Abstract
The potential requirement of either the Prion or Shadoo protein for early mouse embryogenesis was recently suggested. However, the current data did not allow to precise the developmental process that was affected in the absence of both proteins and that led to the observed early lethal phenotype. In the present study, using various Prnp transgenic mouse lines and lentiviral vectors expressing shRNAs that target the Shadoo-encoding mRNA, we further demonstrate the specific requirement of at least one of these two PrP-related proteins at early developmental stages. Histological analysis reveals developmental defect of the ectoplacental cone and important hemorrhage surrounding the Prnp-knockout-Sprn-knockdown E7.5 embryos. By restricting the RNA interference to the trophoblastic cell lineages, the observed lethal phenotype could be attributed to the sole role of these proteins in this trophectoderm-derived compartment. RNAseq analysis performed on early embryos of various Prnp and Sprn genotypes indicated that the simultaneous down-regulation of these two proteins affects cell-adhesion and inflammatory pathways as well as the expression of ectoplacental-specific genes. Overall, our data provide biological clues in favor of a crucial and complementary embryonic role of the prion protein family in Eutherians and emphasizes the need to further evaluate its implication in normal and pathological human placenta biology.
Introduction
The Prion protein, PrP, is the best known member of the prion protein family due to its pivotal role in transmissible spongiform encephalopathies [1–3 for reviews]. However, the physiological function of this ubiquitously expressed protein is still unclear, and the same is largely true for the related Shadoo and Doppel proteins. Various roles in neuroprotection, cellular homeostasis, response to oxidative stress, cell proliferation and differentiation, synaptic function and signal transduction have been proposed for PrP [4]–[7]. Shadoo was recently shown to possess neuro- and stress-protective properties [8]–[10] whereas inactivation of the Doppel-encoding gene in mice resulted in male infertility associated with strain-related variable sperm maturation defects [11], [12]. The difficulty to define a precise role for PrP partially comes from the observation that PrP-encoding gene-knockout (Prnp KO) mice [13], [14], cattle [15] and goat [16] suffer from no drastic developmental phenotype. It was hypothesized that another host-encoded protein is able to compensate for the lack of PrP [17]. Shadoo, which shares some spatial regulation and properties with PrP, appears to be a good candidate for being this hypothetical, host-encoded PrP-like protein [18].
The developmental regulation of the mouse Prnp gene suggested a possible involvement of PrP in embryogenesis [19]–[22]. The two other prion-related proteins are also expressed in early developmental stages according to the available EST databases and to recent reports [23], [24]. The hypothesis of an embryonic role of the PrP protein family was recently reinforced by the observation that PrP and Shadoo are required for early mouse embryogenesis, as lethality was observed around E10.5 in Sprn-(Shadoo-encoding gene-)knockdown, Prnp-knockout embryos [25]. These data also suggested that the physiological role of these proteins may have to be investigated at early developmental stages.
The aim of this study was to get further insight into the biological function of the prion protein family during early embryogenesis using transcriptomic analysis and cell lineage-specific gene targeting.
Results
Shadoo-knockdown (SprnKD)-induced Embryonic Lethality Requires the Lack of PrP Proteins
We have previously reported that the injections of two independent sh-RNA lentiviral solutions (LSI : SIGMA TRCN0000179960 and LS2 : SIGMA TRCN0000184740), both targeting the mouse Sprn transcript, induce embryonic lethality in FVB/N Prnp KO embryos that was not detected on an FVB/N genetic background [25]. However, since then, evidence has been published highlighting other differences than Prnp between these two genetic backgrounds. Prnp-physically linked 129-derived loci were reported to be conserved in FVB/N Prnp KO mice alongside the Prnp locus itself [26]. Although unlikely, such non-Prnp loci could be involved in the observed lethality associated with Sprn knockdown in FVB/N Prnp KO embryos. RNAi is known to potentially induce off-target effects ([27] for recent review), and such off-target events might interfere with the expression of some of the 129-derived genes indirectly selected during the Prnp knockout experiment. Alternatively, alleles of these genes might differentially modulate specific pathways that are involved during the knockdown process, leading to a lethal phenotype. To assess how specific the previously described phenotype is to the double knockout/knockdown and the potential involvement of additional genes coming along with the Prnp knockout allele, we took advantage of our recent derivation, following micro-injection of a transgene based on the phgPrP-vector in FVB/N Prnp KO eggs, of a homozygous P10 transgenic mouse line that expresses the ovine PrP protein at physiological levels. In this transgenic line, all the 129-additional genes of the FVB/N Prnp KO mice are present alongside a functional ovine Prnp transgene. Injection of the sh-RNA lentiviral solution targeting FoxL2 in P10 mice gave results statistically similar to those previously observed for FVB/N or FVB/N Prnp KO eggs (Table 1 and [25]), arguing that the P10 genetic background is not associated with an unusual susceptibility to lentiviral infection and/or RNA interference. Injection in P10 mouse eggs of the shRNA targeting Sprn lentiviral solution (LS2 in [25]) resulted in an embryonic survival rate similar to that observed in FVB/N and significantly higher from that previously detected for FVB/N Prnp KO mice (Table 1, p<0.05). The induced LS2 lethality is thus dependent on the presence or not of a functional PrP-encoding gene and not on other 129 Prnp KO-associated loci.
