Abstract
We previously reported that some L1 family (KpnI family) members are closely associated with the Alu family sequence. To understand the details of the L1-Alu association, the structure of a L1-Alu unit downstream from the beta-globin gene was compared between human and primates. The results revealed that the L1-Alu-associated sequence was formed by the insertion of the L1 sequence, T beta G41, into the 3' poly A tract of the preexisting Alu family sequence. It was estimated that the T beta G41 sequence was inserted after the divergence of Old World monkeys and hominoids and before the divergence of orang-utan and common ancestor of other higher hominoids. From the calculation of the mutation rates of L1 sequences, it was suggested that the T beta G41 was derived from an active L1 sequence which was able to encode reverse transcriptase-related protein.
Full text
PDF



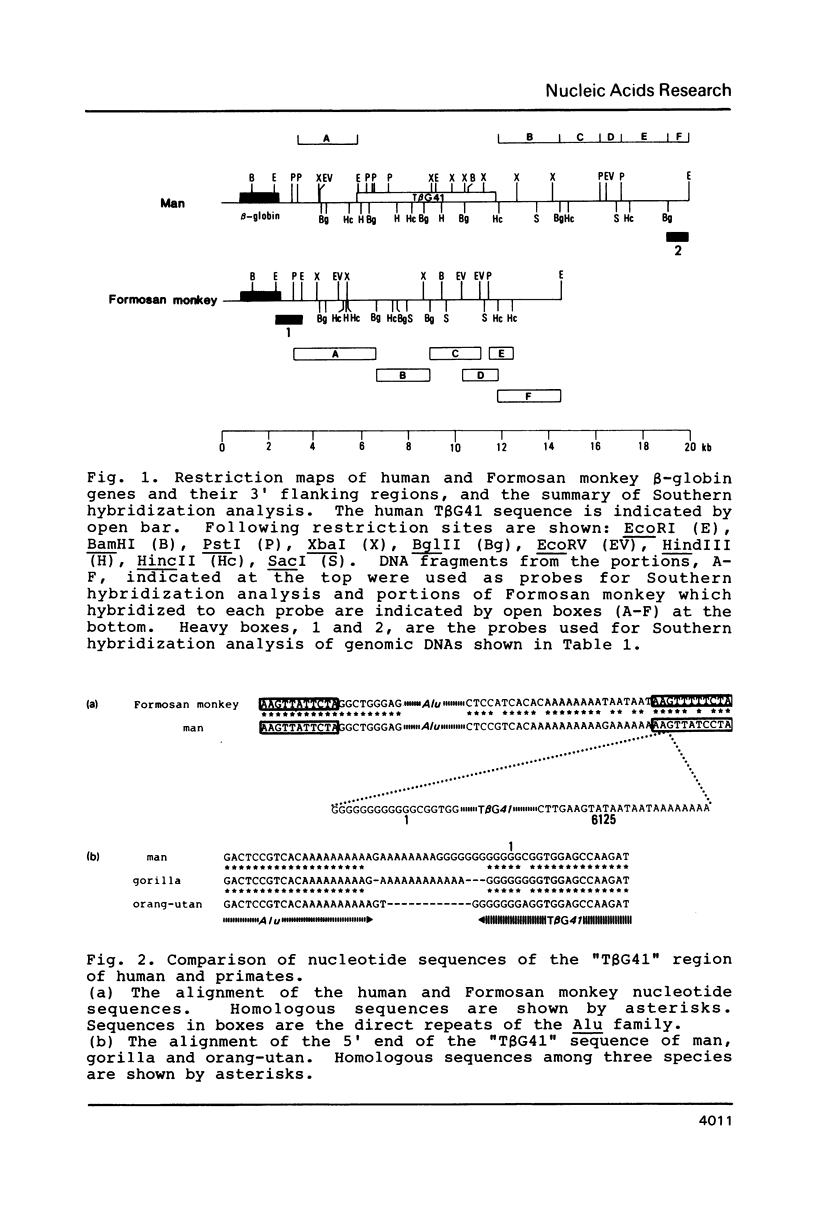


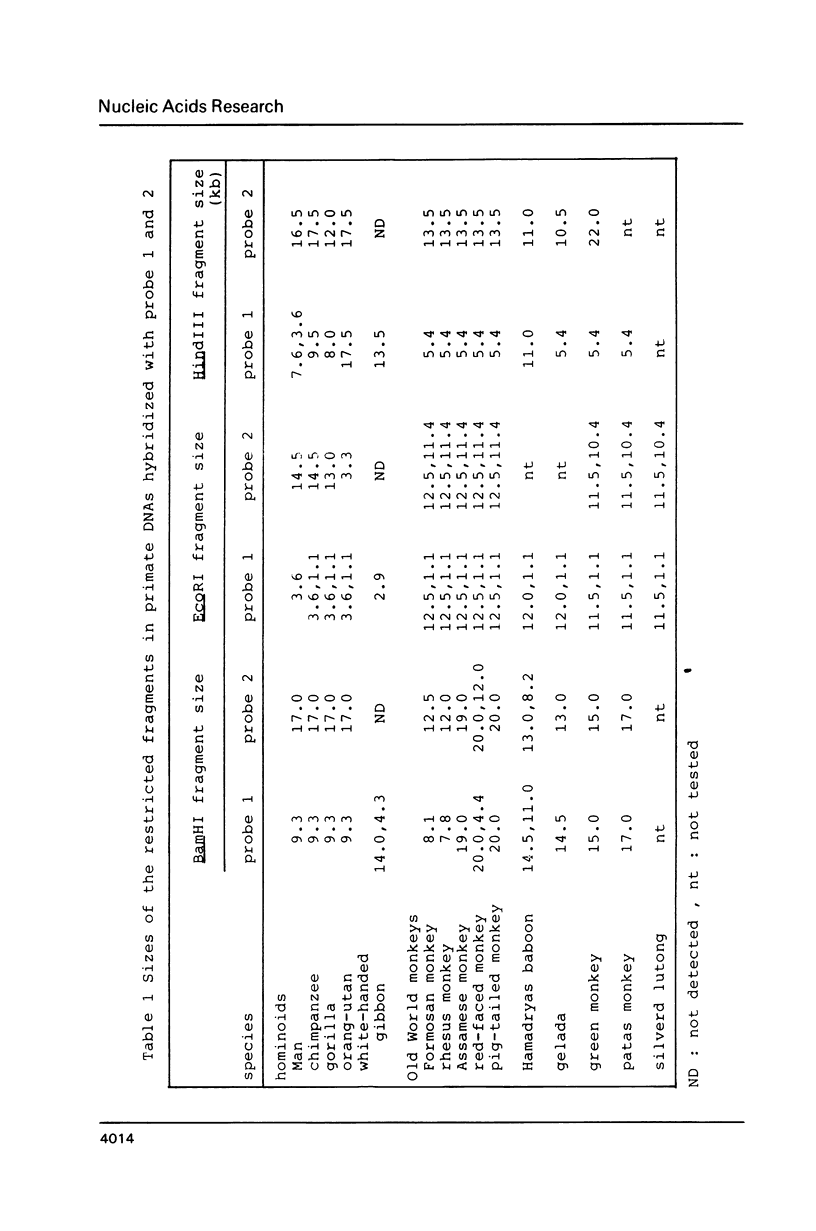






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adams J. W., Kaufman R. E., Kretschmer P. J., Harrison M., Nienhuis A. W. A family of long reiterated DNA sequences, one copy of which is next to the human beta globin gene. Nucleic Acids Res. 1980 Dec 20;8(24):6113–6128. doi: 10.1093/nar/8.24.6113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrie P. A., Jeffreys A. J., Scott A. F. Evolution of the beta-globin gene cluster in man and the primates. J Mol Biol. 1981 Jul 5;149(3):319–336. doi: 10.1016/0022-2836(81)90476-9. [DOI] [PubMed] [Google Scholar]
- Bell G. I., Karam J. H., Rutter W. J. Polymorphic DNA region adjacent to the 5' end of the human insulin gene. Proc Natl Acad Sci U S A. 1981 Sep;78(9):5759–5763. doi: 10.1073/pnas.78.9.5759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton F. H., Loeb D. D., Voliva C. F., Martin S. L., Edgell M. H., Hutchison C. A., 3rd Conservation throughout mammalia and extensive protein-encoding capacity of the highly repeated DNA long interspersed sequence one. J Mol Biol. 1986 Jan 20;187(2):291–304. doi: 10.1016/0022-2836(86)90235-4. [DOI] [PubMed] [Google Scholar]
