A gain-of-function study showed that miR-7a promoted the generation of oligodendrocytes (OL) and retained the cells in their precursor stage. Inhibiting miR-7a reduced oligodendrogenesis but expanded neuronal population. miR-7a might exert these effects by repressing the expression of proneural genes and regulators for OL differentiation.
Abstract
The generation of myelinating cells from multipotential neural stem cells in the CNS requires the initiation of specific gene expression programs in oligodendrocytes (OLs). We reasoned that microRNAs (miRNAs) could play an important role in this process by regulating genes crucial for OL development. Here we identified miR-7a as one of the highly enriched miRNAs in oligodendrocyte precursor cells (OPCs), overexpression of which in either neural progenitor cells (NPCs) or embryonic mouse cortex promoted the generation of OL lineage cells. Blocking the function of miR-7a in differentiating NPCs led to a reduction in OL number and an expansion of neuronal populations simultaneously. We also found that overexpression of this miRNA in purified OPC cultures promoted cell proliferation and inhibited further maturation. In addition, miR-7a might exert the effects just mentioned partially by directly repressing proneuronal differentiation factors including Pax6 and NeuroD4, or proOL genes involved in oligodendrocyte maturation. These results suggest that miRNA pathway is essential in determining cell fate commitment for OLs and thus providing a new strategy for modulating this process in OL loss diseases.
INTRODUCTION
Oligodendrocytes (OLs) play a critical role in the CNS by producing insulating protein membranes that ensheath axons. Significant damages to OLs result in demyelination and hinder effective communication among neurons. Correspondingly, CNS demyelinating conditions, such as spinal cord injury and multiple sclerosis, will result in severe motor, sensory, and cognitive impairment. To obtain proper remyelination, the knowledge regarding the regulators of OL development has been a major focus in understanding the mechanisms promoting differentiation of oligodendrocyte precursor cells (OPCs).
Myelinating OLs are derived from multipotential neural progenitor cells (NPCs). The process of creating a properly functional OL, including cell fate specification and OPC migration, maturation, and myelination, is regulated by the dynamic interplay between transcription factors, epigenetic factors, microRNA (miRNA) regulators, and other cell-extrinsic signals (Emery, 2010; Yu et al., 2010). miRNAs are small, noncoding RNAs that regulate gene expression by posttranscriptional targeting RNA-induced silencing complex (RISC) to cognate mRNA (Bartel, 2004). Studies using a Nestin-Cre line to delete the miRNA-processing enzyme Dicer in the entire CNS show that Dicer function or the maturation of miRNA is required for the proper specification and development of both neurons and OLs during embryonic stages (Kawase-Koga et al., 2009). Moreover, disruption of Dicer function in specified OPCs and OLs by expressing Cre from the Olig1 or Olig2 promoter induces the failure of normal OL differentiation, as well as myelin formation (Dugas et al., 2010; Zhao et al., 2010; Zheng et al., 2010). Therefore these studies indicate that mature miRNA activity is required at various stages of OL development, including the initial production of fate-specified OPCs, the differentiation of mature OLs, the generation of compact CNS myelin during development, and the maintenance of functional myelin sheaths in adult animals (Dugas and Notterpek, 2011).
Several groups have investigated the roles of individual miRNA in promoting functional CNS myelination. miR-219 and miR-338 are two of the most highly induced miRNAs in differentiating OLs, and they are individually necessary and sufficient to promote normal OPC differentiation into OLs in vitro and in vivo (Dugas et al., 2010; Zhao et al., 2010); miR-23a and miR-23b have been identified as miRNAs that are induced approximately fivefold during OL maturation, and overexpression of either one can enhance OL differentiation, probably by reducing the expression of a gene that inhibits normal OL maturation (Lin and Fu, 2009); miR-138 is also induced in differentiating OLs, and it specifically promotes the early stages of OL differentiation, while simultaneously suppressing the later stage of OL differentiation (Dugas et al., 2010; Zhao et al., 2010). In contrast to the identification of miRNAs in promoting OL differentiation, a set of OPC-enriched miRNAs has also been identified (Lau et al., 2008; Shin et al., 2009; Letzen et al., 2010), among which members of the miR-17-92 cluster were found to be both necessary and sufficient to enhance OPC proliferation in vivo and in vitro (Budde et al., 2010). Given the critical role of miRNAs in neurogenesis (Cheng et al., 2009; Kawase-Koga et al., 2009; Shi et al., 2010; Yoo et al., 2011), we hypothesize that oligodendroglial lineage specification might be guided in part by miRNA pathways.
In Lau's investigation, miR-7a, together with several other miRNAs, showed a higher level in A2B5+/GC− OPCs than in A2B5−/GC+ mature OLs sorted from postnatal rat brain (Lau et al., 2008). Because miR-7a shared a similar temporal expression pattern with the miR-17-92 cluster (Miska et al., 2004), we speculated that miR-7a might also play a role in OPC self-renewal and differentiation. In the current study, through in vitro and in utero gain-of-function experiments, we investigated the effect of miR-7a in regulating the generation of OL from multipotential NPCs as well as from embryonic mouse cortex. Also, inhibiting the function of miR-7a in NPC cultures enhanced our conclusion about miR-7a in oligodendrogenesis. Furthermore, we showed that miR-7a controlled oligodendrogenesis by directly targeting proneural genes such as Pax6 and NeuroD4, and it might impede OL differentiation by targeting regulators for this process. Our present study provides insight into the regulation of OL specification and proliferation by miRNAs. This work further suggests that switching off certain neuronal differentiation factors by miRNAs is a critical mechanism for the generation of OLs.
RESULTS
miR-7a expression in different brain cell types in vitro
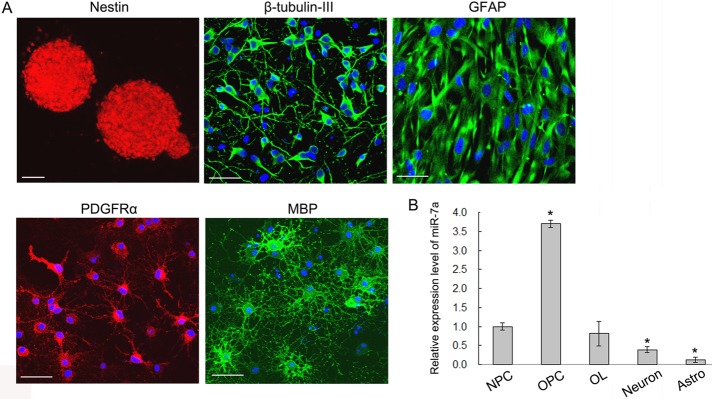
Previous studies indicated that miR-7 was one of the top miRNAs highly expressed in FACS-sorted rat A2B5+/GalC− OPCs, and it was found down-regulated during OL differentiation in vivo using miRNA microarray (Lau et al., 2008). To validate the results, we examined the expression level of mature miR-7a by quantitative real-time PCR (qRT-PCR) with the TaqMan MicroRNA Assay kit. Purified OPCs, differentiated mature OLs, together with primary neurosphere cultures, astrocytes, and cortical neurons, were first identified by immunostaining with specific markers (Figure 1A), and the purity of individual cultures was [mt]95%. Then total RNAs from each cell type were collected, and qRT-PCR showed that miR-7a expression was higher in OL lineage cells than in other neural cell types, with the highest level at the OPC stage (Figure 1B). This result confirmed previous microarray data and suggested that miR-7a might be involved in the generation of OLs.
FIGURE 1:
miR-7a expression in different brain cell types. (A) Representative images of purified neurospheres (Nestin+), neurons (β-tubulin-III+), astrocytes (GFAP+), OPCs (PDGFRα+), and mature OLs (MBP+) from embryonic or neonatal rat cortex used for studying the expression level of miR-7a. Scale bars, 50 μm. (B) The TaqMan MicroRNA Assay was applied to analyze the relative expression level of miR-7a from different cell cultures as indicated. U6 RNA was used as internal control. Data are from three independent experiments and represent mean ± SEM. *p < 0.05 (Student's t test).
Overexpression of miR-7a in differentiating NPCs expanded PDGFRα+ and Olig2+ OPC populations
To investigate whether miR-7a is involved in the differentiation of NPCs, we induced transient overexpression of miR-7a by transfection of miRNA mimics. An increase in the intracellular miR-7a level was confirmed by qRT-PCR (Supplemental Figure 1A).
