Sec16 is an essential factor in “ER exit site” formation, as well as in COPII-mediated traffic in vivo. The results indicate that purified Sec16 alone can self-assemble into homo-oligomeric complexes on a planar lipid membrane and plays an important role in regulating Sar1 GTPase activity at the late steps of COPII vesicle formation.
Abstract
COPII-coated buds are formed at endoplasmic reticulum exit sites (ERES) to mediate ER-to-Golgi transport. Sec16 is an essential factor in ERES formation, as well as in COPII-mediated traffic in vivo. Sec16 interacts with multiple COPII proteins, although the functional significance of these interactions remains unknown. Here we present evidence that full-length Sec16 plays an important role in regulating Sar1 GTPase activity at the late steps of COPII vesicle formation. We show that Sec16 interacts with Sec23 and Sar1 through its C-terminal conserved region and hinders the ability of Sec31 to stimulate Sec23 GAP activity toward Sar1. We also find that purified Sec16 alone can self-assemble into homo-oligomeric complexes on a planar lipid membrane. These features ensure prolonged COPII coat association within a preformed Sec16 cluster, which may lead to the formation of ERES. Our results indicate a mechanistic relationship between COPII coat assembly and ERES formation.
INTRODUCTION
Eukaryotic cells use membrane-bound vesicles or carrier intermediates for protein trafficking between organelles. Transport from the endoplasmic reticulum (ER) to the Golgi is mediated by COPII-coated carriers (Dancourt and Barlowe, 2010; Zanetti et al., 2011). The minimal machinery to drive COPII coat formation involves the small GTPase Sar1, the inner coat complex Sec23/24, and the outer coat complex Sec13/31, and these components sequentially bind to the cytoplasmic surface of the ER membrane. Assembly of the COPII coat on the ER membrane is initiated by GDP–GTP exchange on Sar1 catalyzed by the ER-localized transmembrane guanine nucleotide exchange factor Sec12 (Nakano et al., 1988; Nakano and Muramatsu, 1989; Barlowe and Schekman, 1993). GTP binding induces a conformational change of Sar1 such that it inserts into the ER membrane. Membrane-bound Sar1p-GTP recruits the Sec23/24 complex via its Sec23 portion, and Sec24 captures the cytoplasmically exposed ER export signal of the transmembrane cargo to form a prebudding complex (Bi et al., 2002; Mossessova et al., 2003). Subsequently, the prebudding complex recruits a Sec13/31 complex by interacting with the catalytic motif of Sec23 and the proline-rich domain of Sec31, which then polymerizes the prebudding complexes to form COPII vesicles (Shaywitz et al., 1997; Bi et al., 2007; Tabata et al., 2009). The COPII vesicles destined for the Golgi are at least partially uncoated because TRAPPI complexes on the cis-Golgi membrane tether the vesicles by interacting with Sec23 subunits (Cai et al., 2007). The Sec23 subunit is the GTPase-activating protein (GAP) for Sar1 and therefore stimulates Sar1 GTP hydrolysis upon binding to Sar1, leading to disassembly of the prebudding complex (Yoshihisa et al., 1993; Antonny et al., 2001). However, Sar1-dependent association of Sec23/24 to membranes is stabilized through interactions with transmembrane cargo proteins and repeated cycles of Sec12-dependent GTP loading of Sar1, which facilitate proper and efficient cargo sorting into COPII vesicles (Futai et al., 2004; Sato and Nakano, 2005). In addition, the rate of GTP hydrolysis by Sar1 is further accelerated through the binding of Sec13/31 to the prebudding complex, which enhances the Sec23-mediated GAP activity by an order of magnitude. Such activity has been reported to trigger rapid disassembly of the fully assembled coat in a minimal system (Antonny et al., 2001). However, the significance of this activity remains to be determined. Additional factors are believed to regulate the Sar1 GTPase activity to prevent premature coat disassembly.
COPII vesicles are formed at specific sites of the ER known as ER exit sites (ERES). Sec16 is crucially involved in the organization of ERES because its inactivation causes a loss of ERES in several species (Connerly et al., 2005; Watson et al., 2006; Bhattacharyya and Glick, 2007; Ivan et al., 2008; Castillon et al., 2009). There are independent binding domains for each of the COPII proteins that have been identified in Sec16, the functional significance of which is unknown (Gimeno et al., 1996; Shaywitz et al., 1997). This is because in vitro experiments with purified full-length Sec16 have been much less extensive than those with other COPII coat proteins, and the only biochemical effect observed with full-length Sec16 is its ability to promote efficient assembly of COPII coat proteins on synthetic liposomes (Supek et al., 2002). It has been suggested that Sec16 has no catalytic effect on Sar1 GTPase activity, but it has been implicated to act as a scaffold to stabilize COPII subunits on the ER membrane and to facilitate COPII vesicle formation (Supek et al., 2002).
In this study, we report that full-length Sec16 not only serves as a scaffold for COPII assembly, but it also plays an important role in negatively regulating Sar1 GTPase activity by modulating the interaction with COPII coat components. In addition, we show that purified Sec16 has the ability to self-assemble into homo-oligomeric complexes on the artificial lipid bilayer membrane, which may be the basis of the ERES structure. Our results not only provide potential biochemical and molecular explanations for how Sec16 acts in concert with COPII coat components in the ER, but they also answer a long-standing question about the mechanistic relationship between COPII coat assembly and ER exit site formation.
RESULTS
Localization of the COPII components to the ERES relies on Sec16
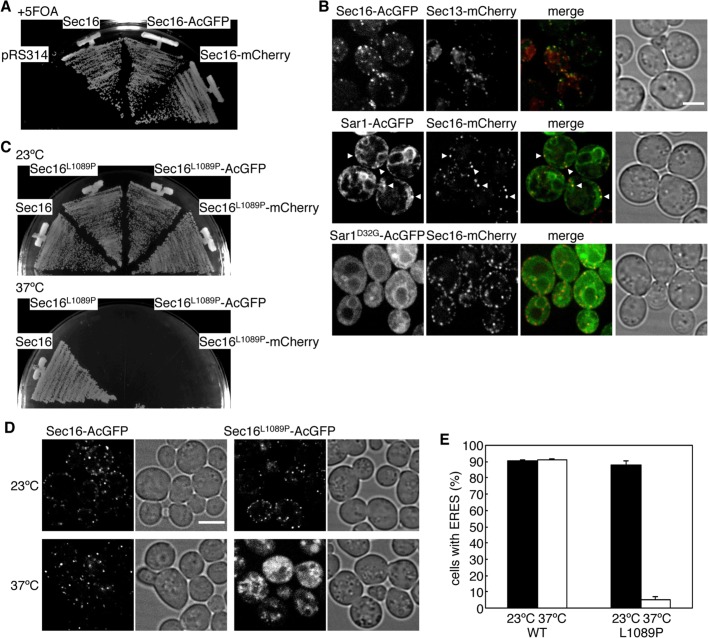
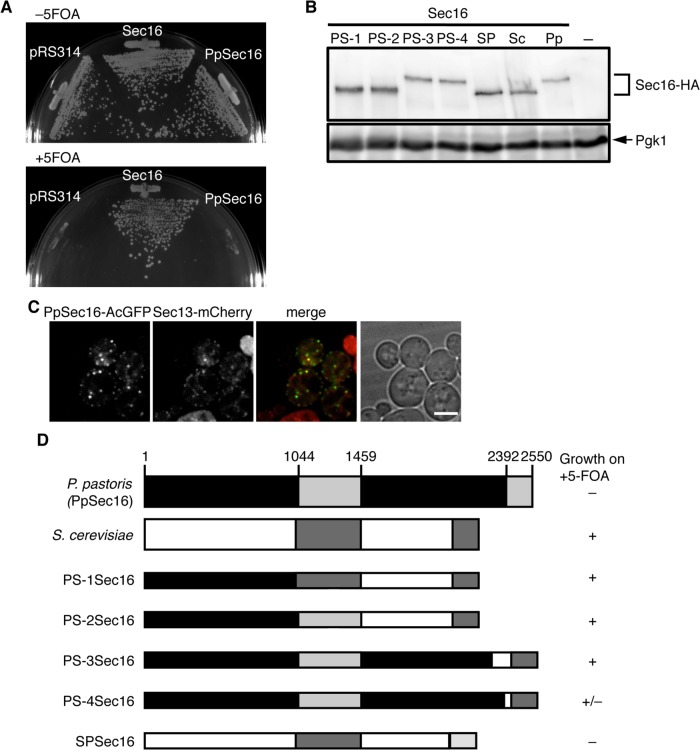
Sec16 localizes as punctate structures at the ERES, specific sites of the ER that are highly conserved among species (Connerly et al., 2005; Bhattacharyya and Glick, 2007; Ivan et al., 2008; Witte et al., 2011). However, the colocalization behavior of Sec16 with COPII components has not been studied extensively in Saccharomyces cerevisiae. To visualize Sec16 in S. cerevisiae cells without interference from the original chromosomal copy of this gene, we fused fluorescent proteins to the C-terminus of Sec16 and expressed it in sec16∆ cells. We found that sec16∆ cells expressing either AcGFP- or mCherry-fused Sec16 (Sec16-AcGFP and Sec16-mCherry, respectively) grew comparably with those expressing untagged Sec16 on plates containing 5-fluoroorotic acid (5-FOA), indicating that fusion Sec16 proteins are functional for growth (Figure 1A). Then we observed cells expressing Sec16-AcGFP or Sec16-mCherry with florescent protein–tagged COPII proteins by confocal florescence microscopy (Figure 1B). As also observed before (Connerly et al., 2005; Castillon et al., 2009; Shindiapina and Barlowe, 2010; Okamoto et al., 2012), Sec13-mCherry appeared as small, multiple dots and colocalized well with Sec16-AcGFP at the ERES. We next examined subcellular localization of Sar1-AcGFP along with Sec16-mCherry. In S. cerevisiae, Sec12 has been reported to localize throughout the entire ER and is not significantly concentrated at the ERES (Soderholm et al., 2004). However, it has not been determined whether Sar1 localizes at the ERES. When coexpressed with Sec16-mCherry, Sar1-AcGFP showed the localization pattern of the whole ER, with some accumulation in punctate structures. We found that the accumulated Sar1-AcGFP colocalized well with Sec16-mCherry, indicating that Sar1-AcGFP is also concentrated at the ERES. To further examine the ERES localization of Sar1, we expressed a GDP-locked form of Sar1D32G-AcGFP and observed it with Sec16-mCherry. Sar1D32G-AcGFP displayed faint ER staining with higher cytosolic signals than the wild type, and no punctate colocalization was observed with Sec16-mCherry. These results suggest that the GTP-form of Sar1 is preferably concentrated at the ERES. Note that both Sec13-mCherry and Sar1-AcGFP complemented the corresponding temperature-sensitive phenotypes (unpublished data).
