Abstract
Eukaryotic cells contain many different membrane compartments with characteristic shapes, lipid compositions, and dynamics. A large fraction of cytoplasmic proteins associate with these membrane compartments. Such protein–lipid interactions, which regulate the subcellular localizations and activities of peripheral membrane proteins, are fundamentally important for a variety of cell biological processes ranging from cytoskeletal dynamics and membrane trafficking to intracellular signaling. Reciprocally, many membrane-associated proteins can modulate the shape, lipid composition, and dynamics of cellular membranes. Determining the exact mechanisms by which these proteins interact with membranes will be essential to understanding their biological functions. In this Technical Perspective, we provide a brief introduction to selected biochemical methods that can be applied to study protein–lipid interactions. We also discuss how important it is to choose proper lipid composition, type of model membrane, and biochemical assay to obtain reliable and informative data from the lipid-interaction mechanism of a protein of interest.
INTRODUCTION
Eukaryotic cells are composed of multiple lipid compartments that display distinct morphologies, dynamics, lipid composition, and sets of interacting proteins. For example, the plasma membrane is enriched with phosphatidylinositol 4,5-biphosphate (PI(4,5)P2), whereas phosphatidylinositol 4-phosphate (PI(4)P) is the predominant phosphoinositide in the Golgi complex. Consequently, the protein compositions of these two membrane compartments vary significantly, giving rise to their distinct morphologies and functional properties (Di Paolo and De Camilli, 2006).
It has been estimated that approximately one-fourth of the genes in the human genome encode for integral membrane proteins, such as transmembrane transporters and channels, as well as for many enzymes and receptors that associate with membranes through transmembrane α-helical anchors (von Heijne, 2006). However, the structural/functional diversity of peripheral membrane proteins is even more widespread. Membrane interactions regulate the subcellular localizations and activities of these proteins, but peripheral membrane proteins can in turn regulate the composition, dynamics, and morphology of cellular membranes. For example, many proteins associated with the regulation of the actin cytoskeleton interact with phosphoinositide-rich membranes, and their activities can be either up-regulated or down-regulated through membrane interactions. Thus transient interactions of actin-binding proteins with phosphoinositide-rich membranes play an important role in regulation of cytoskeletal dynamics (Saarikangas et al., 2010). However, cytoskeleton-associated proteins can also affect the lipid distribution, as well as directly sculpt the plasma membrane. One example is the cytoskeleton-associated Bin–amphiphysin–Rvs (BAR) domain proteins that bind phosphoinositide-rich membranes through curved interfaces and, depending on the geometry of the lipid-binding interface, bend the membrane to generate either plasma membrane protrusions or invaginations (Peter et al., 2004; Frost et al., 2009; Zhao et al., 2011). Thus a rich, bidirectional interplay exists between peripheral membrane proteins and cellular membrane compartments.
Peripheral membrane proteins associate with lipids through many distinct mechanisms. They can interact with a membrane through an unspecific hydrophobic association, for example, by inserting an amphipathic α-helix into the bilayer, and through electrostatic interactions between the protein and lipid head groups. In addition, peripheral membrane proteins may contain a specific binding pocket(s) for certain lipid head groups or may harbor covalently bound lipid anchors, such as palmitoyl, myristoyl, or prenyl modifications. In many cases, the membrane interactions of these proteins simultaneously employ more than one of the above-mentioned mechanisms. Importantly, the mechanism by which a protein interacts with lipids has significant biological consequences, because it determines the specificity and affinity of membrane binding, as well as possible effects of the protein on the morphology and dynamics of the membrane (for a review, see Lemmon et al., 2008).
To reliably examine the interactions of a protein with membranes, one should design a membrane-binding assay that takes into account the subcellular localization and biological function of the protein. Thus the lipid composition and morphology of the model membrane should resemble the biological membrane with which the protein of interest is expected to associate. Furthermore, a large number of different assays exist for studying protein–lipid interactions in vitro, and they provide different types of information concerning the mechanisms of protein–lipid interactions. The choice of an assay therefore depends on the protein of interest and on the type of information needed. In this paper, we will briefly introduce selected biochemical protein–lipid interaction assays and provide some advice on matters that are important to keep in mind when designing and carrying out these experiments. We will focus on assays applied for studying the lipid interactions of cytoplasmic peripheral membrane proteins. Nevertheless, many of the assays presented here can also be applied for extracellular proteins that associate with the outer leaflet of the plasma membrane and for proteins associated with the luminal leaflets of intracellular membrane compartments.
DEFINING THE LIPID COMPOSITION FOR AN ASSAY
Cellular membranes contain a large variety of lipid species that differ in the structure of their lipid head group and in the length and saturation of their hydrophobic acyl chains (Figure 1). Distinct membrane compartments differ both quantitatively and qualitatively in their lipid content, and there are also cell type–specific differences in lipid compositions of individual membrane compartments. For example, the lipid composition of the plasma membrane is different between neuronal and muscle cells, and to add another layer of complexity, many signaling pathways also affect the lipid composition of the plasma membrane. Furthermore, whereas all lipids are quite symmetrically distributed between the two leaflets of the endoplasmic reticulum (ER) membrane bilayer, the Golgi, endosomal organelles, and plasma membrane display an asymmetric lipid distribution, with phosphatidylcholine, sphingomyelin, and glycosphingolipids enriched at the noncytosolic (luminal side), and phosphatidylethanolamine (PE), as well as negatively charged phospholipids, including phosphatidylinositol (PI) and phosphatidylserine, enriched at the cytosolic leaflet (Colbeau et al., 1971; van Meer et al., 2008; van Meer and de Kroon, 2011). The asymmetric distribution of lipids has important functional consequences for many cellular processes, such as vesicle budding and membrane fusion (Zimmerberg and Chernomordik, 1999). In addition to lipid composition, the geometry of membranes also plays an essential role in protein interactions, because many proteins preferentially interact with membranes displaying specific curvature (McMahon et al., 2010; Zhao et al., 2011).
