Abstract
Thrombin receptor, F2R or PAR1 is a G-protein coupled receptor, located in the membrane of endothelial cells. It has been initially found to transduce signals in hemostasis, but recently also known to act in cancer and in vascular development. Mouse embryos lacking PAR1 function die from hemorrhages with varying frequency at midgestation. We have performed a survey of potential PAR1 homologs in the zebrafish genome and identified a teleost ortholog of mammalian PAR1. Knockdown of par1 function in zebrafish embryos demonstrates a requirement for Par1 in cardio-vascular development. Furthermore, we show that function of Par1 requires the presence of a phylogenetically conserved proteolytic cleavage site and a second intracellular domain. Altogether our results demonstrate a high degree of conservation of PAR1 proteins in the vertebrate lineage in respect to amino acid sequence as well as protein function.
Introduction
The protease-activated receptors (PARs) form a class of G-protein-coupled receptors (GPCRs) activated by a unique proteolytic cleavage in the extracellular N-terminus. The PAR family member PAR1, also termed f2r for coagulation factor II receptor, is constitutively expressed at the surface of endothelial cells. It has been shown to be cleaved by a highly selective group of serine proteases that includes thrombin, plasmin [1], coagulation Factor Xa [2] and activated protein C [3]. Recent studies have shown that platelet matrix metalloprotease-1 (MMP-1) also cleaves PAR1, but at a different site [4], [5]. Cleavage unmasks a short peptide sequence that interacts with the second extracellular loop, acting as a tethered ligand for the receptor [6]. The proteolytic activation of PAR1 induces transmembrane signalling to internally located G proteins [7]. MMP-1 creates a longer tethered ligand, which activates a distinct spectrum of G protein pathways [5]. Targeted disruption of Par1 in mice resulted in bleeding at multiple sites, cardiovascular failure and partial embryonic lethality, likely involving endothelial defects that did not impair hemostasis [8], [9]. Whereas vasculogenesis (i.e. the formation of the primitive vascular plexus from endothelial progenitors) was normal, Par1 mutants displayed abnormal yolk sac vascular pattern due to disorganization or delayed remodelling of the primitive vascular network, as well as some breaks in the sinus venous wall. Surprisingly, half of the Par1 −/− embryos survived and became grossly normal adult mice with no bleeding diathesis. Reconstitution of a PAR1 transgene driven by an endothelial-specific promoter rescued the Par1 −/− embryonic lethality [9]. Numerous in vitro data showed that PAR1 contributes to many functions, such as alteration of vascular tone and permeability, angiogenesis, smooth muscle cell proliferation, production of extracellular matrix, and alteration of gene expression including that for adhesion molecules, chemokines, cytokines and recently the EHT (endothelial-to-hematopoietic transition) (for review see [10], [11], [12]. The irreversible N-terminus cleavage and activation of PAR1 necessitates rapid desensitization by phosphorylation, internalization and receptor level recovery from an internal pool of PAR. De novo synthesis is thereafter initiated to restore normal internal level [13], [14].
The zebrafish, due to the facility of observing and manipulating early embryos, is an excellent model for investigating the molecular basis of vertebrate development, including vascular morphogenesis in vivo [15]. The zebrafish vascular system develops in a similar fashion to other vertebrates and because of its small size, the zebrafish embryo can absorb enough oxygen through passive diffusion from water to survive for several days even in the complete absence of a functional vasculature.
In this study, we have examined the possible involvement of Par1 in vascular morphogenesis. We reported the characterization of a zebrafish Par1 protein orthologues to those previously identified in other species. Moreover, zebrafish par1 knockdown led to heart dysfunction and vascular defects. We also demonstrated that the N-terminus and the secondary loop domains of Par1 are necessary for normal cardiovascular development clarifying the role of Par1 during vascular embryonic development.
Results and Discussion
PAR1 is highly conserved throughout evolution
To identify PAR1 homologues in the zebrafish, we performed a BLAST search for a zebrafish homologue of the human PAR1 receptor in the most current zebrafish genome database (Ensembl, Zv9). Similar to previously published [16], we identified a zebrafish homologue, Par1 (ENSDARG00000060012, A2BIP6, BX950872.7). Zebrafish Par1 displays 61.6% consensus and 41.4% identity to human PAR1. As described for other species, zebrafish Par1 hydropathy profile analysis revealed a putative seven transmembrane structure (Fig. S1A–C). Looking for characteristic features of PARs in zebrafish Par1, we identified the sequence R28/SFSGFF, which is very similar to the R41/SFLLRN motif at which human PAR1 is cleaved and activated by thrombin [17]. Whereas the N- and C- terminus are moderately similar, the intra- and extra-cellular loops are highly conserved. In particular, the second extracellular loop (the ligand activation site in other species) and the second intracellular loop (important for signal transduction) are strikingly similar (Fig. S1C). Phylogenetic analysis also showed that zf-Par1 is most closely related to PAR1 of other teleostei and Xenopus (Fig. S1B). Our analysis is consistent with the recently published study from Xu et al [16]. The expression pattern during early zebrafish development reported by this group suggests a role of Par1 in haematopoiesis and vasculogenesis. They described par1 expression in the intermediate cell mass and later in the posterior blood islands (PBI), the heart and veins. Xu et al also found specific expression from 2 days postfertilization (dpf) onward in various organs including the pancreas, the pronephric duct and the gut [16]. Their expression data also indicate maternal contribution of the mRNA, resembling that of the mammalian PAR1 gene.
Additionally to this zebrafish par1 gene, several potential duplicate genes appeared during the search. This issue had previously been addressed, asserting high occurrence of gene duplication for all the PAR family members [18]. A later revision clarified these data as discrepancy between the database versions [16]. We found possible uncharacterized proteins resembling Par1. We performed a phylogenic and extended-homology alignment of the potential duplicate genes versus the zebrafish Par1 (F2r), Par2a (F2rl1.1), Par2b (F2rl1.2) and Par3 (F2rl2) receptors (Fig. S2).
