Abstract
The use of umbilical cord blood (UCB) for allogeneic transplantation has increased dramatically over the past years. However, the limited number of cells available in a single cord blood unit remains a serious obstacle. Here, we wished to establish a nonhuman primate cord blood transplantation model that would allow us to test various hematopoietic stem cell (HSC) expansion and gene therapy strategies. We implemented HOXB4-mediated expansion based on our previous experience with HOXB4 in autologous cells. Cord blood units were divided into two equal parts; half of the cells were transduced with a YFP control vector and cryopreserved, while half were transduced with a HOXB4GFP vector, expanded, and cryopreserved. Both fractions of cells were transplanted into Macaca nemestrina subjects. We found that neutrophil recovery occurred within 19 days in all animals, and both neutrophil and platelet recovery were substantially accelerated compared to human single unit cord blood transplants. In addition, HOXB4-transduced and expanded cells resulted in superior engraftment of all hematopoietic lineages in all animals over non-expanded controls. In conclusion, we have successfully established a nonhuman primate cord blood transplantation model and demonstrated that HOXB4 stimulates expansion and engraftment of repopulating cells. The availability of such a model has significant implications for developing and testing strategies to improve clinical cord blood transplantation, as it will allow comparison of different stem cell expansion methodologies within a single animal. Furthermore, it can be used in long-term follow-up studies to determine how specific expansion techniques affect engraftment of various hematopoietic lineages.
Keywords: hematopoietic stem cell, expansion, nonhuman primate, cord blood transplant, large animal model
INTRODUCTION
Umbilical cord blood (UCB) transplantation provides a treatment option for patients suffering from a wide variety of hematologic and nonhematologic malignancies. Rapid accessibility and a reduced risk of graft-versus-host disease are distinct advantages in the choice of UCB as a source of stem cells for transplantation. However, cell dose is a major issue, especially in adult patients and large pediatric patients, as total nucleated cell (TNC) dose and CD34 cell dose are well documented to be predictors of cord blood transplant success [1]. Therefore, research has focused on overcoming the cell dose barrier, including transplantation of two cord blood units and ex vivo expansion of cells prior to transplant with the goal of generating clinically meaningful cell doses.
The first strategy, double cord blood unit transplantation, is used by most centers currently. Although this technique has helped to overcome cell dose limitations, there continues to be delayed engraftment and immune reconstitution and the potential for increased complications from graft-versus-host disease. In addition, it is typical to see a single unit emerge as the dominant source of long-term hematopoiesis [2]. Furthermore, the cost of cord blood for transplantation ranges between $25,000 and $45,000 per unit; thus, the expenses involved in a double cord blood unit transplant can be considerable. Therefore, even with double unit transplants, there is still a need to achieve faster engraftment and potentially better immune reconstitution to minimize infectious complications. For these reasons, many investigators have looked into novel stem cell expansion strategies.
Unfortunately, expansion strategies that focus solely on the use of cytokines have not shown significant expansion of repopulating cells. In addition, these techniques are associated with an increased rate of differentiation, which leads to a loss of primitive cells. In short, these studies have not translated into improved engraftment in clinical trials [3-5]. Promising new leads to achieve stem cell expansion have emerged from the discovery of self-renewal genes such as HOXB4 [6-8].
We have recently exploited the large animal model to demonstrate a differential effect of HOXB4 overexpression on short- and long-term repopulating cells in vivo. Using a competitive repopulation assay in a large animal model (Macaca nemestrina), we found that HOXB4 overexpression resulted in superior engraftment over non-HOXB4 controls [9]. Interestingly, HOXB4 appears to have the most dramatic effect on short-term repopulating cells, resulting in 56-fold higher short-term engraftment when compared with control-transduced cells. This offers promise in the field of cord blood transplantation; HOXB4 expansion of a portion of a graft may promote short-term engraftment and provide hematopoietic rescue while awaiting engraftment of long-term repopulating cells. Furthermore, we have also demonstrated a differential effect of HOXB4 on cells from different species [10]. In addition, we have recently shown that a combination of HOXB4 and the Notch ligand Delta-1 synergize to yield enhanced generation of cord blood NOD/SCID repopulating cells with higher levels of engraftment of human CD45+, CD34+, CD3+, CD20+, and CD41+ cells compared to either factor used alone [11]. Therefore, by combining other factors with early-acting genes like HOXB4, it is possible to encourage differentiation along lineages that are often underrepresented in populations of hematopoietic stem cells (HSCs) expanded with HOXB4 alone.
Development of an efficient means for expanding stem cells has broad applications, not limited simply to UCB stem cell transplant. For example, for a large percentage of the population, especially minorities, availability of appropriate donors for allogeneic HSC transplantation is limited [12]; thus, alternate sources of HSCs are under investigation, such as umbilical cord blood. Gene therapy is another example of a field that would benefit from these techniques; investigators could increase the number of gene-modified cells in gene therapy protocols and boost cell numbers for transplant following non-myeloablative conditioning.
However, one of the most significant obstacles facing researchers studying ex vivo expansion techniques is the lack of an appropriate model in which to study HSC biology and behavior. The availability of a large animal model would circumvent this limitation and allow the efficient evaluation of these strategies with long-term follow-up. Thus, in the current study, our goal was to establish a nonhuman primate cord blood transplant model. Subsequently, we wished to use this model to determine if nonhuman primate cord blood cells could be expanded to numbers large enough to be of clinical significance and could engraft in a fully myeloablated nonhuman primate recipient.