Table 1. Effect of ShRNA-mediated Sprn knockdown on embryo resorption at E11.5.
| Lentivirus | FoxL2 | FoxL2 | FoxL2 | Shadoo (LS2) | Shadoo (LS2) | Shadoo (LS2) | None |
| Genetic Background | FVB/N | FVB/N Prnp 0/0 | P10 | FVB/N | FVB/N Prnp 0/0 | P10 | P10 |
| Implanted | 56 | 41 | 79 | 101 | 128 | 105 | 28 |
| Resorbed | 28 | 16 | 39 | 49 | 96 | 36 | 5 |
| % Resorbed | 50 | 39 | 49 | 48.5 | 75* | 34.3 | 17.8 |
p<0.05 (x2 test) when compared to any of the FoxL2 results and to LS2 on P10 and FVB/N genetic backgrounds.
These data are a compilation of at least 2 independent experiments. No statistically significant variability was observed between the analyzed litters (3 for P10, more than 4 for the other lentiviral infections).
Histological Analysis of FVB/N PrnpKO SprnKD Embryos Reveals Ectoplacental Cone Defects and Local Hemorrhage
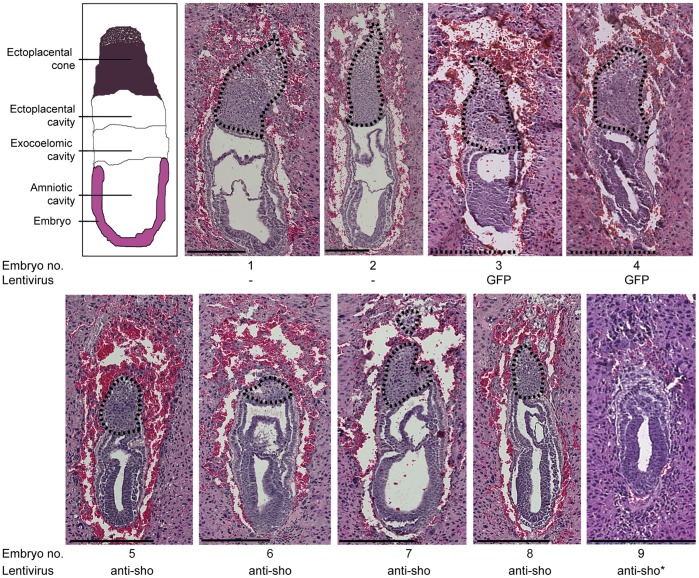
Comparative histological analyses of E7.5 embryos were performed between FVB/N Prnp KO and FVB/N Prnp KO embryos injected at the zygotic stage with either a FG12 lentiviral solution, used as a control as it only encodes GFP (http://www.addgene.org/14884) and could thus allow tthe control of the lineage specific lentiviral infection (see below), or an shRNA targeting Sprn LS2-lentiviral solution [25]. This developmental stage was chosen as a compromise between the early embryonic lethality observed following LS2 injection ([25] and current data) and a developmental timing that could allow the assessment of some embryonic lineage differentiation. FG12-injected embryos (Figure 1, #3 and 4) were found to be slightly developmentally delayed as compared to non-injected embryos (Figure 1, #1 and 2), and some were surrounded by minor hemorrhage, attested by pinkish-red stained red blood cells (#3 for example, 2 out of the 6 analyzed embryos), but otherwise they did not present other major defects (6/6 embryos). LS2-injected embryos were similarly developmentally delayed (7/7 embryos), suggesting that this phenotype is associated with the in vitro manipulation of the eggs, but they were fully comparable in size and developmental stage to FG12-injected controls. Most importantly, in contrast to control embryos (Figure 1), LS2-injected embryos were characterized by reduced ectoplacental cone surfaces. Compared to control embryos, their cones were disorganized, with a notably reduced and even fragmented invasive chorionic trophoblast cell layer (Figure 1, #7 and 8). Such a phenotype was never observed in the above-mentioned control embryos (13/13). In addition, all the seven analyzed embryos were fully surrounded by large hemorrhagic lacunae containing lots of red blood cells (Figure 1, #5 to 8).