The initially identified zebrafish Par1 protein is more closely related to Par1 orthologous in both fish and mammals, however the specific relationship of the potential paralogues to zf Par1 was highly dependent on method of analysis, but always scored higher conservation to Par1 than Par2a/b (Fig. S2A). It is noteworthy that in several simulations, the Par1 paralogues were often grouped with one of the Par1 paralogues in both Takifugu rubripes and Tetraodon nigroviridis. Moreover, highly conserved sequences were found in putative transmembrane domains of the Par1 protein. There is a clear homology between Par1 and Par2a/b, while for the paralogue candidates; N-terminal and C-terminal are different from these three proteins (Fig. S2B). Furthermore, human PAR1 and PAR2 genes are located on the same chromosome (5q13), whereas the here addressed zebrafish par1 and par2(a/b) genes, reside on different chromosomes (5 and 21, respectively). The candidate duplicate genes are located in the same 0.2 Mb region as par2a and par2b (Fig. S2B), further suggesting that they are either coding for Par2-like proteins or that par1 was translocated after duplication. The mentioned putative paralogues needs to be further characterized.
Interestingly, expression analysis of other members of the PAR family in the zebrafish do not indicate that there could be a redundant functional gene of the same family in endothelial cells [16]. An in situ hybridization study demonstrated that zebrafish par1 and par2a-b genes display distinct spatial and temporal expression patterns during development. In addition, par3 was not detected before 3 dpf by RT-PCR [16]. In mammals, four protease-activated receptors constitute the PAR family. In mouse, PAR1 and PAR4 receptors serve at least partially redundant roles in endothelial cells and together are necessary for the measurable thrombin responses [19]. Like others, we failed to identify a zebrafish homolog of the human or mouse PAR4 gene.
Genetic knockdown of par1 disturbs cardio-vascular maturation
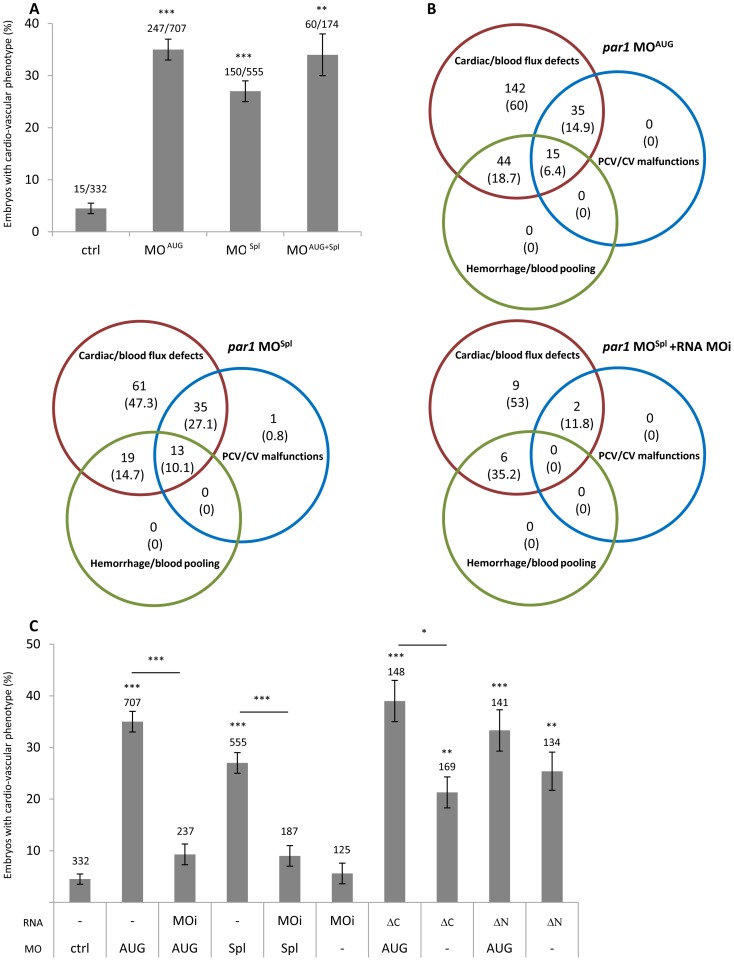
The evolutionary conserved expression of par1 mRNA suggests fundamental functions for this gene. To investigate the function of Par1 in zebrafish during early embryonic development, we used morpholino knockdown as a reverse genetic approach. Our morpholinos target the region surrounding the translation start codon of transcripts (par1 MOAUG) or inhibit the splicing of pre-mRNA between the exon 1 and intron 1 (par1 MOSpl). RT-QPCR, performed on morphants embryos injected with the splice-targeting morpholino, revealed a threefold increase of the non-spliced form. At 48 hours post fertilization (hpf), 35% (MOAUG) and 27% (MOSpl) of par1 morphants displayed a cardio-vascular phenotype ( Fig. 1A ). Our phenotypic analysis of par1 morphants in zebrafish matched well with the partial mouse embryonic phenotype and lethality previously described by others [8], [9]. We observed that par1 morphants with a cardio-vascular phenotype, had weak heart rates associated with slowed to no blood flow. Comparison of par1 morphants showing this phenotype revealed two additional classes of vascular failures ( Fig. 1B ); haemorrhage/blood pooling and PCV/CV malfunctions. We never observed a higher percentage of phenotypes by injecting higher concentration of the morpholino (than 4 ng) or by co-injection of the two morpholinos. Lower concentration of the morpholino revealed concentration-dependent numbers of embryos with a phenotype ( Table 1 ). Co-injections with Rhodamin Dextran indicated no phenotypic variation due to variations in injection volume (data not shown).
Figure 1. par1 knockdown induces cardio-vascular phenotype at 48 hpf.
(A) cardio-vascular phenotype in control and par1 morphants. n values were indicated on the top of each bar of the graph. (B) Venn diagram representation of three classes of vascular failure. n values (% of embryos/embryos with cardio-vascular phenotype) were indicated. (C) cardio-vascular phenotype of morpholino and/or RNA injected embryos. Three par1 mRNA mutants were used. n values were added on the top of each bar of the graph. Average ± error bars from ≥3 independent experiments are presented. * p<0.05; ** p<0.01; *** p<0.001. ctrl, control; MOAUG, par1 MOAUG; MOSpl, par1 MOSpl; MOi, par1 mRNA morpholino-insensitive; ΔC, par1 mRNA morpholino-insensitive lacking intracellular domain; ΔN, par1 mRNA N terminus deleted.