MATERIALS AND METHODS
Experimental Design
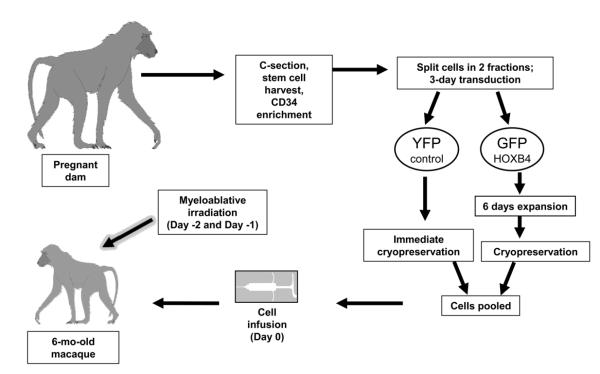
In order to test HOXB4-mediated expansion of cord blood cells in a clinically relevant setting, we developed a nonhuman primate competitive repopulation model (Figure 1). Approximately one week prior to due date, a C-section is performed and cord blood cells are collected from the infant. After processing, the cells are split into two fractions; half are transduced with a control gammaretroviral YFP vector during a 3-day transduction and then frozen. The remaining cells are transduced with a HOXB4GFP vector and expanded for an additional 6 days (for a total of 9 days of ex vivo culture), before being cryopreserved. The use of two different markers, GFP and YFP, allows for a competitive repopulation approach; cells from peripheral blood or bone marrow can easily be analyzed by flow cytometry for the presence of GFP+ and YFP+ cells. Both fractions of cells are maintained in liquid nitrogen for at least 6 months while the infant reaches an appropriate weight. On the day of transplantation, the cells are pooled and intravenously infused into the myeloablated recipient.
Figure 1.
Schematic of experimental design for nonhuman primate autologous cord blood transplant model.
Animal Housing and Care
All pig-tailed macaques (Macaca nemestrina) were housed at the University of Washington National Primate Research Center under conditions approved by the American Association for Accreditation of Laboratory Animal Care. Experimental protocols were approved by the Institutional Animal Care and Use Committee.
For 3 days prior to transplant and continuing through day 56, animals received oral tacrolimus (FK-506) at a dose necessary to maintain serum trough levels between 10 and 15 ng/mL. Over the 2 days prior to transplant, animals were conditioned with fractionated, myeloablative total body irradiation of 1100 cGy [13] from a 6 MV X-ray beam of a single-source linear accelerator located at the Fred Hutchinson Cancer Research Center South Lake Union Facility. The animals underwent a training process to sit calmly in a specially modified cage. The cage provided clear access for the irradiation, while gently restricting excess movement by limiting space. The dose was administered at a rate of 7 cGy/min delivered as a midline tissue dose. The instrument was calibrated weekly to maintain accuracy. Beginning on the day of cell infusion, G-CSF was administered daily until the animals began to engraft, defined as absolute neutrophil count (ANC) > 500/μl for three consecutive days. All animals received standard supportive care, including antibiotics, electrolytes, fluids, and transfusions. Daily complete blood counts (CBC) were used to determine hematopoietic recovery. A total of three macaques were transplanted and followed for this study.
Cord Blood Processing
Red cells from macaque umbilical cord blood were lysed in ammonium chloride red cell lysis buffer. Nucleated cells were incubated for 20 minutes with the 12.8 IgM anti-CD34 antibody, washed, and incubated for another 20 minutes with MACS IgM microbeads (Miltenyi Biotec, Auburn, California). CD34+ cells were enriched via magnetic column separation. Overall, samples ranged in purity from 80–99% CD34+ by flow cytometry.
HOXB4 Transduction and Culture
Transductions were carried out on fibronectin-coated, non-tissue culture-treated plates. Cells were pre-stimulated for 48 hours. After 48 hours, cells were exposed to virus-containing media for two 4-hour transductions (one exposure per day for two consecutive days). Cells were transduced at an MOI of 0.3 (previously determined to cause minimal toxicity). The generation of Phoenix GALV-pseudotyped MSCV-HOXB4-ires-GFP and MSCV-ires-YFP viral vectors has been described previously [9,14,15]. Virus titers were assayed on HT1080 cells, and titers were obtained in the range of 1×105 to 2×105 IU/mL. Vector supernatant was filtered through a 0.45-μm filter and frozen at −80°C until used for transduction.
Cord blood cells were cultured in Iscove’s Modified Dulbecco’s Medium, supplemented with 10% fetal bovine serum and 1% penicillin/streptomycin. The following growth factors were added at a concentration of 100 ng/mL: IL-3, IL-6, TPO, Flt3-L, SCF, and G-CSF. Cultures were split as necessary to maintain cell concentrations in the range of 1 × 105 to 5 × 105 cells/mL.
Flow Cytometry
Flow cytometric data were collected using a Canto I (Becton Dickinson, San Jose, CA) and analyzed using FlowJo software. At least 10,000 events were collected for each sample. Samples were analyzed for expression of the GFP marker (as an indicator of HOXB4 expression) and the YFP marker, as well as for CD3, CD4, CD8, CD13, CD14, CD20, and CD34. Non-transduced cells were used as a control for the gating of HOXB4GFP+ cells and YFP+ cells, and isotype control antibodies were used as a control for gating of positive populations among antibody-labeled cells. All antibodies were purchased from Becton Dickinson.
Colony Forming Unit Assays
Colony forming unit assays were carried out in MethoCult H4230 methylcellulose media (Stem Cell Technologies, Vancouver, Canada) supplemented with 100 ng/mL EPO, IL-3, IL-6, TPO, SCF, G-CSF, and GM-CSF. Plates were incubated at 37°C for 12 to 14 days; after this period of time, colonies of greater than 50 cells were enumerated.
