013e3181a77e9a. [DOI] [PubMed] [Google Scholar]
- 9.Zhou H, Coffin CM, Perkins SL, et al. Malignant peripheral nerve sheath tumor: a comparison of grade, immunophenotype, and cell cycle/growth activation marker expression in sporadic and neurofibromatosis 1-related lesions. The American Journal of Surgical Pathology. 2003;27:1337–1345. doi: 10.1097/00000478-200310000-00006. [DOI] [PubMed] [Google Scholar]
- 10.Xu GF, O'Connell P, Viskochil D, et al. The neurofibromatosis type 1 gene encodes a protein related to GAP. Cell. 1990;62:599–608. doi: 10.1016/0092-8674(90)90024-9. [DOI] [PubMed] [Google Scholar]
- 11.Martin GA, Viskochil D, Bollag G, et al. The GAP-related domain of the neurofibromatosis type 1 gene product interacts with ras p21. Cell. 1990;63:843–849. doi: 10.1016/0092-8674(90)90150-d. [DOI] [PubMed] [Google Scholar]
- 12.Viskochil D. Genetics of neurofibromatosis 1 and the NF1 gene. J Child Neurol. 2002;17:562–570. doi: 10.1177/088307380201700804. discussion 571–562, 646–551. [DOI] [PubMed] [Google Scholar]
- 13.Gottfried ON, Viskochil DH, Couldwell WT. Neurofibromatosis type 1 and tumorigenesis: molecular mechanisms and therapeutic implications. Neurosurgical Focus. 2010;28:E8. doi: 10.3171/2009.11.FOCUS09221. [DOI] [PubMed] [Google Scholar]
- 14.Menon AG, Anderson KM, Riccardi VM, et al. Chromosome 17p deletions and p53 gene mutations associated with the formation of malignant neurofibrosarcomas in von Recklinghausen neurofibromatosis. Proc Natl Acad Sci USA. 1990;87:5435–5439. doi: 10.1073/pnas.87.14.5435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cichowski K, Shih TS, Schmitt E, et al. Mouse models of tumor development in neurofibromatosis type 1. Science. 1999;286:2172–2176. doi: 10.1126/science.286.5447.2172. [DOI] [PubMed] [Google Scholar]
- 16.Kourea HP, Cordon-Cardo C, Dudas M, Leung D, Woodruff JM. Expression of p27(kip) and other cell cycle regulators in malignant peripheral nerve sheath tumors and neurofibromas: the emerging role of p27(kip) in malignant transformation of neurofibromas. Am J Pathol. 1999;155:1885–1891. doi: 10.1016/S0002-9440(10)65508-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Watson MA, Perry A, Tihan T, et al. Gene expression profiling reveals unique molecular subtypes of neurofibromatosis type I–associated and sporadic malignant peripheral nerve sheath tumors. Brain Pathology (Zurich, Switzerland) 2004;14:297–303. doi: 10.1111/j.1750-3639.2004.tb00067.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Badache A, De Vries GH. Neurofibrosarcoma-derived Schwann cells overexpress platelet-derived growth factor (PDGF) receptors and are induced to proliferate by PDGF BB. Journal of Cellular Physiology. 1998;177:334–342. doi: 10.1002/(SICI)1097-4652(199811)177:2<334::AID-JCP15>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 19.Dang I, Nelson JK, DeVries GH. c-Kit receptor expression in normal human Schwann cells and Schwann cell lines derived from neurofibromatosis type 1 tumors. J Neurosci Res. 2005;82:465–471. doi: 10.1002/jnr.20648. [DOI] [PubMed] [Google Scholar]
- 20.Holtkamp N, Atallah I, Okuducu AF, et al. MMP-13 and p53 in the progression of malignant peripheral nerve sheath tumors. Neoplasia (New York, NY) 2007;9:671–677. doi: 10.1593/neo.07304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Widemann BC. Current status of sporadic and neurofibromatosis type 1-associated malignant peripheral nerve sheath tumors. Current Oncology Reports. 2009;11:322–328. doi: 10.1007/s11912-009-0045-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lim LP, Lau NC, Garrett-Engele P, et al. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433:769–773. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- 23.Lee MJ, Su YN, You HL, et al. Identification of forty-five novel and twenty-three known NF1 mutations in Chinese patients with neurofibromatosis type 1. Hum Mutat. 2006;27:832. doi: 10.1002/humu.9446. [DOI] [PubMed] [Google Scholar]
- 24.Lu J, Getz G, Miska EA, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 25.Lee YS, Dutta A. MicroRNAs in cancer. Annual Review of Pathology. 2009;4:199–227. doi: 10.1146/annurev.pathol.4.110807.092222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Subramanian S, Thayanithy V, West RB, et al. Genome-wide transcriptome analyses reveal p53 inactivation mediated loss of miR-34a expression in malignant peripheral nerve sheath tumours. J Pathol. 2009;220:58–70. doi: 10.1002/path.2633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chai G, Liu N, Ma J, et al. MicroRNA-10b regulates tumorigenesis in neurofibromatosis type 1. Cancer Sci. 2010;101:1997–2004. doi: 10.1111/j.1349-7006.2010.01616.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bauer VL, Braselmann H, Henke M, et al. Chromosomal changes characterize head and neck cancer with poor prognosis. Journal of Molecular Medicine (Berlin, Germany) 2008;86:1353–1365. doi: 10.1007/s00109-008-0397-0. [DOI] [PubMed] [Google Scholar]