Table 1. Dose dependence of par1 MOAUG phenotype at 2 dpf.
| Dose (ng) | Embryos (n) | Embryos with phenotype/n (%) | |
| Ctrl MO | 4 | 332 | 4,5±1 |
| par1 MOAUG | 1,3 | 135 | 12,2±2,8 |
| 2 | 106 | 17,2±4,1 | |
| 4 | 332 | 35,0±2,0 |
Intact cleavage site and second intracellular domain are required to rescue par1 knockdown
In order to test the specificity of our antisense strategy, we designed a morpholino-insensitive par1 mRNA construction which is mutated at the MO target, named par1-MOi (Fig. S1D). We observed that co-injection of par1-MOi mRNA with par1-MOAUG or par1-MOSpl was able to rescue the cardio-vascular phenotype, allowing the embryos to develop normally. This provides strong evidence that our observations are specific to par1 knockdown ( Fig. 1B–C and 3A ). Involvement of zebrafish Par1-conserved domains in the cardio-vascular phenotype was evaluated with two mRNA constructs: par1-ΔN, where the N-terminus cleavage site is deleted, and par1-ΔC, lacking the second intracellular domain required for signalling (Fig. S1D). The former cannot be activated by classical cleavage, whereas the latter should have lost the ability to transduce any signal. Co-injection of par1-ΔC or par1-ΔN mRNA with par1-MOAUG was unable to rescue the vascular phenotype, respectively ( Fig. 1C ). These results indicate that the Par1 protein acts during vascular development in the zebrafish embryo and that this activity requires proteolytic cleavage and the intracellular domain as in mammals. Furthermore, overexpression of Par1-ΔC or -ΔN in wild type embryos suggests a dominant negative effect of these truncated receptors.
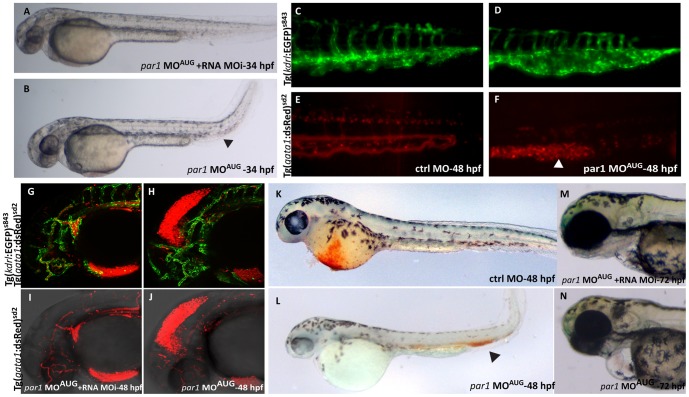
Figure 3. par1 knockdown causes ICM blood pooling, heart edema and head hemorrhage.
(A) 34 hpf rescued embryo with heart beat and blood flow, resembling a ctrl MO embryo at this stage. (B) par1 morphant showing blood pooling (arrowhead) with bend tail and heart edema. (C–F) The posterior part of the embryo at 48 hpf. The ctrl MO embryo has dorsal aorta, CV, ISVs and arteries with expanded lumen with blood flow as seen in the double transgenic background (C and E). The par1 morphant shows ISVs, arteries, dorsal aorta and CV, but the blood cells are pooled in the CV (arrowhead in F) and the vein is bulged and malformed (asterisk in D). (G–J) Head of 48 hpf double transgenic embryos, displaying endothelial cells in green fluorescence and blood cells in red fluorescence (Tg(kdrl:EGFP)s843; Tg(gata1:dsRed)sd2). The blood vessels in the head of both the rescued embryo and the morphant are developed normally, but there is a leakage of blood cells into the mesencephalon of the morphant embryo, causing hemorrhage. Blood flow in the body of both rescued and morphant embryo was normal (not shown). (K–L) O-dianisidine staining at 48 hpf. (K) embryo treated with Ctrl MO displaying normal blood circulation; (L) embryo treated with par1-MOAUG showing reduced level of Hb in the Duct of Cuvier and blood pooling in the tail. (M–N) Embryos at protruding mouth stage (72 hpf). Contrary to the rescue embryo (M), the par1 morphant has a slight cardiac edema, no mouth protrusion and no visible swim bladder (N).
Heart beat weaker and blood flow slower in par1 knockdown
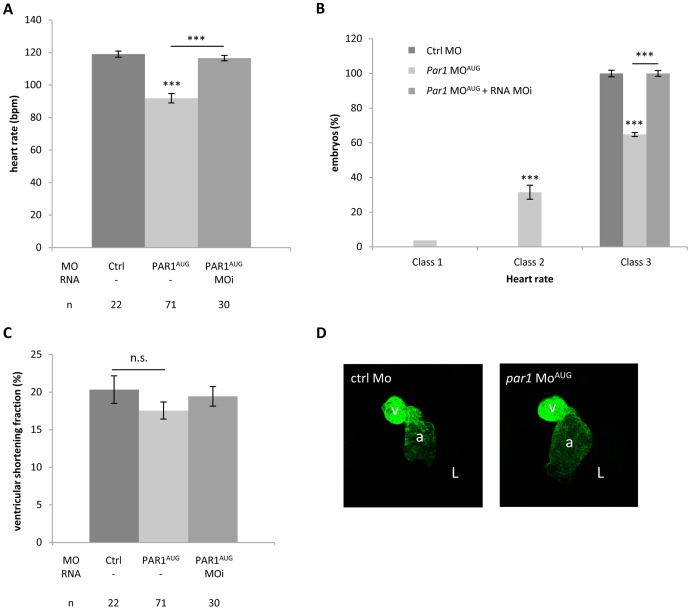
As the heart starts to beat in the one day old zebrafish embryo, no apparent difference was seen before between Ctrl MO and par1 MOAUG injected embryos. At 48 hpf heart rate counts were determined and par1 MOAUG morphants (injected with a dose of 4 ng) displayed significantly lower heart rate compared to controls ( Fig. 2A ). This difference is maintained at 3 dpf. We classified the heart beat rate into three classes; class 1: absent; class 2: heart rate<90 beats per minute (bpm) and class 3: with normal heart rate>90 bpm. Interestingly, injection of par1 MOAUG led to class 1 (3.7%±2) and 2 (31.5%±6 with a mean at 72.9±4.7 bpm) ( Fig. 2B ). Classes 1 and 2 were never observed in Ctrl morphants. Furthermore, par1 morphants heart rate in the normal class where significantly lower (105.4±1.3 bpm) than in controls (117.9±2.8 bpm) (p<0.001). We observed that co-injection of par1-MOi mRNA with par1-MOAUG was able to rescue this heart beat phenotype ( Fig. 2A–B ). Moreover, in embryos with lower cardiac heart rates (<90 bpm), no obvious abnormalities in the heart morphology can be detected before the functional defect appears. We never observed retrograde flow or fibrillation. Ctrl and par1 morphant heart beats regularly and co-ordinately with a 1∶1 ratio of atrial to ventricular beats. Furthermore, anti-myosin heavy chain immunostaining revealed that cardiac looping was not affected by par1 MOAUG injection ( Fig. 2D ). The atrium and the ventricle can be clearly distinguished and constriction was evident at the atrioventricular boundary. In the morphant embryo, the atrium was slightly enlarged. We quantified cardiac contractility by measuring the ventricular shortening fraction (VSF) [20]. VSF of the Ctrl morphant and the par1 MOAUG injected embryo was not significantly different ( Fig. 2C ). Our results demonstrate a requirement for Par1 in cardiac function that is consistent with in vitro studies that have linked PAR1 activation to an increased contractile response in adult cardiomyocytes [21], [22].