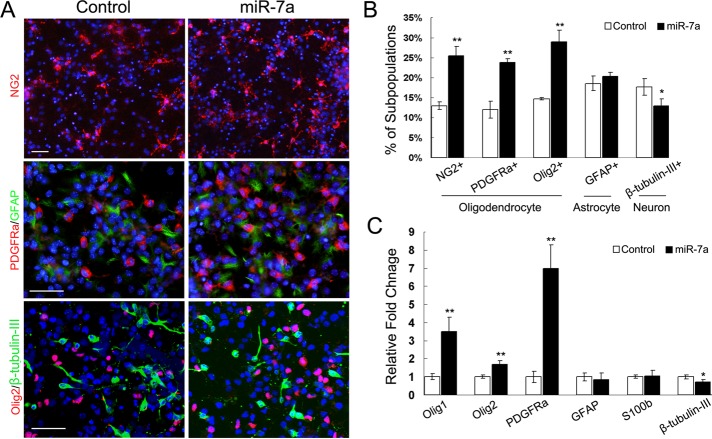
The initial dissociated NPC culture before transfection was a ∼98% Nestin+ population, with <5% GFAP+ cells. β-tubulin-III+ and PDGFRα+ cells were not detected at this time. Another three days' treatment with low concentrations of basic fibroblast growth factor (bFGF) and epidermal growth factor (EGF), together with platelet-derived growth factor (PDGF)-AA, induced the negative control–transfected NPCs differentiating into astrocytes (18.6 ± 1.87%), neurons (17.62 ± 2.07%), and early-stage OLs (12 ± 2.13%, 13.01 ± 1.01%, 14.7 ± 0.34%), as demonstrated by immunostaining for glial fibrillary acidic protein (GFAP), β-tubulin-III, PDGFRα, NG2, and Olig2 (Figure 2, A and B). We did not observe major phenotypic differences between NPCs transfected with control oligonucleotides and nontransfected cells (unpublished data). Transfection of miR-7a mimics increased the percentage of OPCs up to 24∼30%; about twofold that of control transfection, but did not change the percentage of GFAP+ astrocytes. Quantification of β-tubulin-III+ cells showed a slight decrease in the miR-7a transfected group compared with control (Figure 2B).
FIGURE 2:
Overexpression of miR-7a promoted OL generation in vitro. (A) Mouse NPCs were transfected with miR-7a mimics and scrambled miRNA negative control as indicated. The transfected cells were cultured for another 3 d and subjected to immunostaining with antibodies against NG2, PDGFRα, Olig2, GFAP, and β-tubulin-III. Scale bars, 50 μm. (B) Histogram depicts the percentage of NG2+, PDGFRα+, Olig2+, GFAP+, and β-tubulin-III+ cells, which represent three cell types as indicated in differentiated NPCs. Data are from three independent experiments. (C) Relative expression level of neural lineage specific genes was analyzed by qRT-PCR from miR-7a transfected differentiating NPC cultures. GAPDH was used as internal control. Data represent mean ± SEM. *p < 0.05, **p < 0.01 (one-way ANOVA).
Also, the mRNA expression level of several cell lineage-specific genes revealed similar alterations after miR-7a overexpression. The basic helix-loop-helix (bHLH) proteins, Olig1 and Olig2, that have well demonstrated the crucial role in oligodendrogenesis and myelination, together with PDGFRα, were significantly up-regulated by miR-7a transfection in differentiating NPC cultures; expression for GFAP and early formed astrocyte marker S100b showed no obvious differences between the two groups; and the neuron marker β-tubulin-III was decreased in miR-7a–transfected NPC cultures (Figure 2C).
miR-7a induced OL specification in vivo
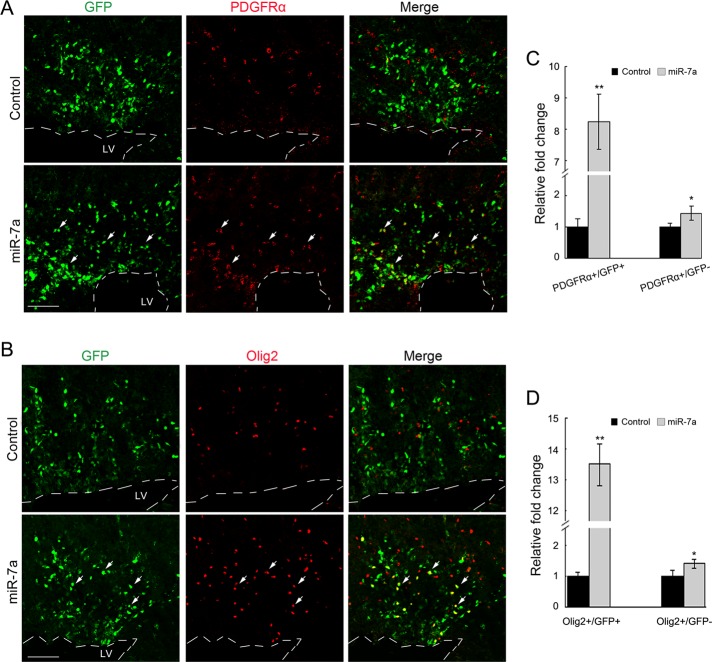
To determine the effect of miR-7a overexpression on OL development in mice, we electroporated an miR-7a–expressing vector into one side of the neocortical ventricular zone of developing embryos at E14.5. The cortices from electroporated embryos were collected 3 d later (E17.5). Blank vector electroporation was included as control. Overexpression of miR-7a induced a significant increase of both PDGFRα+/GFP+ and Olig2+/GFP+ OPCs in the cortex (Figure 3), which suggested that miR-7a could promote OL lineage formation in the developing mouse cortex. Moreover, there was also an increase in the number of either PDGFRα+/GFP− or Olig2+/GFP− cells in the miR-7a–treated group compared with control electroporation, which suggested a possible nonautonomous effect of miR-7a overexpression.
FIGURE 3:
In utero electroporation of miR-7a promoted OL specification in embryonic mouse cortex. (A and B) Mouse embryos at E14.5 were electroporated with expression vector for miR-7a and harvested at E17.5. The sections of electroporated cortices were analyzed by immunostaining with antibodies to PDGFRα and Olig2, respectively. pCIG plasmid electroporation was included as control. Arrows indicate electroporated GFP+ cells colabeling with PDGFRα or Olig2. Dashed line shows the edge of lateral ventricle (LV). Scale bar, 100 μm. (C and D) Quantification of PDGFRα+ or Olig2+ cells in a defined cortical area (0.25 mm2). Y axis indicates the ratio of the number of PDGFRα+/GFP+, PDGFRα+/GFP− and Olig2+/GFP+, Olig2+/GFP− cells in miR-7a–overexpressing cortices to that of the control. Data represent mean ± SEM. **p < 0.01, *p < 0.05 (one-way ANOVA).
Knockdown miR-7a expression repressed NPCs committing to OL lineage
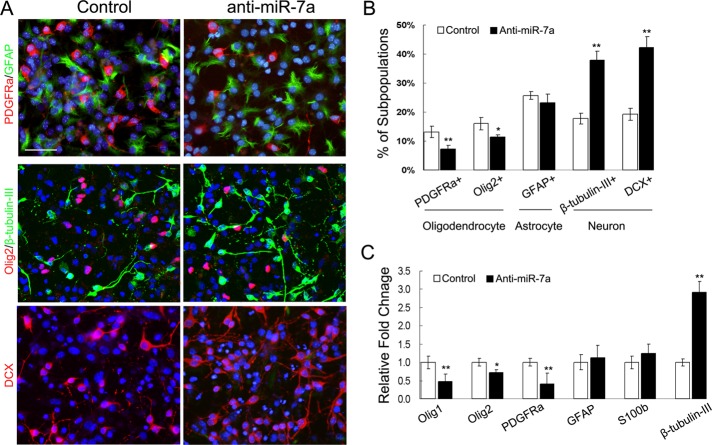
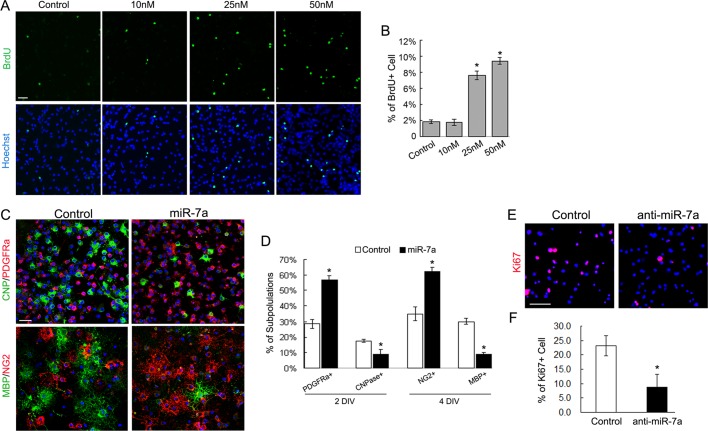
Next we blocked the activity of miR-7a in differentiating NPCs by transfection of hairpin miRNA inhibitors and studied the effect on cell fate commitment. qRT-PCR assay confirmed the efficiency of miRNA inhibitors in reducing the level of mature miR-7a (Supplemental Figure 1B). In general, knocking down the level of miR-7a in differentiated NPCs resulted in an inverse differentiation pattern compared with overexpression study, as revealed by immunostaining and qRT-PCR assay: Inhibiting miR-7a in differentiating NPCs significantly reduced the percentage of OPCs and the mRNA expression of several OPC markers, whereas the generation of β-tubulin-III+ and doublecortin+ (DCX) neurons was greatly promoted. No obvious change in astrocyte maker expression was detected (Figure 4).