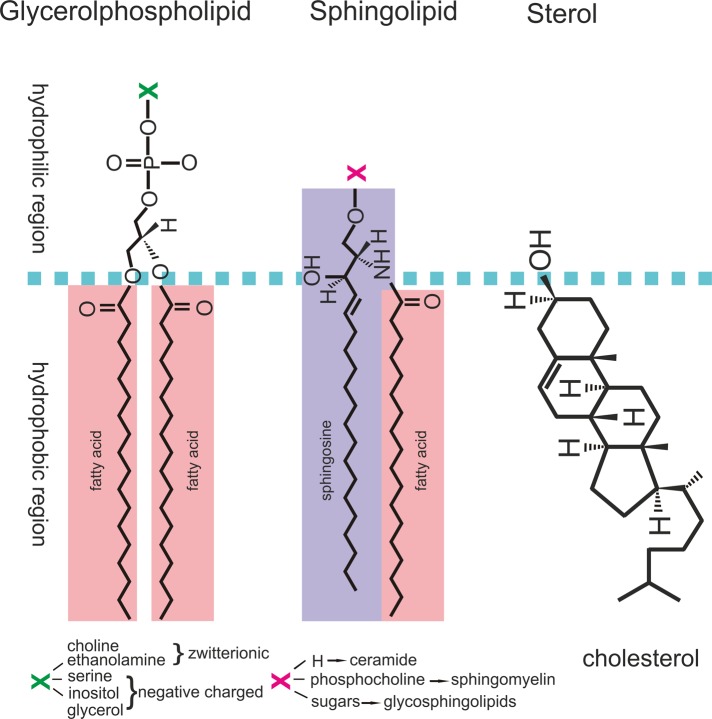
FIGURE 1:
Schematic representation of structures of common lipids. Lipids are hydrophobic or amphiphilic molecules. Lipid molecules are typically composed of two major regions: a hydrophilic region (regions above the dashed line) and hydrophobic tails (regions below the dashed line). Glycerophospholipids are the most abundant lipids in cellular membranes, and they are composed of two fatty acid chains linked to a phosphate and a specific head group. Sphingolipids are composed of a backbone of a sphingoid base, such as sphingosine, connected to a fatty acid chain and a specific head group. Sterol lipids, such as cholesterol (with a small polar head) and its derivatives, are also important components of cellular membranes.
Many factors need to be taken into account when designing an experiment for studying membrane interactions of a specific protein. These include lipid composition, molar ratio of the lipids, membrane curvature, pH, and salt concentration of the buffer. It is important that the lipid composition and other above-mentioned parameters in the assay will resemble as accurately as possible the relevant membrane organelle. For example, if the protein of interest localizes to the cytoplasmic leaflet of the Golgi complex, it is important to include PI(4)P in the vesicles, whereas if the protein of interest localizes to the inner leaflet of the plasma membrane, one should include PI(4,5)P2 at the relevant density in the model membranes.
PREPARATION OF MODEL MEMBRANES
Interactions between proteins and lipids can be studied by using isolated and model membranes in vitro. Isolated membranes display natural compositions of lipids, but isolation of sufficient amounts of specific membrane compartments from cells for biochemical assays is often tedious. Furthermore, isolated biological membranes often display a complex composition that includes membrane proteins. Consequently, the majority of our current knowledge of protein–lipid interactions and their mechanisms are derived from studies on model membranes developed over the last century to enable study of the properties and interactions of biological membranes. The most well-known and common biomimetic systems used for such purposes are lipid vesicles, supported lipid bilayers, and lipid monolayers (Figure 2), which are suitable for distinct biochemical assays.
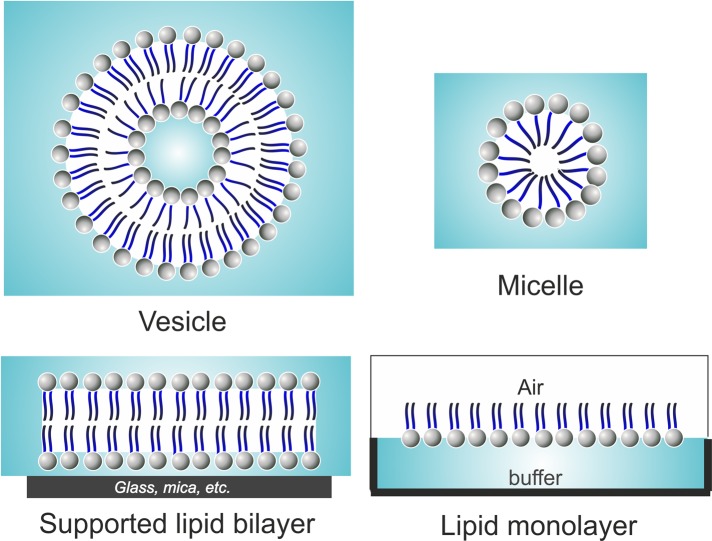
FIGURE 2:
Different model membranes for studying protein–lipid interactions. Lipid vesicles or liposomes are spherical hollow structures composed of a lipid bilayer. Micelles are also spherical, but are formed from a lipid monolayer that contains a hydrophobic core. Planar model membranes include supported lipid bilayers and lipid monolayers. The former are made up of a flat lipid bilayer supported by a solid surface, such as mica, glass, or silicon oxide wafers. Lipid monolayers can be formed by spreading lipid molecules at the surface of a buffer, where the lipid molecules obtain a specific orientation with the polar head groups of phospholipids pointing toward the aqueous subphase and the hydrophobic acyl chains pointing toward the air.
Lipid vesicles
Multilamellar and unilamellar vesicles
Among the model membranes, lipid vesicles (liposomes) of different sizes represent the most widely used tool. This is because liposomes structurally resemble many cellular membrane compartments, and they are suitable for a wide variety of biochemical assays, such as fluorometric and sedimentation/flotation assays. In liposomes, the individual lipid molecules are arranged in a spherical lipid bilayer. Thus liposomes are better suited for most biochemical assays compared with, for example, micelles, which do not have a lipid bilayer structure and display much higher curvature compared with most cellular membrane compartments (see Figure 2). Unilamellar vesicles are composed of a single spherical lipid bilayer, whereas multilamellar vesicles are composed of several spherical lipid bilayers. Multilamellar vesicles can be applied for cosedimentation and coflotation assays, as well as for solid-state nuclear magnetic resonance (NMR) experiments. However, optically clear large unilamellar vesicles are the most commonly used liposome type for biochemical studies.
Hydration of most lipid films of various compositions produces multilamellar vesicles (Φ ≈ 1 μm) that can be downsized by extrusion or sonication into large unilamellar vesicles (Φ ≈ 100–500 nm) or small unilamellar vesicles (Φ < 50 nm), respectively (Szoka and Papahadjopoulos, 1980). Large unilamellar vesicles are typically prepared by force extrusion (Mui et al., 2003). In this technique, a lipid suspension is forced through a polycarbonate filter with a defined pore size to yield vesicles with a diameter distribution near the pore size of the filter. Prior to extrusion through the final pore size, multilamellar vesicle suspensions can be disrupted either by several freeze–thaw cycles or by prefiltering the suspension through a larger pore size (typically 0.2–1.0 μm), which improves the homogeneity of size distribution of the final suspension. It is important to emphasize that, as with all procedures for downsizing lipid dispersions, the extrusion should be performed at a temperature above the highest lipid-phase transition temperature of the lipid mixture. Attempts to extrude below this temperature will be unsuccessful, as the high viscosity of rigid membranes prevents them from passing through the pores and/or results in the formation of unstable vesicles. A detailed description and instructions for preparing large unilamellar vesicles can be found in Mui et al. (2003).