Linear Amplification-Mediated Polymerase Chain Reaction (LAM-PCR)
At periodic intervals following transplantation in L09025, HOXB4GFP+ bone marrow cells were purified by FACS and analyzed by LAM-PCR. The detailed protocol for LAM-PCR has been reported previously [16]. In brief, PCR products were cloned and sequenced. The sequences with legitimate linker and LTR were subjected to BLAT analysis using the University of California, Santa Cruz (UCSC) Genome Browser web site. The rhesus genome from the January 2006 assembly was used for analysis.
NM-PCR for Detection of Retroviral Integration Sites
For analysis of retroviral integration sites in T09214, we used an improved analytical technique involving nebulization mediated (NM)-PCR. Briefly, 300 ng to 3 μg of DNA were nebulized with pressurized nitrogen for 60 seconds. Fragmented DNA was isolated and polished, and modified linkers were ligated following standard procedures (454/Roche-GS 20 DNA Library Preparation Kit, Branford, Connecticut). To amplify the vector-genome junction, dsDNA was amplified in sequential, nested exponential PCR.
Multiplex Pyrosequencing of Retroviral Integration Sites
Successful integration sites amplified by NM-PCR were gel purified to isolate DNA fragments ~800 through ~1,500 bp in length. Gel purified samples were shipped to the University of Illinois at Urbana-Champaign, quality control checked, sequenced on the 454/Roche Titanium system, and FASTA format sequence reads were deposited on a secure server for downstream processing.
Processing of Integration Site Data
For NM-PCR-based amplified vector LTR-chromosome junctions, DNA sequences were processed as previously described [16-18]. The LTR proximal genomic sequences were aligned to the rhesus genome using a stand-alone version of BLAT [19] that generated a BLAST alignment score. Rhesus genome alignments were converted to the human genome position and PERL programs were used to compare localized integration sites to various chromosomal features by using tables available from the University of California at Santa Cruz (UCSC) database as previously described [20].
RESULTS
Development of a Transplant Model
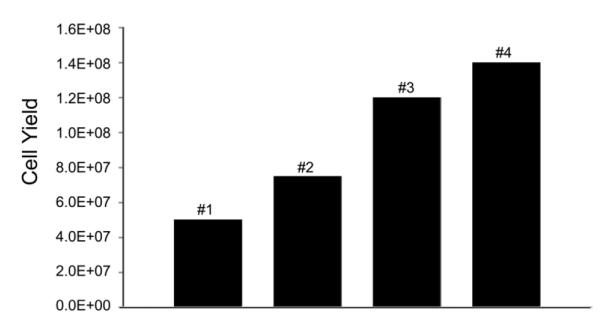
We first wished to establish a cord blood harvest procedure. Over the course of our study, we implemented several improvements to our cord blood collection procedure, including heparin coating of the catheters used to collect blood, use of heparinized saline to flush the vein, massage of the placenta to collect additional cells, as well as other techniques that have been shown to aid in maximizing cell collection during clinical cord blood harvest [21]. By utilizing these practices, we were able to increase cell numbers collected by nearly 3-fold (Figure 2); in addition, successful transduction and expansion of these cells allowed engraftment in our nonhuman primate recipients.
Figure 2. Increasing yields in cord blood harvests.
By making a series of improvements to our cord blood harvest procedure, we were able to increase our harvest yield substantially.
Transduction and Expansion
Next, we wished to determine whether M. nemestrina cord blood cells could be efficiently transduced and expanded. As shown in Table 1, transduction efficiencies ranged from 35% to 46% for the YFP control arm, and from 42% to 46% for the HOXB4GFP arm. Over the three-day transduction period, YFP cells expanded between 3- and 19-fold. HOXB4GFP cells were expanded for an additional 6 days following transduction, for a total of 9 days in culture. These cells expanded between 78- and 204-fold. Transplant doses ranged from 1.0×105 to 7.8×105 YFP+CD34+ cells per kilogram for the control arm, and 2.6×106 to 3.4×106 HOXB4GFP+CD34+ cells per kilogram for the expanded arm. (The difference in YFP+ and GFP+ cell doses is due to the fact that the GFP+ cells were expanded for 6 additional days, thus accounting for an increase in overall cell numbers; however, it is important to reiterate that both experimental arms (GFP+ and YFP+) consisted of identical cell numbers prior to expansion, and thus the HOXB4GFP+ dose consisted of the progeny of an equivalent number of HSCs as the YFP+ dose.) Pre-freeze and post-thaw CFU plating showed that there was no significant loss of repopulating cell viability during cryopreservation (data not shown). These data demonstrate that cord blood cells from macaques can be transduced and expanded to clinically relevant doses.
Table 1.
| L09025 | K09175 | T09214 | |
|---|---|---|---|
| Age | 6 mo. | 7 mo. | 8 mo. |
| Weight | 1.2 kg | 1.0 kg | 1.3 kg |
| Sex | Male | Female | Female |
| Transduction efficiency | 46% (YFP) | 35% (YFP) | 41% (YFP) |
| 46% (GFP) | 45% (GFP) | 42% (GFP) | |
| Fold expansion YFP arm (over 3 days) | 3 | 13 | 19 |
| Fold expansion GFP arm (over 9 days) | 78 | 155 | 204 |
| Transplant Dose | 1.05YFP+CD34+/kg | 1.85YFP+CD34+/kg | 7.85YFP+CD34+/kg |
| 2.66GFP+CD34+/kg | 3.36GFP+CD34+/kg | 3.46GFP+CD34+/kg | |
| Time to ANC > 500/μl (days) | 19 | 7 | 10 |
| Survival post-transplant (days) | 82 | 45 | 270+ |
| Outcome | TTP-like syndrome | Viral pneumonia | Alive |
This table provides a summary of relevant information, including age and weight at time of transplant, transduction efficiencies, fold expansion of YFP and GFP experimental arms, transplant dose, time to neutrophil recovery, and post-transplant survival.