FIGURE 4:
Inhibition of miR-7a repressed oligodendrogenesis and induced neurogenesis in vitro. (A) Mouse NPC cultures were transfected with miR-7a hairpin inhibitor and negative control. Three days after transfection, cells were subjected to immunostaining with antibodies against PDGFRα, Olig2, GFAP, β-tubulin-III, and DCX. Scale bars, 50 μm. (B) Histogram depicts the percentage of PDGFRα+, Olig2+, GFAP+, β-tubulin-III+, and DCX+ cells, which represent three cell types as indicated in differentiated NPCs. Data are from three independent experiments. (C) Relative expression level of several neural lineage specific genes was analyzed by qRT-PCR from miR-7a hairpin inhibitor transfected differentiating NPC cultures. GAPDH was used as internal control. Data represent mean ± SEM. *p < 0.05, **p < 0.01 (one-way ANOVA).
Involvement of miR-7a in OPC proliferation and further differentiation
According to the results just mentioned, we speculated that the increase in the number of OPCs, with the decrease in the percentage of neurons at the same time, from miR-7a–overexpressing NPCs may be due to selective survival, selective stimulation of OPCs generation or proliferation, and/or a direct inhibition effect on neuron generation. As far as selective survival is concerned, no differences in cell death were observed between control NPCs and miR-7a–overexpressing or –silenced NPCs as identified by Caspase3 immunostaining (unpublished data).
To determine whether the increase in the number of OPCs after miR-7a overexpression could be ascribed to a selective miR-7a–mediated stimulation on the proliferation of newly formed OPCs, we examined their proliferative activity with the use of 5-bromodeoxyuridine (BrdU) incorporation analysis. Purified OPC cultures were transfected with miR-7a mimics and grown in differentiation medium. During transfection, to eliminate the possible effect of RNA amount rather than RNA concentration on OPC proliferation, we mixed negative control RNA with miR-7a mimics to make the equal amount of transfected RNA among different miR-7a mimic concentrations. Two days after transfection, a dose-dependent promotion of cell proliferation, as revealed by increased BrdU labeling, was observed in miR-7a mimic-treated cultures (Figure 5A). The percentage of BrdU+ cells was 2.03 ± 0.24% in the control group and 9.76 ± 0.56% in the group treated with 50 nM miR-7a (p = 0.014; Figure 5B). Meanwhile, the cultures were subjected to immunostaining for stage-specific OL markers after treatment for indicated periods. In addition to increased proliferation, a significant expansion of PDGFRα+ and NG2+ OPCs was observed in the miR-7a overexpression group compared to the control group (Figure 5, C and D). Moreover, overexpression of miR-7a resulted in a significant decrease in the number of early differentiated CNP+ OLs (17.52 ± 0.91% in the control group and 8.87 ± 2.98% in the miR-7a group, p = 0.092) 2 d after treatment, as well as a decrease in mature MBP+ OLs (30 ± 1.87% in the control group and 9 ± 0.98% in the miR-7a group, p = 0.017) 4 d after treatment. These results suggest that miR-7a positively regulates OPC proliferation but negatively controls their further differentiation.
FIGURE 5:
Manipulating the expression level of miR-7a in purified OL cultures controlled proliferation and differentiation. (A and B) Purified rat OPCs were transfected with miR-7a mimics at different concentrations as indicated and cultured in OL differentiation medium. Two days after transfection, cultures were subjected to BrdU incorporation assay. Representative images of BrdU immunostaining (A) and quantification of the percentage of BrdU+ cells (B) show that overexpression of miR-7a induced an increase in the number of proliferating OPCs in a dose-dependent manner. (C and D) Stage-specific marker immunostaining indicate that overexpression of miR-7a increased the number of PDGFRα+ and NG2+ OPCs but repressed their further differentiation as shown by CNPase and MBP immunostaining. (E and F) Blocking the activity of miR-7a by inducing hairpin miRNA inhibitors into OPC cultures significantly reduced the number of proliferating cells as indicated by Ki67 staining. Scale bars, 50 μm. Data represent mean ± SEM. *p < 0.05 (one-way ANOVA in B; Student's t test in D and F).
We also tested the effects of blocking miR-7a in proliferating OPC cultures by transfecting hairpin miRNA inhibitors. The immunostaining for mitosis marker Ki67 revealed a significant reduction of proliferating OPC populations in the anti-miR-7a group cultured in the growth medium with mitogens (23.2 ± 3.57% in the control group and 8.67 ± 4.5% in the anti-miR-7a group, p = 0.029; Figure 5, E and F).
We also blocked the effect of miR-7a in differentiating OPCs, but did not observe significant changes in the percentage of OL subpopulations (unpublished data). We presume that because the expression of miR-7a naturally decreases with the differentiation of OL, additional repression is not as effective as in the progenitor stage (when miR-7a is highly expressed) and thus does not induce phenotypic changes.
Target gene identification for miR-7a
To identify the potential physiological targets of miR-7a, we analyzed the computationally predicted targets using TargetScan, PicTar, miRanda, and mirBase prediction algorithms (Krek et al., 2005; Grimson et al., 2007; Bartel, 2009). miR-7a was predicted to target a number of genes involved in neurogenesis, astrogliogenesis, and myelin gene expression (Table 1).
TABLE 1:
Predicted mRNA targets of miR-7a.
| Symbol | Gene name | Functions |
|---|---|---|
| Pax6 | Paired box 6 | Neurogenesis |
| NeuroD4 | Neurogenic differentiation 4 | Neurogenesis |
| Gsk3β | Glycogen synthase kinase 3 beta | Neurogenesis, neuronal migration, neuronal polarization, and axon growth/guidance |
| Mecp2 | Methyl CpG binding protein 2 | Neuronal maturation and synaptogenesis |
| KLF4 | Kruppel-like factor 4 | Indispensible for pluripotency |
| Sp1 | Sp1 transcription factor | Required for myelin gene expression |
| CNP | 2′,3′-cyclical nucleotide 3′ phosphodiesterase | Myelin gene |
| NFib | Nuclear factor I/B | Astrogliogenesis, required for the expression of astrocyte marker GFAP |
| NFic | Nuclear factor I/C | Astrogliogenesis, required for the expression of astrocyte marker GFAP |
| SNCA | Alpha-synuclein | Modulator of oxidative damage |
| NAIF1 | Nuclear apoptosis inducing factor 1 | Involved in cell death |
| Parp1 | Poly(ADP-ribose) polymerase-1 | Mediator of cell death |
Among these genes, Pax6 is a homeo-(HD) and paired-(PD) DNA binding domain containing transcription factor, overexpression of which has been shown to down-regulate Olig2 expression and to promote a neuronal lineage development (Jang and Goldman, 2011); NeuroD4 is a member of the bHLH family of transcription factors that acts as an essential determinant for the cortical projection neuron identity. It has been shown to be a key transcriptional target and cofactor for proneural gene Ngn2, which synergizes with Ngn2 to accelerate target gene transcription in the cortex (Mattar et al., 2008).