It is worth noting that small unilamellar vesicles and micelles can also be applied for many studies, among them circular dichroism (CD) spectroscopy and examination of the structures of membrane proteins (e.g., see Dancea et al., 2008). The most common instrumentation for preparation of small unilamellar vesicles are bath and probe tip sonicators (Tenchov et al., 1985). Micelles, on the other hand, form spontaneously when lipids are added to an aqueous solution at certain concentrations and temperatures that depend on the type of lipid(s) used (Singh et al., 2007; Peric et al., 2006).
Giant unilamellar vesicles
As described above, the large and small unilamellar vesicles are suitable for spectroscopic and sedimentation-based assays. However, they are submicroscopic in size and do not allow light microscopy imaging of membrane dynamics. So-called giant vesicles (Φ ≈ 10–300 μm) therefore represent an excellent model system for microscopic studies on membrane physicochemical properties and protein–membrane interactions. Membrane bending rigidity, stretching elasticity, lipid sorting, and phase separation have been investigated using giant vesicles (Dimova et al., 2006; Bassereau and Goud, 2011), because the size of giant vesicles allows their visualization by light microscopy and micromanipulation of individual vesicles. This permits direct imaging of dynamic processes, including kinetics of protein–lipid interactions, lipid microdomain formation, changes in membrane morphology, and dynamics of lipids (e.g., see Saarikangas et al., 2009; Römer et al., 2010; Bacia et al., 2011; Sorre et al., 2012).
Giant vesicles with the same lipid compositions on both leaflets can be prepared by hydrating a dried lipid film at a temperature above the lipid-phase transition either for a long period of time (up to 36 h, spontaneous swelling method) or in the presence of an external electrical field (the electroformation method). Recently, new techniques have been developed for preparing giant liposomes with asymmetric bilayers by water/oil inverted emulsion and microfluidic jetting (Pautot et al., 2003; Abkarian et al., 2011; Richmond et al., 2011). A detailed discussion concerning methods for preparing giant vesicles can be found in Walde et al. (2010). Among these methods, the electroformation method is most widely used, because it allows good control over vesicle size, shape, and lamellarity (Angelova et al., 1992; Luisi and Walde, 2000). In this method, the lipid mixture is spread on two platinum wires or on an indium tin oxide–coated glass surface dried under a stream of nitrogen. Possible residues of organic solvent are typically further removed by evacuation in a vacuum. An electric field is then applied to generate giant vesicles in a relevant buffer. It is important to note that when using the electroformation method, the giant liposomes generally grow better in low-salt buffer. However, some methods have also been developed to generate giant vesicles in physiological ionic conditions (Estes and Mayer, 2005; Pott et al., 2008).
Supported lipid bilayers
Supported lipid bilayers are made up of a flat lipid bilayer supported on a solid surface, such as mica, glass, or silicon oxide wafers (Figure 2). In such a model system, the polar head groups of the first lipid monolayer are facing toward the support, while their hydrocarbon chains are in contact with the acyl chains of the second monolayer. Supported lipid bilayers offer several advantages over lipid vesicles (Loose and Schwille, 2009). These model membranes can be prepared easily, and they are more stable than lipid vesicles. Importantly, the lipid asymmetry of supported lipid bilayers can be controlled; this is difficult to achieve in vesicular model systems. In addition, as these membrane assemblies are confined to the surface of a solid support, their characterization by surface-sensitive techniques, such as atomic force microscopy, is much easier compared with characterization of free-floating vesicles (Mingeot-Leclercq et al., 2008; Goksu et al., 2009). However, one of the main drawbacks of using classical supported lipid bilayers is the proximity of the lipid bilayer and the solid substrate, which may affect the properties of the system, such as the mobility of membrane components and incorporation of transmembrane proteins. To solve such problems, one can prepare tethered lipid bilayers by introducing spacer molecules between the lipid head groups and the solid surface (Rossi et al., 2007). Another disadvantage of supported lipid bilayers is the frequent presence of defects in the bilayer. Thus the integrity of the supported lipid bilayer should be verified before the addition of proteins of interest.
Different techniques can be applied for preparing supported lipid bilayers, including the Langmuir–Blodgett technique, lipid vesicle fusion, and micellar solutions (for details of these methods, see Tamm and McConnell, 1985; Mingeot-Leclercq et al., 2008; Vacklin et al., 2005).
Lipid monolayers
Phospholipid monolayers, also referred to as Langmuir monolayers, are also useful for certain studies, due to their homogeneity, stability, and planar geometry (Brockman, 1999). Interactions of proteins with phospholipids at the air–water interface of lipid monolayers provide a unique model, for example, for studying membrane phase separation and the mechanisms by which proteins insert into membranes (for a review, see Maget-Dana, 1999). Membrane deformation by proteins has also been studied by using a lipid monolayer combined with electron microscopy (Ford et al., 2002). The two-dimensional lipid density can be controlled by compressing the monolayer, which offers advantages for specific biochemical assays. Lipid monolayers can be prepared by spreading lipid molecules at the surface of a liquid; the lipid molecules will obtain a specific orientation (at an air–water interface, the heads of phospholipids point toward the water and the acyl chains point toward the air).
ASSAYS FOR STUDYING PROTEIN–LIPID INTERACTIONS
To understand the molecular mechanism by which a protein interacts with biological membranes, one needs experimental knowledge of the affinity and specificity of the membrane-binding process and the protein's topology/conformation in the lipid bilayer, as well as possible modulation of physicochemical properties of the membrane by proteins. Thus many methods, including spectroscopy, microscopy, and cosedimentation and coflotation assays, have been developed to aid understanding of the principles of interactions between lipids and proteins.
Vesicle cosedimentation and coflotation assays
The vesicle cosedimentation and coflotation assays are widely applied for studying the affinity and lipid specificity of protein binding to membranes. A cosedimentation assay is based on sedimentation of lipid vesicles and interacting proteins in a buffer by high-speed centrifugation, whereas in the coflotation assay, the vesicles and bound proteins are enriched by density gradient centrifugation (e.g., see Stiasny et al., 2002; Zhao et al., 2010; Pykäläinen et al., 2011). In general, large unilamellar or multilamellar vesicles are the best model membrane systems for these studies. Of the two methods, the cosedimentation assay is faster and more versatile for different buffer conditions and requires smaller amounts of proteins and lipids. However, many membrane-interacting proteins have a tendency to aggregate or oligomerize, which results in their sedimentation in the absence of liposomes. Thus membrane binding of such proteins cannot be reliably examined by vesicle cosedimentation assay. A coflotation assay, which is not that sensitive to protein oligomerization/aggregation, can be used instead. It is important to note, however, that certain proteins can induce vesicle fission or fragmentation. This may cause problems in both vesicle cosedimentation and coflotation assays. To control the integrity of vesicles and ensure the presence of a lipid pellet (in a cosedimentation assay) or a lipid layer (in a coflotation assay), one can include a small amount of labeled (e.g., BODIPY) lipids in the vesicles during their preparation to make them visible to the naked eye.