- DiGiovanni L., Haynes S. R., Misra R., Jelinek W. R. Kpn I family of long-dispersed repeated DNA sequences of man: evidence for entry into genomic DNA of DNA copies of poly(A)-terminated Kpn I RNAs. Proc Natl Acad Sci U S A. 1983 Nov;80(21):6533–6537. doi: 10.1073/pnas.80.21.6533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furano A. V., Somerville C. C., Tsichlis P. N., D'Ambrosio E. Target sites for the transposition of rat long interspersed repeated DNA elements (LINEs) are not random. Nucleic Acids Res. 1986 May 12;14(9):3717–3727. doi: 10.1093/nar/14.9.3717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimaldi G., Singer M. F. Members of the KpnI family of long interspersed repeated sequences join and interrupt alpha-satellite in the monkey genome. Nucleic Acids Res. 1983 Jan 25;11(2):321–338. doi: 10.1093/nar/11.2.321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimaldi G., Skowronski J., Singer M. F. Defining the beginning and end of KpnI family segments. EMBO J. 1984 Aug;3(8):1753–1759. doi: 10.1002/j.1460-2075.1984.tb02042.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasegawa M., Kishino H., Yano T. Dating of the human-ape splitting by a molecular clock of mitochondrial DNA. J Mol Evol. 1985;22(2):160–174. doi: 10.1007/BF02101694. [DOI] [PubMed] [Google Scholar]
- Hattori M., Hidaka S., Sakaki Y. Sequence analysis of a KpnI family member near the 3' end of human beta-globin gene. Nucleic Acids Res. 1985 Nov 11;13(21):7813–7827. doi: 10.1093/nar/13.21.7813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hattori M., Kuhara S., Takenaka O., Sakaki Y. L1 family of repetitive DNA sequences in primates may be derived from a sequence encoding a reverse transcriptase-related protein. Nature. 1986 Jun 5;321(6070):625–628. doi: 10.1038/321625a0. [DOI] [PubMed] [Google Scholar]
- Hattori M., Sakaki Y. Dideoxy sequencing method using denatured plasmid templates. Anal Biochem. 1986 Feb 1;152(2):232–238. doi: 10.1016/0003-2697(86)90403-3. [DOI] [PubMed] [Google Scholar]
- Jones R. S., Potter S. S. L1 sequences in HeLa extrachromosomal circular DNA: evidence for circularization by homologous recombination. Proc Natl Acad Sci U S A. 1985 Apr;82(7):1989–1993. doi: 10.1073/pnas.82.7.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katzir N., Rechavi G., Cohen J. B., Unger T., Simoni F., Segal S., Cohen D., Givol D. "Retroposon" insertion into the cellular oncogene c-myc in canine transmissible venereal tumor. Proc Natl Acad Sci U S A. 1985 Feb;82(4):1054–1058. doi: 10.1073/pnas.82.4.1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol. 1980 Dec;16(2):111–120. doi: 10.1007/BF01731581. [DOI] [PubMed] [Google Scholar]
