An actin-dependent role is shown for Myo1E in the trafficking of newly internalized cargo to early endosomes during CME. The results establish for mammalian cells, similar to budding yeast, interdependence in the recruitment of type I myosins, WIP/WIRE, and N-WASP to endocytic sites to assemble F-actin as endocytic vesicles are being formed.
Abstract
Myosin 1E (Myo1E) is recruited to sites of clathrin-mediated endocytosis coincident with a burst of actin assembly. The recruitment dynamics and lifetime of Myo1E are similar to those of tagged actin polymerization regulatory proteins. Like inhibition of actin assembly, depletion of Myo1E causes reduced transferrin endocytosis and a significant delay in transferrin trafficking to perinuclear compartments, demonstrating an integral role for Myo1E in these actin-mediated steps. Mistargeting of GFP-Myo1E or its src-homology 3 domain to mitochondria results in appearance of WIP, WIRE, N-WASP, and actin filaments at the mitochondria, providing evidence for Myo1E's role in actin assembly regulation. These results suggest for mammalian cells, similar to budding yeast, interdependence in the recruitment of type I myosins, WIP/WIRE, and N-WASP to endocytic sites for Arp2/3 complex activation to assemble F-actin as endocytic vesicles are being formed.
INTRODUCTION
Type I myosins are actin-based motor proteins that are expressed in all eukaryotic cells, from yeasts to mammals (Mooseker and Cheney, 1995; Richards and Cavalier-Smith, 2005). There are eight different subtypes of type I myosin in vertebrates. Every type I myosin has an N-terminal motor domain, a single α-helical neck region, and a C-terminal tail. The C-terminal tail of the type I myosins can be considered either short (Myo1A, 1B, 1C, 1D, 1G, and 1H), consisting of a putative pleckstrin-homology domain within a tail-homology domain (TH1), or long (Myo1E and 1F), with a proline-rich TH2 and a Src-homology 3 (SH3) domain in addition to the TH1 domain (Mooseker and Cheney, 1995; McConnell and Tyska, 2010). Whereas Myo1F is predominately expressed in the hematopoietic tissues, Myo1E is the only “long-tailed” type I myosin that is ubiquitously expressed in mammalian cells. Human Myo1E was previously named myosin-IC, and the rat homologue was called myr3. To minimize confusion, the nomenclature for type I myosins was subsequently standardized (Gillespie et al., 2001).
Clues to Myo1E function have been obtained through studies in frogs and mammals. Green fluorescent protein (GFP)–tagged Myo1E colocalizes with stable F-actin around Xenopus oocyte wounds, suggesting a role coordinating membrane movement and actin-associated forces (Yu and Bement, 2007). In mammalian cells, Myo1E has been found using total internal reflection fluorescence microscopy (TIRFM) to localize to clathrin- and dynamin-containing puncta at the plasma membrane (Krendel et al., 2007). Furthermore, the C-terminal SH3 domain of Myo1E binds to dynamin and synaptojanin-1 in vitro, and expression of the Myo1E tail inhibits transferrin uptake (Krendel et al., 2007). Although the latter results implicate Myo1E in clathrin-mediated endocytosis (CME), its precise role in this process has not been determined.
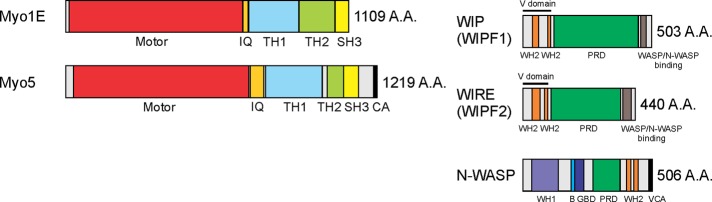
In single-celled eukaryotes such as budding yeast, fission yeast, and Dictyostelium, long-tailed type I myosins directly or indirectly activate the Arp2/3 complex and are involved in actin polymerization and endocytosis (Geli and Riezman, 1996; Jung et al., 1996, 2001; Evangelista et al., 2000; Lechler et al., 2000; Engqvist-Goldstein and Drubin, 2003; Sirotkin et al., 2005; Soulard et al., 2005; Sun et al., 2006). The protein domain structure of human Myo1E is very similar to that of the budding yeast type I myosins Myo3/5, except that it is missing the C-terminal CA domain (Figure 1). The CA domain of Myo3/5, together with verprolin Vrp1, is responsible for activation of the Arp2/3 complex to polymerize actin during the membrane invagination step of budding yeast endocytosis (Geli et al., 2000; Sun et al., 2006). Deletion of the genes encoding the two type I myosins Myo3 and Myo5 completely abolishes endocytic internalization in budding yeast (Sun et al., 2006).
FIGURE 1:
Schematic representation of the domain structure of mammalian type I myosin, Myo1E. Myo1E is composed of an N-terminal motor domain, a neck domain with IQ calmodulin-binding motif, a tail domain with tail-homology (TH) 1 and 2 domains, and a Src-homology (SH3) domain. Budding yeast Myo3/5 proteins have similar domain structures to the mammalian orthologue, Myo1E, except for the addition of a C-terminal (C) and acidic (A) domain. The A and C domains are necessary to activate the yeast Arp2/3 complex for actin polymerization. Mammalian WASP-interacting protein family 1/2 (WIPF1/2), commonly known as WIP and WIRE, related to yeast verprolin (Vrp1), is composed of two WASP-homology 2 (WH2) domains in the N-terminal verprolin-homology domain (V domain), a proline-rich domain (PRD), and a C-terminal WASP/N-WASP–binding domain. Mammalian N-WASP is composed of an N-terminal WIP-binding WASP-homology 1 (WH1) domain, a basic region (B), a GTPase-binding domain (GBD), a PRD, and a C-terminal VCA domain.
In budding yeast, CME has been revealed as a strictly actin-dependent process (Ayscough et al., 1997), in which different endocytic proteins are recruited to clathrin-coated pits (CCPs) with specific timing to perform specific functions (Kaksonen et al., 2005; Sun et al., 2006; Stimpson et al., 2009). However, in mammalian cells, the role of actin in CME has been controversial (Gottlieb et al., 1993; Lamaze et al., 1997; Fujimoto et al., 2000). On one hand, it has been reported that scission takes place when actin polymerization is at its maximum at a CCP (Merrifield et al., 2005), that disruption of actin by latrunculin A (lat-A) affects CCP assembly, constriction, and internalization (Yarar et al., 2005), and that trafficking of nascent endocytic vesicles requires dynamic actin polymerization (Merrifield et al., 1999; Kaksonen et al., 2000). On the other hand, treatment with lat-A and cytochalasin D were reported to inhibit endocytosis in only a subset of cultured cell lines and was dependent on whether the cells were adhering to a substrate or were grown in suspension (Fujimoto et al., 2000). Recent studies suggest that actin assembly forces are most important in mammalian cells under conditions of high membrane tension and when large clathrin “plaques” are internalized (Saffarian et al., 2009; Boulant et al., 2011). Of interest, absence of dynamin-1 and -2 revealed an intrinsic actin assembly mechanism at mammalian endocytic sites similar to that found in yeast cells (Ferguson et al., 2009). It was recently suggested that mammalian CME proteins have a modular organization similar to the one described for yeast cells. In a high time-resolution TIRFM study, Myo1E together with many associated and actin-regulatory proteins were found to have very similar recruitment signatures in NIH 3T3 cells and thus were assigned to the same dynamin/myosin/N-WASP module (Taylor et al., 2011), suggesting a possible functional link between these proteins. Here we provide evidence for a role for Myo1E in coordinating actin assembly and trafficking in CME.
RESULTS
Myo1E is recruited to endocytic sites coincident with recruitment of dynamin, WIP, WIRE, N-WASP, and actin
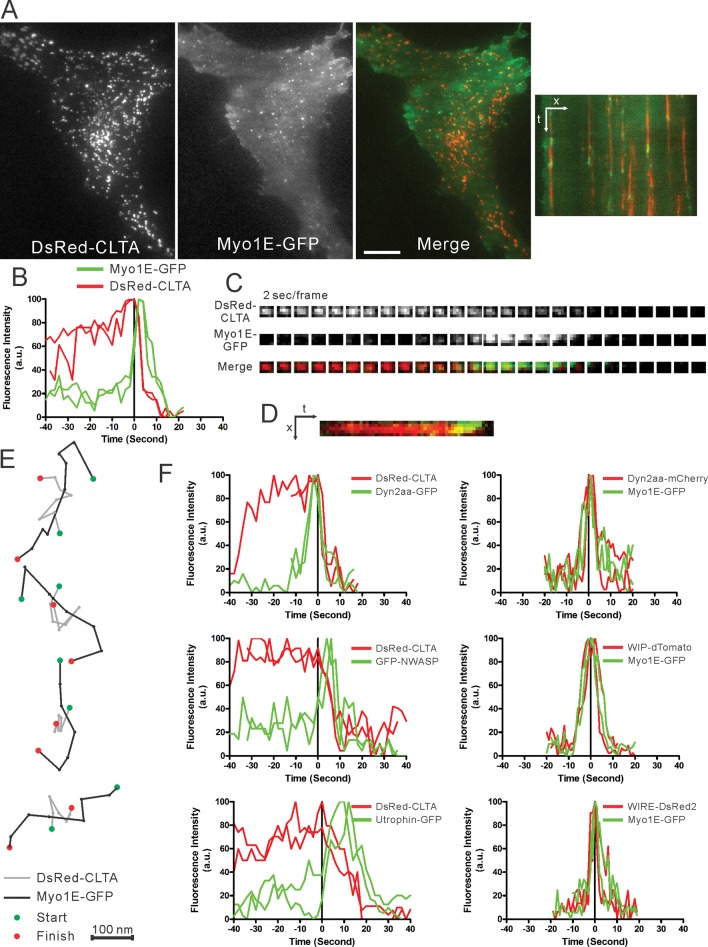
To explore the functions of Myo1E in CME, we first examined in Swiss 3T3 cells the recruitment dynamics of Myo1E using TIRFM (Figure 2A and Supplemental Movie S1). GFP-tagged Myo1E is recruited to CCPs and peaks when DsRed-clathrin light chain a (CLTA) is being internalized (Figure 2B). A representative recruitment profile, a montage overlay, and a kymograph are shown in Figure 2, B–D. Using particle-tracking analysis, we observed that Myo1E-GFP moved in a vectoral manner across the CCPs, whereas DsRed-CLTA was relatively stationary (Figure 2E). This vectoral movement might reflect Myo1E's motor activity driving newly endocytosed vesicles toward the cell interior.