To obtain information about the affinity of a protein to membranes, one should carry out these assays by varying the concentration of one of the components. This is similar to protein–protein interaction assays, in which the concentration of one of the proteins needs to be varied for obtaining the Kd value of the interaction (Pollard, 2010). In lipid-binding assays, it is often more informative to vary the concentration of lipid vesicles rather than protein, because these assays are typically carried out with a large excess of lipids versus proteins. However, it is important to remember that proteins often interact with lipids in a multivalent manner, and the binding may therefore involve variable specificity and affinity interactions between many lipid molecules and amino acids on the surface of the protein. Thus, in contrast to bimolecular protein–protein interactions in which an accurate Kd value can be measured, only an apparent Kd value can be obtained for protein–membrane interactions in most cases.
Besides the vesicle cosedimentation and coflotation assays, other methods such as isothermal titration calorimetry (ITC; Ananthanarayanan et al., 2003) and surface plasmon resonance (SPR; Besenicar et al., 2006) can be considered for measuring the affinity of a protein to membranes. However, due to certain technical limitations (e.g., a need for large quantities of proteins and lipids for ITC, and possible unspecific signals in SPR), these assays have not been widely applied for protein–lipid studies so far.
Fluorescence spectroscopy methods
Approaches based on fluorescence spectroscopy are useful for studying many aspects of lipid–protein interactions due to their intrinsic sensitivity, suitable timescale, and minimal perturbation. Steady-state fluorescence spectroscopy measurements (e.g., fluorescence intensity, spectrum shift, and quenching assays) provide information about molecular interactions within the system and can be applied to determine their affinity. On the other hand, time-resolved fluorescence methods (e.g., fluorescence resonance energy transfer [FRET] and anisotropy) can additionally provide information about the dynamics and topology of the interactions.
The fluorophores for studying protein–lipid interactions can be extrinsic probes, meaning that the lipid and/or protein is conjugated with a fluorescent probe. In both cases, this can be achieved by chemical labeling, whereas in the case of proteins one can also use fluorescent fusion proteins (e.g., green fluorescent protein or its variants). Alternatively, the intrinsic fluorescence of aromatic amino acids of the protein can be applied for studying protein–lipid interactions. Of these, tryptophan fluorescence is the most commonly used, due to its high sensitivity to the local environment. Changes in the intrinsic tryptophan fluorescence of proteins often occur upon conformational changes or ligand binding. In general, the quantum yield of tryptophan fluorescence increases in intensity (sometimes accompanied with a blue shift) when tryptophan is exposed to a more hydrophobic environment, and decreases when it is exposed to an aqueous medium. Thus, if the protein of interest harbors a tryptophan residue that is located at the membrane-binding interface or is sensitive to a possible conformational change induced by membrane-binding, tryptophan fluorescence can be applied for determining the affinity and kinetics of protein–lipid interactions (Kraft et al., 2009).
Membrane probes include fluorescent analogues of natural lipids, as well as lipophilic organic dyes that have little structural resemblance to natural biomolecules. Fluorescent lipids are either head group– or acyl chain–labeled, and special attention should be paid to selection of the probe, as certain fluorescent probes may dramatically affect the properties of the individual lipid molecules and the whole model membrane. The selection of fluorescent probes also relies on the purpose of the study. For example, acyl chain–conjugated BODIPY-PI(4,5)P2 can be applied for studying PI(4,5)P2 microdomain formation (Gambhir et al., 2004; Saarikangas et al., 2009), while nitrobenzoxadiazole PE and N-rhodamine-PE can be used for FRET studies of lipid mixing in membrane fusion (Düzgüneş et al., 2010). Another commonly used fluorescent probe, diphenylhexatriene (DPH), is a hydrophobic molecule that localizes in the acyl chain region of a lipid bilayer; it can be applied for studying the effects of proteins on membrane fluidity (Zaritsky et al., 1985; Saarikangas et al., 2009, Figure 3).
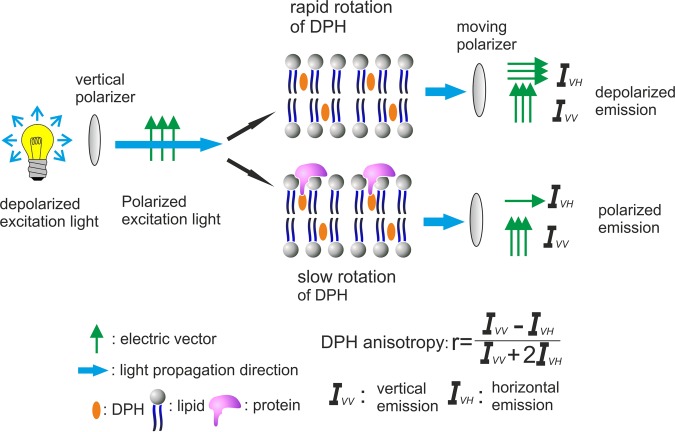
FIGURE 3:
Insertion of proteins into a lipid bilayer increases the lipid acyl chain order, which can be monitored by DPH anisotropy. A linearly polarized excitation beam is generated by a vertical polarizer. The polarized light preferentially excites DPH with transition moments aligned parallel to the incident polarization vector. The resulting fluorescence is collected and directed into two channels through a moving polarizer that measure the intensity of the fluorescence polarized both parallel (IVV) and perpendicular (IVH) to that of the excitation beam. With these two measurements, the fluorescence anisotropy, r, can be calculated. When the membrane is fluid, DPH molecules tumble quickly and have depolarized emission, thus displaying low anisotropy values. However, insertion of proteins into the hydrophobic core of a lipid bilayer diminishes the tumbling of DPH during the excitation state, and the emitted light is polarized in the perpendicular direction, resulting in increased DPH anisotropy.
In fluorescence spectroscopy assays, it is convenient to use a fluorometer with a built-in stirring mechanism, because measurements are commonly initiated with titration. If the instrument does not have this feature, the solution should be rigorously mixed by other methods to avoid trapping air bubbles within the cuvette. In addition, lipid vesicles introduce substantial light scattering at high concentration. Thus it is recommended that a low lipid concentration be used and proper control measurements be included in all assays.
Proteins can exclusively interact with lipid head groups and/or also penetrate the lipid bilayer. Electrostatic interactions of proteins with a membrane surface are salt-sensitive (Saarikangas et al., 2009). Thus carrying out membrane-binding assays (e.g., cosedimentation or fluorometric binding experiments) at different ionic strengths provides general information about whether the protein of interest binds to the membrane in a salt-sensitive or a salt-insensitive manner. The former result would indicate that interaction between the protein and membrane is mainly mediated through electrostatic interactions, while salt-insensitive binding indicates the protein may interact with the acyl chain region of the bilayer.