Figure 2. Slow heart rate in par1 morphants.
(A) Heart rates were determined at 48 hpf and significance was described from an ANOVA-test statistic. The heart rate of par1 morphants was significantly lower than controls. *** p<0.001. n values were indicated. (B) Classification of heart rate. Class 1: heart shows no heart rate; class 2: heart presents lower heart rate <90 bpm; class 3: heart rate seems normal >90 bpm. (C) Ventricular shortening fraction (D) The heart is labeled with anti-myosin heavy chain antibody in 48 hpf embryos. Ventral view with the head on the top. a, atrium; v, ventricle, L; embryo left.
In association with the heart-beat defect, embryos of classes 1 and 2 had a significant slower blood flow, gradually coming to a complete stop in 20% of these embryos. Furthermore, some par1 morphants with cardio-vascular phenotypes had haemorrhages ( Table 2 ) in the head, around the MtA (Metencephalic Artery) and the MCeV (Middle Cerebral Vein) ( Fig. 3J ), blood pooling in the location of the posterior blood island (PBI) ( Fig. 3B and F ) and hemorrhages and oedemas were apparent around the heart itself ( Fig. 3N ). Blood cell pooling can restrain or completely prevent blood flow. In vivo live imaging in the knockdown zebrafish embryo substantiate the internal bleeding in the head, due to haemorrhages from the forming CtA (central arteries) posterior to the MtA (metencephalic artery) and the MCeV (middle cerebral vein), see movie S1, or in the forming CVP (choroidal vascular plexus) anterior to the MtA and the MCeV (data not shown), and all small plexus sprouting from the PHBC (primordial hindbrain channel).
Table 2. Hemorrhages/blood pooling localisation.
| n (% embryos/heart beat defective embryos) | |||||
| Embryos (n) | Embryos with cardio-vascular phenotype (n) | Head | Heart | Body | |
| par1 MOAUG | 707 | 248 | 33 (14) | 44 (18) | 26 (11) |
| par1 MOSpl | 555 | 150 | 24 (16) | 21 (14) | 18 (12) |
Interestingly, we observed that the formation of the vascular system was altered in the caudal vascular plexus ( Fig. 3D and 4B ). Caudal vein (CV) cell rearrangement was in some cases not complete (23% of embryos in grades 1 and 2 with MOAUG and 46% with MOspl). To further characterize this phenotype, we analysed a double transgenic zebrafish strain expressing EGFP under the promoter of the endothelial specific kdrl, and DsRed under the erythroid-specific promoter of gata1. While gata1-positive cells were located throughout the vascular tree upon injection in control morphants ( Fig. 3C and E ), erythroid cells were enclosed by the vascular tissue in the posterior part of the vein in par1 morphants with blood pooling in the location of the PBI (arrowhead in Fig. 3F ). Furthermore, we observed that the CV in this region was dilated and failed to form, as seen with the green fluorescence in Tg(kdrl:EGFP)s843 embryos (asterisk in Fig. 3D ). Blood pooling in the posterior body region of the morphant embryo was confirmed by haemoglobin (Hb) localization using O-dianisidine staining ( Fig. 3K–L ). In control morphants, Hb staining was high in the Duct of Cuvier and weak in the tail ( Fig. 3K ). In par1 morphants with blood clots at PBI, the Hb staining showed that erythrocytes were confined to this region (arrowhead in Fig. 3L ). Bulged similar dilated posterior caudal plexus has already been described in a different study with more or less severe abnormal blood circulation [23], [24]. In virtual cross sections of confocal scans within the posterior cardinal vein double transgenic embryos (Tg(kdrl:EGFP)s843;Tg(gata1:dsRed)sd2) the difference between the morphant and the control embryo became clearer. In control embryos the ISV's are inflated at this stage development (35 hpf), the dorsal aorta can be seen as a single tube and the posterior cardinal vein appears as 2 to 3 tubes in the cross section. In par1 morphants, ISVs are also luminized and the dorsal aorta can be seen as one tube but CV tube does not show a defined number and is somewhat dilated. In comparison in the tnnt2 morphant which has a complete silent heart phenotype [25], the dorsal aorta was small and the intersegmental vessels (ISVs) were collapsed. The caudal vein in the tnnt2 morphant is seen as one big tube filled with all the blood cells that migrate into the vessel [26], [27], and clog, due to lack of blood flow. Furthermore, oedemas in the heart of the par1 morphant were not as severe as in the tnnt2 morphant ( Fig. 4 ). These results suggest a direct requirement for Par1 during posterior cardinal vein remodelling or an indirect function via a slower heart beat rate and a slower blood flow.
Figure 4. par1 knockdown impairs vascular remodeling in the cardinal vein.