FIGURE 2:
Recruitment of Myo1E-GFP when clathrin is being internalized. (A) A low-magnification TIRFM view of Swiss 3T3 cells stably expressing DsRed-clathrin light chain a (CLTA) and transiently expressing Myo1E-GFP. Right, a kymograph of the peripheral region of the same cell, showing recruitment of Myo1E to CCPs at the time of internalization. Bar, 10 μm. (B) Representative temporal recruitment profile of Myo1E-GFP and DsRed-CLTA in Swiss 3T3 cells. Myo1E-GFP is recruited concomitant with clathrin internalization. (C) Montage and (D) kymograph show the recruitment of Myo1E-GFP at the end of the lifetime of DsRed-CLTA. (E) Particle tracking of Myo1E-GFP shows a characteristic vectoral movement at the CCP with reference to CLTA. Bar, 100 nm. (F) Representative temporal recruitment profiles for GFP-tagged dynamin, N-WASP, and actin-marker utrophin (calponin-homology domain) in Swiss 3T3 cells stably expressing DsRed-CLTA (left). Images were captured at 2-s intervals. Representative temporal recruitment profiles of tagged dynamin, WIP, and WIRE in Swiss 3T3 cells expressing Myo1E-GFP (right). Images were captured at 1-s intervals.
CCP scission coincides with maximal displacement of CCPs from the plasma membrane and with peak recruitment of dynamin, F-actin–binding proteins, and actin-assembly regulatory proteins, including N-WASP and Arp3 (Merrifield et al., 2002, 2004, 2005; Merrifield, 2004; Perrais and Merrifield, 2005; Yarar et al., 2005). It is widely accepted that N-WASP is responsible for binding and activating the Arp2/3 complex to polymerize actin. To analyze the relative recruitment timing for these actin-regulatory proteins and Myo1E, we performed a series of temporal recruitment experiments. Consistent with previous reports, dynamin, N-WASP, and the F-actin–binding domain of utrophin are sequentially recruited to the CCPs (Figure 2F, left, and Supplemental Movies S2–S4). The temporal dynamics of Myo1E were very similar to those of dynamin 2 and the two verprolin-family proteins WIP (WASP-interacting protein or WIPF1) and WIRE (WIP-related or WIPF2; Figure 2F, right, and Supplemental Movies S5–S7). These results indicate that Myo1E and the actin-regulatory and -binding proteins arrive at the CCP late in the endocytic pathway and suggest that they may function to coordinate late events such as scission and vesicle trafficking.
Low concentrations of latrunculin A inhibit trafficking of endocytic cargo to perinuclear compartments
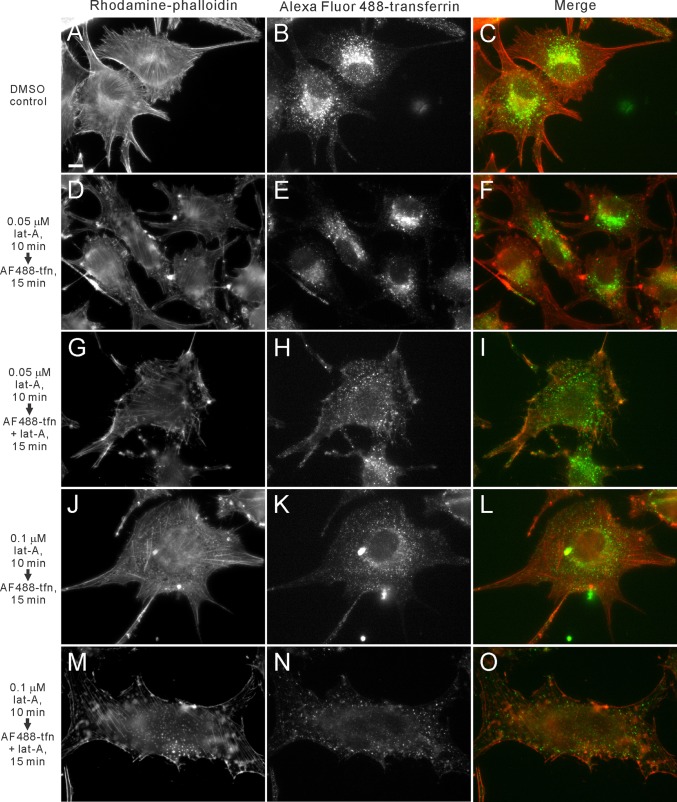
Actin assembly has been implicated in multiple stages of CME, including CCP assembly, constriction and internalization (Yarar et al., 2005). However, the role of actin in postscission trafficking steps has not yet been fully elucidated. Treatment of cells with low concentrations of lat-A has been shown to block events dependent on dynamic actin assembly without disrupting actin stress fibers or cell morphology (Shen and Turner, 2005). Human skin melanoma SK-MEL-28 cells treated with low levels (0.05 or 0.1 μM) of lat-A for 10–25 min showed reduced endocytosis and impaired trafficking of Alexa Fluor 488–transferrin (AF488-tfn) to the perinuclear region (Figure 3). When cells were treated for 10 min with dimethyl sulfoxide (DMSO) or a very low concentration of lat-A (0.05 μM) and were then incubated with AF488-tfn at 37°C for 15 min in the absence of lat-A, similar to the untreated control (Supplemental Figure S1D), the internalized transferrin was trafficked to the perinuclear region in all cells examined (n = 35 and 41; Figure 3, B and E). However, when the cells continued to be treated with 0.05 μM lat-A during incubation with AF488-tfn, 16.2% of the cells showed reduced and scattered localization of transferrin (Figure 3H), 81.1% showed normal perinuclear transferrin localization, and 2.7% displayed abnormal cell shrinkage (n = 37). When the cells were treated at 0.1 μM lat-A for 10 min followed by removal of lat-A and addition of AF488-tfn, the majority of the cells (93.6%) showed scattered AF488-tfn localization (Figure 3K). The remaining 4.3 and 2.1% of the cells showed normal perinuclear localization and abnormal cell shrinkage, respectively (n = 47). Similarly, when these cells were incubated with AF488-tfn in the continued presence of 0.1 μM lat-A, 90.7% of the cells showed scattered AF488-tfn localization (Figure 3N), and the remaining 2.3 and 7% showed normal perinuclear localization and abnormal cell shrinkage, respectively (n = 43). The perinuclear population of internalized transferrin at 10 min colocalized with an early endosome marker, Rab5a-GFP, in untreated cells (Supplemental Figure S2). Of importance, treatment with lat-A does not affect the distribution of Rab5a-GFP at the perinuclear region (Supplemental Figure S3, A–F). When cells were treated with 0.1 μM lat-A for 15 min, the localization of Rab5a-GFP was unchanged despite disruption of F-actin organization (Supplemental Figure S3, E and J). A transferrin-recycling assay showed that the dispersed localization of AF488-tfn in the lat-A–treated cells was not a result of a change in the recycling of transferrin from the endosomal compartments to cell surface. Treatment with lat-A did not affect the recycling of internalized transferrin when compared with the DMSO-treated control cells both qualitatively, by visualizing the internalized transferrin in cells (Supplemental Figure S4), and quantitatively, by a fluorescence-activated cell sorting (FACS)–based analysis (Supplemental Figure S5). These results collectively support the conclusion that actin functions in CME, as previously reported (Merrifield et al., 2002, 2004; Merrifield, 2004; Benesch et al., 2005; Yarar et al., 2005; Le Clainche et al., 2007; Boulant et al., 2011), and additionally in trafficking of internalized cargo.
FIGURE 3:
Low levels of lat-A inhibit endocytic trafficking. (A–O) SK-MEL-28 cells were treated with either DMSO (A–C) or low levels of lat-A for 10 min and subjected to a fluorescent transferrin-uptake assay in the absence (D–F, J–L) or presence (G–I, M–O) of lat-A. (A–C) In the DMSO-treated controls, cells exhibit normal F-actin structure (A) and the Alexa Fluor 488–transferrin was internalized and trafficked to the perinuclear region (B). (C) The merge of A and B. (D–F) In cells treated with 0.05 μM lat-A, despite the mild disruption of F-actin (D), the transferrin uptake (E) is similar to the control (B). (F) The merge of D and E. (G–I) However, when these cells were incubated in the presence of lat-A during fluorescent transferrin-uptake assay, the endocytic trafficking of transferrin is inhibited (H). (I) The merge of G and H. (J–O) In cells treated with 0.1 μM lat-A, either in the absence (J–L) or presence (M–O) of lat-A during the transferrin-uptake assay, significant endocytic and trafficking defects were observed. (D, G, J, M) Cells treated with low levels of lat-A show continued presence of stress fibers. Bar, 10 μm.