Hematopoietic Recovery
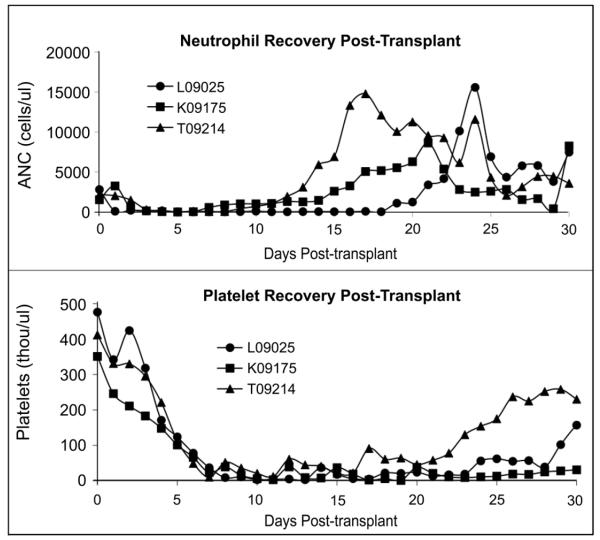
After confirming that we were able to successfully expand cord blood cells ex vivo, we wished to study the hematopoietic recovery kinetics of macaques transplanted with genetically modified cord blood cells. All animals attained neutrophil recovery within the first 3 weeks following transplant. An ANC of greater than or equal to 500 cells per μl was reached by L09025 at day 19, by K09175 at day 7, and by T09214 at day 10. G-CSF was discontinued at day 23 for L09025, day 18 for K09175 and day 15 for T09214. ANC levels remained above 500 cells per μl for the remainder of the study for each animal, though occasional drops in neutrophil counts necessitated an additional dose of G-CSF to boost neutrophil production. The effects of these additional doses can be seen in Figure 3 (top panel) as points when the ANC rises.
Figure 3. Hematopoietic recovery during the first 30 days following autologous cord blood transplant.
Recovery of neutrophils is shown in the top panel, and recovery of platelets is shown in the bottom panel. Neutrophils begin to recover in all three animals after about two weeks. Platelet recovery takes longer, and begins to occur after about 3 weeks.
All animals demonstrated a decline in platelet counts following TBI, and remained thrombocytopenic for at least two to three weeks following transplant. However, Figure 3 (lower panel) shows that T09214 and L09025 began to resume normal thrombopoiesis by the end of the first month post-transplant. K09175 continued to experience thrombocytopenia, and required frequent transfusions over the course of her time on the study. Overall, these data demonstrate that we are able to achieve recovery of neutrophils and platelets in a timely manner following myeloablative conditioning.
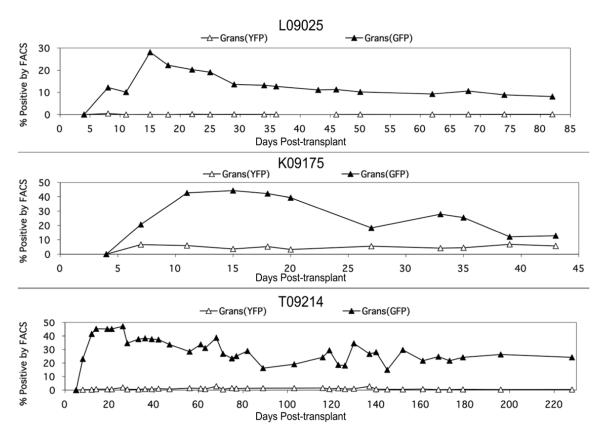
Gene Marking
The inclusion of the GFP or YFP reporter gene in our retroviral vectors allowed us to track the contribution of gene-marked cells in the animals. All animals demonstrated a similar pattern in engraftment of gene-modified granulocytes. This trend is illustrated in Figure 4. In each case, gene marking in the HOXB4GFP arm climbed rapidly over the first two weeks, reached a peak around two to three weeks post-transplant, and then began to decline until eventually stabilizing after approximately the first month. HOXB4GFP marking levels in L09025 and K09175 stabilized around 10%; in T09214, HOXB4GFP marking levels stabilized around 20%. Gene marking in the YFP control arm was detectable in all three animals, though at low levels. In both L09025 and T09214, only about 0.5% to 1.5% of granulocytes were YFP+ by flow cytometric analysis. In K09175, this level was significantly higher and remained relatively consistent at about 5%. Thus, we were able to show improved engraftment kinetics and durability of HOXB4-transduced and expanded cells compared to control-transduced cells.
Figure 4. Percent GFP+ and YFP+ granulocytes in each animal by FACS analysis over the course of the study.
In each case, GFP+ granulocytes reach a peak within the first month post-transplant, and then decline and stabilize. YFP+ granulocytes, though present at very low levels, were detectable at each time point, even in L09025.
Subset Stains
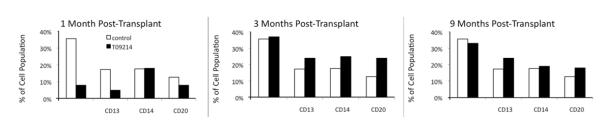
To determine the percentages of, and gene marking within, various subsets contributing to hematopoietic recovery, we performed subset stain assays. Full, comprehensive subset stains were performed on L09025 (approximately three months post-transplant) and T09214 (1, 3, 6, and 9 months post-transplant). Peripheral blood cells were stained for CD3, CD13, CD14, and CD20, and analyzed by flow cytometry to determine the contributions of different subsets to overall hematopoiesis (Figure 5) and the contributions of GFP+/YFP+ cells to different subsets (Figure 6). One month after transplant, T09214 had lower percentages of CD3, CD13, and CD20 cells compared to a non-transplanted control animal. However, by 3 and 9 months post-transplant, T09214 had regained normal hematopoiesis (Figure 5), thus showing that HOXB4-mediated expansion did not lead to any long-term skewing of hematopoiesis.