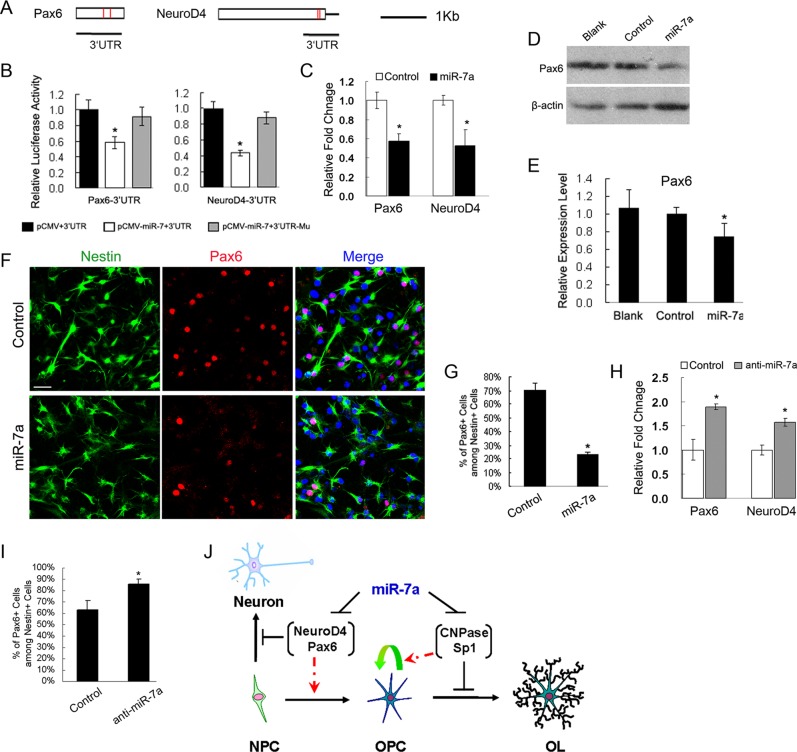
We tested miR-7a–mediated repression on Pax6 and NeuroD4 by placing their 3′ untranslated region (UTR) segments downstream to a cytomegalovirus (CMV)-driven luciferase reporter (Figure 6A) and performed reporter assays in COS-7 cells by cotransfecting with either expression plasmids for miR-7a or a control plasmid. Overexpression of miR-7a was confirmed by qRT-PCR analysis (Supplemental Figure 1C), and it significantly decreased the luciferase activity of reporters carrying Pax6 or NeuroD4 3′ UTR segments with their predicted binding sites, respectively (Figure 6B). Mutation of predicted miR-7a binding sites in the UTR segments for Pax6 and NeuroD4 (Supplemental Figure 2A) resulted in a significant recovery of their luciferase activities in response to miR-7a overexpression (Figure 6B). Thus the mutagenesis studies suggest that there is direct binding between miR-7a and its targets.
FIGURE 6:
miR-7a targeted proneural genes. (A) Sequence analysis for 3′ UTR of mouse Pax6 and NeuroD4 transcripts. The rectangle represents the 3′ UTR of each gene, in which the recognition sites of miR-7a are indicated by red bars. Black lines underneath depict the regions of 3′ UTRs that were cloned into pMir-reporter: Pax6 3′UTR, 1056 base pairs; NeuroD4 3′UTR, 700 base pairs. (B) Luciferase reporter assays for the effects of miR-7a expression on activities of reporters carrying the 3′ UTR segments of Pax6 or NeuroD4, respectively. Their mutant forms with corresponding “seed” sequence mutations were included as negative control. The histogram shows the ratio of the luciferase activity normalized to control expression vector. (C) qRT-PCR analysis of Pax6 and NeuroD4 expression using RNAs isolated from NPCs 3 d after transfection with miR-7a mimics. Scrambled miR transfection is included as control. (D and E) Western blot analysis shows that Pax6 protein level was down-regulated in NPC cultures by miR-7a overexpression compared with either control transfection or no transfection (indicated as “Blank”). β-Actin is included as an internal control. (F and G) Expression of Pax6 in NPC cultures was examined by immunostaining and quantified 3 d after transfection with miR-7a mimics. Nestin staining was used to quantify the percentage of Pax6+ cells. Scale bars, 50 μm. (H) qRT-PCR analysis of Pax6 and NeuroD4 expression using RNAs isolated from differentiating NPC cultures after transfection with anti-miR-7a as well as the scrambled oligonucleotide control as indicated. (I) Quantification of the number of Pax6+ cells among Nestin+ cells in differentiating NPCs transfected with anti-miR-7a revealed by immunostaining. (J) Schematic diagram for the function of miR-7a in regulating OL generation. During the NPC to OPC transition, miR-7a suppresses the expression of proneuronal differentiation genes (such as Pax6 and NeuroD4) to initiate the generation of OL lineage and then suppresses the expression of proOL factors (such as Sp1 and CNPase) to maintain the cells at proliferating precursor stage. Inhibition of expression/stage transition is shown by black lines with bars; promotion of stage transition is shown by red dashed lines with arrow heads. Astrogliogenesis is not involved in the current model due to the lack of confirmed relevance to miR-7a. Data are from three independent experiments. *p < 0.05 (one-way ANOVA).
Next we tested whether miR-7a regulated the expression of Pax6 and NeuroD4. NPCs from rat brain were confirmed to express these genes endogenously by qRT-PCR, both of which showed none or less expression in OL lineage (Supplemental Figure 2B). Then NPC cultures were transfected with miR-7a mimics in parallel with control nucleotide. Three days after transfection, total RNAs isolated from control and miRNA mimic–treated cells were subject to qRT-PCR measurement. The mRNA level of Pax6 was down-regulated ∼40% by miR-7a mimics (Figure 6C), and protein expression was reduced to 75% as shown by Western blot analysis (Figure 6, D and E). Immunostaining with NPC markers showed a significant reduction of Pax6/Nestin double-positive cells after miR-7a overexpression (Figure 6, F and G), which suggested a posttranscriptional inhibition by miR-7a. In addition, mRNA expression of NeuroD4 was drastically decreased after treatment with miR-7a mimics (Figure 6C). In our in utero electroporation study, the number of Pax6+/GFP+ cells was significantly reduced in miR-7a–overexpressing GFP+ cells (Supplemental Figure 3), which further corroborate the effect of miR-7a.
Conversely, knockdown of miR-7a by miRNA inhibitors resulted in an up-regulation of Pax6 and NeuroD4 mRNA level in primary NPC cultures (Figure 6H), and the number of Pax6+ cells was increased in the anti-miR-7a–treated group (Figure 6I). These observations indicate that expression of neuronal genes, such as Pax6 and NeuroD4, can be negatively regulated by miR-7a during the progress of NPC cell fate commitment, especially into oligodendroglial lineage.
Other predicted target genes of miR-7a, such as NFib and NFic (Table 1), are required for astrogliogenesis and are necessary to drive the expression of GFAP (Steele-Perkins et al., 2005; Wilczynska et al., 2009). Because we did not observe alterations in the astrocyte marker expression after miR-7a mimic treatment in differentiating NPCs under current conditions, we supposed that miR-7a did not exert the effect of promoting OL generation by inhibiting astrocyte formation. Therefore we did not test the possibility of these two genes as functional targets for miR-7a.
We also noticed that miR-7a was predicted to target a well-known myelin gene, CNPase and genes associated with OL differentiation, such as Sp1 (Table 1). Although we did not test the binding activity between miR-7a and the 3′ UTR of CNPase, we had shown the down-regulation of CNPase expression in miR-7a–transfected OL cultures (Figure 5, C and D). Additionally, miR-7a could specifically target the 3′ UTR of Sp1 (Supplemental Figure 4, A and B), which is required for the myelin basic protein (MBP) gene expression (Wei et al., 2003, 2004, 2005), and repress its mRNA expression (Supplemental Figure 4C).
In summary, our data suggest that miR-7a may first induce OL specification by negatively regulating proneuronal factors, then maintain newly formed OPCs in their precursor stage by inhibiting proOL differentiation regulators as a second wave of targets for miR-7a (Figure 6J).
DISCUSSION
miR-7a expression in OL lineage cells
Consistent with an earlier investigation of miRNA expression in OL lineage cells from postnatal brain (Lau et al., 2008), we found that the level of miR-7a was four- to fivefold higher in OPCs than in mature OLs identified by the TaqMan MicroRNA Assay, which also revealed its relatively lower expression in neurons and astrocytes purified from postnatal cortex, as well as in neural progenitors from E15 embryonic cortex.