- Kimura M. Estimation of evolutionary distances between homologous nucleotide sequences. Proc Natl Acad Sci U S A. 1981 Jan;78(1):454–458. doi: 10.1073/pnas.78.1.454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhara S., Matsuo F., Futamura S., Fujita A., Shinohara T., Takagi T., Sakaki Y. GENAS: a database system for nucleic acid sequence analysis. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 1):89–99. doi: 10.1093/nar/12.1part1.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laimins L., Holmgren-König M., Khoury G. Transcriptional "silencer" element in rat repetitive sequences associated with the rat insulin 1 gene locus. Proc Natl Acad Sci U S A. 1986 May;83(10):3151–3155. doi: 10.1073/pnas.83.10.3151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerman M. I., Thayer R. E., Singer M. F. Kpn I family of long interspersed repeated DNA sequences in primates: polymorphism of family members and evidence for transcription. Proc Natl Acad Sci U S A. 1983 Jul;80(13):3966–3970. doi: 10.1073/pnas.80.13.3966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W. H., Gojobori T., Nei M. Pseudogenes as a paradigm of neutral evolution. Nature. 1981 Jul 16;292(5820):237–239. doi: 10.1038/292237a0. [DOI] [PubMed] [Google Scholar]
- Loeb D. D., Padgett R. W., Hardies S. C., Shehee W. R., Comer M. B., Edgell M. H., Hutchison C. A., 3rd The sequence of a large L1Md element reveals a tandemly repeated 5' end and several features found in retrotransposons. Mol Cell Biol. 1986 Jan;6(1):168–182. doi: 10.1128/mcb.6.1.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin S. L., Vincent K. A., Wilson A. C. Rise and fall of the delta globin gene. J Mol Biol. 1983 Mar 15;164(4):513–528. doi: 10.1016/0022-2836(83)90048-7. [DOI] [PubMed] [Google Scholar]
- Martin S. L., Voliva C. F., Burton F. H., Edgell M. H., Hutchison C. A., 3rd A large interspersed repeat found in mouse DNA contains a long open reading frame that evolves as if it encodes a protein. Proc Natl Acad Sci U S A. 1984 Apr;81(8):2308–2312. doi: 10.1073/pnas.81.8.2308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyake T., Migita K., Sakaki Y. Some KpnI family members are associated with the Alu family in the human genome. Nucleic Acids Res. 1983 Oct 11;11(19):6837–6846. doi: 