FIGURE 1:
Sec16 localizes with COPII proteins at the ERES. (A) Fluorescent protein–tagged Sec16 is functional. sec16∆ cells expressing Sec16 from pRS316 (URA3) were transformed with pRS314 or pRS314-borne Sec16, or with C-terminally AcGFP- or mCherry-fused Sec16 (Sec16-AcGFP or Sec16-mCherry, respectively). Transformants were streaked on plates containing 5-FOA and incubated at 30°C. (B) Localization of Sec16 with COPII proteins. sec16∆ cells expressing Sec16-AcGFP with Sec13-mCherry, or Sec16-mCherry with Sar1-AcGFP, Sar1D32G-AcGFP, were grown to mid-log phase and observed by fluorescence microscopy. Arrowheads indicate Sar1-AcGFP–concentrated sites overlapping with Sec16-mCherry. (C) Sec16 L1089P mutant shows the temperature-sensitive phenotype in growth. sec16∆ cells expressing wild-type Sec16 or Sec16 P1089L mutant with or without AcGFP or mCherry fusion were streaked on plates and incubated at 23 and 37°C. (D) Sec16 L1089P mutant fails to localize at the ERES at 37°C. sec16∆ cells expressing Sec16-AcGFP or Sec16L1089P-AcGFP were grown for 2 h at 23 or 37°C and observed by fluorescence microscopy. (E) The percentage of cells containing multiple ERES dots is indicated at 23 and 37°C. More than 100 cells were quantified in three individual experiments by fluorescence microscopy, and the error bars represent the SD. Scale bars, 4 μm.
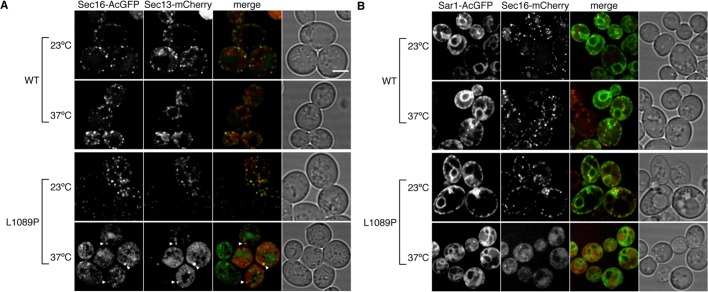
Previous studies showed that localization of COPII proteins is substantially altered in temperature-sensitive sec16-2 mutant after a shift to a nonpermissive temperature (Shindiapina and Barlowe, 2010). Although a point mutation in sec16-2 has been identified at amino acid 1089 with a substitution from leucine to proline (L1089P), the behavior of Sec16 bearing this mutation (Sec16L1089P) under nonpermissive temperatures is not clear. To answer this question, we introduced the L1089P mutation into Sec16-AcGFP or Sec16-mCherry and expressed them in sec16∆ cells. After incubation at a permissive temperature (PT; 23°C) on 5-FOA plates as described in the legend of Figure 1A, sec16∆ cells expressing the indicated Sec16L1089P were obtained, and all were sensitive to incubation at a nonpermissive temperature (NPT; 37°C; Figure 1C). Then we checked the Sec16 localization at the PT and NPT. At the PT, Sec16L1089P-AcGFP localized at the ERES, behaving like wild-type Sec16. After shift to the NPT, Sec16L1089P-AcGFP lost its localization at the ERES, whereas the wild-type control did not show any significant change (Figure 1, D and E). These results indicate that the L1089P mutation causes a loss of ERES localization at the NPT. Next we observed sec16∆ cells coexpressing Sec16L1089P-AcGFP with Sec13-mCherry or Sec16L1089P-mCherry with Sar1-AcGFP (Figure 2). At the PT, Sec16L1089P-AcGFP colocalized with Sec13-mCherry, as did wild-type Sec16-AcGFP. On shifting to the NPT, unlike the wild-type control, Sec13-mCherry completely changed its ERES localization, displaying increased cytosolic signals and intense punctate structures (Figure 2A), as previously reported in the sec16-2 strain (Shindiapina and Barlowe, 2010). Those intense structures colocalized with the aberrant dot structures of Sec16L1089P-AcGFP observed at the NPT. At the PT, Sar1-AcGFP localized throughout the ER, with some accumulation at the ERES colocalizing with Sec16L1089P-mCherry or Sec16-mCherry (Figure 2B). In contrast, Sar1-AcGFP specifically lost the ERES localization in cells coexpressing Sec16L1089P-mCherry at the NPT, although the ER localization was not significantly affected. Sar1-AcGFP was not included in the intense punctate structures as observed with Sec13-mCherry when coexpressed with Sec16L1089P-mCherry at the NPT. These results suggest that correct recruitment of COPII proteins to the ERES relies on the correct localization of Sec16.
FIGURE 2:
Inactivation of Sec16 alters localization of COPII proteins. sec16∆ cells expressing Sec13-mCherry with Sec16-AcGFP or Sec16L1089P-AcGFP (A) or Sar1-AcGFP with Sec16-mCherry or Sec16L1089P-mCherry (B) were grown for 2 h at the PT or the NPT and observed by fluorescence microscopy. Arrowheads indicate abnormal punctate structures of Sec16L1089P-AcGFP and Sec13-mCherry. Scale bars, 4 μm.
Sec16 negatively regulates the acceleration of Sec23 GAP activity by Sec31
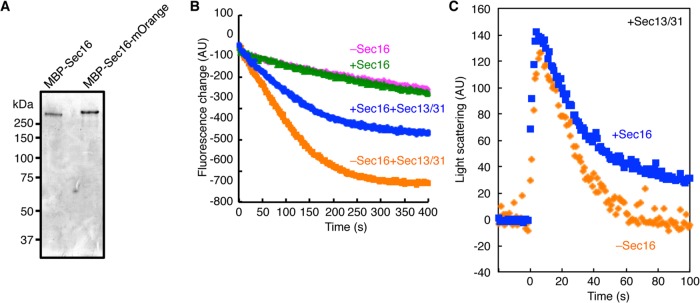
Sec16 has been considered to act mainly as a scaffold for COPII coat assembly at the ERES. This is because the only biochemical activity observed so far with full-length Sec16 is its ability to bind COPII coat components (Supek et al., 2002). To further characterize the biochemical activity of Sec16, we purified full-length Sec16 by using an expression and purification system similar to that reported previously (Supek et al., 2002). We confirmed that maltose-binding protein (MBP)–Sec16 complements sec16∆ cells (Supplemental Figure S1A). When proteins affinity purified by amylose resin from sec16∆ cells carrying the plasmid were subjected to SDS–PAGE, there was a single major band corresponding to MBP-Sec16 (Figure 3A). We next examined Sar1 GTPase activity in the presence of the purified full-length MBP-Sec16. We incubated liposomes preloaded with Sar1-GTP and monitored the decrease in the intrinsic tryptophan fluorescence of Sar1 that accompanies conversion of Sar1-GTP to Sar1-GDP after the addition of Sec23/24 at time 0. No significant difference in the Sec23-mediated GAP activity was observed in the absence or presence of Sec16, as was reported previously (Supek et al., 2002). In contrast, a pronounced difference was detected in the presence of Sec31. The GAP stimulation by Sec31 was markedly diminished in the presence of Sec16, indicating that Sec16 interferes with the Sec31-mediated stimulation of the GAP activity of Sec23 (Figure 3B). These results are in agreement with data reported very recently that indicate that the N-terminally truncated Sec16 mutant displays an inhibitory effect on the ability of Sec31 to stimulate Sec23 GAP activity by preventing the recruitment of Sec31 (Kung et al., 2012). However, we confirmed that the full-length MBP-Sec16 purified here is able to facilitate binding of COPII proteins, including Sec31, on liposomes, as previously reported (Supplemental Figure S1B; Supek et al., 2002). Because COPII coat dissociation from the ER membrane is dependent on GAP-stimulated Sar1 GTP hydrolysis, we next determined whether the GAP inhibition by Sec16 coincides with coat disassembly. To do that, we examined a single round of real-time COPII coat binding and dissociation from liposomes in the presence or absence of Sec16 by light scattering (Figure 3C). When Sec23/24 was added to liposomes preloaded with Sar1-GTP and Sec13/31, an instant increase in light scattering signal was observed, which corresponds to the rapid recruitment of COPII components. Subsequently, the light scattering signal declined (−3.68 ± 0.46 arbitrary units/s) due to disassembly of the COPII coat upon Sar1-GTP hydrolysis. As expected, the presence of Sec16 decelerated the dissociation of the COPII coat (−2.81 ± 0.76 arbitrary units/s). These results provide evidence that Sec16 has a role in regulating Sec31-mediated Sec23 GAP activation, which contributes to the stabilization of the COPII coat.