- 29.Abou-Elhamd KE, Habib TN, Moussa AE, Badawy BS. The role of genetic susceptibility in head and neck squamous cell carcinoma. Eur Arch Otorhinolaryngol. 2008;265:217–222. doi: 10.1007/s00405-007-0436-4. [DOI] [PubMed] [Google Scholar]
- 30.Scully C, Field JK, Tanzawa H. Genetic aberrations in oral or head and neck squamous cell carcinoma 2: chromosomal aberrations. Oral Oncology. 2000;36:311–327. doi: 10.1016/s1368-8375(00)00021-x. [DOI] [PubMed] [Google Scholar]
- 31.Wang FE, Zhang C, Maminishkis A, et al. MicroRNA-204/211 alters epithelial physiology. FASEB J. 2010;24:1552–1571. doi: 10.1096/fj.08-125856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Miller SJ, Rangwala F, Williams J, et al. Large-scale molecular comparison of human schwann cells to malignant peripheral nerve sheath tumor cell lines and tissues. Cancer Res. 2006;66:2584–2591. doi: 10.1158/0008-5472.CAN-05-3330. [DOI] [PubMed] [Google Scholar]
- 33.Barkan B, Starinsky S, Friedman E, Stein R, Kloog Y. The Ras inhibitor farnesylthiosalicylic acid as a potential therapy for neurofibromatosis type 1. Clin Cancer Res. 2006;12:5533–5542. doi: 10.1158/1078-0432.CCR-06-0792. [DOI] [PubMed] [Google Scholar]
- 34.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 35.Sheela S, Riccardi VM, Ratner N. Angiogenic and invasive properties of neurofibroma Schwann cells. J Cell Biol. 1990;111:645–653. doi: 10.1083/jcb.111.2.645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Muir D, Neubauer D, Lim IT, Yachnis AT, Wallace MR. Tumorigenic properties of neurofibromin-deficient neurofibroma Schwann cells. Am J Pathol. 2001;158:501–513. doi: 10.1016/S0002-9440(10)63992-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mahller YY, Vaikunth SS, Currier MA, et al. Oncolytic HSV and erlotinib inhibit tumor growth and angiogenesis in a novel malignant peripheral nerve sheath tumor xenograft model. Mol Ther. 2007;15:279–286. doi: 10.1038/sj.mt.6300038. [DOI] [PubMed] [Google Scholar]
- 38.Mu G, Liu H, Zhou F, et al. Correlation of overexpression of HMGA1 and HMGA2 with poor tumor differentiation, invasion, and proliferation associated with let-7 downregulation in retinoblastomas. Hum Pathol. 2010;41:493–502. doi: 10.1016/j.humpath.2009.08.022. [DOI] [PubMed] [Google Scholar]
- 39.Stenvang J, Silahtaroglu AN, Lindow M, Elmen J, Kauppinen S. The utility of LNA in microRNA-based cancer diagnostics and therapeutics. Semin Cancer Biol. 2008;18:89–102. doi: 10.1016/j.semcancer.2008.01.004. [DOI] [PubMed] [Google Scholar]
- 40.Lagos-Quintana M, Rauhut R, Meyer J, Borkhardt A, Tuschl T. New microRNAs from mouse and human. RNA (New York, NY) 2003;9:175–179. doi: 10.1261/rna.2146903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Huang J, Zhao L, Xing L, Chen D. MicroRNA-204 regulates Runx2 protein expression and mesenchymal progenitor cell differentiation. Stem Cells. 2010;28:357–364. doi: 10.1002/stem.288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Courboulin A, Paulin R, Giguere NJ, et al. Role for miR-204 in human pulmonary arterial hypertension. J Exp Med. 2011;208:535–548. doi: 10.1084/jem.20101812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Conte I, Carrella S, Avellino R, et al. miR-204 is required for lens and retinal development via Meis2 targeting. Proc Natl Acad Sci USA. 2010;107:15491–15496. doi: 10.1073/pnas.0914785107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Natera-Naranjo O, Aschrafi A, Gioio AE, Kaplan BB. Identification and quantitative analyses of microRNAs located in the distal axons of sympathetic neurons. RNA. 2010;16:1516–1529. doi: 10.1261/rna.1833310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Garzon R, Garofalo M, Martelli MP, et al. Distinctive microRNA signature of acute myeloid leukemia bearing cytoplasmic mutated nucleophosmin. Proc Natl Acad Sci USA. 2008;105:3945–3950. doi: 10.1073/pnas.0800135105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lam EK, Wang X, Shin VY, et al. A microRNA contribution to aberrant Ras activation in gastric cancer. Am J Transl Res. 2011;3:209–218. [PMC free article] [PubMed] [Google Scholar]
- 47.Viskochil DH. Disorders of the Ras pathway: an introduction. Am J Med Genet C Semin Med Genet. 2011;157:79–82. doi: 10.1002/ajmg.c.30301. [DOI] [PubMed] [Google Scholar]
- 48.Yan N, Ricca C, Fletcher J, et al. Farnesyltransferase inhibitors block the neurofibromatosis type I (NF1) malignant phenotype. Cancer Research. 1995;55:3569–3575. [PubMed] [Google Scholar]
- 49.Berlingieri MT, Manfioletti G, Santoro M, et al. Inhibition of HMGI-C protein synthesis suppresses retrovirally induced neoplastic transformation of rat thyroid cells. Mol Cell Biol. 1995;15:1545–1553. doi: 10.1128/mcb.15.3.1545. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 50.Lee Y, Yang X, Huang Y, et al. Network modeling identifies molecular functions targeted by miR-204 to suppress head and neck tumor metastasis. PLoS Comput Biol. 2010;6:e1000730. doi: 10.1371/journal.pcbi.1000730. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.