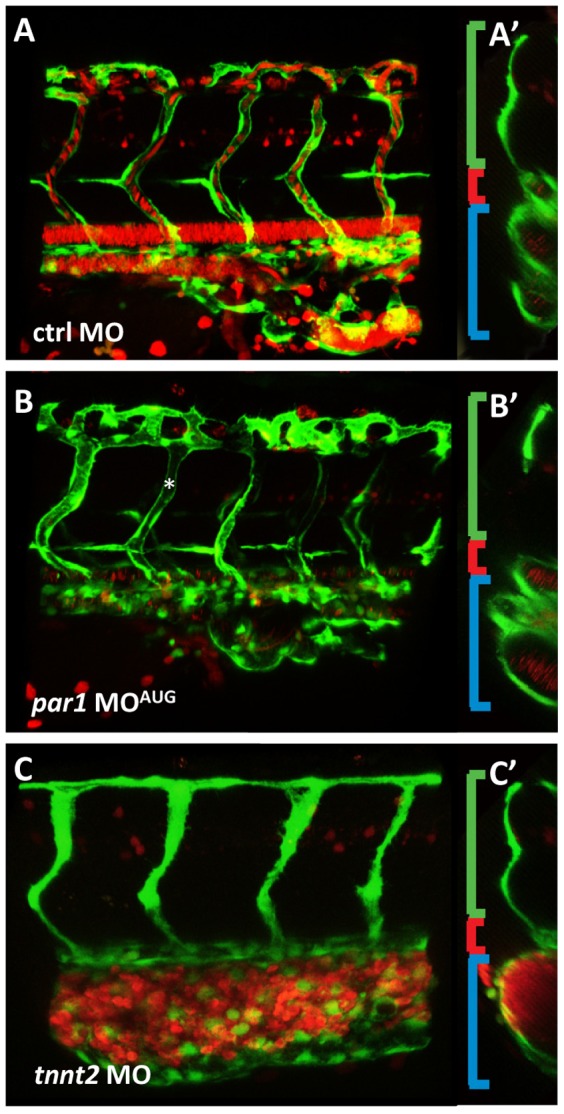
(A–B) Lateral views of double transgenic embryos (Tg(kdrl:EGFP)s843;Tg(gata1:dsRed)sd2) at 35 hpf. (A) The intersegmental vessels are connected to the CV and the one intersegmental artery (most posterior) seen in the figure has blood flowing from the dorsal aorta. (A′) In the mid-cross section, the intersegmental artery is visible (green bracket), the dorsal aorta can be seen as one tube (red bracket), and the CV is seen as two tubes (blue bracket). (B) par1 morphant lateral view. ISVs are lumenized (asterisk) (B′) In cross section, the CV tube does not show a defined number. Due to mislaid vein remodeling the tube is disorganized, cell clusters are apparent instead of a round tube. (C) The tnnt2 knockdown shows no mature inflation of the dorsal aorta and only a minor primary lumen in the ISV. The CV is bulged and full of blood cells in this region. (C′) The small dorsal aorta is a clear difference to the par1 knockdown, additionally to the lack of lumen in the ISVs and blood cells always clogging in the region of origin, PBI.
Finally, considering that PAR1 modulates endothelial barrier integrity through actomyosin contraction and adherens junction modifications [28], we investigated the effect of Par1 knockdown on VE-cadherin-based adherens junctions using antibodies against VE-cadherin. Staining patterns were similar in the par1 morphant and in the control morphant (Fig. S3), This would indicate intact adherens junctions in the endothelial cells of the par1 morphant. Yue et al recently discovered that Par1 is a negative regulator of the hematopoietic development. Zebrafish embryos in their knockdown experiments had increased number of hematopoietic cells and in mESC (mouse embryonic stem cells), both primitive and definitive erythroid number a.m., was increased if the F2 receptor was chemically or genetically blocked [12]. One may speculate that the increasing number of erythrocytes causes strain on the heart and the blood vessel, resulting in heart failure and bleeding. According to Yue et al, in vitro blocking Par1 causes more endothelial cells to undergo endothelial-hematopoetic-transition and abrogation of junctions in the cells. Our analysis of junctional integrity in the endothelial cells, using the VE-cadherin antibody, revealed no difference between par1 MO injected embryos and control embryos.
Conclusion
Our sequence analysis and functional data on Par1 in the zebrafish confirm the protein to be highly conserved in the animal kingdom [8], [9]. The PAR1 receptor is clearly involved in cardiovascular development and vessel maturation. Non-functional PAR1 causes defects in vascular maturation and internal bleeding, as shown with live imaging in the zebrafish knockdown embryo. Finally, we demonstrated that the N-terminus and the secondary loop domains of Par1 are necessary for normal cardiovascular development and that PAR1 function and probably activation and signaling is conserved across vertebrates.
Materials and Methods
Ethics and fish care
All protocols in this study were conducted in strict accordance with the French and Swiss guidelines for the care and use of laboratory animals. No specific ethics approval was required for this project, as all zebrafish (Danio rerio) used in this study were between 0 and 3 days old. Zebrafish embryos were killed with a bleach solution (sodium hypochlorite 6.15%) added to the culture system water at 1 part bleach to 5 parts water. Zebrafish embryos remained in this solution for at least 5 minutes to ensure death. Given the age of the embryos, pain perception has not yet developed at these earlier stages so this is not considered as a painful procedure. For in situ hybridization or immunostaining, zebrafish embryos were killed and fixed on PFA 4%. Zebrafish were maintained and staged according to Westerfield [29]. Experiments were performed using the standard Ab wild type strain. All the transgenic lines used came from zebrafish. The following zebrafish transgenic lines were used: Tg(flia:EGFP)γ1 [30], Tg(kdrl:EGFP)s843 [31], and Tg(gata1:dsRed)sd2 [32].
Zebrafish PAR1 gene identification
The human PAR1 amino acid (aa) sequence (P25116 UniProt) was used to perform BLAST searches in both Ensembl and NCBI databases. No human tissue was used for this study. We obtained the human sequence from a genebank and performed sequence analysis in silico. In silico alignment analysis was done by two different approaches, LALIGN (http://www.ch.embnet.org/software/LALIGN_form.html) and Vector NTI AlignX software, and both were used in the determination of a bona fide zebrafish PAR1 homologous. Multiple alignments were likewise done by AlignX or PRALINE [33]. Proteomic analysis and predictions were made with TMHMM, TMPred and Predictprotein.org (ExPASy portal: http://www.expasy.ch/). The clone corresponding to the sequence identified in silico (# CO935967) was ordered from ImaGenes (Berlin, Germany) and re-sequenced to confirm the cDNA.