Previous studies have suggested the possible role of miR-7a in early CNS development according to its expression patterns: During the earliest stage of rat cortical development, the level of miR-7a increased about fourfold between embryonic day E11 and E13 as identified by qRT-PCR analysis (Nielsen et al., 2009). In mouse brain, the signal of Northern blot for miR-7a peaked in the fetus from E12.5 through E17.5 and gradually decreased after birth (Miska et al., 2004), and displayed a similar temporal expression pattern to miR-9 and miR-17-92 clusters. Recently, the miR-17-92 cluster showed functions in promoting OPC proliferation (Budde et al., 2010), and miR-9 was identified as one of the top miRNAs highly expressed in postnatal A2B5+/GC− OPCs in comparison to A2B5−/GC+ mature OLs (Lau et al., 2008), although its correlation with OL lineage progression has not been illustrated. Apart from these, little is known about miR-7a function in the development of vertebrate nervous system. The expression pattern of miR-7a obtained from different neural cell subtypes indicated that it might play a role in determining the fate commitment of OL lineage, because its expression peaked at the OPC stage during the development of NPCs to mature OL and it was much higher than in neurons and astrocytes.
OL generation from NPCs driven by miR-7a
By manipulating the level of miR-7a in NPC cultures isolated from embryonic mouse brain, we found that miR-7a could promote the generation of OL lineage, but could not accelerate the formation of fully mature OLs in current culture conditions (unpublished data). After overexpression of miR-7a mimics, immunostaining with OPC makers PDGFRα, NG2, and OL linage maker Olig2 revealed a significant increase in the number of OPCs, which was confirmed in qRT-PCR analysis. At the same time, quantification of neuron maker β-tubulin-III revealed a slight reduction compared with control treatment, but neither the percentage of GFAP+ astrocytes nor the mRNA level of GFAP and S100b were influenced. Therefore we speculated that miR-7a might promote oligodendrogenesis from NPCs, at least partially, by inhibiting neurogenesis. In addition, the up-regulation of the mRNA expression level for Olig1 and Olig2 after miR-7a treatment is consistent with their important roles in OL development. Olig1 has been suggested as a key factor for the induction of OPC formation in the brain: Cortical NPC cultures infected with an Olig1-expressing adenovirus yielded a substantial amount of OPCs (Lu et al., 2000); ectopic expression of Olig1 in embryonic mouse forebrain ventricle appears to promote OL formation in all brain regions (Lu et al., 2001); and overexpression of Olig1 initiates the differentiation of NPCs into OPCs both in vitro (Balasubramaniyan et al., 2004) and in vivo (Maire et al., 2010). Meanwhile, Olig2 is required for OPC fate specification from NPCs and subsequent OPC differentiation (Copray et al., 2006; Maire et al., 2010).
In developing mouse cortex, we found that overexpression of miR-7a was able to induce OL generation, defined by PDGFRα and Olig2 immunostaining, which further strengthened the critical function of this miRNA in oligodendrogenesis.
Consistent with overexpression data, blocking the function of miR-7a during NPC differentiation led to reduction in the expression of Olig2 as well as PDGFRα. As presumed, the blockage of OPC generation was accompanied by a conspicuous expansion of neuron population, as identified by early neuronal marker DCX and β-tubulin-III. Because the viability of neural subtypes was not affected by either miR-7a overexpression or knockdown treatment, the opposite effects of miR-7a on OL specification and neuronal differentiation suggested a distinct role of miR-7a in initiating OL lineage formation while repressing neuronal development.
A critical role of miR-7a in maintaining OLs at precursor stage
To investigate the further progression of OPCs in miR-7a mimic treatment, we tested its effect in purified rat OL cultures. Our BrdU incorporation assay revealed that miR-7a could promote OPC proliferation even in the differentiation condition and that the formation of CNP+ and MBP+ OL was repressed by miR-7a overexpression. In contrast, blocking the endogenous activity of miR-7a in OPC cultures greatly reduced their proliferative activity even in the growth medium with mitogens. Here Ki67 immunostaining, instead of the BrdU assay, was applied to show the number of proliferating cells due to the labeling of more positive cells than in the BrdU assay, as reported earlier (Smith et al., 1995). These results suggested a critical role of miR-7a in maintaining newly formed OPCs at their proliferating stage and were consistent with our observations of the miR-7a expression pattern during OL differentiation. Therefore we presume that, in vertebrate brain development, miR-7a might work with other partners, such as miR-219 and miR-338 (Dugas et al., 2010; Zhao et al., 2010), to direct the formation and maturation of OL in cohort. The regulatory network for miR-7a expression remains to be explored further.
We also noticed that miR-7 has shown its various roles in other species: It has proven to be a key player in controlling photoreceptor differentiation in the Drosophila eye (Li and Carthew, 2005); it is expressed in certain cell subtypes in the Drosophila spinal cord (Aboobaker et al., 2005); and its expression in the hypothalamus neurons with sensory or neurosecretory functions has been identified in zebrafish (Tessmar-Raible et al., 2007). Also, during retinoic acid (RA)-induced differentiation of neuroblastoma cells, miR-7 expression is slightly down-regulated, and its overexpression reduces neurite outgrowth of differentiated neuroblastoma (Chen et al., 2010). Moreover, miR-7 has been suggested as a tumor repressor, and it decreases the viability and invasiveness of several cancer cell types, including glioblastoma and schwannoma (Kefas et al., 2008; Webster et al., 2009; Saydam et al., 2011). Thus miR-7a may have pleiotropic effects on different biological processes, and it will promote OL generation in a context and temporally specific manner.
Proneural factors and myelin genes as targets of miR-7a
The timing of OL specification, proliferation, and differentiation is tightly controlled by the balance of activities between promoters and repressors. Proneural genes Pax6 and NeuroD4 could be the physiological targets for miR-7a due to the validation of direct binding between miR-7a and their 3′ UTRs and to the regulation of their expression levels by miR-7a. This conclusion is consistent with their roles in directing neuronal versus glial fate determination in the CNS: Neuronal bHLH genes, NeuroD4/Mash1 double mutants showed two missed longitudinal columns of hindbrain neurons and retinal bipolar cells, and instead, those cells that normally differentiate into neurons adopted glial fate (Ohsawa et al., 2005); Pax6 is a multifunctional player regulating neural NPC proliferation and differentiation through the control of different downstream molecules in a highly context-dependent manner (Sansom et al., 2009). In the developing ventral spinal cord, combinatorial actions of Pax6 together with other factors controlled neurogenesis and gliogenesis: Mash1 itself promoted differentiation of both neurons and OLs; Pax6, however, converted Mash1 to become neurogenic, whereas Olig2 selectively enhanced Mash1-dependent oligodendrogenesis (Sugimori et al., 2007).
After formation of OL lineage cells from NPCs, miR-7a sequentially maintained these cells at their precursor stage. The proliferation-promoting effect of miR-7a in OPC cultures was similar to effects observed previously in other cell types, such as lung carcinoma cells (Cheng et al., 2005), although the mechanism remained unclear. We believe, however, that the lack of further differentiation of OPC cultures transfected with miR-7a may ascribe to its inhibitory effect on myelin gene expression directly or indirectly. CNPase is one of the well-known early myelin genes that predicated as a target of miR-7a, and we showed that its expression was down-regulated in miR-7a–transfected OL cultures. As to the function of another predicted target, Sp1 has been suggested to play an essential role in the regulation of MBP expression through binding to the GC-rich region of MBP promoter (Wei et al., 2003). It competes off the binding of Nkx2.2 to MBP promoter and then reverses the Nkx2.2-mediated repression of the MBP promoter (Wei et al., 2005). Meanwhile, through interaction with Sox10, Sp1 activates and contributes to the tissue-specific expression of MBP in the CNS (Wei et al., 2004). In the current study, we demonstrated that miR-7a could bind to the 3′ UTR of Sp1 (Supplemental Figure 4B), which suggested Sp1 as a physiological target for miR-7a. Also, several other previously reported target genes for miR-7a, such as epidermal growth factor receptor (Kefas et al., 2008; Saydam et al., 2011) and IRS-1/2, upstream regulators of the Akt pathway (Kefas et al., 2008), have also been identified to play roles in OL myelination (Aguirre et al., 2007; Flores et al., 2008). Therefore it is conceivable that miR-7a maintains the proliferative activity of newly formed OPCs partially by inhibiting the expression of myelin genes.
Our observations suggest that an active repression of a cohort of proneuronal differentiation factors and OL differentiation activators by miR-7a can be one of the critical steps necessary for initiating OL specification and maintaining their proliferative, progenitor state (Figure 6J). These results can therefore provide insight into the effective generation of OL lineage cell from NPCs, especially in OL loss therapies such as spinal cord injury.