Myo1E depletion reduces rates of endocytosis and trafficking of endocytic cargo to perinuclear compartments
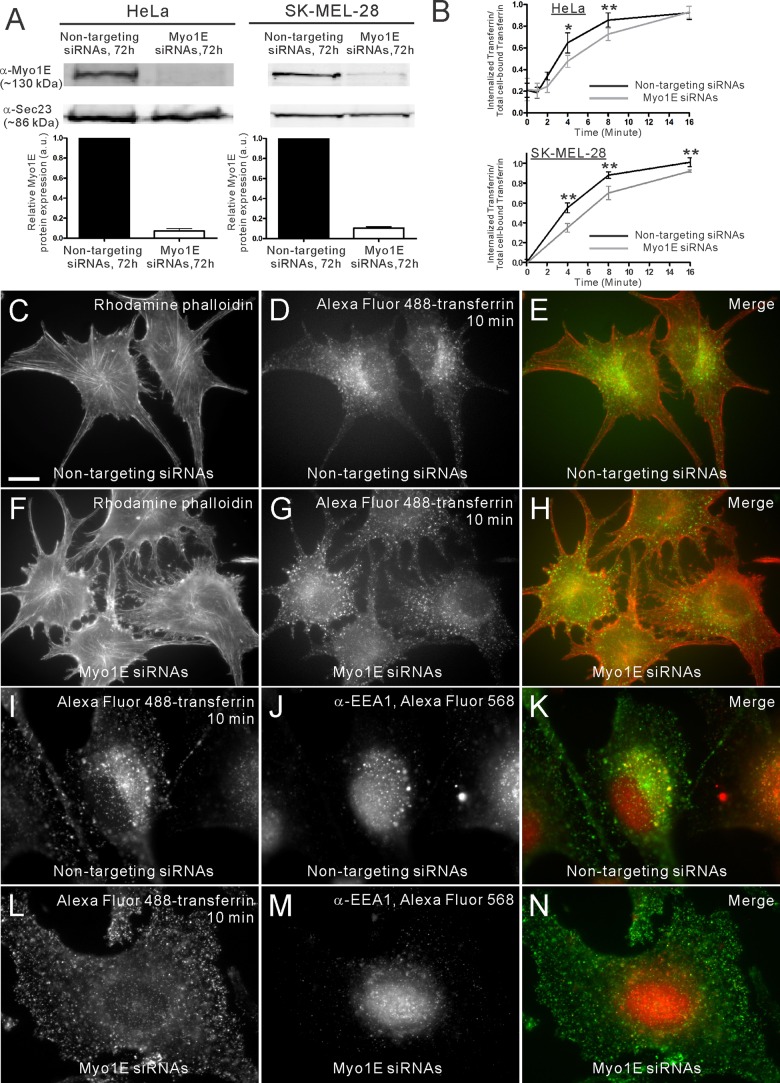
Because the appearance of GFP-tagged Myo1E at endocytic sites is coincident with appearance of actin and its regulators, and since actin assembly inhibition resulted in endocytic and trafficking defects, we next sought to determine the effect of Myo1E depletion on CME. RNA interference–mediated knockdown of human Myo1E was performed in both HeLa and SK-MEL-28 cells. Quantitative Western blotting showed a reduction of 96 and 89% in Myo1E protein levels 72 h posttransfection in HeLa and SK-MEL-28 cells, respectively (Figure 4A). In both cases, Myo1E protein levels were quantified relative to a loading control, Sec23. Quantitative enzyme-linked immunosorbent assay (ELISA)–based transferrin-uptake assays were performed in HeLa cells to assess endocytosis in Myo1E-knockdown (Myo1E-KD) cells. HeLa cells were serum starved, pulse incubated with biotinylated transferrin on ice, and then moved to 31°C, which is permissive for internalization (Engqvist-Goldstein et al., 2004; Zhang et al., 2005). The amount of internalized biotinylated transferrin is measured by the difference between the total transferrin fraction (phosphate-buffered saline [PBS] washed) and the internalized fraction (acid washed). Myo1E-KD cells showed a 27 and 36% reduction, respectively, in the internalized biotinylated transferrin at 4 min (p < 0.05; n = 3) in HeLa and SK-MEL-28 cells (Figure 4B). To capture these subtle kinetic defects during the early stages of CME in the Myo1E-KD cells, it is important to perform the transferrin-uptake assay at a lower temperature, at 31°C, to slow the internalization process in a pulse-chase assay. When the assay was performed at 37°C, the rate of internalization was faster, and it was more difficult to observe endocytic defects in the Myo1E-KD cells (n = 2; Supplemental Figure S6 compared with Figure 4B). To visually assess endocytosis and trafficking in Myo1E-KD cells, we performed fluorescent transferrin-uptake assays. After 10 min of AF488-tfn uptake, internalized transferrin accumulated at the perinuclear region in cells treated with control small interfering RNAs (siRNAs; Figure 4D), similar to Figure 3B. By quantifying the total fluorescence of the cell in the Myo1E-KD and control cells we observed a 51 ± 18% reduction in transferrin uptake (n = 6 and 7 for Myo1E-KD and control cells, respectively) and an inhibition of trafficking to the perinuclear region in the Myo1E-KD cells. Instead of being internalized and trafficked to the perinuclear endosomal structures, the transferrin appeared to have a scattered localization (Figure 4G). Immunofluorescence staining showed that in control cells the internalized transferrin in the perinuclear region colocalizes with the early endosomal marker EEA1 (Figure 4, I–K) but not with the late endosomal marker Rab7 or CHMP4 (Supplemental Figure S7). In contrast, the fluorescent transferrin did not colocalize with EEA1 in the Myo1E-KD cells (Figure 4, L–N). Similar to the lat-A–treated cells, the localization of an early endosomal marker was unchanged in Myo1E-depleted cells (Supplemental Figure S3). These data indicate that in addition to a role in endocytic uptake, Myo1E plays an important role in transferrin trafficking from the cell surface to perinuclear early endosomal structures.
FIGURE 4:
Depletion of Myo1E reduces endocytosis and inhibits downstream endocytic trafficking. (A) Western blots and quantification of Myo1E depletion using siRNAs. Depletion of Myo1E in HeLa and SK-MEL-28 cells after 72 h showed a 96 and 90% reduction in Myo1E protein levels, respectively. Sec23 serves as a loading control. (B) ELISA-based transferrin-uptake assay showed a 27 and 36% reduction in internalization of biotinylated transferrin at 4-min in Myo1E-depleted HeLa and SK-MEL-28 cells compared with control, respectively. *p < 0.05 and **p < 0.01. (C, F) F-actin staining and (D, G) fluorescent transferrin uptake in control and Myo1E-depleted SK-MEL-28 cells, respectively. Ten minutes after internalization, Alexa Fluor 488-transferrin–containing puncta were concentrated at the perinuclear region in the control cells (D). (D, G) By quantifying the total fluorescence of the cell, there is a 51 ± 18% reduction of the internalized transferrin in Myo1E-depleted cells compared with the control cells (cells = 6 and 7 for Myo1E-depleted and control cells, respectively). In addition, the internalized puncta were localized at the cell periphery rather than at the perinuclear region. (I–K) Immunofluorescence shows colocalization of the perinuclear-concentrated Alexa Fluor 488–transferrin puncta and an early endosome marker EEA1 in the control cells. However, in the Myo1E-depleted cells (L–N), the puncta were dispersely localized and did not colocalize with EEA1, which retains its perinuclear localization. Bar, 10 μm.
Myo1E depletion increases clathrin and dynamin lifetimes at CCPs
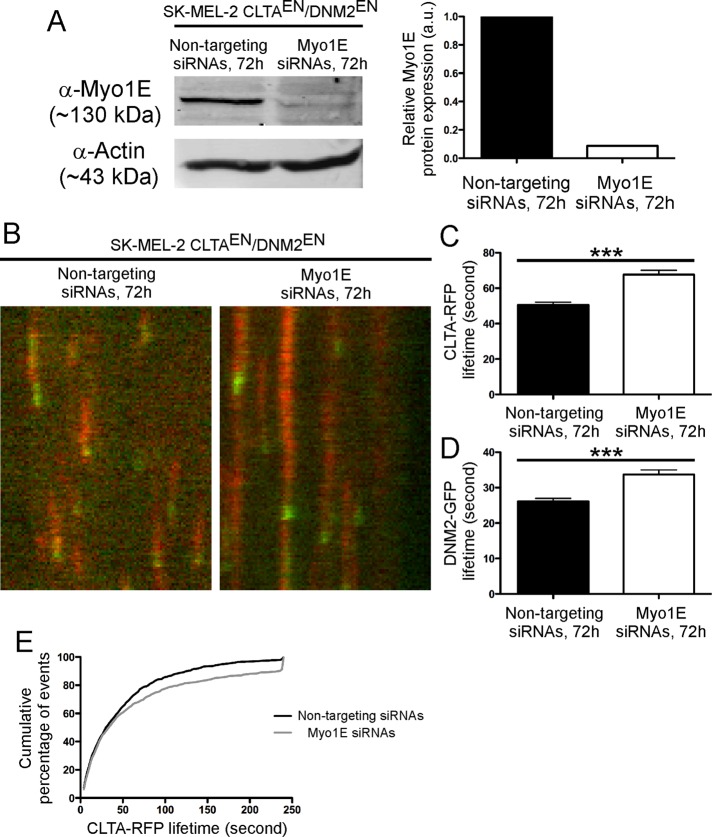
To study the effect of Myo1E depletion on CCP dynamics, we used genome-edited SK-MEL-2 CLTAEN/DNM2EN cells. These cells express clathrin light chain–red fluorescent protein (RFP) and dynamin-GFP at endogenous levels and display more homogeneous CCP dynamics than cells overexpressing these constructs (Doyon et al., 2011). Myo1E was depleted by siRNA, and the cells were imaged using TIRFM. Quantitative Western blotting showed a 91% reduction in Myo1E protein levels 72 h posttransfection (Figure 5A). CLTA-RFP and DNM2-GFP puncta were detected and tracked by using Imaris software (see Materials and Methods).
FIGURE 5:
Depletion of Myo1E in genome-edited CLTAEN/DNM2EN SK-MEL-2 cells increased the average lifetime of CLTA-RFP and DNM2-GFP. (A) Western blots and quantification of Myo1E depletion using siRNAs after 72 h showed a 91% reduction of Myo1E protein expression. (B) Representative examples of kymograph showing the lifetime of CLTA-RFP (red) and DNM2-GFP (green) in CLTAEN/DNM2EN cells treated with control or Myo1E siRNAs. (C) The average lifetime of CLTA-RFP in Myo1E-depleted cells (61.9 ± 2.4 s; tracks, 982; cells, 8) is significantly longer than that in the control cells (48.6 ± 2.1 s; tracks, 1293; cells, 8). (D) The average lifetime of DNM2-GFP in Myo1E-depleted cells (33.7 ± 1.3 s; tracks, 998; cells, 8) is also significantly longer than in the control cells (26.1 ± 0.85 s; tracks, 934; cells, 8). Data are presented as means ± SEM. ***p < 0.0001. (E) Cumulative percentage of frequencies of CLTA-RFP lifetime in the analysis.
Kymograph analysis of typical CME events in a control cell showed CLTA-RFP tracks terminated by a short burst of DNM2-GFP (Figure 5B, left, and Supplemental Movie S8). The average lifetimes for CLTA-RFP and DNM2-GFP were 48.6 ± 2.1 s (tracks, 1293; cells, 8) and 26.1 ± 0.85 s (tracks, 934; cells, 8), respectively. These average lifetimes were consistent with the ones we measured previously (Doyon et al., 2011). However, in cells depleted of Myo1E, the lifetimes of CLTA-RFP and DNM2-GFP appeared to be significantly longer (p < 0.0001; Figure 5B, right, and Supplemental Movie S9). In contrast to the control cells, the average lifetimes for CLTA-RFP and DNM2-GFP in Myo1E-depleted cells were 61.9 ± 2.4 s (tracks, 982; cells, 8) and 33.7 ± 1.3 s (tracks, 998; cells, 8), respectively (Figure 5, B–D). Most dramatically, 10.2% of CLTA-RFP tracks in Myo1E-depleted cells persisted throughout the entire length of the 4-min movie, as compared with only 0.9% in the control cells (Figure 5, B and E).