Several fluorescence spectroscopy methods can be applied for studying the interaction of proteins with the acyl chain region of the lipid bilayer. One method is fluorescence anisotropy studies of the membrane-embedded hydrophobic probe DPH. This rigid hydrophobic probe spontaneously inserts into the lipid bilayer and is oriented, on average, parallel to the acyl chains. The fluorescence anisotropy for DPH has been widely used to monitor changes in the rotational diffusion of acyl chains in the membrane interior (Zaritsky et al., 1985). Insertion of proteins or protein motifs, such as α-helices, into the lipid bilayer, is expected to decrease the rotational mobility of DPH, and thus increase the DPH anisotropy (Figure 3). Therefore DPH anisotropy is a relatively simple method for examining whether a protein of interest penetrates into the acyl chain region of the lipid bilayer. For more detailed studies concerning the membrane penetration and depth of membrane insertion, one can take advantage of lipids brominated at different positions along the acyl chains. Bromine-labeled lipids are collisional quenchers that can be applied to measure the depth of membrane insertion of a tryptophan residue of a protein. For more details on this method, see Ladokhin (1999), Zhao and Kinnunen (2002), and Saarikangas et al. (2009). In addition to fluorometric methods, such as the ones described above, lipid monolayers can be applied to study protein penetration into membranes, because membrane insertion of proteins causes augmented surface pressure (Zhao et al., 2006; Jones et al., 2012).
It is important to note that membrane binding may also induce significant conformational changes in the protein. For example, an unstructured region of a peripheral membrane protein can fold into an amphiphilic α-helix when penetrating into the lipid bilayers (e.g., see Gallop et al., 2006). Both micelles and liposomes can be applied in studies of the secondary and tertiary structures of membrane-bound proteins by CD spectroscopy, NMR, and x-ray crystallography (Zhao and Kinnunen, 2002; Dancea et al., 2008; Chill and Naider, 2011; Hanson et al., 2012).
THE EFFECTS OF PROTEIN BINDING ON THE PHYSICOCHEMICAL PROPERTIES OF THE MEMBRANES
Proteins have a limited number of interaction partners and often display their functions through bimolecular interactions. However, biological membranes display physical properties that cannot be explained at the single-molecule level. For example, membranes display trans-bilayer coupling, phase behavior, and elasticity. The chemical composition of the bilayer affects its mechanical properties and, conversely, membranes adapt their compositions such that the physical properties are maintained when external conditions change (Janmey and Kinnunen, 2006). Peripheral membrane proteins can cause changes in the physicochemical properties of the membranes, such as alterations in membrane fluidity and phase behavior, and can induce formation of lipid microdomains.
Protein-induced lipid clustering can be studied by fluorescence spectroscopy using an acyl chain–labeled lipid molecule such as BODIPY-PI(4,5)P2 (Gambhir et al., 2004). This fluorescent probe has highly superimposable absorption and emission spectra and exhibits self-quenching properties when two or more molecules are brought into proximity. If protein binding induces the formation of PI(4,5)P2 microdomains, BODIPY-PI(4,5)P2 molecules are clustered together, resulting in intramolecular self-quenching (Figure 4). In addition to BODIPY-labeled lipids, pyrene-labeled phospholipids can be applied to study lipid microdomain formation upon protein binding (Pap et al., 1995; Kinnunen and Holopainen, 2000). It is important to note that fluorometric lipid-clustering assays are very sensitive to small environmental changes; for example, divalent cations induce phosphoinositide clustering in model membranes (Wang et al., 2012). Thus such assays should always be carried out with proper controls and special attention should be paid to ensure no changes in the buffer conditions occur during the assay.
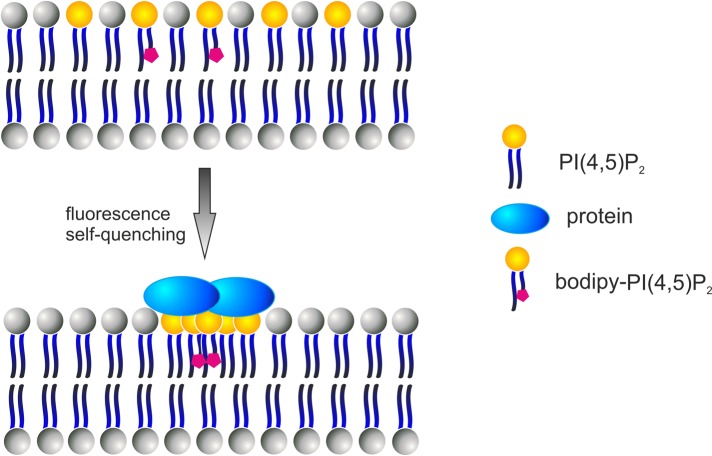
FIGURE 4:
Detection of PI(4,5)P2 microdomain formation by a fluorometric assay using BODIPY-PI(4,5)P2.Interaction of a protein in a multivalent manner with specific lipids (e.g., PI(4,5)P2) may induce lipid clustering. Furthermore, oligomerization of membrane-binding proteins may induce clustering of specific lipid molecules. The BODIPY fluorescent probe has highly superimposable absorption and emission spectra and exhibits self-quenching properties when two or more molecules are brought into proximity. Upon lipid clustering, BODIPY-labeled lipids (e.g., PI(4,5)P2) form excimers, which results in intramolecular self-quenching of the BODIPY fluorescence. This change in BODIPY fluorescence can be applied for studying the effects of a protein on clustering of specific BODIPY-conjugated lipid species.
Other lipid probes, such as laurdan, prodan, and pyrene, are extensively used in studies concerning structural and dynamical properties of membranes (e.g., phase behavior, lipid order, membrane fusion). These probes can be incorporated into the lipid bilayer either alone or covalently linked to fatty acids. They locate at different depths in the bilayer and display distinct photophysical properties. Pyrene-labeled phospholipids are commonly used for studying lateral diffusion and segregation of lipids, as well as for membrane fusion assays (Kinnunen and Holopainen, 2000). Laurdan and prodan probes are sensitive to the physical state of the surrounding lipids, and are thus applied to study changes in lipid order and membrane-phase behavior (Parasassi et al., 1998; Krasnowska et al., 1998; Kaiser et al., 2009).
MEMBRANE MORPHOLOGY ANALYSIS
The shape and dynamics of the plasma membrane can be modulated by proteins that directly bind membranes and sculpt them into desired shapes (McMahon and Gallop, 2005). Besides in vivo studies, membrane-sculpting processes can be examined by electron microscopy and live imaging of vesicles. Commonly used electron microscopy methods for membrane morphology analysis are negative staining of protein–vesicle mixtures and Epon-embedded thin sections of uranyl acetate–stained protein–vesicle samples (e.g., see Peter et al., 2004; Mattila et al., 2007). Structural features of the protein-induced changes in membrane topology can be examined in more detail using electron tomography analysis of Epon-embedded liposome samples (Mattila et al., 2007; Saarikangas et al., 2009). In the electron microscopy studies, it is important to use a vesicle size that is large enough for the protein to bend the membrane. For example, if the protein is expected to generate membrane tubules with a diameter of ∼200 nm, one should use vesicles with an average diameter of at least 500 nm.