Figure 1. Histological analysis of E7.5 embryos.
E7.5 embryos were fixed and stained by hematoxylin, eosin, and saffron. Top left: schematic representation of a mouse E7.5 embryo. 1,2: FVB/N PrnpKO embryos. 3,4: FG12-injected FVB/N PrnpKO embryos. 5–8: LS2-injected FVB/N PrnpKO embryos. 3–8: embryos that were injected at the zygotic stage. 9: LS2-infected FVB/N PrnpKO embryos. *: infection performed at the blastocyst stage. Interesting features include i) the size differences between injected and non-injected embryos, ii) the relatively important hemorrhagic tissue that is totally surrounding the LS2-injected FVB/N PrnpKO embryos 5, 6 and 8, iii) the developmental defect of the ectoplacental cone (area surrounded using a dashed line ) of all the LS2-injected FVB/N PrnpKO embryos (5–8) that even leads to its nearly complete disappearance in embryo 6 and iv) the important developmental delay and the total disorganization of the extra-embryonic ectoderm and of the ectoplacental cone of embryo 9. Scale: 250 µm. Sho: Sprn.
Trophoblastic-restricted RNA Interference Induces Embryonic Lethality
The embryonic lethality reported for FVB/N Prnp KO Sprn KD embryos occurred at a developmental stage at which trophoblastic lineage cells proliferate and differentiate to provide adequate nutriments and metabolic exchanges to the fetus via a developing, expanding and maturing placenta. Histological analysis of the embryos clearly pointed to a defect in the ectoplacental cone development (Figure 1). The ectoplacental cone derives from the outer trophectoderm. Separation of the trophectoderm from the inner cell mass is the first lineage distinction that occurs during mammalian development at the blastocyst stage. To assess whether the lethal phenotype could be associated with a developmental defect restricted to the embryo or to trophectoderm-derived, extra-embryonic tissues, we took advantage of the capacity of lentiviral vectors to specifically infect the trophoblast when used on blastocysts ([28], [29], Figure S1).
The obtained results showed no statistical difference in the percentage of surviving embryos when lentiviral solutions of ShRNA-targeting FoxL2 on either genetic background or of ShRNA LS2 targeting Sprn on FVB/N embryos were used (Table 2). These results highlighted that i) this targeted lentivirus delivery protocol leads to identical survival rate compared to zygotic injection when a gene known to have no obvious function in the placental development such as FoxL2 is downregulated and ii) that Srpn downregulation does not appear to interfere with the placenta development of FVB/N embryos or at least with no detectable incidence on the survival rate. The ShRNA-targeting Sprn LS2 lentiviral solution applied on FVB/N Prnp KO blastocysts led to a strikingly different output with a statistically higher resorption rate (100% versus <50%, Table 2, p<0.05). A similar trend was observed when the ShRNA-targeting Sprn LS1 lentiviral solution was used to infect FVB/N Prnp KO blastocysts, resulting in a resorption rate above 60% as compared with less than 40% with the ShRNA-targeting FoxL2 lentiviral solution (Table 2). The lower rate observed with LS1 is consistent with the previously reported overall lower capacity of this lentiviral solution to induce a phenotype as compared with LS2 [25] likely to be in relation with its apparent lower efficiency to knockdown Sprn (see Fig. 1 in [25]). Overall, these results were not statistically different from those for zygotic lentiviral infections (Tables 1 and 2).