Myo1E SH3 domain is necessary and sufficient to recruit WIP, WIRE, and N-WASP in vivo
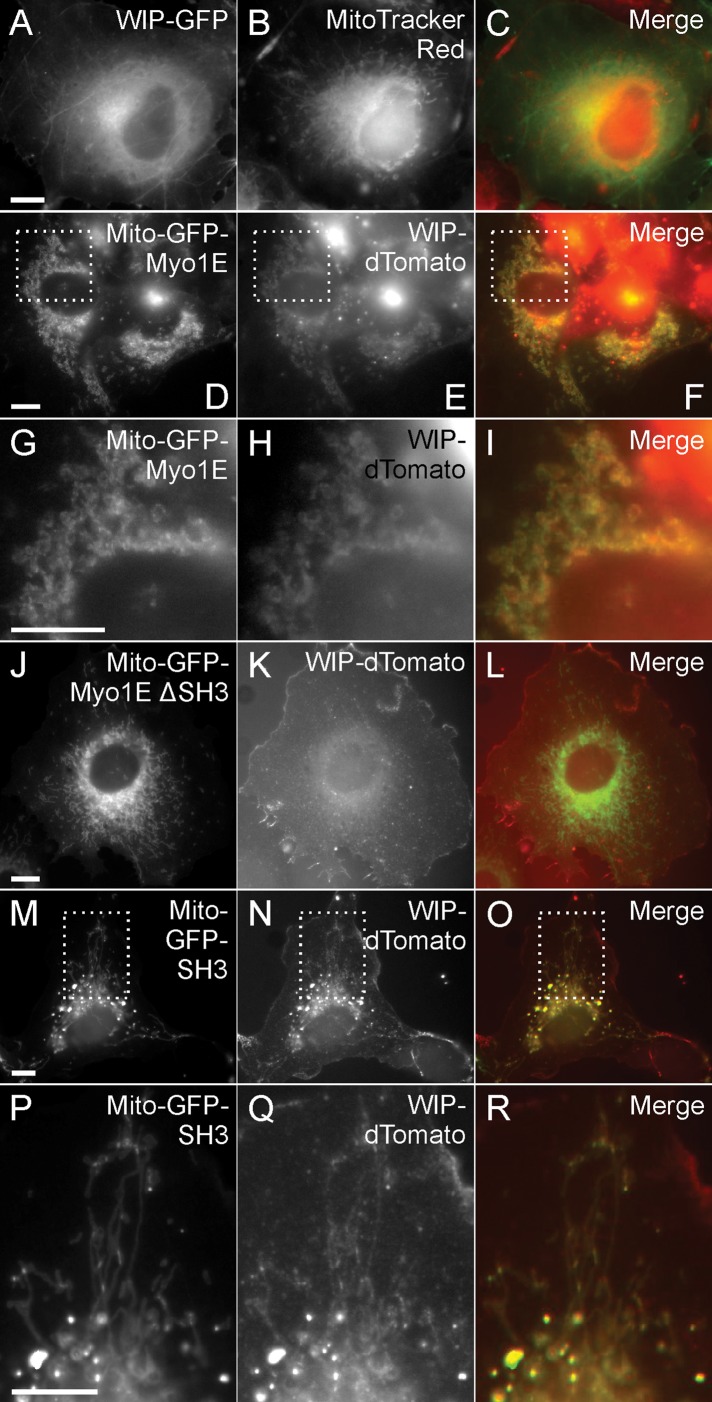
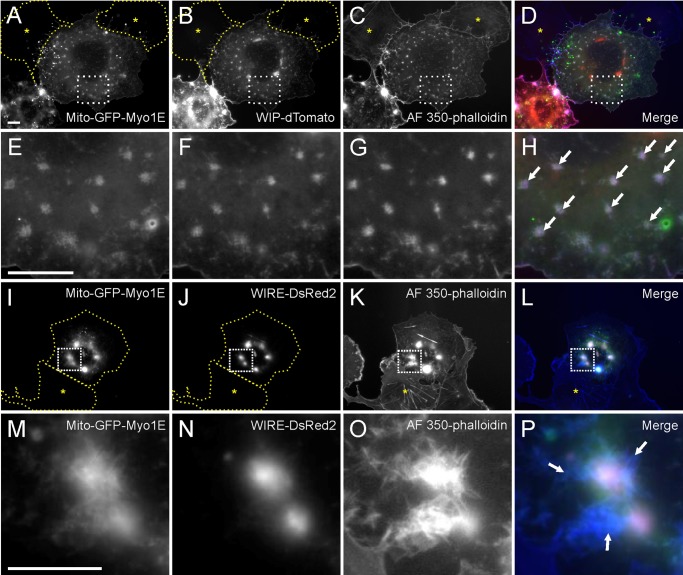
Our live cell imaging showed that Myo1E and other actin-regulatory proteins are recruited to the CCPs with similar lifetimes and dynamics. To test for functional interrelationships among these proteins, we N-terminally tagged full-length Myo1E with a mitochondria outer membrane-localizing (Mito) signal (Pinyol et al., 2007) and GFP. The Mito-GFP-Myo1E colocalized with the mitochondria marker MitoTracker Red (Supplemental Figure S8, A–C) but not with DsRed-CLTA in Cos-7 cells (Supplemental Figure S8, D–F). Cells were cotransfected with Mito-GFP-Myo1E, and fluorescent protein–tagged WIP, WIRE, or N-WASP to determine whether Mito-GFP-Myo1E is able to recruit these proteins to the mitochondria in vivo. In control cells transfected with either WIP-GFP (Figure 6, A–C) or WIRE-GFP (Supplemental Figure S9, A–C) alone, localization to mitochondria was not observed. However, when GFP-Myo1E was targeted to the mitochondrial outer membrane, both WIP-dTomato and WIRE-DsRed2 were recruited to the mitochondria as well (Figure 6, D–I, and Supplemental Figure S9, D–F). The recruitment of WIP and WIRE required the SH3 domain of Myo1E. When Mito-GFP-Myo1E with the SH3 domain deleted (∆SH3) was coexpressed with either WIP-dTomato or WIRE-DsRed2, neither WIP nor WIRE was recruited to mitochondria (Figure 6, J–L, and Supplemental Figure S9, G–I). However, recruitment of WIP and WIRE was observed when only the SH3 domain of Myo1E was mistargeted to the mitochondria (Figure 6, M–R, and Supplemental Figure S9, J–L). Thus, the SH3 domain of Myo1E is necessary and sufficient to recruit WIP and WIRE in vivo. When cells were cotransfected with Mito-GFP-Myo1E and either WIP-dTomato (Figure 7, A–H) or WIRE-DsRed (Figure 7, I–P) and then labeled with AF350-conjugated phalloidin, filamentous actin was detected at the mitochondria (white arrows). When WIP or WIRE was not cotransfected with Mito-GFP-Myo1E, actin could still be observed around mitochondria but less prominently (unpublished data). By contrast, untransfected cells displayed normal actin organization with no aberrant actin structures (yellow asterisks). In addition, targeting of GFP-Myo1E full length to mitochondria resulted in recruitment of N-WASP-dTomato to mitochondria (Supplemental Figure S9, M–R). It was previously demonstrated that actin polymerization on mitochondria is detected when N-WASP is mistargeted to the mitochondria (Pinyol et al., 2007). Consistently, when we mistargeted GFP-tagged N-WASP to mitochondria and labeled the cells with rhodamine–phalloidin, we detected prominent actin structures at the GFP-N-WASP–decorated mitochondria (Supplemental Figure S10, A–D). Our results collectively suggest that the SH3 domain of Myo1E may be responsible for recruiting the actin-regulatory proteins WIP, WIRE, and their interacting partner N-WASP to initiate Arp2/3 complex–based actin polymerization.
FIGURE 6:
Myo1E SH3 domain is necessary and sufficient to recruit WIP. (A–D) Transient expression of WIP-GFP (A) in Cos-7 control cells labeled with the mitochondria marker MitoTracker Red (B). WIP-GFP normally does not colocalize with MitoTracker Red. (C) The merge of A and B. (E, F) In cells cotransfected with mitochondria-targeted Mito-GFP-Myo1E and WIP-dTomato, WIP-dTomato is recruited to mitochondria. (G–I) High magnification of E and F showing the colocalization of GFP-Myo1E–decorated mitochondria and WIP-dTomato. (I) The merge of G and H. (J–R) SH3 domain of Myo1E is necessary and sufficient to recruit WIP-dTomato. (J–L) In cells cotransfected with Mito-GFP-Myo1E ΔSH3 and WIP-dTomato, WIP-dTomato is not recruited to the GFP-Myo1E-decorated mitochondria. (M–R) In cells cotransfected with Mito-GFP-SH3 and WIP-dTomato, the recruitment of WIP-dTomato is restored. (O) The merge of M and N. (P–R) High magnification of M–O shows the colocalization of GFP-SH3–decorated mitochondria and WIP-dTomato. (R) The merge of P and Q. Bars, 10 μm.
FIGURE 7:
Actin assembly at the GFP-Myo1E and WIP-dTomato or WIRE-DsRed2-decorated mitochondria. Cotransfection of Mito-GFP-Myo1E and WIP-dTomato (A–H) or WIRE-DsRed2 (I–P) results in actin assembly at the mitochondria. Yellow asterisk marks an untransfected cell that has no aberrant actin structures in the cytoplasm. Arrows indicate actin meshwork assembled on the GFP-Myo1E and WIP-dTomato or WIRE-DsRed-decorated mitochondria. Bar, 10 μm.