Microinjection of morpholino antisense oligonucleotides (MOs)
MOs were purchased from Gene Tools (LLC, Philomath, OR, USA). For the experiments 4 ng of each morpholino were used for injection into one- to two-cell stage zebrafish embryos [34]. Par1-specific MOAUG (5′-CCGTCACCAACAGAACCCGCAACAT-3′ ) was used to inhibit Par1 protein synthesis (translational initiation codon is underlined). We also performed some experiments with a splice site directed morpholino MOspl (5′-GAAAGTCTGTAAGCGTCTTACCGTT-3′) that inhibited the splicing of pre-mRNA between the exon 1 and intron 1. (5′-CCGTCAGGAAGACAACGCGCAAGAT-3′) was used as a control MO. For a complete knockdown of the heart beat, an antisense morpholino oligonucleotide targeting the cardiac troponin T translation start codon and flanking 5′ sequence was injected (5′-CATGTTTGCTCTGATCTGACACGCA-3′) [25].
Quantitative RT-PCR
cDNA synthesis and RT-PCR reaction were performed as previously described [35]. Gene expression level was normalized to RPL13. The Rpl13a primers were previously described in [36]. Each sample was tested in duplicate. The par1 primers were fw 5′-TGTTACAATGATTAAATGCTGCAA-3′, rv 5′-CATCAGTGACGGTGAGGAAA-3′.
Heart rate counts and ventricular contractibility analysis
Heart rates (beats per minute) were counted manually under Leica stereo microscope M80 for control MO and par1 MOAUG injected zebrafish embryos at 48 hpf. Embryos were anesthetized 10 sec in tricaine, transferred to a recording chamber, mounted and perfused with embryo medium. The ventricular shorthening fraction (VSF) was measured according to Wang et al [20]. The heart rate (beats/min) and the VSF were recorded by the same operator after a recovery in embryo medium of at least 10 min.
Hemoglobin staining
In situ Hb staining was carried out using O-dianisidine as described by Kwan et al [37]. The presence of yellow-brown coloration indicates the presence of hemoglobin in zebrafish embryos.
Constructions and mRNA injection
cDNA from clone # CO935967 was cloned in pCS2+ vector. Directed mutagenesis allowed us to construct a cDNA mutated at the MO target (MOi clone for MO insensitive RNA) as well as two deletion mutants ΔC (lacking the 69 C-terminal aa) and ΔN (lacking the first 79 aa). A detailed description of the plasmids is available upon request. mRNA synthesis was performed using the mMESSAGE mMACHINE high yield capped RNA Transcription Kit (Ambion Inc., Austin, TX, USA). 150 ng/ml of transcribed mRNA was injected into one-cell stage embryos. For the rescue experiments, mRNA and morpholino were co-injected.
Immunofluorescence
Zebrafish anti-Ve-cadherin antibody was used diluted 1∶500 [38]. Alexa 688 goat anti-rabbit IgG were diluted 1∶1000. Anti-myosin heavy chain antibody (Millipore, Temecula, CA, USA) was used diluted 1∶200 to visualize the heart at 48 hpf.
Microscope imaging
Living zebrafish embryos were mounted in methyl-cellulose on a slide (with slope) and imaged with a LeicaDFC 420C camera on a Leica MZF III binocular. Photos were processed with Adobe Photoshop CS4.
Confocal imaging
Living zebrafish embryos were mounted in 0.8% low melting agarose in a small Petri dish with cover slip bottom, covered with egg water, and imaged on the Leica TCS SP5 confocal microscope. Objective ×20 with 2 times zoom or ×63 were used. Fixed embryos were mounted in fluorescent mounting medium and imaged on a slide with a cover slip on the same Leica SP5. All images were analysed with Imaris 7.1.1 (Bitplane) Software. All images are compressed z-stalk of an embryo. For overnight imaging, embryos were mounted as described above and kept in a chamber (The Box) with constant temperature of 28°C.
Statistical analysis
For Fig. 1, all values are expressed as means ± standard error. Error bars are standard errors estimated as √(p(1−p)/n) where p is the proportion of embryos exhibiting a phenotype and n the total number of embryos investigated (or 1/n when p = 0 or 1). Comparisons were performed by chi-squared test. For Fig. 2, comparisons between multiple groups were performed by 1-way ANOVA followed by Tukey's tests. Significance level was set at P<0.05.
Supporting Information
Par1 is highly conserved throughout evolution. (A) Alignment of PAR1 protein sequences; black background: identical amino acid; grey background: conservative change; transmembrane segments and functional domains are indicated. (B) phylogenetic tree (C) Secondary structure prediction for zebrafish Par1 conforms to a seven-pass membrane receptor structure. Alignment and comparison between functional domains of human PAR1 and zebrafish Par1; black box: thrombin cleavage site; green letters: tethered ligand domain; underlined: hirudin–like sequence; TM1: transmembrane domain 1. (D) Three par1 mRNA mutants were designed: par1-MOi, morpholino-insensitive; par1-ΔN N-terminus deleted; and par1-ΔC, a par1-MOi lacking the intracellular domain. Asterisks indicate the morpholino-insensitive sequence on par1 mRNA.
(TIF)
Par1 putative paralogues. (A) Ensembl Phylogenetic tree. (B) Location and sequence analysis (C) Hierarchical multiple sequence alignment (PRALINE) of zebrafish PARs and putative paralogues.
(TIF)
The endothelial adherens junctions appear intact in par1 knockdown. (A–C) Lateral views after labelling of adherens junctions in the ISVs with anti ve-cadherin antibody (green) in a (Tg(kdrl:EGFP)s843) (red) transgenic embryo. (D–F) Immunolocalization of ve-cadherin alone. (E–F) Ve-cadherin labelled junctions appeared normal at 33 hpf in par1 morphants.