Figure 5. Hematopoietic composition of T09214 at various time points post-transplant, compared to a non-transplanted control animal.
At 1 month post-transplant, T09214 has lower percentages of CD3, CD13, and CD20 cells compared to a non-transplanted control animal. By 3 and 9 months post-transplant, T09214 has regained relatively normal hematopoietic reconstitution.
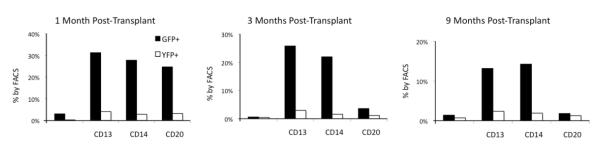
Figure 6. Percent of GFP+ and YFP+ cells among hematopoietic lineages at various time points post-transplant.
Over time, the percentage of GFP+ cells is dropping among all lineages, showing that HOXB4GFP+ cells are conferred with a short-term, but not long-term, repopulating advantage. At each time point, GFP+ cells outnumber YFP+ cells in all 4 lineages; this effect is most dramatic among myeloid populations.
The percentage of gene marking within individual subsets is represented in Figure 6. Here, we show that, at each time point, the percentage of GFP+ cells is higher than the percentage of YFP+ cells within each subset. However, this effect is much more exaggerated among the myeloid lineages (a 10–30 percentage point difference) than among the lymphoid lineages (typically a 1–3 percentage point difference). Thus, we show that the HOXB4-transduced and -expanded cells are more prevalent than control-transduced cells among multiple lineages, with the most pronounced effect on myeloid populations.
Retroviral Integration Site Analysis
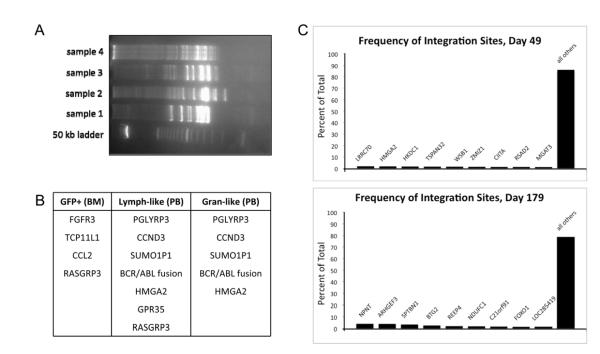
Retroviral integration site analysis was performed on L09025 at day 82 post-transplant using a LAM-PCR based approach. We used this technique to analyze three separate populations of cells (GFP-sorted cells from the bone marrow, lymphocyte-like cells from the peripheral blood, and granulocyte-like cells from the peripheral blood). The presence of multiple bands on the acrylamide gel shown in Figure 7A indicates a polyclonal population. Furthermore, the table in Figure 7B lists several of the most frequently identified integration sites, and it is noted that several of these show up among more than one of the three populations studied, thus indicating that true hematopoietic repopulating cells have been transduced and expanded.
Figure 7. Integration site analysis.
Retroviral integration site analysis was performed at day 82 in L09025 (A and B) and day 49 and 179 in T09214 (C). For L09025, three separate populations were analyzed: GFP-sorted cells from bone marrow (BM), lymphocyte-like cells from peripheral blood (PB), and granulocyte-like cells from PB. The gel (A) shows numerous bands indicating a polyclonal population. The table (B) lists the integration sites that were found in each population (integration was either in or within 125,000 base pairs of each gene listed.) Several of the sites turned up among multiple populations, thus indicating that true repopulating cells were expanded. Analysis of PB in T09214 (C) at day 49 and day 179 revealed nearly 300 unique integration sites at each time point; the graphs show the frequency of the top 9 most common sites and indicate relatively uniform site distribution with no evidence of clonal outgrowth.
When analyzing retroviral integration sites in T09214, we modified our approach and opted to use an improved technique involving NM-PCR (detailed above) followed by 454/Roche pyrosequencing. This improved method allows for the capture of exponentially more valid integration sites, and thus we elected to switch to this technique for future analyses. Based on this method, we investigated integration sites in T09214 at two time points post-transplant, day 49 and day 179. These analyses were highly efficient, and we were able to capture over 2,200 valid integration sites at day 49, of which 298 were unique. Analysis at day 179 was similarly efficient; we were able to collect over 2,700 valid integration sites, with 263 of these being unique. In Figure 7C, the results are illustrated graphically, showing the frequency of the nine most commonly captured integration sites alongside the combined contribution of all other sites.
Outcome
L09025 survived for 82 days post-transplant. This animal was eventually euthanized due to complications from TTP (thrombotic thrombocytopenic purpura)-like syndrome, evidenced by pancytopenia and acute renal failure. A bone marrow biopsy was performed at time of euthanasia, and it showed a fragmented and focally hemorrhagic marrow with a cellularity of 80–90%. Erythroid, myeloid and megakaryocytic lineages were all present with apparent normal maturation. Lymphocytes and plasma cells were also readily apparent. Immature cells, such as blasts, did not appear increased in number, and no fibrosis was noted.
K09175 survived for 45 days post-transplant before succumbing to viral pneumonia. A bone marrow biopsy taken soon after death indicated the presence of a few lymphocytes that showed cytotoxic granules in the cytoplasm; in other words, large granular lymphocytes were observed, which tend to be increased in infectious/inflammatory processes. There was no evidence of blasts or of any malignant cell morphology.