Dynamin recruits Myo1E independent of its C-terminal proline-rich domain in vivo
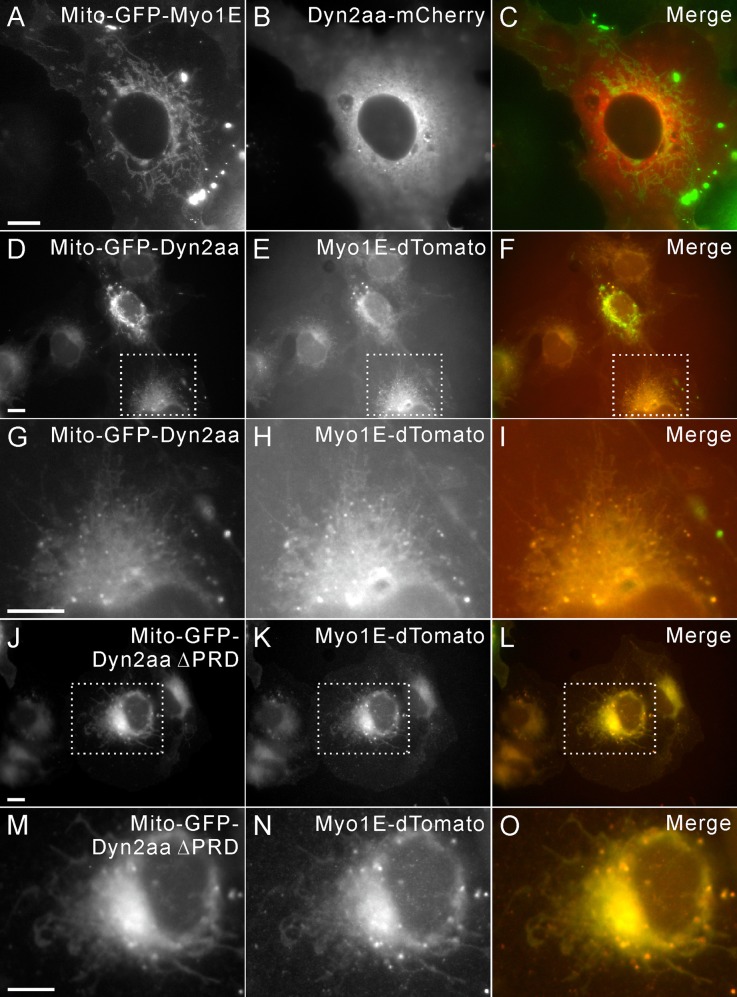
It has been shown by immunoprecipitation that the SH3 domain of Myo1E interacts with dynamin, possibly via dynamin's proline-rich domain (PRD; Krendel et al., 2007). However, mistargeting of GFP-Myo1E did not result in recruitment of dynamin to mitochondria (Figure 7, A and B). Because live cell imaging analyses showed that dynamin is recruited to the CCP during late steps of the endocytic pathway (Merrifield et al., 2002; Figure 2F) but just before Myo1E (Figure 2F), we asked whether dynamin can recruit Myo1E. When GFP-tagged dynamin was targeted to mitochondria, Myo1E-dTomato was efficiently recruited (Figure 8, D–I). Myo1E recruitment to the GFP-dynamin–decorated mitochondria was not dependent on dynamin's PRD. When GFP-dynamin with its C-terminal PRD deleted (∆PRD) was targeted to mitochondria, Myo1E was still efficiently recruited to mitochondria (Figure 8, J–O). The recruitment of Myo1E to mitochondria by GFP-dynamin ∆PRD could be because of the oligomerization of endogenous dynamin with the ectopically expressed protein. However, a similar high efficiency of Myo1E recruitment is observed in both the presence and absence of the PRD domain (Figure 8, F and L). These results are consistent with the previous findings that localization of Myo1E to either CCPs in CME or actin coats in Xenopus oocyte cortical granules is dependent on Myo1E's TH2 domain (Krendel et al., 2007; Yu and Bement, 2007) and suggest that dynamin might recruit Myo1E to endocytic sites independent of a PRD–SH3 domain interaction.
FIGURE 8:
Dynamin recruits Myo1E independent of its proline-rich domain. (A, B) Cotransfection of Mito-GFP-Myo1E and dynamin-mCherry shows that the latter is not recruited to the mitochondria. (C) The merge of A and B. (D–F) Cotransfection of Mito-GFP-dynamin and Myo1E-dTomato shows that Myo1E-dTomato is efficiently recruited to the GFP-dynamin–decorated mitochondria. (F) The merge of D and E. (G–I) High magnification of D–F showing the colocalization of GFP-dynamin–decorated mitochondria and Myo1E-dTomato. (I) The merge of G and H. (J–L) Cotransfection of Mito-GFP–dynamin ΔPRD and Myo1E-dTomato shows efficient recruitment of Myo1E-dTomato to the mitochondria. (L) The merge of J and K. (M–O) High magnification of J–L showing the colocalization of GFP-dynamin ΔPRD–decorated mitochondria and Myo1E-dTomato. (O) The merge of M and N. Bars, 10 μm.
DISCUSSION
Myo1E function in mammalian endocytosis
Clathrin-mediated endocytosis is a multistep process, which involves invagination of the plasma membrane, scission, and trafficking of internalized vesicles to early endosomes. It has been suggested that actin is involved in CCP assembly, CCP lateral motility, CCP constriction, and possibly scission and trafficking of the nascent vesicle away from the plasma membrane (Yarar et al., 2005). Understanding how actin functions in endocytosis requires analysis of the roles of proteins that function with and regulate actin. Because actin and type I myosin are required to invaginate the plasma membrane during budding yeast endocytosis, we investigated the endocytic role of Myo1E in mammalian cell endocytosis.
Here we presented evidence for roles of Myo1E in facilitating normal clathrin and dynamin dynamics, recruiting actin regulatory and polymerizing factors to CCPs during the late stages of CME, and facilitating cargo trafficking from the plasma membrane to perinuclear early endosomes. Inhibition of actin assembly by lat-A caused reduced transferrin endocytosis and a significant delay in its trafficking to perinuclear early endosomal compartments. Depletion of Myo1E resulted in similar endocytic defects, suggesting that it has an integral role in these actin-dependent steps. Depletion of Myo1E resulted in a relatively mild reduction—27% in HeLa cells and 36% in SK-MEL-28 cells—in the kinetics of internalization of the cargo transferrin as measured using a quantitative ELISA. Our results suggest that Myo1E may play a role in both cargo internalization and the transport of the newly internalized vesicles to their target endosomal compartments. This conclusion is consistent with our live cell imaging data showing that Myo1E is recruited to endocytic sites at the time when the clathrin-coated structure is internalizing into the cell. At this point, we do not know whether the defect in transferrin trafficking from the cell surface to the perinuclear early endosomes results from defects in CCP budding off from the plasma membrane or from a failure in a later vesicular transport step for the internalized vesicles. In a genome-edited cell line in which both endogenous CLTA and DNM2 loci had been fused to sequences encoding RFP and GFP, respectively, Myo1E depletion resulted in a significant increase in CLTA-RFP and DNM2-GFP lifetimes. In addition, there was a >11-fold increase in the number of static CLTA-RFP puncta in the Myo1E-depleted cells compared with the control cells. Recruitment of DNM2-GFP to these CLTA-RFP puncta was not affected, although the average DNM2-GFP lifetime was longer in the Myo1E-depleted cells. These results suggest that when Myo1E is depleted, CCV formation is impaired, leading to longer clathrin and dynamin lifetimes.
In yeasts and Dictyostelium, type I myosins coordinate actin assembly with endocytosis. Here targeting GFP-Myo1E to mitochondria resulted in recruitment of the verprolin-family proteins (WIP and WIRE) and N-WASP, providing evidence that Myo1E may also play a role in actin assembly regulation. The recruitment of WIP and WIRE depends on the Myo1E SH3 domain. In budding and fission yeast, type I myosin has been reported to interact with verprolin and to be involved in WASP-Arp2/3 complex–based actin polymerization (Anderson et al., 1998; Geli et al., 2000; Sirotkin et al., 2005; Sun et al., 2006). Similar to results for type I myosin in budding yeast, our studies provide evidence for interdependence in recruitment to endocytic sites of mammalian Myo1E with WIP, WIRE, and N-WASP, thus activating the Arp2/3 complex to assemble F-actin as endocytic vesicles are being formed. This conclusion is also consistent with a role for actin in propelling nascent endocytic vesicles and endosomes (Merrifield et al., 1999; Kaksonen et al., 2000).
Myo1E recruitment to endocytic sites
The assignment of endocytic proteins to distinct modules in budding yeast helped to identify subsets of proteins that interact and function together during endocytosis (Kaksonen et al., 2005; Sun et al., 2006, 2007; Okreglak and Drubin, 2007; Toshima et al., 2007; Toret et al., 2008; Liu et al., 2009, 2010; Stimpson et al., 2009). On the basis of a similar approach in mammalian cells, nine proteins were recently assigned to the dynamin/myosin/N-WASP module based on their temporal recruitment signatures at endocytic sites (Taylor et al., 2011). These proteins include Myo1E, dynamin 1 and 2, synaptojanin 2β1, N-WASP, Hip1R, myo6, Synd2, and Eps8. Myo1E SH3 domain interacts with the PRD-containing proteins dynamin 1 and 2 and synaptojanin 1 (Krendel et al., 2007). The SH3 domain of human Myo1E is required, although not sufficient, to localize Myo1E to endocytic sites in Swiss 3T3 cells (Krendel et al., 2007). Therefore interactions between the Myo1E SH3 domain and PRD-containing endocytic proteins are hypothesized to be responsible for the recruitment of Myo1E to the endocytic sites (Krendel et al., 2007). Consistent with the previous findings, we showed that Myo1E is recruited by Mito-GFP-dynamin 2aa to mitochondria but found that the reverse is not true and that recruitment of DNM2-GFP to endocytic sites was not affected when Myo1E was depleted in genome-edited SK-MEL-2 cells. However, since both Myo1E and actin localized at the tubulated CCPs in primary fibroblast cells derived from dynamin 1 and 2–knockout mice (Ferguson et al., 2009), we propose that additional factors, perhaps other PRD-containing proteins, also contribute to Myo1E recruitment to endocytic sites.
Conserved roles of type I myosin in actin assembly and endocytosis
Accumulating evidence points to a role for type I myosins in actin assembly and endocytosis in diverse organisms. In budding yeast, type I myosin is responsible for actin nucleation necessary for endocytic internalization (Sun et al., 2006). In neutrophils, depletion of another “long-tail” type I myosin, Myo1F, leads to a reduction of actin polymerization in the cortical actin network (Kim et al., 2006). In the African trypanosome Trypanosoma brucei, depletion of the only type I myosin, TbMyo1, results in ∼80% reduction of tomato lectin uptake (Spitznagel et al., 2010). On the basis of this study, TbMyo1 was suggested to be involved in vesicle scission and movement into the cytoplasm (Spitznagel et al., 2010). Both the mammalian and the budding yeast type I myosins Myo1E and Myo3/5, respectively, have very similar protein domain structures. It is therefore logical to speculate that they would have similar biological functions. However, the additional C-terminal CA domain in budding yeast Myo3/5, together with verprolin Vrp1, directly activates the Arp2/3 complex to trigger actin polymerization and is crucial for yeast endocytosis. Here we provide evidence to suggest a conserved role for Myo1E and Myo3/5 in actin assembly and in facilitating endocytosis. In contrast to the direct interaction between Myo3/5 and Arp2/3 complex, the recruitment by Myo1E of actin-regulatory and polymerization factors to assemble actin may enable additional temporal and spatial regulation. A role for type I myosin function in actin regulation at endocytic sites from yeast to mammals fits with the recent appreciation of similarities across species in actin assembly integration with endocytic vesicle formation (Ferguson et al., 2009; Liu et al., 2009).