MATERIALS AND METHODS
Primary culture of mouse cortical NPCs and rat OPCs
All animal experiment protocols were approved by the Animal Care and Use Committee of the Fourth Military Medical University and were conducted in accordance with the guidelines for the care and use of laboratory animals.
For mouse cortical NPC cultures, cortical precursors were isolated from C57/BL6 mouse embryos at E15.5 as described previously (Chen et al., 2007). Briefly, each cortex was cut into two or three pieces, transferred to ice-cold neurosphere growth medium (DMEM/F12 supplemented with N2 and 20 ng/ml EGF + 20 ng/ml bFGF), and dissociated by mechanical trituration with a fire-polished glass Pasteur pipette. The cell suspension was passed through a 50-μm nylon pouch, and 5 × 104 cells/ml were plated into a 100-mm dish. After 5 d, neurospheres were formed, and, to induce the differentiation of NPCs, dissociated secondary neurospheres were plated onto poly-d-lysine–coated dishes or coverslips in medium with 5 ng/ml EGF, 5 ng/ml bFGF, and 10 ng/ml PDGF-AA as previously described (Balasubramaniyan et al., 2004).
Isolation and culture of rat OPCs followed protocol as previously described (Zhao et al., 2007, 2010). Briefly, brains were removed from P2 Sprague Dawley rat pups, and the cortices were dissected. Cortical pieces were enzymatically digested followed by mechanical dissociation. Cells were resuspended in DMEM with 10% fetal bovine serum and plated onto T75 flasks. The resulting mixed glial cultures were maintained for 7–10 d. Purified OPCs were prepared by differential shaking and were seeded onto poly-l-ornithine–coated 35-mm dishes at the density of 3 × 104 cells/cm2 in OL growth medium (DMEM/F12 supplemented with 0.1% bovine serum albumin, 10 nM d-biotin, 5 μg/ml insulin, 5 ng/ml sodium selenium, 50 μg/ml apo-transferrin, 10 nM hydrocortisone, 1 mM sodium pyruvate, 100 U/ml penicillin, 10 ng/ml fibroblast growth factors, and 10 ng/ml platelet-derived growth factor AA). OPCs were amplified in growth medium for ∼4 d and passaged with isolation medium (0.2% DNase I + 5 μg/ml insulin + 0.04% EDTA). To initiate differentiation, OPCs were seeded onto poly-d-lysine–coated dishes or coverslips in differentiation medium with 30 ng/ml triiodothyronine (T3), 10 ng/ml ciliary neurotrophic factor, and 5 μg/ml N-acetyl-l-cysteine.
Duplex miRNA mimics and hairpin miRNA inhibitor transfection
For our in vitro transfection study, dissociated NPCs or purified OPCs were transfected with 50 nM miRIDIAN miRNA mimics (C-310591-07; Dharmacon, Lafayette, CO) or hairpin inhibitors (IH-310591-08; Dharmacon) using Lipofectamine 2000 (Invitrogen, Carlsbad, CA) 1 d after plating. For both transfections, either miRIDIAN miRNA mimic negative control (CN-001000-01) or hairpin inhibitor negative control (IN-001005-01) transfection was included, respectively. The transfection efficiency was evaluated by qRT-PCR with the TaqMan MicroRNA Assay (Applied Biosystems, Carlsbad, CA). As indicated, 3 or 5 d posttransfection, cultures were harvested for immunocytochemistry and qRT-PCR assay.
RNA extraction and qRT-PCR
Total RNAs were purified from tissue or cell cultures using TRIzol reagent according to the manufacturer's instruction (Invitrogen). RNA was transcribed to cDNA with the PrimeScript II 1st Strand cDNA Synthesis Kit (Takara Bio, Shiga, Japan). qRT-PCR was performed using the CFX96 Touch Real-Time PCR Detection System (Bio-Rad Laboratories, Hercules, CA), and the relative gene expression was normalized to internal control Gapdh. Primer sequences for SybrGreen probes of target genes are as follows. Rat and mouse Gapdh: acaagatggtgaaggtcggtgtga and agcttcccattctcagccttgact; mouse Pdgfrα: ggagactcaag taaccttgcac and tcagttctgacgttgctttcaa; mouse Olig2: gggaggtcatgccttacgc and ctcc agcgagttggtgagc; mouse Olig1: gcagccacctatctcctcatc and cgagtagggtaggataacttcgc; mouse Gfap: tctcgaatgactcctccactc and aagctccgcctggtagacat; mouse β-tubulin-III: cccagcggcaactatgtagg and ccagaccgaacactgtcca; rat and mouse Pax6: aagagtggcgac tccagaagttgt and tttatcatacatgccgtctgcgcc; rat and mouse NeuroD4: tgcccagagactgtg gtactgaaa and agagctcagacctttgtccatcca; rat and mouse Sp1: agtgccctaagcgtttcatgagg a and ctgcatgacgttgatgccactgtt.
For mature miRNA expression analysis, cDNA was synthesized using the TaqMan MicroRNA Reverse Transcription kit (Applied Biosystems) and 10 ng of total RNA along with miR-7a specific stem loop RT primer supplied in the miR-7a TaqMan MicroRNA Assay (000268; Applied Biosystems). qRT-PCR was performed with TaqMan PCR master mix according to the manufacturer's instruction, and a U6 snRNA TaqMan probe (001973; Applied Biosystems) was used as endogenous control.
Cell proliferation assay
A BrdU incorporation assay was used to assess cell proliferation. OPC cultures were exposed to 20 uM BrdU for 12 h and fixed in 4% formaldehyde. Cells were subjected to DNA denaturation with 2 M hydrochloric acid for 30 min and then neutralized with 0.1 M sodium borate at pH 8.5 for 10 min. After permeabilization with 0.03% Triton X-100 in 5% normal donkey serum for 15 min, cultures were incubated with primary antibody against BrdU (1:200; Sigma) for 1 h at room temperature followed by donkey anti-mouse secondary antibody conjugated to Alexa 594 (1:1000; Molecular Probes, Eugene, OR) for another hour. Hoechst was used to label nuclei. BrdU incorporation was evaluated by examining five random fields per coverslip, and the percentage of cells positive for BrdU was compared with that of the control condition. Experiments were replicated using cells from three different primary cultures.
In utero electroporation
For in utero electroporation, DNA solution (1 μl) in phosphate-buffered saline containing 0.01% fast green was injected into the lateral ventricle of the C57 mice embryos at E14.5. After injection, electroporation (five 50-ms square pulses of 35 V with 990-ms intervals) was carried out. Plasmid DNAs (1 mg/ml) used for electroporation were pCIG-miR-7a and control pCIG. Embryos were harvested 72 h after electroporation and processed for immunohistology. At least five embryos from three to four female mice with the expression of each vector were analyzed and characterized. To compare the effect of miR-7a and control vector treatment on OL generation, the number of PDGFRα+/GFP+, PDGFRα+/GFP− and Olig2+/GFP+, Olig2+/GFP− were counted in defined areas (0.25 mm2).
Immunostaining and Western blot assay
Immunostaining was performed on tissue or cell cultures using the standard protocols just mentioned along with fluorescent secondary antibodies (Millipore, Billerica, MA). Hoechst was used to stain nuclei and determine the percentage of immunopositive cells. The primary antibodies were: mouse anti-CNPase (1:300; Sigma, St. Louis, MO), rabbit anti-NG2 (1:300; Millipore), mouse anti-MBP (1:1000; Abcam, Cambridge, UK), rabbit anti-PDGFRα (1:500; Abcam), rabbit anti-Olig2 (1:200; Abcam), rabbit anti-Pax6 (1:400; Proteintech, Chicago, IL), mouse anti-Nestin (1:100; Chemicon, Temecula, CA), mouse anti–β-tubulin-III (1:500; R&D Systems, Minneapolis, MN), mouse anti-GFAP (1:4000; Sigma), and rabbit anti-Ki67 (1:1000; Abcam).
