Abstract
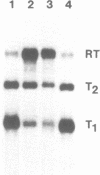
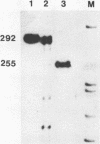
The rat rDNA transcription unit extends 560-565 bp into the spacer downstream of the 28S rRNA coding region. The site of 3′ end formation is located in front of a conserved 18 bp sequence element which is repeated eight times in the 3′ spacer between nucleotides +582 and +1767 relative to the 3′ terminus of 28S rRNA. These sequence motifs are almost identical to the RNA polymerase I transcription termination signal (the Sal I box) that has previously been identified in the 3′ terminal spacer of mouse rDNA. Interestingly, each of the single rat elements contains one or more base substitutions as compared to the murine Sal I box. Individual rat Sal I boxes were cloned and tested for their ability to interact with the murine termination factor and to direct transcription termination. It is shown that five of the eight boxes represent genuine transcription terminators, while three elements contain certain point mutations which are not recognized by the nuclear Sal I box-binding protein and therefore are functionally inactive.
Full text
PDF











Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Clos J., Normann A., Ohrlein A., Grummt I. The core promoter of mouse rDNA consists of two functionally distinct domains. Nucleic Acids Res. 1986 Oct 10;14(19):7581–7595. doi: 10.1093/nar/14.19.7581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dignam J. D., Lebovitz R. M., Roeder R. G. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983 Mar 11;11(5):1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dover G. A., Flavell R. B. Molecular coevolution: DNA divergence and the maintenance of function. Cell. 1984 Oct;38(3):622–623. doi: 10.1016/0092-8674(84)90255-1. [DOI] [PubMed] [Google Scholar]
- Erickson J. M., Schmickel R. D. A molecular basis for discrete size variation in human ribosomal DNA. Am J Hum Genet. 1985 Mar;37(2):311–325. [PMC free article] [PubMed] [Google Scholar]
- Financsek I., Mizumoto K., Mishima Y., Muramatsu M. Human ribosomal RNA gene: nucleotide sequence of the transcription initiation region and comparison of three mammalian genes. Proc Natl Acad Sci U S A. 1982 May;79(10):3092–3096. doi: 10.1073/pnas.79.10.3092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grummt I., Kuhn A., Bartsch I., Rosenbauer H. A transcription terminator located upstream of the mouse rDNA initiation site affects rRNA synthesis. Cell. 1986 Dec 26;47(6):901–911. doi: 10.1016/0092-8674(86)90805-6. [DOI] [PubMed] [Google Scholar]
- Grummt I., Maier U., Ohrlein A., Hassouna N., Bachellerie J. P. Transcription of mouse rDNA terminates downstream of the 3' end of 28S RNA and involves interaction of factors with repeated sequences in the 3' spacer. Cell. 1985 Dec;43(3 Pt 2):801–810. doi: 10.1016/0092-8674(85)90253-3. [DOI] [PubMed] [Google Scholar]
- Grummt I., Rosenbauer H., Niedermeyer I., Maier U., Ohrlein A. A repeated 18 bp sequence motif in the mouse rDNA spacer mediates binding of a nuclear factor and transcription termination. Cell. 1986 Jun 20;45(6):837–846. doi: 10.1016/0092-8674(86)90558-1. [DOI] [PubMed] [Google Scholar]
- Grummt I., Roth E., Paule M. R. Ribosomal RNA transcription in vitro is species specific. Nature. 1982 Mar 11;296(5853):173–174. doi: 10.1038/296173a0. [DOI] [PubMed] [Google Scholar]
- Grummt I. Specific transcription of mouse ribosomal DNA in a cell-free system that mimics control in vivo. Proc Natl Acad Sci U S A. 1981 Feb;78(2):727–731. doi: 10.1073/pnas.78.2.727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson S., Sollner-Webb B. A transcriptional terminator is a novel element of the promoter of the mouse ribosomal RNA gene. Cell. 1986 Dec 26;47(6):891–900. doi: 10.1016/0092-8674(86)90804-4. [DOI] [PubMed] [Google Scholar]
- Kempers-Veenstra A. E., Oliemans J., Offenberg H., Dekker A. F., Piper P. W., Planta R. J., Klootwijk J. 3'-End formation of transcripts from the yeast rRNA operon. EMBO J. 1986 Oct;5(10):2703–2710. doi: 10.1002/j.1460-2075.1986.tb04554.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Volpe A., Simeone A., D'Esposito M., Scotto L., Fidanza V., de Falco A., Boncinelli E. Molecular analysis of the heterogeneity region of the human ribosomal spacer. J Mol Biol. 1985 May 25;183(2):213–223. doi: 10.1016/0022-2836(85)90214-1. [DOI] [PubMed] [Google Scholar]
- Labhart P., Reeder R. H. Characterization of three sites of RNA 3' end formation in the Xenopus ribosomal gene spacer. Cell. 1986 May 9;45(3):431–443. doi: 10.1016/0092-8674(86)90329-6. [DOI] [PubMed] [Google Scholar]
- McStay B., Reeder R. H. A termination site for Xenopus RNA polymerase I also acts as an element of an adjacent promoter. Cell. 1986 Dec 26;47(6):913–920. doi: 10.1016/0092-8674(86)90806-8. [DOI] [PubMed] [Google Scholar]
- Miesfeld R., Arnheim N. Species-specific rDNA transcription is due to promoter-specific binding factors. Mol Cell Biol. 1984 Feb;4(2):221–227. doi: 10.1128/mcb.4.2.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishima Y., Financsek I., Kominami R., Muramatsu M. Fractionation and reconstitution of factors required for accurate transcription of mammalian ribosomal RNA genes: identification of a species-dependent initiation factor. Nucleic Acids Res. 1982 Nov 11;10(21):6659–6670. doi: 10.1093/nar/10.21.6659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moss T., Mitchelson K., de Winter R. The promotion of ribosomal transcription in eukaryotes. Oxf Surv Eukaryot Genes. 1985;2:207–250. [PubMed] [Google Scholar]
- Simeone A., La Volpe A., Boncinelli E. Nucleotide sequence of a complete ribosomal spacer of D. melanogaster. Nucleic Acids Res. 1985 Feb 25;13(4):1089–1101. doi: 10.1093/nar/13.4.1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommerville J. RNA polymerase I promoters and transcription factors. Nature. 1984 Jul 19;310(5974):189–190. doi: 10.1038/310189a0. [DOI] [PubMed] [Google Scholar]
- Tautz D., Dover G. A. Transcription of the tandem array of ribosomal DNA in Drosophila melanogaster does not terminate at any fixed point. EMBO J. 1986 Jun;5(6):1267–1273. doi: 10.1002/j.1460-2075.1986.tb04356.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C. An exonuclease protection assay reveals heat-shock element and TATA box DNA-binding proteins in crude nuclear extracts. Nature. 1985 Sep 5;317(6032):84–87. doi: 10.1038/317084a0. [DOI] [PubMed] [Google Scholar]
- Yavachev L. P., Georgiev O. I., Braga E. A., Avdonina T. A., Bogomolova A. E., Zhurkin V. B., Nosikov V. V., Hadjiolov A. A. Nucleotide sequence analysis of the spacer regions flanking the rat rRNA transcription unit and identification of repetitive elements. Nucleic Acids Res. 1986 Mar 25;14(6):2799–2810. doi: 10.1093/nar/14.6.2799. [DOI] [PMC free article] [PubMed] [Google Scholar]