Giant liposomes containing fluorescently labeled lipids and/or proteins can also be applied to study the effects of proteins on membrane topology. The dynamics of protein membrane interaction, membrane microdomain formation, and changes in the shape of the liposome can be readily observed with fluorescence microscopy methods (e.g., see Saarikangas et al., 2009; Wollert et al., 2009; Römer et al., 2007, 2010; Fyfe et al., 2011; Sorre et al., 2012). Although the spatial resolution of such assays is significantly lower compared with the above-mentioned electron microscopy methods, information obtained with this system from the dynamics of membrane deformation by proteins can be highly valuable. However, when carrying out live imaging of giant unilamellar vesicles, it is important to keep in mind that they are very sensitive to small perturbations (e.g., osmolarity) of the environment. Thus osmolarity should be precisely controlled, and all assays should be accompanied by appropriate control experiments in the absence of protein.
CONCLUSIONS
A number of fluorometric, centrifugation, and microscopic assays are available for studying the molecular mechanisms by which proteins interact with membranes and how these interactions affect the conformation and dynamics of both proteins and lipid bilayers. Each of these assays provides a distinct type of information and has specific technical limitations. It is therefore important to choose an assay that is suitable for the specific structural/chemical properties of the protein of interest and to make sure the particular assay will provide the type of information needed. Because lipid compositions vary significantly between different cell types, their membrane compartments, and the two leaflets of individual membranes, it is also important to choose the lipid composition of the model membrane such that it resembles as accurately as possible the native membrane compartment of interest. We hope that the information provided in this Perspective, and links to the more detailed references concerning various methods, will help you in designing and performing protein–lipid assays in a way that will generate reliable and important information about these interactions.
Acknowledgments
We thank Minna Poukkula and Maria Vartiainen for valuable comments on the manuscript. P.L. and H.Z. were supported by grants from the Academy of Finland and the Finnish Cancer Research Foundation.
Abbreviations used:
- CD
circular dichroism
- DPH
diphenylhexatriene
- ER
endoplasmic reticulum
- FRET
fluorescence resonance energy transfer
- ITC
isothermal titration calorimetry
- NMR
nuclear magnetic resonance
- PE
phosphatidylethanolamine
- PI
phosphatidylinositol
- PI(4)P
phosphatidylinositol 4-phosphate
- PI(4,5)P2
phosphatidylinositol 4,5-biphosphate
- PS
phosphatidylserine
- SPR
surface plasmon resonance
Footnotes
REFERENCES
- Abkarian M, Loiseau E, Massiera G. Continuous droplet interface crossing encapsulation (cDICE) for high throughput monodisperse vesicle design. Soft Matter. 2011;7:4610–4614. [Google Scholar]
- Ananthanarayanan B, Stahelin RV, Digman MA, Cho W. Activation mechanisms of conventional protein kinase C isoforms are determined by the ligand affinity and conformational flexibility of their C1 domains. J Biol Chem. 2003;278:46886–46894. doi: 10.1074/jbc.M307853200. [DOI] [PubMed] [Google Scholar]
- Angelova MI, Soleau S, Meleard PH, Faucon JF, Bothorel F. Preparation of giant vesicles by external AC electric fields. Kinetics and applications. Progr Colloid Polymer Sci. 1992;89:127–131. [Google Scholar]
- Bacia K, Futai E, Prinz S, Meister A, Daum S, Glatte D, Briggs JA, Schekman R. Multibudded tubules formed by COPII on artificial liposomes. Sci Rep. 2011:17. doi: 10.1038/srep00017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassereau P, Goud B. Physics, biology and the right chemistry. F1000 Biol Rep. 2011;3:7. doi: 10.3410/B3-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besenicar M, Macek P, Lakey JH, Anderluh G. Surface plasmon resonance in protein-membrane interactions. Chem Phys Lipids. 2006;141:169–178. doi: 10.1016/j.chemphyslip.2006.02.010. [DOI] [PubMed] [Google Scholar]
- Brockman H. Lipid monolayers: why use half a membrane to characterize protein-membrane interactions? Curr Opin Struct Biol. 1999;9:438–443. doi: 10.1016/S0959-440X(99)80061-X. [DOI] [PubMed] [Google Scholar]
- Chill JH, Naider F. A solution NMR view of protein dynamics in the biological membrane. Curr Opin Struct Biol. 2011;21:627–633. doi: 10.1016/j.sbi.2011.07.003. [DOI] [PubMed] [Google Scholar]
- Colbeau A, Nachbaur J, Vignais PM. Enzymic characterization and lipid composition of rat liver subcellular membranes. Biochim Biophys Acta. 1971;249:462–492. doi: 10.1016/0005-2736(71)90123-4. [DOI] [PubMed] [Google Scholar]
- Dancea F, Kami K, Overduin M. Lipid interaction networks of peripheral membrane proteins revealed by data-driven micelle docking. Biophys J. 2008;94:515–524. doi: 10.1529/biophysj.107.115923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimova R, Aranda S, Bezlyepkina N, Nikolov V, Riske KA, Lipowsky R. A practical guide to giant vesicles. Probing the membrane nanoregime via optical microscopy. J Phys Condens Matter. 2006;18:S1151–S1176. doi: 10.1088/0953-8984/18/28/S04. [DOI] [PubMed] [Google Scholar]
- Di Paolo G, De Camilli P. Phosphoinositides in cell regulation and membrane dynamics. Nature. 2006;443:651–657. doi: 10.1038/nature05185. [DOI] [PubMed] [Google Scholar]
- Düzgüneş N, Faneca H, Lima MC. Methods to monitor liposome fusion, permeability, and interaction with cells. Methods Mol Biol. 2010;606:209–232. doi: 10.1007/978-1-60761-447-0_16. [DOI] [PubMed] [Google Scholar]
- Estes DJ, Mayer M. Giant liposomes in physiological buffer using electroformation in a flow chamber. Biochim Biophys Acta. 2005;1712:152–160. doi: 10.1016/j.bbamem.2005.03.012. [DOI] [PubMed] [Google Scholar]
- Ford MGJ, Mills IG, Peter BJ, Vallis Y, Praefcke GJ, Evans PR, McMahon HT. Curvature of clathrin-coated pits driven by epsin. Nature. 2002;419:361–366. doi: 10.1038/nature01020. [DOI] [PubMed] [Google Scholar]