For Western blot analysis, protein lysates were resolved by SDS–PAGE and blotted using standard procedures. Antibodies used were rabbit anti-Pax6 (Proteintech), mouse anti–β-actin (Sigma), and horseradish peroxidase–conjugated secondary antibodies (Jackson ImmunoResearch, West Grove, PA). Signals were revealed by chemiluminescence with the ECL kit (Pierce, Rockford, IL), according to the manufacturer's instruction.
miRNA expression vectors and luciferase reporter assays
The mouse miR-7a locus on chromosome 13 with its ∼500–base pair flanking sequences was PCR amplified from genomic DNA and inserted into pCMV6 or pCIG expression vector (Zhao et al., 2010). The expression of mature miRNA was verified by qRT-PCR with the TaqMan MicroRNA assay. Segments carrying the putative miR-7a binding sites in 3′ UTR of Pax6, NeuroD4, and Sp1 were cloned into pMir-REPORT vector (Ambion, Grand Island, NY). Primer sequences for cloning target genes are: mouse miR-7a, ccatggtgtttccgtcacttgg and tagacagtagtagtgcagggtt; mouse Pax6-3′UTR, ctcatttcacctggagtgtcag and tacaaa ggctttggcatggt; mouse NeuroD4-3′UTR, tagcctcaaagcttcactggga and tccatccttctatggc caacct; mouse Sp1-3′UTR-segment1, atgggattgtggcacccaatgt and tcccatgattgtgtgcttc cct; and mouse Sp1-3′UTR-segment2, atcacactgtgccttctccact and agggcatcatgcacaactcag a.
Using site-directed mutagenesis based on overlap extension PCR (Ho et al., 1989), the predicted miR-7a binding sites in pMir-reporter-Pax6-UTR were changed from TTTTC C to ATAAGG and from TCTTCC to TGAAGG, respectively; the predicted binding sites in pMir-reporter-NeuroD4-UTR were changed from GTCTTC to CAG AAG and from TTTTCC to CTAAGG, respectively; the three predicted binding sites in pMir-reporter-Sp1-segment1 were changed from GTCTT to CAGAA, from TCTTTC to AGCATG, and from CTTCC to GAAGG.
For luciferase reporter assays, 10 ng of luciferase reporter construct DNA was cotransfected with 200 ng of vector expressing miR-7a into COS-7 cells by Lipofectamine 2000. The pRSV-renilla luciferase plasmid was included to control the transfection efficiencies. Luciferase activity was assayed 48 h after transfection using the dual-luciferase reporter assay system (Promega, Madison, WI). Three transfection assays were performed to obtain statistically significant data.
Statistical analysis
Quantifications were performed from at least three independent experiments, and data were presented as mean ± SEM in the graphs. Student's t test was used to compare two sets of data, and one-way analysis of variance (ANOVA) was applied in multiple comparisons. A value of p < 0.05 was considered statistically significant.
Supplementary Material
Acknowledgments
We thank Haifeng Zhang for help with confocal laser scanning microscopy and Lingling Fei and Jianyong Qiu for technical support. We also thank Jian Wang and Yazhou Wang for discussion of the results. This work was supported by grants from the National Natural Science Foundation of China (30800325), the Scientific Research Foundation for the Returned Overseas Chinese Scholars from the State Education Ministry, and the Outstanding Young Researcher Foundation of the Fourth Military Medical University.
Abbreviations used:
- ANOVA
analysis of variance
- bFGF
basic fibroblast growth factor
- bHLH
basic helix-loop-helix
- BrdU
5-bromodeoxyuridine
- CMV
cytomegalovirus
- DCS
doublecortin+
- EGF
epidermal growth factor
- GFAP
glial fibrillary acidic protein
- MBP
myelin basic protein
- miRNA
microRNA
- NPC
neural progenitor cell
- OL
oligodendrocyte
- OPC
oligodendrocyte precursor cell
- PDGF
platelet-derived growth factor
- qRT-PCR
quantitative real-time PCR
- RISC
RNA-induced silencing complex
- UTR
untranslated region
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E12-04-0270) on June 13, 2012.
REFERENCES
- Aboobaker AA, Tomancak P, Patel N, Rubin GM, Lai EC. Drosophila microRNAs exhibit diverse spatial expression patterns during embryonic development. Proc Natl Acad Sci USA. 2005;102:18017–18022. doi: 10.1073/pnas.0508823102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aguirre A, Dupree JL, Mangin JM, Gallo V. A functional role for EGFR signaling in myelination and remyelination. Nat Neurosci. 2007;10:990–1002. doi: 10.1038/nn1938. [DOI] [PubMed] [Google Scholar]
- Balasubramaniyan V, Timmer N, Kust B, Boddeke E, Copray S. Transient expression of Olig1 initiates the differentiation of neural stem cells into oligodendrocyte progenitor cells. Stem Cells. 2004;22:878–882. doi: 10.1634/stemcells.22-6-878. [DOI] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budde H, Schmitt S, Fitzner D, Opitz L, Salinas-Riester G, Simons M. Control of oligodendroglial cell number by the miR-17-92 cluster. Development. 2010;137:2127–2132. doi: 10.1242/dev.050633. [DOI] [PubMed] [Google Scholar]
- Chen H, et al. miR-7 and miR-214 are specifically expressed during neuroblastoma differentiation, cortical development and embryonic stem cells differentiation, and control neurite outgrowth in vitro. Biochem Biophys Res Commun. 2010;394:921–927. doi: 10.1016/j.bbrc.2010.03.076. [DOI] [PubMed] [Google Scholar]
- Chen Y, Balasubramaniyan V, Peng J, Hurlock EC, Tallquist M, Li J, Lu QR. Isolation and culture of rat and mouse oligodendrocyte precursor cells. Nat Protoc. 2007;2:1044–1051. doi: 10.1038/nprot.2007.149. [DOI] [PubMed] [Google Scholar]
- Cheng AM, Byrom MW, Shelton J, Ford LP. Antisense inhibition of human miRNAs and indications for an involvement of miRNA in cell growth and apoptosis. Nucleic Acids Res. 2005;33:1290–1297. doi: 10.1093/nar/gki200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng LC, Pastrana E, Tavazoie M, Doetsch F. miR-124 regulates adult neurogenesis in the subventricular zone stem cell niche. Nat Neurosci. 2009;12:399–408. doi: 10.1038/nn.2294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Copray S, Balasubramaniyan V, Levenga J, de Bruijn J, Liem R, Boddeke E. Olig2 overexpression induces the in vitro differentiation of neural stem cells into mature oligodendrocytes. Stem Cells. 2006;24:1001–1010. doi: 10.1634/stemcells.2005-0239. [DOI] [PubMed] [Google Scholar]
- Dugas JC, Cuellar TL, Scholze A, Ason B, Ibrahim A, Emery B, Zamanian JL, Foo LC, McManus MT, Barres BA. Dicer1 and miR-219 are required for normal oligodendrocyte differentiation and myelination. Neuron. 2010;65:597–611. doi: 10.1016/j.neuron.2010.01.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dugas JC, Notterpek L. MicroRNAs in oligodendrocyte and Schwann cell differentiation. Dev Neurosci. 2011;33:14–20. doi: 10.1159/000323919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emery B. Transcriptional and post-transcriptional control of CNS myelination. Curr Opin Neurobiol. 2010;20:601–607. doi: 10.1016/j.conb.2010.05.005. [DOI] [PubMed] [Google Scholar]
- Flores AI, Narayanan SP, Morse EN, Shick HE, Yin X, Kidd G, Avila RL, Kirschner DA, Macklin WB. Constitutively active Akt induces enhanced myelination in the CNS. J Neurosci. 2008;28:7174–7183. doi: 10.1523/JNEUROSCI.0150-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimson A, Farh KK, Johnston WK, Garrett-Engele P, Lim LP, Bartel DP. MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol Cell. 2007;27:91–105. doi: 10.1016/j.molcel.2007.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho SN, Hunt HD, Horton RM, Pullen JK, Pease LR. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene. 1989;77:51–59. doi: 10.1016/0378-1119(89)90358-2. [DOI] [PubMed] [Google Scholar]
- Jang ES, Goldman JE. Pax6 expression is sufficient to induce a neurogenic fate in glial progenitors of the neonatal subventricular zone. PLoS One. 2011;6:e20894. doi: 10.1371/journal.pone.0020894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawase-Koga Y, Otaegi G, Sun T. Different timings of Dicer deletion affect neurogenesis and gliogenesis in the developing mouse central nervous system. Dev Dyn. 2009;238:2800–2812. doi: 10.1002/dvdy.22109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kefas B, et al. microRNA-7 inhibits the epidermal growth factor receptor and the Akt pathway and is down-regulated in glioblastoma. Cancer Res. 2008;68:3566–3572. doi: 10.1158/0008-5472.CAN-07-6639. [DOI] [PubMed] [Google Scholar]