T09214 continues to survive at 12 months post-transplant and demonstrates normal health and behavior. Complete blood counts, blood chemistries, appetite, weight gain, and social interaction are all commensurate with a normal, healthy, untransplanted macaque. Regular, periodic blood cultures show no signs of fungal or bacterial infection. The animal continues to be closely monitored by CBC and blood chemistry every two weeks, gene marking analysis once per month, comprehensive veterinary exam once per month, full subset stains every three months, and integration analysis every three months.
DISCUSSION
Here, we have established a nonhuman primate cord blood transplant model and demonstrated that this model allows the evaluation of stem cell expansion strategies. Furthermore, using a HOXB4-mediated expansion technique, we have shown sustained engraftment of gene-modified, expanded, autologous cord blood cells in three macaques. Our findings show that HOXB4-expanded cells result in superior engraftment of cells of multiple lineages, compared to control non-expanded cells. To our knowledge, this paper describes the first successful cord blood transplants performed in the nonhuman primate model.
In this study, macaques were transplanted with autologous cord blood CD34+ cells approximately 6–8 months after C-section delivery and cord blood collection. Half of the cells in the harvested graft were transduced with a YFP control vector and cryopreserved, while the other half were transduced with a HOXB4GFP vector and expanded for an additional 6 days (Figure 1). Post-transplant follow-up included CBC, blood chemistry, gene marking analysis, subset stains, and integration analysis.
We have previously demonstrated that HOXB4-transduced and expanded cells exhibit improved engraftment over HOXB4-transduced, non-expanded cells [9]. In this prior work, we also showed that control-transduced (no HOXB4) and expanded cells resulted in very low engraftment levels. This is because, in the absence of HOXB4, cells in culture tend to favor differentiation in lieu of self-renewal, thus resulting in a loss of the primitive phenotype. Therefore, an expanded graft of cells not containing HOXB4 has a low repopulating potential. Consequently, in the current work we elected to elaborate upon our previous findings and utilize an experimental design in which we compared engraftment of HOXB4-expanded cells and control, non-expanded cells.
All three macaques attained neutrophil engraftment within 19 days post-transplant, and began to recover platelet counts within the first month post-transplant (Figure 3). This is a drastic improvement over trends in human cord blood transplantation using non-expanded cells, in which neutrophil engraftment takes an average of 3–4 weeks, even in the double cord blood unit setting [22], and platelet recovery takes an average of sixty days [23]. Therefore, our results indicate that HOXB4-transduced and expanded cells substantially abbreviate the period of pancytopenia following cord blood transplantation. We cannot entirely disregard the possibility that differences in engraftment kinetics between humans and macaques also contribute to this shortened period of pancytopenia; however, we believe this is unlikely given the fact that cells in our studies were cultured and manipulated, which generally leads to a loss of stem cells and decrease in engraftment, unless conditions are used that could maintain or expand stem cells.
For comparison purposes, we also evaluated the number of days of severe neutropenia (ANC < 100/μl) following HOXB4-mediated bone marrow transplant and HOXB4-mediated cord blood transplant. Macaques receiving HOXB4-transduced and -expanded bone marrow cells (detailed in previously published studies from our laboratory) showed an average of 2 days of severe neutropenia [9]. In contrast, macaques in our current study receiving HOXB4-transduced and -expanded cord blood cells demonstrated an average of 7 days of severe neutropenia. This difference in post-transplant kinetics is not unanticipated however, as it is well established that recovery after cord blood transplant is substantially delayed compared to recovery after bone marrow transplant [24].
All animals demonstrated a similar trend in gene marking levels, with the HOXB4GFP arm increasing to 30–50% within the first three weeks, and stabilizing around 10–20% thereafter (Figure 4). This characteristic shape of the gene marking graphs was observed in our previous work involving HOXB4-mediated bone marrow transplant as well [9]. These findings support the conclusion that, at least under the currently used in vitro conditions, HOXB4 has a larger impact on short-term repopulating cells than on long-term repopulating cells. Within the first 30 days, when short-term repopulating cells are beginning to emerge, it is observed that the difference between percent GFP+ cells and percent YFP+ cells is, on average, between 25- and 30-fold. However, note that the difference in doses of HOXB4GFP+ cells and YFP+ cells infused is, on average, only between 15- and 20-fold (Table 1). This disparity between the number of GFP+/YFP+ cells infused and the percentage of GFP+/YFP+ cells within the first month post-transplant provides evidence that the HOXB4-transduced cells have been conferred with a competitive repopulating advantage.
Further evidence that HOXB4 affects short-term repopulating cells more profoundly than long-term repopulating cells can be seen in Figure 6. This figure shows the GFP+/YFP+ composition of individual subsets at three time points for T09214: 1, 3, and 9 months post-transplant. In general, the prevalence of HOXBGFP+ cells is dropping over time among all lineages. In fact, at 9 months post-transplant, the only lineages in which GFP+ cells are present at greater than 2% are CD13 and CD14 cells (in these subsets, GFP+ cells are present at 13.2% and 14.3%, respectively). Furthermore, subset stain results for both L09025 (not shown) and T09214 (Figure 6) consistently demonstrate that GFP+ percentages are higher than YFP+ percentages among both myeloid and lymphoid cells, but that this effect is most dramatic among myeloid populations, indicating that HOXB4 preferentially expands cells of the myeloid lineage. In addition, retroviral integration site analysis on macaques L09025 and T09214 show that hematopoietic repopulation is polyclonal, thus indicating that there was no outgrowth of clonal cells during in vitro expansion.