MATERIALS AND METHODS
Cell culture and transfection
Swiss 3T3 cells and Swiss 3T3 cells stably expressing DsRed-clathrin were kindly provided by Wolfhard Almers (Oregon Health and Science University, Portland, OR). COS-7, HeLa, and SK-MEL-28 cells were purchased from the American Type Culture Collection (Manassas, VA). The cells were grown in DMEM containing 10% fetal bovine serum (FBS). Swiss 3T3 cells stably expressing DsRed-clathrin were cultured in DMEM containing 10% FBS and 0.4 mg/ml G-418. Swiss 3T3, HeLa, and SK-MEL-28 cells were transfected using Lipofectamine 2000 (Invitrogen, Carlsbad, CA), and COS-7 cells were transfected using FuGENE-6 transfection reagent (Roche, Basel, Switzerland) following standard protocols.
DNA constructs, antibodies, and reagents
Full-length mouse and human Myo1E sequences were amplified from Swiss 3T3 and SK-MEL-28 cDNAs and cloned into the pEGFP-N1 vector (Clontech, Mountain View, CA). A Mito-Flag-GFP–tag empty vector with additional multiple cloning sites (pMito-Flag-GFP-JC3) was made from pCMV-Tag2B Mito-Flag-GFP-syndapin II plasmid (Kessels and Qualmann, 2006). Mito-Flag-GFP–tagged rat dynamin 2aa was made by replacing the syndapin II sequence with the full-length dynamin 2aa sequence. Mito-GFP-dynamin 2aa ΔPRD was made by deleting the PRD domain in the construct. Mito-Flag-GFP–tagged mouse Myo1E was made by cloning the full-length sequence into the JC3 vector. Mito-Flag-GFP mouse Myo1E deletion constructs, ΔSH3 and SH3 domain alone, were made from pMito-Flag-GFP-JC3 mouse Myo1E. The pRSET-B dTomato and mCherry were kindly provided by Roger Tsien. The pdTomato-N1 and pmCherry-N1 vectors were made by replacing EGFP with the dTomato and mCherry sequences in the pEGFP-N1 vector. pEGFP-N1-rat dynamin 2aa was kindly provided by Mark McNiven. pmCherry N1-dynamin 2aa was made by subcloning rat dynamin 2aa into the pmCherry-N1 vector. pDsRed2-C1-human WIRE was kindly provided by Pontus Aspenström. The GFP-utrophin (amino acids 1–261) was kindly provided by William Bement. WIP-dTomato was made by subcloning WIP from CB6-GFP-WIP (kindly provided by Michael Way) into the pdTomato-N1 vector. The pdTomato-N1-N-WASP was made by subcloning the N-WASP sequence from emerald GFP-Flag-rat N-WASP (kindly provided by Matthew Welch) into the pdTomato-N1 vector. The pEGFP-C1-human clathrin LCa was kindly provided by Lois Greene.
The polyclonal goat anti-Myo1E antibody (N13) was purchased from Santa Cruz Biotechnology (Santa Cruz, CA). The polyclonal rabbit anti-human Sec23 antibody was kindly provided by Randy Schekman. The polyclonal sheep anti-human transferrin antibody (PC070) was purchased from The Binding Site (Birmingham, United Kingdom). The polyclonal rabbit anti-EEA1 antibody (2411) was purchased from Cell Signaling Technology (Danvers, MA). The MitoTracker Red CMXRos (M-7512), rhodamine–phalloidin (R415), Alexa Fluor 350–phalloidin (A22281), Alexa Fluor 488–conjugated human transferrin (T-13342), Alexa Fluor 568–conjugated donkey anti–rabbit immunoglobulin G (IgG; A10042), and CellLight BacMam early endosomes-GFP (C10586) were purchased from Invitrogen. The Odyssey blocking buffer (927-40000), IRDye 680 donkey anti–rabbit IgG (926-32223), and IRDye 800CW donkey anti–goat IgG (926-32214) were purchased from LI-COR Biosciences (Lincoln, NE). The polyclonal rabbit anti-Rab7 and anti-CHMP4 antibodies were kindly provided by Suzanne Pfeffer and Phyllis Hanson, respectively.
Live cell imaging and image analysis
In all experiments, cells were plated on precleaned borosilicate glass coverslips (25 mm, number 1; Thermo Fisher Scientific, Waltham, MA). Shortly before imaging, medium was replaced with an imaging buffer containing phenol red–free DMEM (Invitrogen), l-glutamine, 10 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES; Invitrogen) at pH 7.4, and 5% FBS. Cells were imaged in an environmental control system set to 37°C (Precision Control, Fall City, WA). Experiments were performed using a TIRF inverted microscope (IX81; Olympus, Tokyo, Japan) equipped with a 60×/1.45 numerical aperture Plan Apo Objective (Olympus, Center Valley, PA) and fully controlled by MetaMorph, version 7 Molecular Devices, Sunnyvale, CA). Two solid-state laser lines (488 and 568 nm; CVI Melles Griot, Albuquerque, NM) were coupled to a TIRF condenser through two optical fibers. The two channels were simultaneously imaged through a dual-view beam splitter (Photometrics, Tucson, AZ) to separate the green and red emission signals to two sides of the camera using a 565-nm dichroic mirror and 530/30 and 630/50 nm emission filters. Images were collected with a charge-coupled device camera OrcaER2 (1024 × 1024, 14-bit; Hamamatsu, Bridgewater, NJ). Cells were typically imaged at 0.5 Hz for 120 frames, without binning and using a time exposure between 0.4 and 0.9 s.
Analysis was performed using Imaris software, version 7.1 (Bitplane, Saint Paul, MN). CCSs were detected within a 100-μm2 region of interest (ROI) using the spot module. Object segmentation was performed using an estimated size of 350 nm and a manually adjusted quality filter. Objects detected were then tracked using the tracking module and Brownian motion algorithm. To avoid transient breaks in trajectory and lifetime, an estimated displacement of 500 nm and a gap closing of 7 frames were incorporated. Finally, the fidelity of the tracking results was assessed visually by overlapping detected tracks with the original image. Tracks touching the edges of the ROI were excluded from the analysis. Mean lifetime was calculated using the average of individual track lifetime.
Two-color TIRFM movies were also analyzed using ImageJ (National Institutes of Health, Bethesda, MD; Le Clainche et al., 2007). The GFP and DsRed channels were aligned with an image of fluorescent beads. The two channels were then merged. The maximum fluorescence intensity of GFP or DsRed at each CCP was measured and plotted against time. The different CCP tracks were aligned at time zero when the fluorescence of DsRed-clathrin is at maximum before internalization occurs.
Transferrin-uptake assays
For the fluorescent transferrin-uptake assay, SK-MEL-28 cells were grown in six-well plates and treated with siRNAs as described. After 48 h, the cells were seeded on 25-mm round glass coverslips (0.17 mm in thickness) in six-well plates overnight. The cells were serum starved at 37°C for 1 h in starvation medium. They were then incubated with 25 μg/ml human transferrin conjugated to Alexa Fluor 488 in starvation medium (DMEM containing 20 mM HEPES, pH 7.4, and 5 mg/ml BSA) at 37°C for 15 min. The coverslips were then fixed in 4% paraformaldehyde (PFA) at room temperature for 20 min and mounted on glass slides using ProLong Gold antifade reagent with 4′,6-diamidino-2-phenylindole (DAPI; Invitrogen). The internalized fluorescent transferrin was quantified by measuring the average fluorescence intensity of the whole cell using ImageJ software.
For the quantitative ELISA-based transferrin-uptake assay, HeLa cells were grown in six-well plates and treated with siRNAs as described. After 72 h, the cells were washed two times with starvation medium and incubated at 37°C for 1 h. They were then put on ice in a 4°C cold room, and the medium was replaced with 4°C starvation medium containing 2 μg/ml biotinylated-human transferrin. The cells were incubated for 1 h on ice in the cold room. They were then washed two times with ice-cold starvation medium and moved immediately to a 31 or 37°C water bath for various durations (0, 4, 8, and 16 min) to allow transferrin internalization. To measure internalized transferrin, surface-bound transferrin was stripped by adding 2 ml of ice-cold stripping solution (10 mM HCl, 150 mM NaCl, pH 2.0) to the cells for 2 min, followed by a wash with 10 ml of ice-cold PBS. This stripping and washing steps were repeated once. To measure total transferrin bound, cells were washed in 10 ml of ice-cold PBS (Engqvist-Goldstein et al., 2004). Cells were then lysed in lysis buffer (PBS containing 50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 5 mM EDTA, 1% NP-40, and 0.1% SDS) containing complete Mini Protease Inhibitor Cocktail (Roche) and 1 mM phenylmethylsulfonyl fluoride. An ELISA was used to quantify the amount of biotinylated transferrin in the cell lysate (Buss et al., 2001). Briefly, ELISA plates were coated with anti-transferrin antibody (1:1000; The Binding Site) diluted in 50 mM NaHCO3, pH 9.6, overnight at 4°C. The plates were washed twice with PBS and blocked at 37°C for 1 h with blocking solution (10 mM Tris-HCl, pH 7.4, 50 mM NaCl, 1 mM EDTA, 1% Triton X-100, 0.1% SDS, and 0.2% BSA). Protein concentrations of cell lysates were measured using the BCA protein assay (Pierce, Thermo Scientific, Pittsburgh, PA) and diluted with the blocking solution. The diluted cell lysates were added to the ELISA plates and incubated at 4°C overnight. The plates were then washed two times with PBS and incubated with blocking solution at room temperature for 5 min. The ELISA plates were then incubated with streptavidin-conjugated horseradish peroxidase (1:1000; BioLegend, San Diego, CA) at room temperature for 1 h, washed three times with PBS, and incubated with 0.4 mg/ml o-phenylenediamine dihydrochloride (Sigma-Aldrich, St. Louis, MO) in 0.1 M NaH2PO4, pH 5.0, containing 0.01% H2O2 at room temperature for 5–10 min. The absorbance was quantified at 492 nm using an ELISA plate reader.
Treatment of HeLa and SK-MEL-28 cells with siRNA
ON-TARGETplus siRNA pools against human Myo1E and nontargeting siRNA #4 (Dharmacon, Lafayette, CO) were diluted to 20 μM with 1× siRNA buffer (Dharmacon), aliquoted, and frozen at −20°C. HeLa or SK-MEL-28 cells were seeded in six-well plates 1 d before transfection. On the day of transfection, cells were ∼30% confluent. Five microliters of siRNA solution (20 μM) was added to 250 μl of OptiMEM (Invitrogen) in tube 1. In tube 2, 10 μl of Lipofectamine RNAiMAX (Invitrogen) was added to 240 μl of OptiMEM. Tubes 1 and 2 were incubated at room temperature for 5 min before the two solutions were combined. The mixture was gently mixed and incubated at room temperature for another 25 min. It was then added to cells grown in 2 ml of DMEM with 10% FBS and incubated for 3 d.