- Frost A, Unger VM, de Camili P. The BAR domain superfamily: membrane-molding macromolecules. Cell. 2009;137:191–196. doi: 10.1016/j.cell.2009.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fyfe I, Schuh AL, Edwardson JM, Audhya A. Association of the endosomal sorting complex ESCRT-II with the Vps20 subunit of ESCRT-III generates a curvature-sensitive complex capable of nucleating ESCRT-III filaments. J Biol Chem. 2011;286:34262–34270. doi: 10.1074/jbc.M111.266411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallop JL, Jao CC, Kent HM, Butler PJ, Evans PR, Langen R, McMahon HT. Mechanism of endophilin N-BAR domain-mediated membrane curvature. EMBO J. 2006;25:2898–2910. doi: 10.1038/sj.emboj.7601174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gambhir A, Hangyas-Mihalyne G, Zaitseva I, Cafiso DS, Wang J, Murray D, Pentyala SN, Smith SO, McLaughlin S. Electrostatic sequestration of PIP2 on phospholipid membranes by basic/aromatic regions of proteins. Biophys J. 2004;86:2188–2207. doi: 10.1016/S0006-3495(04)74278-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goksu EI, Vanegas JM, Blanchette CD, Lin WC, Longo ML. AFM for structure and dynamics of biomembranes. Biochim Biophys Acta. 2009;1788:254–266. doi: 10.1016/j.bbamem.2008.08.021. [DOI] [PubMed] [Google Scholar]
- Hanson MA, et al. Crystal structure of a lipid G protein-coupled receptor. Science. 2012;335:851–855. doi: 10.1126/science.1215904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janmey PA, Kinnunen PK. Biophysical properties of lipids and dynamic membranes. Trends Cell Biol. 2006;16:538–546. doi: 10.1016/j.tcb.2006.08.009. [DOI] [PubMed] [Google Scholar]
- Jones EM, Dubey M, Camp PJ, Vernon BC, Biernat J, Mandelkow E, Majewski J, Chi EY. Interaction of tau protein with model lipid membranes induces tau structural compaction and membrane disruption. Biochemistry. 2012;51:2539–2550. doi: 10.1021/bi201857v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser HJ, Lingwood D, Levental I, Sampaio JL, Kalvodova L, Rajendran L, Simons K. Order of lipid phases in model and plasma membranes. Proc Natl Acad Sci USA. 2009;106:16645–16650. doi: 10.1073/pnas.0908987106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinnunen PK, Holopainen JM. Mechanisms of initiation of membrane fusion: role of lipids. Biosci Rep. 2000;20:465–482. doi: 10.1023/a:1010402819509. [DOI] [PubMed] [Google Scholar]
- Kraft CA, Garrido JL, Leiva-Vega L, Romero G. Quantitative analysis of protein-lipid interactions using tryptophan fluorescence. Sci Signal. 2009;2:pl4. doi: 10.1126/scisignal.299pl4. [DOI] [PubMed] [Google Scholar]
- Krasnowska EK, Gratton E, Parasassi T. Prodan as a membrane surface fluorescence probe: partitioning between water and phospholipid phases. Biophys J. 1998;74:1984–1993. doi: 10.1016/S0006-3495(98)77905-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ladokhin AS. Analysis of protein and peptide penetration into membranes by depth-dependent fluorescence quenching: theoretical considerations. Biophys J. 1999;76:946–955. doi: 10.1016/S0006-3495(99)77258-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemmon MA. Membrane recognition by phospholipid-binding domains. Nat Rev Mol Cell Biol. 2008;9:99–111. doi: 10.1038/nrm2328. [DOI] [PubMed] [Google Scholar]
- Loose M, Schwille P. Biomimetic membrane systems to study cellular organization. J Struct Biol. 2009;168:143–151. doi: 10.1016/j.jsb.2009.03.016. [DOI] [PubMed] [Google Scholar]
- Luisi PL, Walde P. Giant Vesicles: Perspectives in Supramolecular Chemistry. Chichester, UK: John Wiley & Sons; 2000. [Google Scholar]
- Maget-Dana R. The monolayer technique: a potent tool for studying the interfacial properties of antimicrobial and membrane-lytic peptides and their interactions with lipid membranes. Biochim Biophys Acta. 1999;1462:109–140. doi: 10.1016/s0005-2736(99)00203-5. [DOI] [PubMed] [Google Scholar]
- Mattila PK, Pykäläinen A, Saarikangas J, Paavilainen VO, Vihinen H, Jokitalo E, Lappalainen P. Missing-in-metastasis and IRSp53 deform PI(4,5)P2-rich membranes by an inverse BAR domain-like mechanism. J Cell Biol. 2007;176:953–964. doi: 10.1083/jcb.200609176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMahon HT, Gallop JL. Membrane curvature and mechanisms of dynamic cell membrane remodelling. Nature. 2005;438:590–596. doi: 10.1038/nature04396. [DOI] [PubMed] [Google Scholar]
- McMahon HT, Kozlov MM, Martens S. Membrane curvature in synaptic vesicle fusion and beyond. Cell. 2010;140:601–605. doi: 10.1016/j.cell.2010.02.017. [DOI] [PubMed] [Google Scholar]
- Mingeot-Leclercq MP, Deleu M, Brasseur R, Dufrêne YF. Atomic force microscopy of supported lipid bilayers. Nat Protoc. 2008;3:1654–1659. doi: 10.1038/nprot.2008.149. [DOI] [PubMed] [Google Scholar]
- Mui B, Chow L, Hope MJ. Extrusion technique to generate liposomes of defined size. Methods Enzymol. 2003;367:3–14. doi: 10.1016/S0076-6879(03)67001-1. [DOI] [PubMed] [Google Scholar]
- Pap EHW, Hanicak A, van Hoek A, Wirtz KWA, Visser AJWG. Quantitative analysis of lipid-lipid and lipid-protein interactions in membranes by use of pyrene-labeled phosphoinositides. Biochemistry. 1995;34:9118–9125. doi: 10.1021/bi00028a022. [DOI] [PubMed] [Google Scholar]
- Parasassi T, Krasnowska EK, Bagatolli LA, Gratton E. Laurdan and Prodan as polarity sensitive fluorescence membrane probes. J Fluorescence. 1998;8:365–373. [Google Scholar]
- Pautot S, Frisken BJ, Weitz DA. Engineering asymmetric vesicles. Proc Natl Acad Sci USA. 2003;100:10718–10721. doi: 10.1073/pnas.1931005100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peric M, Alves M, Bales BL. Combining precision spin-probe partitioning with time-resolved fluorescence quenching to study micelles. Application to micelles of pure lysomyristoylphosphatidylcholine (LMPC) and LMPC mixed with sodium dodecyl sulfate. Chem Phys Lipids. 2006;142:1–13. doi: 10.1016/j.chemphyslip.2006.02.003. [DOI] [PubMed] [Google Scholar]
- Peter BJ, Kent HM, Mills IG, Vallis Y, Butler PJ, Evans PR, McMahon HT. BAR domains as sensors of membrane curvature: the amphiphysin BAR structure. Science. 2004;303:495–499. doi: 10.1126/science.1092586. [DOI] [PubMed] [Google Scholar]