Table 2. Effect of trophoblastic-restricted ShRNA- mediated Sprn knockdown on embryo resorption at E13.5.
| Lentivirus | FoxL2 | FoxL2 | Shadoo (LS2) | Shadoo (LS2) | Shadoo (LS1) |
| Genetic Background | FVB/N | FVB/N Prnp 0/0 | FVB/N | FVB/N Prnp 0/0 | FVB/N Prnp 0/0 |
| Implanted | 37 | 24 | 44 | 40 | 66 |
| Resorbed | 19 | 9 | 17 | 40 | 40 |
| % Resorbed | 51.3 | 37.5 | 38.6 | 100* | 60.6 |
p<0.05 (x2 test) when compared to any of the other results.
These data are a compilation of at least 2 independent experiments. No statistically significant variability was observed between the analyzed litters (more than 4 for each lentiviral infections).
Histological analyses performed on E7.5 FVB/N Prnp KO embryos that had been infected at the blastocyst stage with either lentiviral LS2 or FG12 solutions revealed that while FG12 did not interfere with the developmental process of the embryo (data not shown), the use of LS2 resulted in i) an overall developmental delay associated with no other obvious major defects and ii) a disorganization of the extra-embryonic ectoderm and ectoplacental cone, independently of the observed retarded developmental stage with a nearly complete disappearance of the invasive chorionic trophoblast cell layer (Figure 1, #9). This phenotype is thus similar, although more pronounced, to that observed after zygotic infection. It should be mentioned that these embryos were not surrounded by significant hemorrhage, suggesting that this phenotype is either associated with the zygotic stage of injection or, most probably, not yet apparent due to the developmental delay and the much reduced decidualisation. Altogether, these observations indicate that a dysfunction of the trophoblastic lineage induced by the knockdown of Sprn in the absence of PrP is sufficient to induce a rate of embryonic lethality similar to the one associated with the Sprn KD Prnp KO genetic background.
Analysis of SprnKD Embryos Reveals Differential Expression of Few Genes Involved in Major Developmental Processes along Prnp
RNASeq analysis was performed on pools of E6.5 and E7.5 FVB/N and FVB/N Prnp KO embryos that were injected at the pronuclear stage with either LS1 or LS2 ShRNA targeting Sprn lentiviral solutions, as previously described [25]. These two developmental time-points were chosen according to the lethality observed in FVB/N Prnp KO Sprn KD embryos that was found to be already substantial at E8.5, refining the timescale previously described [25]. Genes were considered to be differentially expressed when their deregulation was observed for both LS1 and LS2. Genes that were deregulated by only one of these ShRNAs were considered to result either from RNA-interference off-target effect or from variation between biological replicates. Of note is the observation that all the differentially expressed genes were similarly either upregulated or downregulated by both ShRNA when compared to FVB/N wild-type embryos (Table S1).
Validation by PCR analysis of RNASeq data obtained during this experiment has been previously reported [30]. Sprn itself was not detected as a differentially expressed gene due to the too low number of reads for its transcript (<5 reads). This quantitative approach confirmed that this locus is only expressed at low levels in early embryos, at least 100-fold less than Prnp. However, assessment of the Sprn-down-regulation through LS2-induced RNA interference had already been reported [25] and confirmed in the present experiment by RT-PCR and QPCR, with an overall >65% level of inhibition (Figure S2).