Latrunculin A treatment
Latrunculin A powder (Invitrogen) was dissolved in DMSO at a stock concentration of 1 mM and stored at −80°C. SK-MEL-28 cells on coverslips were treated with 0.05–0.25 μM lat-A or equivalent concentration of DMSO diluted in starvation medium for 10 or 15 min. They were then analyzed by a fluorescent transferrin-uptake assay either in the absence or presence of lat-A. The coverslips were then fixed in 4% PFA at room temperature for 20 min and processed for immunofluorescence.
Transduction with CellLight early endosomes-GFP BacMam baculovirus
Ten thousand cells were grown overnight on coverslips in sic-well dishes. BacMam reagents were added (20 μl) to each of the well on the next day. The cells were incubated with the BacMam reagents overnight at 37°C and then processed for fixation and imaging.
Recycling transferrin assay
SK-MEL-28 cells were grown overnight on coverslips and serum starved at 37°C for 1 h in starvation medium. The cells were then incubated with 10 μg/ml human transferrin conjugated to Alexa Fluor 488 in starvation medium at 37°C for 1 h. The cells were washed with cold starvation buffer and cooled on ice. The cells were then washed with cold acidic buffer (DMEM containing 0.5% acetic acid and 500 mM NaCl) for 40 s to remove surface-bound fluorescent transferrin. The cells were washed five times with cold starvation buffer. They were then treated with either 0.01% DMSO or 0.1 μM lat-A for 15 min. The cells were acid washed again with the acid buffer and fixed with 4% PFA. To assay the recycling of internalized transferrin, the cells were incubated at 37°C for 15 min before the acid wash. The fixed cells were labeled with 6.6 nM rhodamine–phalloidin for 1 h and mounted on glass slide using ProLong Gold antifade reagent with DAPI.
Western blotting and quantitative analysis
For Western blotting, cells treated with siRNAs were washed once with PBS in six-well plates. The cells were dissociated by incubation in 1 ml of 0.5 mM EDTA for 5 min. The dissociated cells were transferred to 1.5-ml tubes and centrifuged briefly at 3000 rpm for 5 min. The supernatant was removed while leaving the cell pellet intact. To lyse the cells, 150 μl of boiling 2× protein sample buffer (125 mM Tris-HCl, pH 6.8, 10% glycerol, 10% SDS, 130 mM dithiothreitol, 0.05% bromophenol blue, 12.5% β-mercaptoethanol) was used to resuspend the pellet quickly by pipeting up and down. The tube was incubated in a 95°C hot block for 5 min. The cell lysates were loaded on SDS–PAGE and analyzed by immunoblotting.
For immunoblotting, protein samples on the gel were transferred to a polyvinylidene fluoride (PVDF) Immobilon-FL transfer membrane (IPFL-00010; Millipore, Billerica, MA) in transfer buffer (25 mM Trizma base, 200 mM glycine, 20% methanol, 0.025% SDS) at 50-V constant voltage in a 4°C cold room for 1 h. After the protein transfer, the PVDF membrane was rinsed briefly in TBS and then incubated in Odyssey blocking buffer (LI-COR Biosciences) diluted at 1:1 in PBS at room temperature for 1 h. All the incubation and washing steps were performed on a rotator. The membrane was incubated overnight at 4°C in primary antibodies, anti-Myo1E (1:200) and anti-Sec23 (1:200), diluted in Odyssey blocking buffer/PBS solution. After three 5-min washes with TBST (Tris-buffered saline with 0.1% Tween-20), the membrane was incubated with IRDye 680 or 800CW antibodies (LI-COR Biosciences) diluted at 1:5000 in Odyssey blocking buffer/PBS solution containing 0.1% Tween-20 at room temperature in the dark for 1 h. After three 5-min washes of TBST, the membrane was incubated in TBS and scanned in a LI-COR Odyssey infrared imaging system (LI-COR Biosciences). The protein expression levels were quantified using the Odyssey application software.
Quantitative transferrin-recycling assay and FACS analysis
Pulse labeling with 20 μg/ml Alexa Fluor 647–conjugated transferrin was performed at 37°C for 1 h in DMEM supplemented with 1% BSA (DMEM-BSA) to allow endocytosis. Cells were then transferred on ice, and surface-bound Alexa Fluor 647–transferrin was removed using ice-cold acid buffer (DMEM, 0.5% acetic acid, and 0.5 M NaCl) for 45 s, and the cells were then neutralized by extensive washes of DMEM-BSA. Next the cells were incubated on ice with DMEM-BSA and 0.1 μM lat-A (or DMSO as a control) for 20 min. To allow the efflux of internal Alexa Fluor 647–transferrin, cells were transferred to 37°C in DMEM-BSA in the presence of 0.1 μM lat–A (or DMSO) and nonfluorescent transferrin. The chase was performed using the following time course: 0, 20, and 40 min. Finally, the cells were transferred to ice, transferrin exposed extracellularly on the plasma membrane was removed using cold acid buffer, and cells were collected using trypsin-EDTA. After cold fixation in 1% PFA, intracellular fluorescence of Alexa Fluor 647–transferrin was quantified by FACS (Beckman-Coulter FC500). The results are expressed as a percentage of the intracellular fluorescence measured at time zero.
To measure the surface expression of transferrin receptor, cells were incubated on ice with DMEM-BSA and 0.1 μM lat-A (or DMSO as a control) for 20 min. Then, Alexa Fluor 647–conjugated transferrin (in the continuous presence of lat-A or DMSO) was provided for 40 min on ice. The cells were then washed extensively with PBS, collected, and fixed with 1% PFA. The fluorescence intensity was measured by FACS.
Supplementary Material
Acknowledgments
We thank Ann Fischer and Michelle Richner for help with cell culture. We thank W. Almers, M. Kessels and B. Qualmann, R. Tsien, M. McNiven, P. Aspenström, M. Way, M. Welch, L. Greene, S. Pfeffer, P. Hanson, and R. Schekman for providing the Swiss 3T3 DsRed-clathrin cell line, Mito-Flag-syndapin II plasmid, pRSET-B dTomato and mCherry plasmids, pEGFP-N1-rat dynamin 2aa plasmid, GFP-utrophin calponin homology–domain plasmid, CB6-GFP-WIP plasmid, emerald GFP-Flag-rat N-WASP plasmid, pEGFP-C1-human clathrin LCa plasmid, and anti-Rab7, anti-CHMP4, and anti-Sec23 antibodies, respectively. This work was supported by a Croucher Foundation Postdoctoral Fellowship to J.C. and National Institutes of Health Grant R01 GM65462 to D.G.D.
Abbreviations used:
- CCPs
clathrin-coated pits
- CLTA
clathrin light chain a
- CME
clathrin-mediated endocytosis
- lat-A
latrunculin A
- Myo1E
myosin 1E
- PRD
proline-rich domain
- SH3
src-homology 3
- TIRFM
total internal reflection fluorescence microscopy
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E11-04-0383) on June 6, 2012.
REFERENCES
- Anderson BL, Boldogh I, Evangelista M, Boone C, Greene LA, Pon LA. The Src homology domain 3 (SH3) of a yeast type I myosin, Myo5p, binds to verprolin and is required for targeting to sites of actin polarization. J Cell Biol. 1998;141:1357–1370. doi: 10.1083/jcb.141.6.1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayscough KR, Stryker J, Pokala N, Sanders M, Crews P, Drubin DG. High rates of actin filament turnover in budding yeast and roles for actin in establishment and maintenance of cell polarity revealed using the actin inhibitor latrunculin-A. J Cell Biol. 1997;137:399–416. doi: 10.1083/jcb.137.2.399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benesch S, Polo S, Lai FP, Anderson KI, Stradal TE, Wehland J, Rottner K. N-WASP deficiency impairs EGF internalization and actin assembly at clathrin-coated pits. J Cell Sci. 2005;118:3103–3115. doi: 10.1242/jcs.02444. [DOI] [PubMed] [Google Scholar]
- Boulant S, Kural C, Zeeh JC, Ubelmann F, Kirchhausen T. Actin dynamics counteract membrane tension during clathrin-mediated endocytosis. Nat Cell Biol. 2011;13:1124–1131. doi: 10.1038/ncb2307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buss F, Arden SD, Lindsay M, Luzio JP, Kendrick-Jones J. Myosin VI isoform localized to clathrin-coated vesicles with a role in clathrin-mediated endocytosis. EMBO J. 2001;20:3676–3684. doi: 10.1093/emboj/20.14.3676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyon JB, et al. Rapid and efficient clathrin-mediated endocytosis revealed in genome-edited mammalian cells. Nat Cell Biol. 2011;13:331–337. doi: 10.1038/ncb2175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engqvist-Goldstein AE, Drubin DG. Actin assembly and endocytosis: from yeast to mammals. Annu Rev Cell Dev Biol. 2003;19:287–332. doi: 10.1146/annurev.cellbio.19.111401.093127. [DOI] [PubMed] [Google Scholar]
- Engqvist-Goldstein AE, Zhang CX, Carreno S, Barroso C, Heuser JE, Drubin DG. RNAi-mediated Hip1R silencing results in stable association between the endocytic machinery and the actin assembly machinery. Mol Biol Cell. 2004;15:1666–1679. doi: 10.1091/mbc.E03-09-0639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evangelista M, Klebl BM, Tong AH, Webb BA, Leeuw T, Leberer E, Whiteway M, Thomas DY, Boone C. A role for myosin-I in actin assembly through interactions with Vrp1p, Bee1p, and the Arp2/3 complex. J Cell Biol. 2000;148:353–362. doi: 10.1083/jcb.148.2.353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson SM, et al. Coordinated actions of actin and BAR proteins upstream of dynamin at endocytic clathrin-coated pits. Dev Cell. 2009;17:811–822. doi: 10.1016/j.devcel.2009.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimoto LM, Roth R, Heuser JE, Schmid SL. Actin assembly plays a variable, but not obligatory role in receptor-mediated endocytosis in mammalian cells. Traffic. 2000;1:161–171. doi: 10.1034/j.1600-0854.2000.010208.x. [DOI] [PubMed] [Google Scholar]