- Pollard TD. A guide to simple and informative binding assays. Mol Biol Cell. 2010;21:4061–4067. doi: 10.1091/mbc.E10-08-0683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pott T, Bouvrais H, Meleard P. Giant unilamellar vesicle formation under physiologically relevant conditions. Chem Phys Lipids. 2008;154:115–119. doi: 10.1016/j.chemphyslip.2008.03.008. [DOI] [PubMed] [Google Scholar]
- Pykäläinen A, et al. Pinkbar is an epithelial-specific BAR domain protein that generates planar membrane structures. Nat Struct Mol Biol. 2011;18:902–907. doi: 10.1038/nsmb.2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richmond DL, Schmid EM, Martens S, Stachowiak JC, Liska N, Fletcher DA. Forming giant vesicles with controlled membrane composition, asymmetry, and contents. Proc Natl Acad Sci USA. 2011;108:9431–9436. doi: 10.1073/pnas.1016410108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Römer W, et al. Shiga toxin induces tubular membrane invaginations for its uptake into cells. Nature. 2007;450:670–675. doi: 10.1038/nature05996. [DOI] [PubMed] [Google Scholar]
- Römer W, et al. Actin dynamics drive membrane reorganization and scission in clathrin-independent endocytosis. Cell. 2010;140:540–553. doi: 10.1016/j.cell.2010.01.010. [DOI] [PubMed] [Google Scholar]
- Rossi C, Briand E, Parot P, Odorico M, Chopineau J. Surface response methodology for the study of supported membrane formation. J Phys Chem B. 2007;111:7567–7576. doi: 10.1021/jp0686792. [DOI] [PubMed] [Google Scholar]
- Saarikangas J, Zhao H, Lappalainen P. Regulation of the actin cytoskeleton-plasma membrane interplay by phosphoinositides. Physiol Rev. 2010;90:259–289. doi: 10.1152/physrev.00036.2009. [DOI] [PubMed] [Google Scholar]
- Saarikangas J, Zhao H, Pykäläinen A, Laurinmaki P, Mattila PK, Kinnunen PK, Butcher SJ, Lappalainen P. Molecular mechanisms of membrane deformation by I-BAR domain proteins. Curr Biol. 2009;19:95–107. doi: 10.1016/j.cub.2008.12.029. [DOI] [PubMed] [Google Scholar]
- Singh J, Miller J, Ranganathan R. Physicochemical characterization of phospholipid solubilized mixed micelles and a hydrodynamic model of interfacial fluorescence quenching. J Phys Chem B. 2007;111:9317–9324. doi: 10.1021/jp0720340. [DOI] [PubMed] [Google Scholar]
- Sorre B, Callan-Jones A, Manzi J, Goud B, Prost J, Bassereau P, Roux A. Nature of curvature coupling of amphiphysin with membranes depends on its bound density. Proc Natl Acad Sci USA. 2012;109:173–178. doi: 10.1073/pnas.1103594108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stiasny K, Allison SL, Schalich J, Heinz FX. Membrane interactions of the tick-borne encephalitis virus fusion protein E at low pH. J Virol. 2002;76:3784–3790. doi: 10.1128/JVI.76.8.3784-3790.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szoka F, Jr., Papahadjopoulos D. Comparative properties and methods of preparation of lipid vesicles (liposomes) Annu Rev Biophys Bioeng. 1980;9:467–508. doi: 10.1146/annurev.bb.09.060180.002343. [DOI] [PubMed] [Google Scholar]
- Tamm LK, McConnell HM. Supported phospholipid bilayers. Biophys J. 1985;47:105–113. doi: 10.1016/S0006-3495(85)83882-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenchov BG, Yanev TK, Tihova MG, Koynova RD. A probability concept about size distributions of sonicated lipid vesicles. Biochim Biophys Acta. 1985;816:122–130. doi: 10.1016/0005-2736(85)90400-6. [DOI] [PubMed] [Google Scholar]
- Vacklin HP, Tiberg F, Thomas RK. Formation of supported phospholipid bilayers via co-adsorption with β-d-dodecyl maltoside. Biochim Biophys Acta. 2005;1668:17–24. doi: 10.1016/j.bbamem.2004.11.001. [DOI] [PubMed] [Google Scholar]
- van Meer G, de Kroon AI. Lipid map of the mammalian cell. J Cell Sci. 2011;124:5–8. doi: 10.1242/jcs.071233. [DOI] [PubMed] [Google Scholar]
- van Meer G, Voelker DR, Feigenson GW. Membrane lipids: where they are and how they behave. Nat Rev Mol Cell Biol. 2008;9:112–124. doi: 10.1038/nrm2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Heijne G. Membrane-protein topology. Nat Rev Mol Cell Biol. 2006;12:909–918. doi: 10.1038/nrm2063. [DOI] [PubMed] [Google Scholar]
- Walde P, Cosentino K, Engel H, Stano P. Giant vesicles: preparations and applications. Chembiochem. 2010;11:848–865. doi: 10.1002/cbic.201000010. [DOI] [PubMed] [Google Scholar]
- Wang YH, Collins A, Guo L, Smith-Dupont KB, Gai F, Svitkina T, Janmey PA. Divalent cation-induced cluster formation by polyphosphoinositides in model membranes. J Am Chem Soc. 2012;134:3387–3395. doi: 10.1021/ja208640t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wollert T, Wunder C, Lippincott-Schwartz J, Hurley JH. Membrane scission by the ESCRT-III complex. Nature. 2009;458:172–177. doi: 10.1038/nature07836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaritsky A, Parola AH, Abdah M, Masalha H. Homeoviscous adaptation, growth rate, and morphogenesis in bacteria. Biophys J. 1985;2:337–339. doi: 10.1016/S0006-3495(85)83788-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H, Hakala M, Lappalainen P. ADF/cofilin binds phosphoinositides in a multivalent manner to act as a PIP(2)-density sensor. Biophys J. 2010;98:2327–2336. doi: 10.1016/j.bpj.2010.01.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H, Kinnunen PK. Binding of the antimicrobial peptide temporin L to liposomes assessed by Trp fluorescence. J Biol Chem. 2002;277:25170–25177. doi: 10.1074/jbc.M203186200. [DOI] [PubMed] [Google Scholar]
- Zhao H, Pykäläinen A, Lappalainen P. I-BAR domain proteins: linking actin and plasma membrane dynamics. Curr Opin Cell Biol. 2011;23:14–21. doi: 10.1016/j.ceb.2010.10.005. [DOI] [PubMed] [Google Scholar]
- Zhao H, Sood R, Jutila A, Bose S, Fimland G, Nissen-Meyer J, Kinnunen PK. Interaction of the antimicrobial peptide pheromone Plantaricin A with model membranes: implications for a novel mechanism of action. Biochim Biophys Acta. 2006;1758:1461–1474. doi: 10.1016/j.bbamem.2006.03.037. [DOI] [PubMed] [Google Scholar]
- Zimmerberg J, Chernomordik LV. Membrane fusion. Adv Drug Deliv Rev. 1999;38:197–205. doi: 10.1016/s0169-409x(99)00029-0. [DOI] [PubMed] [Google Scholar]