- Krek A, et al. Combinatorial microRNA target predictions. Nat Genet. 2005;37:495–500. doi: 10.1038/ng1536. [DOI] [PubMed] [Google Scholar]
- Lau P, Verrier JD, Nielsen JA, Johnson KR, Notterpek L, Hudson LD. Identification of dynamically regulated microRNA and mRNA networks in developing oligodendrocytes. J Neurosci. 2008;28:11720–11730. doi: 10.1523/JNEUROSCI.1932-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letzen BS, Liu C, Thakor NV, Gearhart JD, All AH, Kerr CL. MicroRNA expression profiling of oligodendrocyte differentiation from human embryonic stem cells. PLoS One. 2010;5:e10480. doi: 10.1371/journal.pone.0010480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Carthew RW. A microRNA mediates EGF receptor signaling and promotes photoreceptor differentiation in the Drosophila eye. Cell. 2005;123:1267–1277. doi: 10.1016/j.cell.2005.10.040. [DOI] [PubMed] [Google Scholar]
- Lin ST, Fu YH. miR-23 regulation of lamin B1 is crucial for oligodendrocyte development and myelination. Dis Model Mech. 2009;2:178–188. doi: 10.1242/dmm.001065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu QR, Cai L, Rowitch D, Cepko CL, Stiles CD. Ectopic expression of Olig1 promotes oligodendrocyte formation and reduces neuronal survival in developing mouse cortex. Nat Neurosci. 2001;4:973–974. doi: 10.1038/nn718. [DOI] [PubMed] [Google Scholar]
- Lu QR, Yuk D, Alberta JA, Zhu Z, Pawlitzky I, Chan J, McMahon AP, Stiles CD, Rowitch DH. Sonic hedgehog–regulated oligodendrocyte lineage genes encoding bHLH proteins in the mammalian central nervous system. Neuron. 2000;25:317–329. doi: 10.1016/s0896-6273(00)80897-1. [DOI] [PubMed] [Google Scholar]
- Maire CL, Wegener A, Kerninon C, Nait Oumesmar B. Gain-of-function of Olig transcription factors enhances oligodendrogenesis and myelination. Stem Cells. 2010;28:1611–1622. doi: 10.1002/stem.480. [DOI] [PubMed] [Google Scholar]
- Mattar P, Langevin LM, Markham K, Klenin N, Shivji S, Zinyk D, Schuurmans C. Basic helix-loop-helix transcription factors cooperate to specify a cortical projection neuron identity. Mol Cell Biol. 2008;28:1456–1469. doi: 10.1128/MCB.01510-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miska EA, Alvarez-Saavedra E, Townsend M, Yoshii A, Sestan N, Rakic P, Constantine-Paton M, Horvitz HR. Microarray analysis of microRNA expression in the developing mammalian brain. Genome Biol. 2004;5:R68. doi: 10.1186/gb-2004-5-9-r68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen JA, Lau P, Maric D, Barker JL, Hudson LD. Integrating microRNA and mRNA expression profiles of neuronal progenitors to identify regulatory networks underlying the onset of cortical neurogenesis. BMC Neurosci. 2009;10:98. doi: 10.1186/1471-2202-10-98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohsawa R, Ohtsuka T, Kageyama R. Mash1 and Math3 are required for development of branchiomotor neurons and maintenance of neural progenitors. J Neurosci. 2005;25:5857–5865. doi: 10.1523/JNEUROSCI.4621-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sansom SN, Griffiths DS, Faedo A, Kleinjan DJ, Ruan Y, Smith J, van Heyningen V, Rubenstein JL, Livesey FJ. The level of the transcription factor Pax6 is essential for controlling the balance between neural stem cell self-renewal and neurogenesis. PLoS Genet. 2009;5:e1000511. doi: 10.1371/journal.pgen.1000511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saydam O, et al. miRNA-7 attenuation in Schwannoma tumors stimulates growth by upregulating three oncogenic signaling pathways. Cancer Res. 2011;71:852–861. doi: 10.1158/0008-5472.CAN-10-1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y, Zhao X, Hsieh J, Wichterle H, Impey S, Banerjee S, Neveu P, Kosik KS. MicroRNA regulation of neural stem cells and neurogenesis. J Neurosci. 2010;30:14931–14936. doi: 10.1523/JNEUROSCI.4280-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin D, Shin JY, McManus MT, Ptacek LJ, Fu YH. Dicer ablation in oligodendrocytes provokes neuronal impairment in mice. Ann Neurol. 2009;66:843–857. doi: 10.1002/ana.21927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith MD, Healy E, Thompson V, Morley A, Rees JL. Use of in situ detection of histone mRNA in the assessment of epidermal proliferation: comparison with the Ki67 antigen and BrdU incorporation. Br J Dermatol. 1995;132:359–366. doi: 10.1111/j.1365-2133.1995.tb08668.x. [DOI] [PubMed] [Google Scholar]
- Steele-Perkins G, Plachez C, Butz KG, Yang G, Bachurski CJ, Kinsman SL, Litwack ED, Richards LJ, Gronostajski RM. The transcription factor gene Nfib is essential for both lung maturation and brain development. Mol Cell Biol. 2005;25:685–698. doi: 10.1128/MCB.25.2.685-698.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugimori M, Nagao M, Bertrand N, Parras CM, Guillemot F, Nakafuku M. Combinatorial actions of patterning and HLH transcription factors in the spatiotemporal control of neurogenesis and gliogenesis in the developing spinal cord. Development. 2007;134:1617–1629. doi: 10.1242/dev.001255. [DOI] [PubMed] [Google Scholar]
- Tessmar-Raible K, Raible F, Christodoulou F, Guy K, Rembold M, Hausen H, Arendt D. Conserved sensory-neurosecretory cell types in annelid and fish forebrain: insights into hypothalamus evolution. Cell. 2007;129:1389–1400. doi: 10.1016/j.cell.2007.04.041. [DOI] [PubMed] [Google Scholar]
- Webster RJ, Giles KM, Price KJ, Zhang PM, Mattick JS, Leedman PJ. Regulation of epidermal growth factor receptor signaling in human cancer cells by microRNA-7. J Biol Chem. 2009;284:5731–5741. doi: 10.1074/jbc.M804280200. [DOI] [PubMed] [Google Scholar]
- Wei Q, Miskimins WK, Miskimins R. The Sp1 family of transcription factors is involved in p27(Kip1)-mediated activation of myelin basic protein gene expression. Mol Cell Biol. 2003;23:4035–4045. doi: 10.1128/MCB.23.12.4035-4045.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Q, Miskimins WK, Miskimins R. Sox10 acts as a tissue-specific transcription factor enhancing activation of the myelin basic protein gene promoter by p27Kip1 and Sp1. J Neurosci Res. 2004;78:796–802. doi: 10.1002/jnr.20342. [DOI] [PubMed] [Google Scholar]
- Wei Q, Miskimins WK, Miskimins R. Stage-specific expression of myelin basic protein in oligodendrocytes involves Nkx2.2-mediated repression that is relieved by the Sp1 transcription factor. J Biol Chem. 2005;280:16284–16294. doi: 10.1074/jbc.M500491200. [DOI] [PubMed] [Google Scholar]
- Wilczynska KM, Singh SK, Adams B, Bryan L, Rao RR, Valerie K, Wright S, Griswold-Prenner I, Kordula T. Nuclear factor I isoforms regulate gene expression during the differentiation of human neural progenitors to astrocytes. Stem Cells. 2009;27:1173–1181. doi: 10.1002/stem.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo AS, Sun AX, Li L, Shcheglovitov A, Portmann T, Li Y, Lee-Messer C, Dolmetsch RE, Tsien RW, Crabtree GR. MicroRNA-mediated conversion of human fibroblasts to neurons. Nature. 2011;476:228–231. doi: 10.1038/nature10323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y, Casaccia P, Lu QR. Shaping the oligodendrocyte identity by epigenetic control. Epigenetics. 2010;5:124–128. doi: 10.4161/epi.5.2.11160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X, et al. MicroRNA-mediated control of oligodendrocyte differentiation. Neuron. 2010;65:612–626. doi: 10.1016/j.neuron.2010.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao XH, Jin WL, Ju G. An in vitro study on the involvement of LINGO-1 and Rho GTPases in Nogo-A regulated differentiation of oligodendrocyte precursor cells. Mol Cell Neurosci. 2007;36:260–269. doi: 10.1016/j.mcn.2007.07.008. [DOI] [PubMed] [Google Scholar]
- Zheng K, Li H, Zhu Y, Zhu Q, Qiu M. MicroRNAs are essential for the developmental switch from neurogenesis to gliogenesis in the developing spinal cord. J Neurosci. 2010;30:8245–8250. doi: 10.1523/JNEUROSCI.1169-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.