- Geli MI, Lombardi R, Schmelzl B, Riezman H. An intact SH3 domain is required for myosin I-induced actin polymerization. EMBO J. 2000;19:4281–4291. doi: 10.1093/emboj/19.16.4281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geli MI, Riezman H. Role of type I myosins in receptor-mediated endocytosis in yeast. Science. 1996;272:533–535. doi: 10.1126/science.272.5261.533. [DOI] [PubMed] [Google Scholar]
- Gillespie PG, et al. Myosin-I nomenclature. J Cell Biol. 2001;155:703–704. doi: 10.1083/jcb.200110032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottlieb TA, Ivanov IE, Adesnik M, Sabatini DD. Actin microfilaments play a critical role in endocytosis at the apical but not the basolateral surface of polarized epithelial cells. J Cell Biol. 1993;120:695–710. doi: 10.1083/jcb.120.3.695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung G, Remmert K, Wu X, Volosky JM, Hammer JA 3rd. The Dictyostelium CARMIL protein links capping protein and the Arp2/3 complex to type I myosins through their SH3 domains. J Cell Biol. 2001;153:1479–1497. doi: 10.1083/jcb.153.7.1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung G, Wu X, Hammer JA 3rd. Dictyostelium mutants lacking multiple classic myosin I isoforms reveal combinations of shared and distinct functions. J Cell Biol. 1996;133:305–323. doi: 10.1083/jcb.133.2.305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaksonen M, Peng HB, Rauvala H. Association of cortactin with dynamic actin in lamellipodia and on endosomal vesicles. J Cell Sci. 2000;113:4421–4426. doi: 10.1242/jcs.113.24.4421. [DOI] [PubMed] [Google Scholar]
- Kaksonen M, Toret CP, Drubin DG. A modular design for the clathrin- and actin-mediated endocytosis machinery. Cell. 2005;123:305–320. doi: 10.1016/j.cell.2005.09.024. [DOI] [PubMed] [Google Scholar]
- Kessels MM, Qualmann B. Syndapin oligomers interconnect the machineries for endocytic vesicle formation and actin polymerization. J Biol Chem. 2006;281:13285–13299. doi: 10.1074/jbc.M510226200. [DOI] [PubMed] [Google Scholar]
- Kim SV, Mehal WZ, Dong X, Heinrich V, Pypaert M, Mellman I, Dembo M, Mooseker MS, Wu D, Flavell RA. Modulation of cell adhesion and motility in the immune system by Myo1f. Science. 2006;314:136–139. doi: 10.1126/science.1131920. [DOI] [PubMed] [Google Scholar]
- Krendel M, Osterweil EK, Mooseker MS. Myosin 1E interacts with synaptojanin-1 and dynamin and is involved in endocytosis. FEBS Lett. 2007;581:644–650. doi: 10.1016/j.febslet.2007.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamaze C, Fujimoto LM, Yin HL, Schmid SL. The actin cytoskeleton is required for receptor-mediated endocytosis in mammalian cells. J Biol Chem. 1997;272:20332–20335. doi: 10.1074/jbc.272.33.20332. [DOI] [PubMed] [Google Scholar]
- Le Clainche C, Pauly BS, Zhang CX, Engqvist-Goldstein AE, Cunningham K, Drubin DG. A Hip1R-cortactin complex negatively regulates actin assembly associated with endocytosis. EMBO J. 2007;26:1199–1210. doi: 10.1038/sj.emboj.7601576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lechler T, Shevchenko A, Li R. Direct involvement of yeast type I myosins in Cdc42-dependent actin polymerization. J Cell Biol. 2000;148:363–373. doi: 10.1083/jcb.148.2.363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Sun Y, Drubin DG, Oster GF. The mechanochemistry of endocytosis. PLoS Biol. 2009;7:e1000204. doi: 10.1371/journal.pbio.1000204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Sun Y, Oster GF, Drubin DG. Mechanochemical crosstalk during endocytic vesicle formation. Curr Opin Cell Biol. 2010;22:36–43. doi: 10.1016/j.ceb.2009.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McConnell RE, Tyska MJ. Leveraging the membrane–cytoskeleton interface with myosin-1. Trends Cell Biol. 2010;20:418–426. doi: 10.1016/j.tcb.2010.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merrifield CJ. Seeing is believing: imaging actin dynamics at single sites of endocytosis. Trends Cell Biol. 2004;14:352–358. doi: 10.1016/j.tcb.2004.05.008. [DOI] [PubMed] [Google Scholar]
- Merrifield CJ, Feldman ME, Wan L, Almers W. Imaging actin and dynamin recruitment during invagination of single clathrin-coated pits. Nat Cell Biol. 2002;4:691–698. doi: 10.1038/ncb837. [DOI] [PubMed] [Google Scholar]
- Merrifield CJ, Moss SE, Ballestrem C, Imhof BA, Giese G, Wunderlich I, Almers W. Endocytic vesicles move at the tips of actin tails in cultured mast cells. Nat Cell Biol. 1999;1:72–74. doi: 10.1038/9048. [DOI] [PubMed] [Google Scholar]
- Merrifield CJ, Perrais D, Zenisek D. Coupling between clathrin-coated-pit invagination, cortactin recruitment, and membrane scission observed in live cells. Cell. 2005;121:593–606. doi: 10.1016/j.cell.2005.03.015. [DOI] [PubMed] [Google Scholar]
- Merrifield CJ, Qualmann B, Kessels MM, Almers W. Neural Wiskott Aldrich syndrome protein (N-WASP) and the Arp2/3 complex are recruited to sites of clathrin-mediated endocytosis in cultured fibroblasts. Eur J Cell Biol. 2004;83:13–18. doi: 10.1078/0171-9335-00356. [DOI] [PubMed] [Google Scholar]
- Mooseker MS, Cheney RE. Unconventional myosins. Annu Rev Cell Dev Biol. 1995;11:633–675. doi: 10.1146/annurev.cb.11.110195.003221. [DOI] [PubMed] [Google Scholar]
- Okreglak V, Drubin DG. Cofilin recruitment and function during actin-mediated endocytosis dictated by actin nucleotide state. J Cell Biol. 2007;178:1251–1264. doi: 10.1083/jcb.200703092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrais D, Merrifield CJ. Dynamics of endocytic vesicle creation. Dev Cell. 2005;9:581–592. doi: 10.1016/j.devcel.2005.10.002. [DOI] [PubMed] [Google Scholar]
- Pinyol R, Haeckel A, Ritter A, Qualmann B, Kessels MM. Regulation of N-WASP and the Arp2/3 complex by Abp1 controls neuronal morphology. PLoS One. 2007;2:e400. doi: 10.1371/journal.pone.0000400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards TA, Cavalier-Smith T. Myosin domain evolution and the primary divergence of eukaryotes. Nature. 2005;436:1113–1118. doi: 10.1038/nature03949. [DOI] [PubMed] [Google Scholar]
- Saffarian S, Cocucci E, Kirchhausen T. Distinct dynamics of endocytic clathrin-coated pits and coated plaques. PLoS Biol. 2009;7:e1000191. doi: 10.1371/journal.pbio.1000191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen L, Turner JR. Actin depolymerization disrupts tight junctions via caveolae-mediated endocytosis. Mol Biol Cell. 2005;16:3919–3936. doi: 10.1091/mbc.E04-12-1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirotkin V, Beltzner CC, Marchand JB, Pollard TD. Interactions of WASp, myosin-I, and verprolin with Arp2/3 complex during actin patch assembly in fission yeast. J Cell Biol. 2005;170:637–648. doi: 10.1083/jcb.200502053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soulard A, Friant S, Fitterer C, Orange C, Kaneva G, Mirey G, Winsor B. The WASP/Las17p-interacting protein Bzz1p functions with Myo5p in an early stage of endocytosis. Protoplasma. 2005;226:89–101. doi: 10.1007/s00709-005-0108-4. [DOI] [PubMed] [Google Scholar]
- Spitznagel D, O'Rourke JF, Leddy N, Hanrahan O, Nolan DP. Identification and characterization of an unusual class I myosin involved in vesicle traffic in Trypanosoma brucei. PLoS One. 2010;5:e12282. doi: 10.1371/journal.pone.0012282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stimpson HE, Toret CP, Cheng AT, Pauly BS, Drubin DG. Early-arriving Syp1p and Ede1p function in endocytic site placement and formation in budding yeast. Mol Biol Cell. 2009;20:4640–4651. doi: 10.1091/mbc.E09-05-0429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y, Carroll S, Kaksonen M, Toshima JY, Drubin DG. PtdIns(4,5)P2 turnover is required for multiple stages during clathrin- and actin-dependent endocytic internalization. J Cell Biol. 2007;177:355–367. doi: 10.1083/jcb.200611011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y, Martin AC, Drubin DG. Endocytic internalization in budding yeast requires coordinated actin nucleation and myosin motor activity. Dev Cell. 2006;11:33–46. doi: 10.1016/j.devcel.2006.05.008. [DOI] [PubMed] [Google Scholar]
- Taylor MJ, Perrais D, Merrifield CJ. A high precision survey of the molecular dynamics of mammalian clathrin-mediated endocytosis. PLoS Biol. 2011;9:e1000604. doi: 10.1371/journal.pbio.1000604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toret CP, Lee L, Sekiya-Kawasaki M, Drubin DG. Multiple pathways regulate endocytic coat disassembly in Saccharomyces cerevisiae for optimal downstream trafficking. Traffic. 2008;9:848–859. doi: 10.1111/j.1600-0854.2008.00726.x. [DOI] [PubMed] [Google Scholar]
- Toshima J, Toshima JY, Duncan MC, Cope MJ, Sun Y, Martin AC, Anderson S, Yates JR, 3rd, Mizuno K, Drubin DG. Negative regulation of yeast Eps15-like Arp2/3 complex activator, Pan1p, by the Hip1R-related protein, Sla2p, during endocytosis. Mol Biol Cell. 2007;18:658–668. doi: 10.1091/mbc.E06-09-0788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yarar D, Waterman-Storer CM, Schmid SL. A dynamic actin cytoskeleton functions at multiple stages of clathrin-mediated endocytosis. Mol Biol Cell. 2005;16:964–975. doi: 10.1091/mbc.E04-09-0774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu HY, Bement WM. Multiple myosins are required to coordinate actin assembly with coat compression during compensatory endocytosis. Mol Biol Cell. 2007;18:4096–4105. doi: 10.1091/mbc.E06-11-0993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang CX, Engqvist-Goldstein AE, Carreno S, Owen DJ, Smythe E, Drubin DG. Multiple roles for cyclin G-associated kinase in clathrin-mediated sorting events. Traffic. 2005;6:1103–1113. doi: 10.1111/j.1600-0854.2005.00346.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.