10.1093/nar/11.19.6837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyata T., Hayashida H. Extraordinarily high evolutionary rate of pseudogenes: evidence for the presence of selective pressure against changes between synonymous codons. Proc Natl Acad Sci U S A. 1981 Sep;78(9):5739–5743. doi: 10.1073/pnas.78.9.5739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyata T., Yasunaga T. Rapidly evolving mouse alpha-globin-related pseudo gene and its evolutionary history. Proc Natl Acad Sci U S A. 1981 Jan;78(1):450–453. doi: 10.1073/pnas.78.1.450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potter S. S. Rearranged sequences of a human Kpn I element. Proc Natl Acad Sci U S A. 1984 Feb;81(4):1012–1016. doi: 10.1073/pnas.81.4.1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindler C. W., Rush M. G. Discrete size classes of monkey extrachromosomal circular DNA containing the L1 family of long interspersed nucleotide sequences are produced by a general non-sequence specific mechanism. Nucleic Acids Res. 1985 Nov 25;13(22):8247–8258. doi: 10.1093/nar/13.22.8247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shafit-Zagardo B., Brown F. L., Maio J. J., Adams J. W. KpnI families of long, interspersed repetitive DNAs associated with the human beta-globin gene cluster. Gene. 1982 Dec;20(3):397–407. doi: 10.1016/0378-1119(82)90208-6. [DOI] [PubMed] [Google Scholar]
- Shafit-Zagardo B., Brown F. L., Zavodny P. J., Maio J. J. Transcription of the KpnI families of long interspersed DNAs in human cells. Nature. 1983 Jul 21;304(5923):277–280. doi: 10.1038/304277a0. [DOI] [PubMed] [Google Scholar]
- Singer M. F. SINEs and LINEs: highly repeated short and long interspersed sequences in mammalian genomes. Cell. 1982 Mar;28(3):433–434. doi: 10.1016/0092-8674(82)90194-5. [DOI] [PubMed] [Google Scholar]
- Skowronski J., Singer M. F. Expression of a cytoplasmic LINE-1 transcript is regulated in a human teratocarcinoma cell line. Proc Natl Acad Sci U S A. 1985 Sep;82(18):6050–6054. doi: 10.1073/pnas.82.18.6050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voliva C. F., Martin S. L., Hutchison C. A., 3rd, Edgell M. H. Dispersal process associated with the L1 family of interspersed repetitive DNA sequences. J Mol Biol. 1984 Oct 5;178(4):795–813. doi: 10.1016/0022-2836(84)90312-7. [DOI] [PubMed] [Google Scholar]