(TIF)
Hemorrhage in the head of a par1 morphant. Live imaging of a rescued double transgenic embryo (Tg(kdrl:EGFP)s843;Tg(gata1:dsRed)sd2, injected with MOpar1 and RNA MOi) to the left, and par1 morphant to the right. Live imaging on a SP5 Leica confocal, overnight imaging from 34 hpf to 46 hpf, z-stalks taken every 20 minutes, objective ×10, zoom ×2. Notice how fast the blood cells leak out of the CtA (central artery) in the morphant.
(AVI)
Acknowledgments
We sincerely thank Dr. Michel Volovitch, Dr. Emmanuelle Tillet, Dr. Philippe Huber, Helen Preiss and Dr. Heinz-Georg Belting for helpful discussions and critical reading of the manuscript. We also thank Kumuthini Kalendra for fish care.
Funding Statement
This work has been supported by the Cantons Basel-Stadt and Basel-Land, the Swiss National Science Foundation, and by a Network of Excellence grant “Cells into Organs” from the FP6 of the European Community. Carole Gauron is supported by ANR Proteophane 2008. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Kuliopulos A, Covic L, Seeley SK, Sheridan PJ, Helin J, et al. (1999) Plasmin desensitization of the PAR1 thrombin receptor: kinetics, sites of truncation, and implications for thrombolytic therapy. Biochemistry 38: 4572–4585. [DOI] [PubMed] [Google Scholar]
- 2. Riewald M, Kravchenko VV, Petrovan RJ, O'Brien PJ, Brass LF, et al. (2001) Gene induction by coagulation factor Xa is mediated by activation of protease-activated receptor 1. Blood 97: 3109–3116. [DOI] [PubMed] [Google Scholar]
- 3. Riewald M, Petrovan RJ, Donner A, Mueller BM, Ruf W (2002) Activation of endothelial cell protease activated receptor 1 by the protein C pathway. Science 296: 1880–1882. [DOI] [PubMed] [Google Scholar]
- 4. Boire A, Covic L, Agarwal A, Jacques S, Sherifi S, et al. (2005) PAR1 is a matrix metalloprotease-1 receptor that promotes invasion and tumorigenesis of breast cancer cells. Cell 120: 303–313. [DOI] [PubMed] [Google Scholar]
- 5. Trivedi V, Boire A, Tchernychev B, Kaneider NC, Leger AJ, et al. (2009) Platelet matrix metalloprotease-1 mediates thrombogenesis by activating PAR1 at a cryptic ligand site. Cell 137: 332–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Macfarlane SR, Seatter MJ, Kanke T, Hunter GD, Plevin R (2001) Proteinase-activated receptors. Pharmacol Rev 53: 245–282. [PubMed] [Google Scholar]
- 7. Seeley S, Covic L, Jacques SL, Sudmeier J, Baleja JD, et al. (2003) Structural basis for thrombin activation of a protease-activated receptor: inhibition of intramolecular liganding. Chem Biol 10: 1033–1041. [DOI] [PubMed] [Google Scholar]
- 8. Connolly AJ, Ishihara H, Kahn ML, Farese RV Jr, Coughlin SR (1996) Role of the thrombin receptor in development and evidence for a second receptor. Nature 381: 516–519. [DOI] [PubMed] [Google Scholar]
- 9. Griffin CT, Srinivasan Y, Zheng YW, Huang W, Coughlin SR (2001) A role for thrombin receptor signaling in endothelial cells during embryonic development. Science 293: 1666–1670. [DOI] [PubMed] [Google Scholar]
- 10. Hirano K, Kanaide H (2003) Role of protease-activated receptors in the vascular system. J Atheroscler Thromb 10: 211–225. [DOI] [PubMed] [Google Scholar]
- 11. Hirano K (2007) The roles of proteinase-activated receptors in the vascular physiology and pathophysiology. Arterioscler Thromb Vasc Biol 27: 27–36. [DOI] [PubMed] [Google Scholar]
- 12. Yue R, Li H, Liu H, Li Y, Wei B, et al. (2012) Thrombin Receptor Regulates Hematopoiesis and Endothelial-to-Hematopoietic Transition. Dev Cell 22: 1092–1100. [DOI] [PubMed] [Google Scholar]
- 13. Ishii K, Chen J, Ishii M, Koch WJ, Freedman NJ, et al. (1994) Inhibition of thrombin receptor signaling by a G-protein coupled receptor kinase. Functional specificity among G-protein coupled receptor kinases. J Biol Chem 269: 1125–1130. [PubMed] [Google Scholar]
- 14. Paing MM, Stutts AB, Kohout TA, Lefkowitz RJ, Trejo J (2002) beta -Arrestins regulate protease-activated receptor-1 desensitization but not internalization or Down-regulation. J Biol Chem 277: 1292–1300. [DOI] [PubMed] [Google Scholar]
- 15. McKinney MC, Weinstein BM (2008) Chapter 4. Using the zebrafish to study vessel formation. Methods Enzymol 444: 65–97. [DOI] [PubMed] [Google Scholar]
- 16. Xu H, Echemendia N, Chen S, Lin F (2011) Identification and expression patterns of members of the protease-activated receptor (PAR) gene family during zebrafish development. Dev Dyn 240: 278–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Vu TK, Wheaton VI, Hung DT, Charo I, Coughlin SR (1991) Domains specifying thrombin-receptor interaction. Nature 353: 674–677. [DOI] [PubMed] [Google Scholar]
- 18. Kim S, Carrillo M, Kulkarni V, Jagadeeswaran P (2009) Evolution of primary hemostasis in early vertebrates. PLoS One 4: e8403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kataoka H, Hamilton JR, McKemy DD, Camerer E, Zheng YW, et al. (2003) Protease-activated receptors 1 and 4 mediate thrombin signaling in endothelial cells. Blood 102: 3224–3231. [DOI] [PubMed] [Google Scholar]
- 20. Wang YX, Qian LX, Yu Z, Jiang Q, Dong YX, et al. (2005) Requirements of myocyte-specific enhancer factor 2A in zebrafish cardiac contractility. FEBS Lett 579: 4843–4850. [DOI] [PubMed] [Google Scholar]
- 21. Jiang T, Danilo P Jr, Steinberg SF (1998) The thrombin receptor elevates intracellular calcium in adult rat ventricular myocytes. J Mol Cell Cardiol 30: 2193–2199. [DOI] [PubMed] [Google Scholar]