FIGURE 3:
Sec16 negatively regulates the Sar1 GTPase activation of the COPII protein. (A) MBP-Sec16 and MBP-Sec16-mOrange are purified from yeast cells. Proteins from sec16∆ cells expressing MBP-Sec16 or MBP-Sec16-mOrange were purified by amylose resin and subjected to SDS–PAGE followed by Coomassie brilliant blue staining. (B) Activation of Sar1 GTPase was examined with Sec16. After preincubation of Sar1 (800 nM), GTP (100 μM), and synthetic liposomes (100 μg/ml) with or without MBP-Sec16 (70 nM) in the presence or absence of Sec13/31 (50 nM), Sec23/24 (50 nM) was added at 0 s to start the reaction. The subsequent decrease in tryptophan fluorescence signal was monitored over time at 340 nm. (C) Coat disassembly is monitored by light scattering assay. Light scattering signals represent the state of the coat-assembled/disassembled liposomes. After preincubation of Sar1 (800 nM), GTP (100 μM), and liposomes (100 μg/ml) with Sec13/31 (75 nM), Sec23/24 (75 nM) was injected into the mixture at 0 s. Where indicated, MBP-Sec16 (60 nM) was added in the starting mixture. The subsequent decrease in change of light scattering signal was monitored over time. The data represent the average of three independent experiments.
The regions of Sec16 required for binding with COPII coat proteins
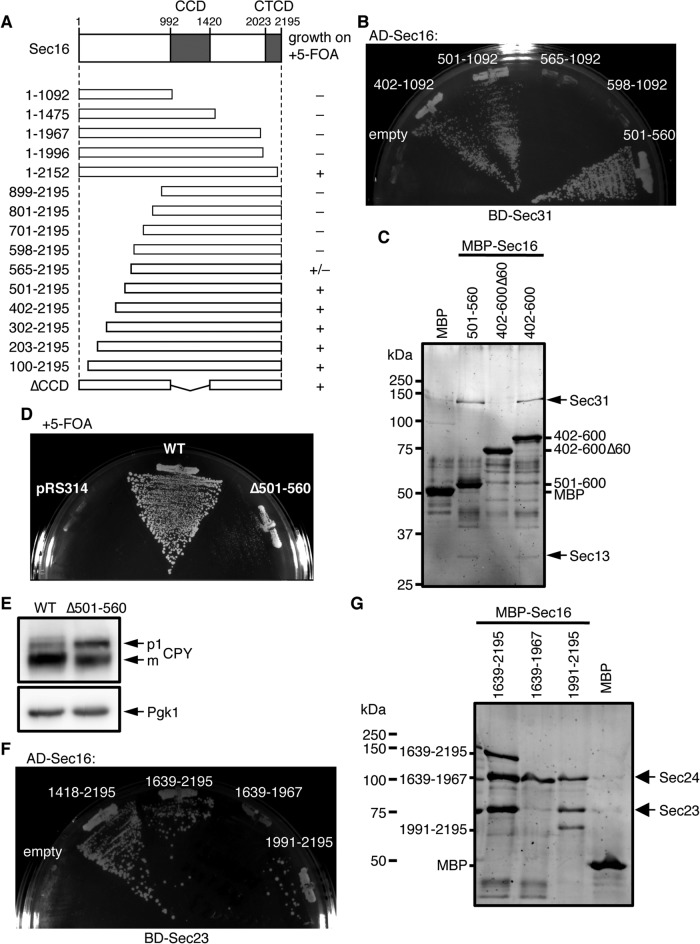
Sec16 has two conserved domains: the central conserved domain (CCD) and the C-terminal conserved domain (CTCD). The L1089P mutation is located in the CCD, which is well conserved among Sec16 homologues in various species. In agreement with an earlier study (Whittle and Schwartz, 2010), Sec16 lacking CCD was still functional in sec16∆ cells based on a growth assay on 5-FOA plates (Figure 4A). This result suggests that the CCD is not absolutely necessary for Sec16 function. Therefore, to analyze the mechanism of GAP activity control, we first mapped the functional regions of Sec16. We made a series of N-terminal- and C-terminal-truncated mutants of Sec16 and performed complementation assays in sec16∆ cells (Figure 4A). Sec161-1996 with truncation of the C-terminal 200 amino acids residues, including the CTCD, no longer showed growth in sec16∆ cells on 5-FOA plates. On the other hand, sec16∆ cells expressing the Sec16501-2195 mutant lacking 500 amino acid residues at the N-terminal region still grew on 5-FOA plates, comparable to cells transformed with the full-length wild type. Previous study showed that when expressed from a 2 μ plasmid, Sec16 lacking the N-terminal 564 amino acids complements a SEC16-null strain (Espenshade et al., 1995). However, the same mutant expressed from a CEN plasmid allowed sec16∆ cells to grow only very slowly, and mutants with longer truncations resulted in a loss of viability. These results suggest that both the upstream region of CCD and the C-terminal region of Sec16 are essential for its function. It should be noted that all the truncated forms of Sec16 examined in this study were expressed in S. cerevisiae cells (Supplemental Figure S2A). The regions in Sec16 that were previously identified as the binding regions for Sec23 and Sec31 were amino acid residues 1639–2195 (corresponding to the C-terminal region) and 447–1043 (corresponding to the upstream region of CCD), respectively (Gimeno et al., 1996; Shaywitz et al., 1997).
FIGURE 4:
Mapping of the binding regions of Sec23 and Sec31 within Sec16. (A) The effect of Sec16 truncations on cell growth. A schematic drawing of Sec16 is shown indicating the location of two conserved domains. The ability of each Sec16 truncation mutant to support the growth of sec16∆ cells was tested on 5-FOA plates as described in the legend of Figure 1A. (B) Mapping of the Sec31-binding region within Sec16 by a yeast two-hybrid assay. The PJ69-4A strain was transformed with plasmids containing the binding domain (BD)–fused Sec31 and the activation domain (AD)–fused Sec16 fragment, and transformants were grown on −histidine plates at 30ºC for 5 d. (C) Sec31 interacts with the 501–560 region of Sec16. MBP and indicated MBP-Sec16 purified from E. coli were immobilized on amylose resin and incubated with Sec13/31. Affinity-isolated proteins were subjected to SDS–PAGE and stained with Sypro Orange. (D) Sec16 lacking the Sec31-binding region does not fully support growth of sec16∆ cells. The ability of Sec16 lacking the 501–560 region to support the growth of sec16∆ cells was tested on 5-FOA plates as described in the legend of Figure 1A. (E) The Sec31-binding region is important for Sec16 function to facilitate ER exit of secretory proteins. Total cell extracts of sec16∆ cells expressing wild-type Sec16 or Sec16∆501-560 were separated by SDS–PAGE followed by immunoblotting with anti-CPY or anti-Pgk1 antiserum. (F) Yeast two-hybrid assay between Sec23 and the C-terminal region of Sec16. The Pj69-4A strain was transformed with plasmids containing BD-fused Sec23 and AD-fused Sec16 fragments, and transformants were grown on −histidine plates at 30ºC for 5 d. (G) Sec23 interacts with the C-terminal region including the CTCD of Sec16. MBP and indicated MBP-Sec16 purified from E. coli were immobilized on amylose resin and incubated with Sec23/24. Affinity-isolated proteins were subjected to SDS–PAGE and stained with Sypro Orange. Sec24 and MBP-Sec161639-1967 have roughly the same apparent mobility on the gel.