These studies offer promise in improving outcome following cord blood transplant. HOXB4-mediated expansion of a portion of a graft may improve engraftment of short-term repopulating cells, thus providing hematopoietic recovery/rescue while longer-term repopulating cells emerge. This would abbreviate the period of pancytopenia and reduce susceptibility to infection following cord blood transplant, thus enhancing survival. The large animal model that we have developed here can also be used for studying alternative expansion strategies, such as HOXB4 protein-mediated expansion. A recombinant TAT-HOXB4 protein has been developed [8] with proven efficacy in expanding hematopoietic stem cells; the utilization of this protein may offer a means of expanding cord blood cells without the accompanying risks of genetic modification. This model will also provide an important platform for new and potentially more potent stem cell expansion agents such as NUP98-HOXA10hd [25] and the small molecule SR1 [26], alone or in combination, to achieve clinically useful expansion of CB derived stem cells.
Furthermore, the development of this model is a valuable contribution to the field of pre-clinical modeling. It is often difficult to study stem cell expansion techniques because of the challenge of finding a suitable in vivo assay. The murine model has proven to be convenient for studying certain aspects of cell expansion and gene therapy, such as transduction efficiency of repopulating cells, short-term engraftment kinetics, and multi-lineage repopulation capability. However, there are many shortcomings of the murine model that often interfere with the ability to translate these findings into human patients [27]. Therefore, our nonhuman primate model is highly valuable for efficient evaluation of expansion strategies, with direct translation to the clinic. Likewise, major histocompatibility complex (MHC) typing is established for nonhuman primates [28,29]; thus, allogeneic transplantation can be performed and evaluated in this setting, thereby offering a significant advantage. In addition, the overwhelming homology between humans and nonhuman primates allows for many of the same cytokines, growth factors, drugs, and therapeutic regimens to be used. Furthermore, one of the most frequently studied diseases, HIV, can be modeled in the nonhuman primate, offering the potential to study this disease using gene-modified cord blood cells.
CONCLUSIONS
Here, we have established a large-animal cord blood transplant model which can be utilized in a variety of ways in order to test different cord blood expansion techniques as a precursor to clinical trials. In addition, we have shown that HOXB4 transduction and expansion of cord blood stem cells leads to improved engraftment of short-term repopulating cells. Furthermore, we have demonstrated that HOXB4 is capable of expanding cord blood cells to doses high enough to be of clinical relevance.
ACKNOWLEDGMENTS
This work was supported in part by NIH grants P30 DK056465, R01 HL084345, RL1 HL092554, and R01 HL098489. We thank Bonnie Larson and Helen Crawford for help in preparing the manuscript. H.P. Kiem is a Markey Molecular Medicine Investigator and the recipient of the José Carreras/E. Donnall Thomas Endowed Chair for Cancer Research.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
There are no financial interests/relationships to disclose.
REFERENCES
- 1.Gluckman E, Rocha V, Arcese W, et al. Factors associated with outcomes of unrelated cord blood transplant: guidelines for donor choice. Exp Hematol. 2004;32:397–407. doi: 10.1016/j.exphem.2004.01.002. [DOI] [PubMed] [Google Scholar]
- 2.Ballen KK, Spitzer TR, Yeap BY, et al. Double unrelated reduced-intensity umbilical cord blood transplantation in adults. Biol Blood Marrow Transplant. 2007;13:82–89. doi: 10.1016/j.bbmt.2006.08.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jaroscak J, Goltry K, Smith A, et al. Augmentation of umbilical cord blood (UCB) transplantation with ex vivo-expanded UCB cells: results of a phase 1 trial using the AastromReplicell System. Blood. 2003;101:5061–5067. doi: 10.1182/blood-2001-12-0290. [DOI] [PubMed] [Google Scholar]
- 4.Pecora AL, Stiff P, Jennis A, et al. Prompt and durable engraftment in two older adult patients with high risk chronic myelogenous leukemia (CML) using ex vivo expanded and unmanipulated unrelated umbilical cord blood. Bone Marrow Transplant. 2000;25:797–799. doi: 10.1038/sj.bmt.1702222. [DOI] [PubMed] [Google Scholar]
- 5.Shpall EJ, Quinones R, Giller R, et al. Transplantation of ex vivo expanded cord blood. Biol Blood Marrow Transplant. 2002;8:368–376. doi: 10.1053/bbmt.2002.v8.pm12171483. [DOI] [PubMed] [Google Scholar]
- 6.Sauvageau G, Thorsteinsdottir U, Eaves CJ, et al. Overexpression of HOXB4 in hematopoietic cells causes the selective expansion of more primitive populations in vitro and in vivo. Genes Dev. 1995;9:1753–1765. doi: 10.1101/gad.9.14.1753. [DOI] [PubMed] [Google Scholar]
- 7.Antonchuk J, Sauvageau G, Humphries RK. HOXB4-induced expansion of adult hematopoietic stem cells ex vivo. Cell. 2002;109:39–45. doi: 10.1016/s0092-8674(02)00697-9. [DOI] [PubMed] [Google Scholar]
- 8.Krosl J, Austin P, Beslu N, Kroon E, Humphries RK, Sauvageau G. In vitro expansion of hematopoietic stem cells by recombinant TAT-HOXB4 protein. Nat Med. 2003;9:1428–1432. doi: 10.1038/nm951. [DOI] [PubMed] [Google Scholar]