- 22. Yasutake M, Haworth RS, King A, Avkiran M (1996) Thrombin activates the sarcolemmal Na(+)-H+ exchanger. Evidence for a receptor-mediated mechanism involving protein kinase C. Circ Res 79: 705–715. [DOI] [PubMed] [Google Scholar]
- 23. Chen E, Larson JD, Ekker SC (2006) Functional analysis of zebrafish microfibril-associated glycoprotein-1 (Magp1) in vivo reveals roles for microfibrils in vascular development and function. Blood 107: 4364–4374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Song M, Yang H, Yao S, Ma F, Li Z, et al. (2007) A critical role of vascular endothelial growth factor D in zebrafish embryonic vasculogenesis and angiogenesis. Biochem Biophys Res Commun 357: 924–930. [DOI] [PubMed] [Google Scholar]
- 25. Sehnert AJ, Huq A, Weinstein BM, Walker C, Fishman M, et al. (2002) Cardiac troponin T is essential in sarcomere assembly and cardiac contractility. Nat Genet 31: 106–110. [DOI] [PubMed] [Google Scholar]
- 26. Herbert SP, Huisken J, Kim TN, Feldman ME, Houseman BT, et al. (2009) Arterial-venous segregation by selective cell sprouting: an alternative mode of blood vessel formation. Science 326: 294–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Iida A, Sakaguchi K, Sato K, Sakurai H, Nishimura D, et al. (2010) Metalloprotease-dependent onset of blood circulation in zebrafish. Curr Biol 20: 1110–1116. [DOI] [PubMed] [Google Scholar]
- 28. Feistritzer C, Riewald M (2005) Endothelial barrier protection by activated protein C through PAR1-dependent sphingosine 1-phosphate receptor-1 crossactivation. Blood 105: 3178–3184. [DOI] [PubMed] [Google Scholar]
- 29.Westerfield M (1993) The zebrafish book a guide for the laboratory use of zebrafish Danio (Brachydanio) rerio. Eugene, OR: Institute of Neuroscience University of Oregon.
- 30. Lawson ND, Weinstein BM (2002) In vivo imaging of embryonic vascular development using transgenic zebrafish. Dev Biol 248: 307–318. [DOI] [PubMed] [Google Scholar]
- 31. Jin SW, Beis D, Mitchell T, Chen JN, Stainier DY (2005) Cellular and molecular analyses of vascular tube and lumen formation in zebrafish. Development 132: 5199–5209. [DOI] [PubMed] [Google Scholar]
- 32. Traver D, Paw BH, Poss KD, Penberthy WT, Lin S, et al. (2003) Transplantation and in vivo imaging of multilineage engraftment in zebrafish bloodless mutants. Nat Immunol 4: 1238–1246. [DOI] [PubMed] [Google Scholar]
- 33. Simossis VA, Heringa J (2005) PRALINE: a multiple sequence alignment toolbox that integrates homology-extended and secondary structure information. Nucleic Acids Res 33: W289–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Nasevicius A, Ekker SC (2000) Effective targeted gene ‘knockdown’ in zebrafish. Nat Genet 26: 216–220. [DOI] [PubMed] [Google Scholar]
- 35. Bouzaffour M, Dufourcq P, Lecaudey V, Haas P, Vriz S (2009) Fgf and Sdf-1 pathways interact during zebrafish fin regeneration. PLoS One 4: e5824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Tang H, Low B, Rutherford SA, Hao Q (2005) Thrombin induces endocytosis of endoglin and type-II TGF-beta receptor and down-regulation of TGF-beta signaling in endothelial cells. Blood 105: 1977–1985. [DOI] [PubMed] [Google Scholar]
- 37. Kwan TT, Liang R, Verfaillie CM, Ekker SC, Chan LC, et al. (2006) Regulation of primitive hematopoiesis in zebrafish embryos by the death receptor gene. Exp Hematol 34: 27–34. [DOI] [PubMed] [Google Scholar]
- 38. Blum Y, Belting HG, Ellertsdottir E, Herwig L, Luders F, et al. (2008) Complex cell rearrangements during intersegmental vessel sprouting and vessel fusion in the zebrafish embryo. Dev Biol 316: 312–322. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Par1 is highly conserved throughout evolution. (A) Alignment of PAR1 protein sequences; black background: identical amino acid; grey background: conservative change; transmembrane segments and functional domains are indicated. (B) phylogenetic tree (C) Secondary structure prediction for zebrafish Par1 conforms to a seven-pass membrane receptor structure. Alignment and comparison between functional domains of human PAR1 and zebrafish Par1; black box: thrombin cleavage site; green letters: tethered ligand domain; underlined: hirudin–like sequence; TM1: transmembrane domain 1. (D) Three par1 mRNA mutants were designed: par1-MOi, morpholino-insensitive; par1-ΔN N-terminus deleted; and par1-ΔC, a par1-MOi lacking the intracellular domain. Asterisks indicate the morpholino-insensitive sequence on par1 mRNA.
(TIF)
Par1 putative paralogues. (A) Ensembl Phylogenetic tree. (B) Location and sequence analysis (C) Hierarchical multiple sequence alignment (PRALINE) of zebrafish PARs and putative paralogues.
(TIF)
The endothelial adherens junctions appear intact in par1 knockdown. (A–C) Lateral views after labelling of adherens junctions in the ISVs with anti ve-cadherin antibody (green) in a (Tg(kdrl:EGFP)s843) (red) transgenic embryo. (D–F) Immunolocalization of ve-cadherin alone. (E–F) Ve-cadherin labelled junctions appeared normal at 33 hpf in par1 morphants.
(TIF)
Hemorrhage in the head of a par1 morphant. Live imaging of a rescued double transgenic embryo (Tg(kdrl:EGFP)s843;Tg(gata1:dsRed)sd2, injected with MOpar1 and RNA MOi) to the left, and par1 morphant to the right. Live imaging on a SP5 Leica confocal, overnight imaging from 34 hpf to 46 hpf, z-stalks taken every 20 minutes, objective ×10, zoom ×2. Notice how fast the blood cells leak out of the CtA (central artery) in the morphant.
(AVI)