Therefore we next asked whether the Sec16 truncation mutants unable to support growth of sec16∆ cells determined here have the ability to interact with COPII components. First, we used yeast two-hybrid analysis to examine the interaction between the fragments of the upstream region of the Sec16 CCD and Sec31 (Figure 4B). Sec16 fragments of amino acid residues 402–1092 and 501–1092 both interacted with Sec31, whereas the fragments of amino acid residues 565–1092 and 598–1092 did not, which agrees well with the result of the growth assay shown in Figure 4A. These were further examined by in vitro binding assays. We incubated purified Sec13/31 with MBP-fused Sec16 fragments purified from Escherichia coli cells (Supplemental Figure S2B), and assessed their interactions by MBP pull-down assays (Figure 4C). In agreement with the results of the yeast two-hybrid interaction analysis, MBP-Sec16402-600 and MBP-Sec16501-560 pulled down similar amounts of Sec31. On the other hand, Sec31 was not precipitated with MBP alone and with MBP-Sec16402-600∆60 in which MBP was fused with the Sec16 fragment of amino acids 402–600 but lacking the 60 amino acids from 501 to 560. We also examined the interaction between MBP-Sec16501-560 and Sec23/24, and we were not able to detect any binding activity between the two proteins (Supplemental Figure S2C). Taking the results together, we conclude that Sec16 has a specific binding site for Sec31 within amino acids 501–560 of Sec16. To test whether this region is important for Sec16 function, we generated sec16∆ cells expressing the Sec16∆501-560 mutant, which lacks the residues 501–560, and incubated them on 5-FOA plates as described earlier (Figure 4D). Sec16∆501-560 allowed sec16∆ cells to grow only very slowly compared with cells transformed with the full-length wild type. To determine whether this growth defect is due to impaired intracellular transport, we monitored transport of the vacuolar protease carboxypeptidase Y (CPY) as a marker for early events in the secretory pathway. As shown in Figure 4E, the ER form of CPY (p1) accumulated within sec16∆ cells expressing Sec16∆501-560, indicating that the deletion of residues 501–560 in Sec16 indeed caused a block in ER-to-Golgi transport. These results show that Sec31 binding to Sec16 is a critical requirement for ER-to-Golgi transport.
Next, since the binding region for Sec23 in Sec16 was shown to include the 557 amino acids from the C-terminal end, we tried to narrow down the Sec23-binding region of Sec16. As shown in Figure 4F, yeast two-hybrid analysis showed that deletion constructs of Sec161418-2195 and Sec161639-2195 interact with Sec23. In addition, the Sec161991-2195 fragment, which includes the CTCD, interacted with Sec23, whereas Sec161639-1967, which lacks the CTCD, did not. To further verify the binding site of Sec16 for Sec23, we carried out in vitro binding assays. The MBP-fused C-terminal fragments of Sec16 expressed and purified from E. coli (Supplemental Figure S2B) were mixed with Sec23/24 and incubated with amylose resin (Figure 4G). MBP-Sec161991-2195, as well as MBP-Sec161639-2195, successfully pulled down the Sec23/24 complex, whereas MBP-Sec161639-1967 failed to interact with the Sec23/24 complex. These results are in good agreement with those obtained from yeast two-hybrid assays (Figure 4F), and we conclude that the interaction with Sec23 occurs at the C-terminal end, including the CTCD of Sec16.
Chimeras between S. cerevisiae Sec16 and Pichia pastoris Sec16 are functional in S. cerevisiae
It has been shown that when expressed in S. cerevisiae sec12∆ cells, Pichia pastoris Sec12 fully rescues its lethality phenotype (Soderholm et al., 2004). We examined whether this is also the case with Sec16. We expressed P. pastoris Sec16 (Ppsec16) from the S. cerevisiae SEC16 promoter in S. cerevisiae sec16∆ cells and performed complementation assays on 5-FOA plates (Figure 5A). Expression of PpSec16 did not impair the growth of the S. cerevisiae sec16∆ cells simultaneously expressing S. cerevisiae Sec16. However, sec16∆ cells with PpSec16 were not able to grow on 5-FOA plates, indicating that PpSec16 is not functionally interchangeable with the S. cerevisiae Sec16. Immunoblotting analysis of hemagglutinin (HA)-tagged versions of the PpSec16 revealed that they can indeed be expressed in S. cerevisiae cells (Figure 5B). It has been shown that organization of the ERES in P. pastoris observed by fluorescence microscopy is different from that of S. cerevisiae; ERES of PpSec16 appear as discrete puncta, whereas the ERES of S. cerevisiae appear as small spots (Connerly et al., 2005). Therefore, to examine whether the nonfunctionality of PpSec16 in S. cerevisiae cells is related to the difference in the spatial organization of the ERES, we fused PpSec16 with AcGFP (PpSec16-AcGFP) at the C-terminal end as described previously (Connerly et al., 2005) and observed PpSec16-AcGFP in S. cerevisiae cells by fluorescence microscopy (Figure 5C). When expressed in cells with a normal chromosomal copy of SEC16, PpSec16-AcGFP colocalized well with Sec13-mCherry at the ERES. These results suggest that PpSec16 is able to localize at the ERES of S. cerevisiae but is not fully functional. Next we tried to identify which part of PpSec16 is functionally replaceable with the corresponding region of the S. cerevisiae Sec16. We made a series of chimeric Sec16 by exchanging C-terminal segments from PpSec16 with the corresponding segments from the S. cerevisiae Sec16 (Figure 5D). When expressed in S. cerevisiae sec16∆ cells, chimeras of PS-1, PS-2, and PS-3 allowed the host cells to grow on 5-FOA plates at levels comparable to sec16∆ cells transformed with the wild-type S. cerevisiae Sec16, and PS-4 also showed partial complementation of the sec16∆ growth defect, which all involve the CTCD of the S. cerevisiae Sec16. In contrast, a chimera of SPSec16, which consists largely of the S. cerevisiae Sec16 domain followed by the C-terminal segment of PpSec16 including its CTCD, did not rescue the lethal sec16∆ phenotype. We confirmed that when tagged with the HA epitope, all of the chimeric Sec16 examined here were expressed in S. cerevisiae cells (Figure 5B). These results suggest that CTCD of Sec16 plays a key role in Sec16 function, which is in reasonable agreement with the results shown in Figure 4.
FIGURE 5:
The CTCD is critical for Sec16 function. (A) The ability of PpSec16 to support growth of sec16∆ cells was tested on 5-FOA plates as described in the legend of Figure 1A. (B) Total cell extracts of S. cerevisiae cells (SEY6210) expressing HA-tagged S. cerevisiae (Sc), P. pastoris (Pp), or chimeric Sec16 indicated were separated by SDS–PAGE followed by immunoblotting with anti-HA or anti-Pgk1 antiserum. Chimeras examined are depicted in D. (C) PpSec16 localizes at the ERES in S. cerevisiae cells. S. cerevisiae cells (SEY6210) expressing PpSec16-AcGFP with Sec13-mCherry were grown to mid-log phase and examined by fluorescence microscopy. Scale bar, 4 μm. (D) Chimeras between S. cerevisiae (ScSec16) and P. pastoris (PpSec16) Sec16. A schematic diagram shows chimeric forms of Sec16. The sequence derived from PpSec16 is depicted as black and light gray bars, and the sequence from ScSec16 is shown as white and dark gray bars. Gray bars display the CDC and CTCD. PS-1Sec16; PpSec161-1010 and ScSec16960-2195, PS-2Sec16; PpSec161-1461 and ScSec161423-2195, PS-3Sec16; PpSec161-2266 and ScSec161895-2195, PS-4Sec16; PpSec161-2361 and ScSec161991-2195, SPSec16; and ScSec161-1996 and PpSec162369-2550. The ability of each Sec16 chimera to support the growth of sec16∆ cells was tested on 5-FOA plates as described in the legend of Figure 1A.