- 9.Zhang X-B, Beard BC, Beebe K, Storer B, Humphries RK, Kiem H-P. Differential effects of HOXB4 on nonhuman primate short- and long-term repopulating cells. PLoS Medicine. 2006;3 doi: 10.1371/journal.pmed.0030173. DOI: 10.1371/journal.pmed.0030173- [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang X-B, Schwartz JL, Humphries RK, Kiem H-P. Effects of HOXB4 overexpression on ex vivo expansion and immortalization of hematopoietic cells from different species. Stem Cells. 2007;25:2074–2081. doi: 10.1634/stemcells.2006-0742. [DOI] [PubMed] [Google Scholar]
- 11.Watts KL, Delaney C, Humphries RK, Bernstein I, Kiem H-P. Combination of HOXB4 and delta-1 ligand improves expansion of cord blood cells. Blood. 2010;116:5859–5866. doi: 10.1182/blood-2010-05-286062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Johansen KA, Schneider JF, McCaffree MA, Woods GL. Efforts of the United States’ National Marrow Donor Program and Registry to improve utilization and representation of minority donors (Review) Transfusion Medicine. 2008;18:250–259. doi: 10.1111/j.1365-3148.2008.00865.x. [DOI] [PubMed] [Google Scholar]
- 13.Watts KL, Beard BC, Wood BL, Kiem HP. Myeloablative irradiation in non-human primates. Journal of Medical Primatology. 2009;38:425–432. doi: 10.1111/j.1600-0684.2009.00368.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Antonchuk J, Sauvageau G, Humphries RK. HOXB4 overexpression mediates very rapid stem cell regeneration and competitive hematopoietic repopulation. Exp Hematol. 2001;29:1125–1134. doi: 10.1016/s0301-472x(01)00681-6. [DOI] [PubMed] [Google Scholar]
- 15.Pineault N, Abramovich C, Ohta H, Humphries RK. Differential and common leukemogenic potentials of multiple NUP98-Hox fusion proteins alone or with Meis1. Mol Cell Biol. 2004;24:1907–1917. doi: 10.1128/MCB.24.5.1907-1917.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Beard BC, Keyser KA, Trobridge GD, et al. Unique integration profiles in a canine model of long-term repopulating cells transduced with gammaretrovirus, lentivirus, and foamy virus. Hum Gene Ther. 2007;18:423–434. doi: 10.1089/hum.2007.011. [DOI] [PubMed] [Google Scholar]
- 17.Beard BC, Dickerson D, Beebe K, et al. Comparison of HIV-derived lentiviral and MLV-based gammaretroviral vector integration sites in primate repopulating cells. Molecular Therapy. 2007;15:1356–1365. doi: 10.1038/sj.mt.6300159. [DOI] [PubMed] [Google Scholar]
- 18.Trobridge GD, Miller DG, Jacobs MA, et al. Foamy virus vector integration sites in normal human cells. PNAS. 2006;103:1498–1503. doi: 10.1073/pnas.0510046103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kent WJ. BLAT--the BLAST-like alignment tool. Genome Res. 2002;12:656–664. doi: 10.1101/gr.229202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Karolchik D, Baertsch R, Diekhans M, et al. The UCSC Genome Browser Database. Nucleic Acids Res. 2003;31:51–54. doi: 10.1093/nar/gkg129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bornstein R, Flores AI, Montalban MA, del Rey MJ, de la SJ, Gilsanz F. A modified cord blood collection method achieves sufficient cell levels for transplantation in most adult patients. Stem Cells. 2005;23:324–334. doi: 10.1634/stemcells.2004-0047. [DOI] [PubMed] [Google Scholar]
- 22.Barker JN, Weisdorf DJ, Defor TE, et al. Transplantation of 2 partially HLA-matched umbilical cord blood units to enhance engraftment in adults with hematologic malignancy. Blood. 2005;105:1343–1347. doi: 10.1182/blood-2004-07-2717. [DOI] [PubMed] [Google Scholar]
- 23.Laughlin MJ, Eapen M, Rubinstein P, et al. Outcomes after transplantation of cord blood or bone marrow from unrelated donors in adults with leukemia. N Engl J Med. 2004;351:2265–2275. doi: 10.1056/NEJMoa041276. [DOI] [PubMed] [Google Scholar]
- 24.Petropoulou AD, Rocha V. Risk factors and options to improve engraftment in unrelated cord blood transplantation. Stem Cells International. 2011;2011:610514. doi: 10.4061/2011/610514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sekulovic S, Gasparetto M, Lecault V, et al. Ontogeny stage-independent and high-level clonal expansion in vitro of mouse hematopoietic stem cells stimulated by an engineered NUP98-HOX fusion transcription factor. Blood. :9999. doi: 10.1182/blood-2011-04-350066. prepublished online August 24, 2011; doi:10.1182/blood-2011-04-350066- [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Boitano AE, Wang J, Romeo R, et al. Aryl hydrocarbon receptor antagonists promote the expansion of human hematopoietic stem cells. Science. 2010;329:1345–1348. doi: 10.1126/science.1191536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Horn PA, Thomasson BM, Wood BL, Andrews RG, Morris JC, Kiem H-P. Distinct hematopoietic stem/progenitor cell populations are responsible for repopulating NOD/SCID mice compared with nonhuman primates. Blood. 2003;102:4329–4335. doi: 10.1182/blood-2003-01-0082. [DOI] [PubMed] [Google Scholar]
- 28.Kaizu M, Borchardt GJ, Glidden CE, et al. Molecular typing of major histocompatibility complex class I alleles in the Indian rhesus macaque which restrict SIV CD8+ T cell epitopes. Immunogenetics. 2007;59:693–703. doi: 10.1007/s00251-007-0233-7. [DOI] [PubMed] [Google Scholar]
- 29.Tanaka-Takahashi Y, Yasunami M, Naruse T, et al. Reference strand-mediated conformation analysis-based typing of multiple alleles in the rhesus macaque MHC class I Mamu-A and Mamu-B loci. Electrophoresis. 2007;28:918–924. doi: 10.1002/elps.200600586. [DOI] [PubMed] [Google Scholar]