The C-terminal region of Sec16 modulates the assembly of COPII proteins
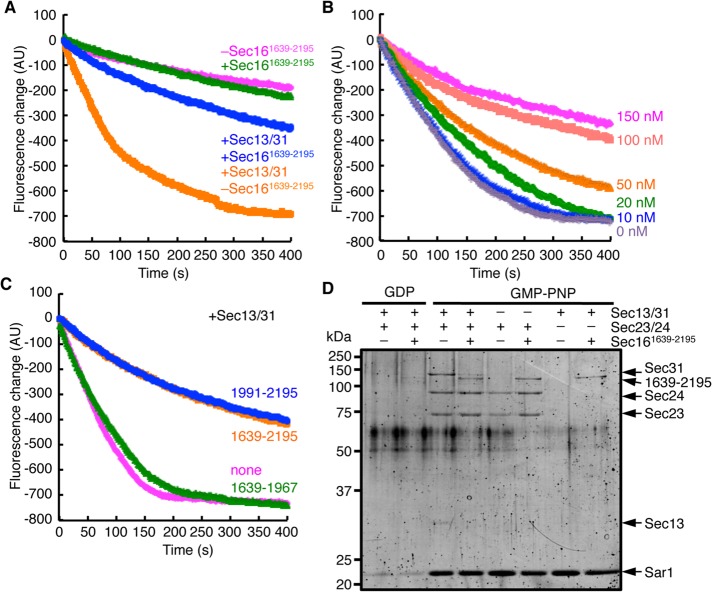
Having established that the C-terminal region of Sec16 is important for its function, we considered it essential to obtain a better understanding of the functional significance of the interaction between Sec16 and Sec23. We examined the ability of the C-terminal fragment of MBP-Sec161639-2195, which includes the Sec23-binding core region, to influence the Sar1 GTPase activity, using a tryptophan-based fluorescence assay (Figure 6A). Consistent with the results of full-length Sec16 in Figure 3B, the presence of MBP-Sec161639-2195 did not cause a significant difference in the GAP activity. In addition, in the presence of MBP-Sec161639-2195, acceleration of the Sec23 GAP activity by Sec31 was significantly reduced (Figure 6, A and B). Sec31 also failed to stimulate the GAP activity for Sar1 in the presence of MBP-Sec161991-2195, whereas MBP-Sec161639-1967 did show GAP activation by Sec31 to the same extent as the control (Figure 6C). These results led us to examine whether Sec31 is properly recruited onto the Sar1/Sec23/24 complex in the presence of the C-terminal fragment of Sec16. To test this, we carried out an in vitro flotation assay with liposomes. Purified Sar1, Sec23/24, and Sec13/31 were incubated with liposomes and GDP or the nonhydrolyzable GTP analogue GMP-PNP in the presence or absence of MBP-Sec161639-2195. The liposomes were then floated on a sucrose density gradient, and liposome-bound proteins recovered in the top fraction were analyzed as described previously (Supek et al., 2002). COPII proteins were allowed to bind to liposomes in a GMP-PNP–dependent manner in the absence of MBP-Sec161639-2195. However, in reactions with MBP-Sec161639-2195, which migrated immediately below the Sec31 band, there was a marked decrease in the amount of Sec31 bound to liposomes (Figure 6D). Of interest, MBP-Sec161639-2195 was also detected in the floating fraction in conditions with Sar1-GMP-PNP regardless of the presence or absence of Sec23/24. These results demonstrate that the C-terminal region of Sec16 including the CTCD binds the Sar1-GTP/Sec23 complex and sterically prevents Sar1/Sec23 association with Sec31, which limits Sec31-stimulated Sec23 GAP activity.
FIGURE 6:
The CTCD alters the interaction between Sec23 and Sec31. (A) The GTPase activity of Sar1 was examined with MBP-Sec161639-2195 (150 nM) as in Figure 3B. Where indicated, Sec13/31 was added in the starting mixture. (B) Sar1 GTPase activity was monitored with the indicated concentrations of MBP-Sec161639-2195 in the presence of Sec13/31 as in A. (C) Sar1 GTPase activity was monitored with indicated MBP-Sec16 in the presence of Sec13/31 as in A. (D) Liposome-binding assay of COPII proteins with MBP-Sec161639-2195. Sar1 (800 nM), Sec23/24 (100 nM), Sec13/31 (150 nM), and MBP-Sec161639-2195 (300 nM) were incubated with GDP or GMP-PNP (100 μM) and synthetic liposomes (100 μg/ml) subjected to flotation on a sucrose density gradient. Float fractions were subjected to SDS–PAGE and stained with Sypro Orange.
Sec16 forms a homo-oligomer via autoassembly
Sec16 is peripherally bound to the ER membrane and included in the ERES. Although we showed that the C-terminal fragment of Sec16 can be recruited to membranes via membrane-bound Sar1, Sar1 recruitment to membranes is not a critical determinant of the membrane localization of full-length Sec16, as shown with the Sec16L1089P mutant (Figure 2B). We next investigated how Sec16 localizes to particular patches of the ER membrane. Several lines of evidence, including experiments using truncated versions of Sec16, suggest that Sec16 forms at least a homodimer (Bhattacharyya and Glick, 2007; Ivan et al., 2008). To test whether full-length Sec16 is associated with itself in S. cerevisiae, we expressed MBP-Sec16 and Sec16-AcGFP simultaneously in yeast cells and carried out MBP pull-down by amylose resin. As shown in Figure 7A, Sec16-AcGFP was coisolated with MBP-Sec16, whereas no Sec16-AcGFP binding was detectable in the absence of MBP-Sec16. These results indicate that Sec16 is associated with itself.
FIGURE 7:
Sec16 self-assembles on the membrane. (A) Sec16 self-assembles. Salt extracts of sec16∆ cells simultaneously expressing MBP-Sec16 and Sec16-AcGFP were incubated with amylose resin. Total lysates and eluted proteins were subjected to SDS–PAGE followed by immunoblotting with anti-MBP or anti-GFP antibody. (B) Imaging of MBP-Sec16-mOrange molecules on a planar lipid bilayer. Right, the system for observing the horizontal planar lipid membrane under a microscope. An artificial planar lipid bilayer was formed across the hole on the bottom of the upper chamber and observed under evanescent field illumination before and after injection of MBP-Sec16-mOrange (200 pM). Scale bar, 10 μm. (C) Distribution of the fluorescence intensity of MBP-Sec16-mOrange on the membrane. A histogram of the fluorescence intensity was fitted to a single-Gaussian distribution (gray curve). (D) Intensity profile showing multiple-step photobleaching. Two representative examples are shown in which four bleaching steps are clearly discernible.
To examine the molecular assembly of Sec16 in aqueous solution, size exclusion chromatography is difficult to apply to Sec16, because an unphysiologically high salt concentration is required to keep the purified MBP-Sec16 soluble and prevent nonspecific aggregation. Thus, to analyze the native molecular forms of membrane-associated Sec16, we used a quantitative imaging approach. As we reported previously, a horizontal planar lipid bilayer membrane competent for COPII vesicle budding can be formed across a small hole in a thin plastic film (Tabata et al., 2009). We purified fluorescently labeled Sec16 (MBP-Sec16-mOrange) from sec16∆ cells (Figure 3A), which was confirmed to complement sec16∆ cells (Supplemental Figure S1A). We added Sec16-mOrange onto the planar membrane and visualized the behavior of membrane-adsorbed Sec16-mOrange using an objective-type total internal reflection fluorescence microscope designed for single-molecule detection (Figure 7B). When Sec16-mOrange on the membrane was monitored with evanescent field illumination, dispersed fluorescent spots were observed. The distribution of the fluorescence intensities of these spots fitted well with a single-Gaussian distribution (Figure 7C), suggesting that these small spots consisted of uniformly self-assembled Sec16-mOrange instead of random aggregation of the monomers. To determine the number of Sec16 molecules contained in each self-assembled complex, we investigated the photobleaching characteristics of individual spots. The fluorescence of Sec16-mOrange disappeared in multiple steps, suggesting that the membrane-bound Sec16-mOrange existed as a multimer. Two examples of the photobleaching characteristics are shown in Figure 7D. The majority of the spots displayed between four and six bleaching steps. It is difficult to determine the exact number of Sec16 contained in each spot because the fluorescent protein moieties (mOrange) of fusion proteins are not always fluorescently active, due to failure of chromophore formation. Taken together, these data suggest that Sec16 alone can self-assemble into multimeric units on the membrane.
DISCUSSION
The GTPase activity of Sar1 is incrementally stimulated in two steps: first by the association with Sec23 and then by the binding of Sec31 to the Sec23/Sar1 complex, resulting in further enhancement of the GAP activity of Sec23. The Sec23 GAP activity is involved in the fidelity of cargo sorting and concentration into COPII vesicles (Sato and Nakano, 2005), whereas the significance of the increase in GAP activity occurring on Sec31 binding is not readily apparent. The acceleration of the GAP activity by Sec31 has been reported to drive rapid disassembly of the fully assembled coat in a minimal system (Antonny et al., 2001). Apparently, additional layers of regulation must be present to avoid self-destruction of the outer COPII cage during vesicle formation. Here we showed that Sec16 interferes with Sec31-mediated Sec23 GAP activation but does not directly affect the GAP activity of Sec23 itself (Figure 3B). This effect leads to a stabilization of the COPII coat complex (Figure 3C), which could allow for a sufficient amount of time for completion of the coat assembly. This conclusion is highly consistent with the previous observation that, although COPII vesicle formation is driven by either GTP or GMP-PNP, the stimulation of budding efficiency by Sec16 is dependent only on GTP (Supek et al., 2002). In addition, we demonstrated that Sec16 alone has the ability to uniformly self-assemble on the membrane surface (Figure 7C), which may serve as the basis of the ERES structure. We propose that these features contribute to spatially restrict the vesicle budding to the ERES, whereby prolonged association of COPII coat within a preformed Sec16 cluster permits the formation of specific subdomains in the ER that is highly active for COPII vesicle formation. In other words, the significance of GAP stimulation by Sec13/31 may be to prevent COPII coat assembly at any sites of the ER other than the ER exit sites.
The mechanism for recruitment of Sec16 to the ER membrane remains to be elucidated. Direct interaction with charged phospholipids is required for Sec16 to bind to the membrane because Sec16 does not bind to liposomes composed of only neutral phospholipids (Supek et al., 2002). In higher eukaryotes, the mechanism for assembly of Sec16 to the ERES involves TFG-1 (TRK-fused gene) as a direct interactor of Sec16 (Witte et al., 2011). However, no TFG-1 homologue is evident in yeast, suggesting that it does not represent the core COPII machinery. Mapping of the ERES targeting domain of Sec16 in mammals and Drosophila identified the N-terminal nonconserved region enriched in charged residues and the CCD (Bhattacharyya and Glick, 2007; Ivan et al., 2008; Hughes et al., 2009). The L1089P mutation in the S. cerevisiae sec16-2 mutant, which we showed here abolishes ERES localization (Figure 1D), is located in the CCD. In P. pastoris, the temperature-sensitive mutation disrupting the ERES has also been identified in the CCD of PpSec16 (Connerly et al., 2005). Therefore this domain is likely to somehow play a critical role in membrane targeting of Sec16. The CCD of Sec16 is found to contain a conserved fold, the ancestral coatomer element 1 (ACE1), and to form a complex with Sec13 in a way similar to the Sec13–Sec31 interaction (Whittle and Schwartz, 2010). It remains unclear what the functional significance of Sec13 association with Sec16 would be, because the CCD is dispensable for Sec16 function in yeast. One possible explanation for this is that since we detected direct association between the C-terminal domain of Sec16 and Sec23 (Figure 4, F and G) and Sec31 binds to the upstream portion of the CCD (Figure 4, B and C), it is possible that Sec23/24 and Sec13/31 act as linkers for connecting the adjacent Sec16 even in the absence of CCD. In fact, we found that Sec16∆CCD has the ability to localize to the ERES (Supplemental Figure S3, A and B). Our finding uncovers a direct contact between Sar1 and the C-terminal region of Sec16. We showed here that membrane-bound Sar1 is sufficient to recruit the C-terminal fragment of Sec16 to liposomal membranes (Figure 6D). However, we also observed that the GDP-locked mutant Sar1D32G failed to localize at the ERES (Figure 1B), and wild-type Sar1 lost its ERES localization upon inactivation of Sec16L1089P at the NPT (Figure 2B). These results suggest that successful assembly of Sec16 on the ER membrane is a prerequisite for Sar1 recruitment to the ERES. This idea is supported by the previous observation that Drosophila Sec16 interacts only with the GTP-locked form of Sar1 and not with the GDP-locked form (Ivan et al., 2008).
After binding to the membranes, Sec16 should act as a scaffold to recruit COPII coat proteins. It has been shown that there are distinct binding sites on Sec16 for each COPII subunit (Gimeno et al., 1996; Shaywitz et al., 1997). Our study provides significant additional details on COPII-binding sites: residues 501–560 for Sec31 and the C-terminal region including the CTCD for Sec23 and Sar1. Our results demonstrate that the binding region (residues 501–560) for Sec31 is critical for Sec16 function (Figure 4, D and E). It has been shown that Sec16 binds to the C-terminal α-solenoid domain of Sec31 but not to the catalytically important proline-rich domain (Shaywitz et al., 1997), suggesting that Sec31 has distinct binding sites for the Sar1/Sec23 complex and Sec16. Our yeast two-hybrid analysis showed that the Sec31A1239V mutant is defective in its interaction with MBP-Sec16501-560 (Supplemental Figure S4). The A1239V mutation corresponds to a mutation in the temperature-sensitive sec31-1 allele, which is located in the C-terminal α-solenoid domain (Salama et al., 1997). This result further supports the idea that the Sec31 interaction with Sec16 and Sar1/Sec23 occurs in separate domains. In contrast to Sec16∆501-560, Sec16 mutants lacking the C-terminal region including the CTCD did not support sec16∆ growth on 5-FOA plates at all (Figure 4A). In addition, the results from the S. cerevisiae–P. pastoris chimera of Sec16 also strengthen the functional importance of the C-terminal region of Sec16 (Figure 4C). The data also indicate that the ERES in P. pastoris are functionally equivalent to those marked by Sec16 in S. cerevisiae, although the organization of the ERES differs between these species.
It is not clear whether Sec16 is included in budded COPII vesicles. Our findings, as well as those from prior studies, indicate that Sec16 associates with multiple COPII subunits and thereby could be part of the COPII coat structure. Indeed, a fraction of Sec16 has been shown to be present in in vitro–generated COPII vesicles in S. cerevisiae (Espenshade et al., 1995), and Sec16 in Drosophila was reported to localize to a tubulovesicular compartment distinct from the ER (Ivan et al., 2008). However, Sec16 seems to be present at a significantly lower stoichiometry relative to other COPII components (Connerly et al., 2005). Moreover, fluorescence recovery after photobleaching analysis showed that Sec16 has a longer half-life on the ER membrane than other COPII components with a substantial immobile fraction (Hughes et al., 2009). Our data showed that the association of the C-terminal Sec16 domain with the Sec23/Sar1-GTP complex hinders the Sec31–Sec23 interaction (Figure 6D). The proline-rich region of Sec31 is involved in the interaction with the Sar1-GTP/Sec23 complex (Bi et al., 2007), and this region seems to be the only anchorage point where Sec13/31 can link to the prebudding complex for polymerization. Hence the presence of Sec16 could prevent prebudding complex polymerization. It is therefore possible that Sec16 is not a stoichiometric subunit of the COPII coat on the vesicles but instead may mainly act on the ER membrane to initiate COPII coat assembly.
While our manuscript was in preparation, Kung et al. (2012) reported results similar to those we present here. They showed that purified Sec16 fragment lacking the N-terminal 564 amino acid residues robustly inhibits the GAP activation by Sec31 without affecting the GAP activity itself, which was partially abrogated by Sec24 mutant, which lacks the ability to interact with Sec16. Although the detailed mechanism of how the Sec16 fragment exerts such an effect remained unclear, the inhibitory effect was found to be due to prevention of Sec31 recruitment to the Sec23/Sar1 complex on membranes. According to our results, it is now clear that the Sec16 fragment used in their study lacks the Sec31-binding domain (i.e., residues 501–560 in Sec16) and thus only partially complements sec16∆ cells (Figure 4). In addition, we showed that the CTCD of Sec16 is involved in the prevention of Sec31 binding to the Sec23/Sar1 complex (Figure 6). However, there is a critical difference between their study and ours regarding the activity of full-length Sec16. They showed that full-length Sec16 has little or no activity to regulate the Sar1 GTPase activation of the COPII coat proteins (Kung et al., 2012). One possible reason for the contradiction between their results and ours might be the different expression system and procedures for preparation of Sec16: they incubated yeast lysates with nickel-nitriloacetic acid overnight at 4°C to isolate histidine-tagged Sec16, whereas we isolated it by incubation with amylose resin for 1.5 h at 4°C. These differences might influence the full-length Sec16 activity. Coupled with the results on the N-terminally truncated fragment and full-length Sec16, they proposed that the N-terminal domain of Sec16 acts as an autoinhibitory domain to counteract prevention of the Sec31 assembly into COPII complex on the membrane (Kung et al., 2012). Accordingly, our purified full-length Sec16 might have the conformation released from its autoinhibitory status and thereby serve as inhibitor of GAP stimulation. Alternatively, one might expect that MBP fusion to the N-terminus of Sec16 could sterically interfere with an autoinhibitory function of this N-terminal region. However, this is clearly ruled out because MBP-fused Sec16 supported the growth of sec16∆ cells as well as the wild-type Sec16, whereas the corresponding truncated mutant (Sec16565-2195) showed only partial growth (Figure 4A and Supplemental S1). In addition, unlike their N-terminally truncated mutant, MBP-Sec16 is indeed capable of facilitating recruitment of COPII coat, including Sec31, to liposomal membranes, as reported earlier by the same group (Supek et al., 2002). There is no direct evidence that Sec16 has different conformational states. As reported with mammalian Sec16 (Farhan et al., 2010; Zacharogianni et al., 2011), protein modification such as phosphorylation could induce the conformational change of Sec16, although this remains to be studied further.
In summary, the present study sheds light on both the structural and the regulatory functions of Sec16. Our data explain at least in part the mechanism for how Sec16 functions in relation to the COPII coat proteins in the ER. Further studies should be directed at understanding how COPII subunit assembly correlates with ERES formation.
MATERIALS AND METHODS
Strains and media
The strains used here are listed in Supplemental Table S1. Yeast strains were grown or incubated in synthetic medium (0.67% yeast nitrogen base and 2% glucose, with auxotrophic supplements). Gene deletions were performed by a PCR-based procedure. For SEC16 deletion, SEY6210 and CB023 strains were transformed with URA3-based plasmid expressing SEC16 (pTYY4), and then the entire coding region of SEC16 was replaced with E. coli kanr by a PCR-based procedure (Gueldener et al., 2002). PCR verified chromosomal SEC16 knockout. If necessary, 5-FOA was used at a final concentration of 0.1%.
Plasmids
To construct pTYY1, 620 base pairs of the upstream region of SEC16 were amplified from the S. cerevisiae genome by PCR and cloned into the SacI and BamHI sites of pRS316. The plasmid pTYY4 was generated by ligating a PCR-amplified SEC16 gene and 640 base pairs of its downstream region into the BamHI and SalI sites of pTYY1. To generate pTYY41, SEC16 genes including upstream and downstream regions were cloned into the SacI and XhoI sites of pRS314, and the BamHI and XhoI restriction sites were introduced just before the stop codon of each gene. For fusion of fluorescent protein at the C-terminus, the DNA fragments of two tandemly fused AcGFP or mCherry were inserted into the BamHI and XhoI sites of pTYY41 to create plasmids pTYY42 (Sec16-AcGFP) and pTYY43 (Sec16-mCherry). The Cu2+-inducible plasmid to express N-terminally MBP-fused Sec16 was generated as described previously (Supek et al., 2002). To generate pTYY44, the PCR-amplified CUP1 promoter was cloned into the SacI and BamHI sites of pRS314, and then the SEC16 gene including its downstream region was ligated into the BamHI and SalI sites. To construct pTYY45 for fusion of MBP at the N-terminus of Sec16, the PCR-amplified DNA fragment of MBP was introduced into the BamHI site of pTYY44. For yeast two-hybrid analysis, SEC31, SEC23, and truncations of SEC16 genes were amplified by PCR from yeast genomic DNA. SEC13 and SAR1 genes flanked with upstream and downstream regions were amplified by PCR from S. cerevisiae genomic DNA and cloned into the SacI and XhoI sites of pRS316. PpSEC16 was amplified by PCR from P. pastoris genomic DNA and cloned into the SmaI and XhoI sites of a plasmid carrying the S. cerevisiae SEC16 promoter and terminator on pRS314. For fusion of fluorescent protein at the C-terminus, BamHI (for SEC13), BglII (for SAR1), or NheI/XhoI (for PpSEC16) restriction sites were introduced just before the stop codon of each gene. To generate plasmids for yeast two-hybrid analysis, SEC16 truncations and SEC23 were cloned into the BamHI and SalI sites of pGAD-C1 and pGBDU-C1, respectively, and SEC31 was ligated into pGBDU-C1 by using BamHI and PstI sites (James et al., 1996). PCR-based site-directed mutagenesis was carried out to generate mutations of L1089P in SEC16, D32G in SAR1, and A1239V in SEC31. For purification of Sec16 proteins from E. coli, PCR-amplified DNA fragments of each SEC16 truncate were cloned into the BamHI and SalI sites of pMALc2x (New England BioLabs, Ipswich, MA).
Fluorescence microscopy
Yeast cells expressing fluorescent protein–fused proteins were grown to mid-log phase. Fluorescence microscopy observation was carried out using an Olympus IX71 microscope (Olympus, Tokyo, Japan) equipped with a CSU10 spinning-disk confocal scanner (Yokogawa Electric Corporation, Tokyo, Japan) and an electron-multiplying charge-coupled device camera (iXon, DV897; Andor Technology, South Windsor, CT). In this setting, a 473-nm solid-state laser (J050BS; Showa Optronics, Tokyo, Japan) was used to excite AcGFP and mCherry at 561 nm (Jive; Cobolt, Solna, Sweden).
MBP-Sec16-mOrange was viewed on a planar synthetic bilayer as described previously. Synthetic lipid bilayers composed of a major–minor mix dissolved in n-decane were formed horizontally at the bottom of the upper chamber in buffer (20 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid [HEPES]–KOH, pH 6.8, 160 mM KoAc, 1 mM CaCl2). Purified MBP-Sec16-mOrange was added from the top of the upper chamber. After 10 min of incubation, excess proteins were removed by a glass pipette, and then MBP-Sec16-mOrange bound to the lipid bilayer was observed with an objective-type total internal reflection fluorescence microscope that was constructed on an inverted microscope (IX71; Olympus). An oil immersion objective lens (PlanApo, 100×/1.45 numerical aperture; Olympus) was located just below the lower chamber. Bilayers were illuminated by an evanescent wave with a 532-nm solid-state laser (Compass 215M-75; Coherent, Santa Clara, CA) as described before (Tabata et al., 2009). The acquired images were analyzed by Andor iQ (Andor Technology).
Protein preparation
Sar1, Sec23/24, and Sec13/31 were purified as described previously (Barlowe et al., 1994; Salama et al., 1997). Full-length Sec16 was purified from yeast basically as described previously (Supek et al., 2002). YTY020 cells were grown at 30°C. At an OD600 of 0.5, CuSO4 was added to a final concentration of 0.4 mM. After 12 h of induction, cells were centrifuged, washed with water, and then resuspended in suspension buffer (20 mM HEPES-KOH, pH 7.4, 1 M KoAc, 5 mM EDTA) containing 5× protease inhibitor cocktail (PIC; Roche, Indianapolis, IN). The cell suspension was frozen as drops by pouring into liquid nitrogen and stored at −80°C. Sixty-four liters of culture typically yielded up to 150 g of cells. The frozen cells were blended and thawed on ice. After dilution with B-1 buffer (20 mM HEPES-KOH, pH 7.4, 1 mM EDTA, 1 mM dithiothreitol [DTT]) containing 1× PIC, KoAc was added into lysate to a final concentration of 500 mM, and the mixture was incubated for 15 min on ice. From the supernatant by 15 min centrifugation at 10,000 × g, the stripped peripheral proteins were recovered by ultracentrifugation at 100,000 × g for 1 h. The supernatant was incubated with amylose resin (New England BioLabs) for 1.5 h at 4°C. The resin was washed with B-2 buffer (20 mM HEPES-KOH, pH 7.4, 500 mM KoAc, 1 mM EDTA, 1 mM DTT) and then with B-3 buffer (20 mM HEPES-KOH, pH 7.4, 500 mM KoAc, 10% glycerol). MBP-Sec16 was eluted with B-4 buffer (20 mM HEPES-KOH, pH 6.8, 450 mM KoAc, 10 mM maltose, 10% glycerol). The fragments of Sec16 were purified from E. coli. The E. coli cells were incubated in LB medium at 30°C. At an OD of around 0.5, isopropyl-β-d-thiogalactoside was added into the culture to a final concentration of 1 mM. After further 4-h incubation, cells were collected, washed with buffer (20 mM HEPES, pH 7.4, 160 mM KoAc), and then resuspended in sonication buffer (20 mM HEPES, pH 7.4, 160 mM KoAc, 5 mM EDTA) containing 2× PIC. Cells were broken by sonication. After unbroken cells were removed by centrifugation at 10,000 × g for 10 min, the supernatant was incubated with amylose resin for 1.5 h at 4°C. The resin was washed with wash buffer (20 mM HEPES, pH 7.4, 160 mM KoAc, 1 mM EDTA), and MBP-Sec16 fragments were eluted with elution buffer (20 mM HEPES, pH 6.8, 160 mM KoAc, 10 mM maltose).
In vitro binding assay
MBP and MBP-Sec16 fragments (100 nM) were immobilized on amylose resin in binding buffer (20 mM HEPES-KOH, pH 6.8, 160 mM KoAc) and incubated with Sec23/24 (100 nM) or Sec13/31 (100 nM) for 1.5 h at 4°C. Beads were collected and washed twice with binding buffer. Proteins were eluted with elution buffer (20 mM HEPES, pH 6.8, 160 mM KoAc, 10 mM maltose) and subjected to SDS–PAGE followed by Sypro Orange staining.
Liposome-binding assay, fluorometric GTPase assay, and light scattering analysis
Liposome-binding assay, the Sar1 GTPase assay, and light scattering assay were carried out as described previously (Antonny et al., 2001; Sato and Nakano, 2005). All experiments were performed with synthetic major–minor mix liposomes (Matsuoka et al., 1998).
Supplementary Material
Acknowledgments
We are grateful to the Yeast Genetic Resource Center of Japan for strains. We also thank members of the Sato laboratory for helpful discussions. This work was supported by a Grant-in-Aid for Scientific Research of the Japan Society for the Promotion of Science (T.Y. and K.S.) and in part by the Targeted Proteins Research Program of the Ministry of Education, Culture, Sports, Science and Technology of Japan (K.S.).
Abbreviations used:
- CCD
central conserved domain
- CTCD
C-terminal conserved domain
- ERES
endoplasmic reticulum exit sites
- 5-FOA
5-fluoroorotic acid
- GAP
GTPase-activating protein
- PT
permissive temperature
- NPT
nonpermissive temperature
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E12-05-0356) on June 6, 2012.
REFERENCES
- Antonny B, Madden D, Hamamoto S, Orci L, Schekman R. Dynamics of the COPII coat with GTP and stable analogues. Nat Cell Biol. 2001;3:531–537. doi: 10.1038/35078500. [DOI] [PubMed] [Google Scholar]
- Barlowe C, Orci L, Yeung T, Hosobuchi M, Hamamoto S, Salama N, Rexach MF, Ravazzola M, Amherdt M, Schekman R. COPII: a membrane coat formed by Sec proteins that drive vesicle budding from the endoplasmic reticulum. Cell. 1994;77:895–907. doi: 10.1016/0092-8674(94)90138-4. [DOI] [PubMed] [Google Scholar]
- Barlowe C, Schekman R. SEC12 encodes a guanine-nucleotide-exchange factor essential for transport vesicle budding from the ER. Nature. 1993;365:347–349. doi: 10.1038/365347a0. [DOI] [PubMed] [Google Scholar]
- Bhattacharyya D, Glick BS. Two mammalian Sec16 homologues have nonredundant functions in endoplasmic reticulum (ER) export and transitional ER organization. Mol Biol Cell. 2007;18:839–849. doi: 10.1091/mbc.E06-08-0707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi X, Corpina RA, Goldberg J. Structure of the Sec23/24–Sar1 pre-budding complex of the COPII vesicle coat. Nature. 2002;419:271–277. doi: 10.1038/nature01040. [DOI] [PubMed] [Google Scholar]
- Bi X, Mancias JD, Goldberg J. Insights into COPII coat nucleation from the structure of Sec23•Sar1 complexed with the active fragment of Sec31. Dev Cell. 2007;13:635–645. doi: 10.1016/j.devcel.2007.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai H, Yu S, Menon S, Cai Y, Lazarova D, Fu C, Reinisch K, Hay JC, Ferro-Novick S. TRAPPI tethers COPII vesicles by binding the coat subunit Sec23. Nature. 2007;445:941–944. doi: 10.1038/nature05527. [DOI] [PubMed] [Google Scholar]
- Castillon GA, Watanabe R, Taylor M, Schwabe TME, Riezman H. Concentration of GPI-anchored proteins upon ER exit in yeast. Traffic. 2009;10:186–200. doi: 10.1111/j.1600-0854.2008.00857.x. [DOI] [PubMed] [Google Scholar]
- Connerly PL, Esaki M, Montegna EA, Strongin DE, Levi S, Soderholm J, Glick BS. Sec16 is a determinant of transitional ER organization. Curr Biol. 2005;15:1439–1447. doi: 10.1016/j.cub.2005.06.065. [DOI] [PubMed] [Google Scholar]
- Dancourt J, Barlowe C. Protein sorting receptors in the early secretory pathway. Annu Rev Biochem. 2010;79:777–802. doi: 10.1146/annurev-biochem-061608-091319. [DOI] [PubMed] [Google Scholar]
- Espenshade P, Gimeno RE, Holzmacher E, Teung P, Kaiser CA. Yeast SEC16 gene encodes a multidomain vesicle coat protein that interacts with Sec23p. J Cell Biol. 1995;131:311–324. doi: 10.1083/jcb.131.2.311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farhan H, Wendeler MW, Mitrovic S, Fava E, Silberberg Y, Sharan R, Zerial M, Hauri HP. MAPK signaling to the early secretory pathway revealed by kinase/phosphatase functional screening. J Cell Biol. 2010;189:997–1011. doi: 10.1083/jcb.200912082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Futai F, Hamamoto S, Orci L, Schekman R. GTP/GDP exchange by Sec12p enables COPII vesicle bud formation on synthetic liposomes. EMBO J. 2004;23:4146–4155. doi: 10.1038/sj.emboj.7600428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gimeno RE, Espenshade P, Kaiser CA. COPII coat subunit interactions: Sec24p and Sec23p bind to adjacent regions of Secl6p. Mol Biol Cell. 1996;7:1815–1823. doi: 10.1091/mbc.7.11.1815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gueldener U, Heinisch J, Koehler GJ, Voss D, Hegemann JH. A second set of loxP marker cassettes for Cre-mediated multiple gene knockouts in budding yeast. Nucleic Acids Res. 2002;30:e23. doi: 10.1093/nar/30.6.e23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes H, et al. Organisation of human ER-exit sites: requirements for the localisation of Sec16 to transitional ER. J Cell Sci. 2009;122:2924–2934. doi: 10.1242/jcs.044032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivan V, de Voer G, Xanthakis D, Spoorendonk KM, Kondylis V, Rabouille C. Drosophila Sec16 mediates the biogenesis of tER sites upstream of Sar1 through an arginine-rich motif. Mol Biol Cell. 2008;19:4352–4365. doi: 10.1091/mbc.E08-03-0246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James P, Halladay J, Craig EA. Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics. 1996;144:1425–1436. doi: 10.1093/genetics/144.4.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kung LF, et al. Sec24p and Sec16p cooperate to regulate the GTP cycle of the COPII coat. EMBO J. 2012;31:1014–1027. doi: 10.1038/emboj.2011.444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuoka K, Orci L, Amherdt M, Bednarek SY, Hamamoto S, Schekman R, Yeung T. COPII-coated vesicle formation reconstituted with purified coat proteins and chemically defined liposomes. Cell. 1998;93:263–275. doi: 10.1016/s0092-8674(00)81577-9. [DOI] [PubMed] [Google Scholar]
- Mossessova E, Bickford LC, Goldberg J. SNARE selectivity of the COPII coat. Cell. 2003;114:483–495. doi: 10.1016/s0092-8674(03)00608-1. [DOI] [PubMed] [Google Scholar]
- Nakano A, Brada D, Schekman R. A membrane glycoprotein, Sec12p, required for protein transport from the endoplasmic reticulum to the Golgi apparatus in yeast. J Cell Biol. 1988;107:851–863. doi: 10.1083/jcb.107.3.851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano A, Muramatsu M. A novel GTP-binding protein, Sar1p, is involved in transport from the endoplasmic reticulum to the Golgi apparatus. J Cell Biol. 1989;109:2677–2691. doi: 10.1083/jcb.109.6.2677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto M, Kurokawa K, Matsuura-Tokita K, Saito C, Hirata R, Nakano A. High-curvature domains of the ER are important for the organization of ER exit sites in Saccharomyces cerevisiae. J Cell Sci. 2012 doi: 10.1242/jcs.100065. doi: 10.1242/jcs.100065. [DOI] [PubMed] [Google Scholar]
- Salama NR, Chuang JS, Schekman RW. SEC31 encodes an essential component of the COPII coat required for transport vesicle budding from the endoplasmic reticulum. Mol Biol Cell. 1997;8:205–217. doi: 10.1091/mbc.8.2.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato K, Nakano A. Dissection of COPII subunit-cargo assembly and disassembly kinetics during Sar1p-GTP hydrolysis. Nat Struct Mol Biol. 2005;12:167–174. doi: 10.1038/nsmb893. [DOI] [PubMed] [Google Scholar]
- Shaywitz DA, Espenshade PJ, Gimeno RE, Kaiser CA. COPII subunit interactions in the assembly of the vesicle coat. J Biol Chem. 1997;272:25413–25416. doi: 10.1074/jbc.272.41.25413. [DOI] [PubMed] [Google Scholar]
- Shindiapina P, Barlowe C. Requirements for transitional endoplasmic reticulum site structure and function in Saccharomyces cerevisiae. Mol Biol Cell. 2010;21:1530–1545. doi: 10.1091/mbc.E09-07-0605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soderholm J, Bhattacharyya D, Strongin D, Markovitz V, Connerly PL, Reinke CA, Glick BS. The transitional ER localization mechanism of Pichia pastoris Sec12. Dev Cell. 2004;6:649–659. doi: 10.1016/s1534-5807(04)00129-7. [DOI] [PubMed] [Google Scholar]
- Supek F, Madden DT, Hamamoto S, Orci L, Schekman R. Sec16p potentiates the action of COPII proteins to bud transport vesicles. J Cell Biol. 2002;158:1029–1038. doi: 10.1083/jcb.200207053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabata KV, Sato K, Ide T, Nishizaka T, Nakano A, Noji H. Visualization of cargo concentration by COPII minimal machinery in a planar lipid membrane. EMBO J. 2009;28:3279–3289. doi: 10.1038/emboj.2009.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson P, Townley AK, Koka P, Palmer KJ, Stephens DJ. Sec16 defines endoplasmic reticulum exit sites and is required for secretory cargo export in mammalian cells. Traffic. 2006;7:1678–1687. doi: 10.1111/j.1600-0854.2006.00493.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whittle JRR, Schwartz TU. Structure of the Sec13–Sec16 edge element, a template for assembly of the COPII vesicle coat. J Cell Biol. 2010;190:347–361. doi: 10.1083/jcb.201003092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witte K, Schuh AL, Hegermann J, Sarkeshik A, Mayers JR, Schwarze K, Yates JR, 3rd, Eimer S, Audhya A. TFG-1 function in protein secretion and oncogenesis. Nat Cell Biol. 2011;13:550–558. doi: 10.1038/ncb2225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshihisa T, Barlowe C, Schekman R. Requirement for a GTPase-activating protein in vesicle budding from the endoplasmic reticulum. Science. 1993;259:1466–1468. doi: 10.1126/science.8451644. [DOI] [PubMed] [Google Scholar]
- Zacharogianni M, Kondylis V, Tang Y, Farhan H, Xanthakis D, Fuchs F, Boutros M, Rabouille C. ERK7 is a negative regulator of protein secretion in response to amino-acid starvation by modulating Sec16 membrane association. EMBO J. 2011;30:3684–3700. doi: 10.1038/emboj.2011.253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanetti G, Pahuja KB, Studer S, Shim S, Schekman R. COPII and the regulation of protein sorting in mammals. Nat Cell Biol. 2011;14:20–28. doi: 10.1038/ncb2390. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.