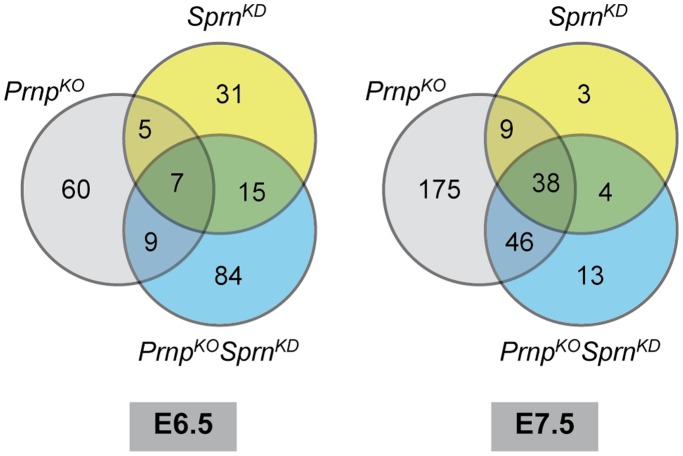
Sprn-downregulation on a FVB/N genetic background induces the differential expression of 58 and 54 transcripts at E6.5 and E7.5, respectively, of which, strikingly, only 5 were found at both developmental stages (Figure 2 and Table S1). This total represents less than 0.14% of all the identified transcripts. Of note is the deregulation in SprnKD embryos of genes specifically expressed in the ectoplacental cone, such as the prolactin-related Prl2C5, Prl2a1 [31]–[34], and/or in inflammatory response, such as interleukin 15 (IL15), complement factor H, granzymes and transmembrane serine protease members, Prss28 and 29 [35], in good correlation with the histological phenotype. Whereas at E6.5, 12 (22%) of the differentially expressed genes were also differentially expressed in Prnp KO embryos ([30] and Figure 2), at E7.5, 47 (87%) of the genes differentially expressed in SprnKD embryos were also differentially expressed in E7.5 Prnp KO embryos ([30] and Figure 2).
Figure 2. Venn diagram of the number of differentially expressed genes.
A venn diagram of the number of genes differentially expressed between PrnpKO, Sprn KD, Prnp KO Sprn KD embryos and their wild type counterparts is given at E6.5 and E7.5.
The same analysis performed on FVB/N Prnp KO genetic background reveals the differential expression of 115 and 101 genes at E6.5 and 7.5, respectively when compared to FVB/N embryos, of which only two were found at both developmental stages (Figure 2 and Table S1). At E6.5, 84 genes (73%) were found to be specifically deregulated in Sprn KD Prnp KO embryos compared to their Prnp KO or Srpn KD counterparts whereas this number drops to 13 (13%) at E7.5. This observation highlights that at early developmental stages, the simultaneous down-regulation of these two genes appears to impact specific biochemical pathways that are not disturbed or to a much lesser extent in embryos invalidated for only one of them. Within these affected genes, several were also specifically expressed in the ectoplacental cone, such as the prolactin-related Prl2C1 and Prl2C3 loci.
In silico analysis of the observed transcriptomic alterations identified few pathways that encompass more than 10 of the differentially expressed genes (Table 3). The top functions associated with these pathways, molecular transport, hematological system development, cardiovascular system development, skeletal and muscular system developments, hair and skin developments, were identical to those recently reported for similar transcriptomic analysis of FVB/N Prnp KO embryos [30] and/or for PrP function in adult tissues [4]–[7]. Interestingly, the above-mentioned genes that were specifically deregulated in Sprn KD Prnp KO E6.5 embryos were identified as being involved in functions such as developmental and genetic disorders, cellular movement and development (Table 3 and Table S1). Many of these genes encode for proteins involved in the extracellular matrix, such as collagens, its remodeling, such as metalloproteinases and the biglycan (Bgn) locus, and cell-cell interactions such as cadherins. Since trophectoderm is the first differentiated tissue to form with cells needing adhesive structures, this observation is in accordance with the histological observed defect of the ectoplacental cone development.
Table 3. List of top-function clusters for differentially expressed genes in Sprn-knockdown embryos.
| Developmental Stage/Genetic Background | Top Functions | Genes |
| E6.5 FVB/N Sprn KD versus WT | Cell Signaling. Cellular Growth and Differentiation. Hematological System | ABCA1, ADAMTS1, ANGPT2, CDH22, CIDEA, CTSK, CXCL13, DIO2, IL15, INHBA, LMCD1, PCSK5, TIMP3, TNNC1, TPSAB1 |
| Developmental and Genetic Disorder | RMRP, RPPH1 | |
| E6.5 FVB/N Prnp KO Sprn KD versus WT | Developmental and Genetic Disorder, Metabolic Disease | ADH1C, C3, C1QB, CD74, CDH11, CFH, COL1A1, COL1A2, CTSK, HLA-DRB1, IL15, LGALS3BP, MMP2, MMP238, RHOJ, SERPINA1 |
| Lipid metabolism, Molecular transport | CCDC80,CDH11, CHODL, EAF2, GJA4, ITIH5, KIAA1324, SDCBP2, SEMA5A, SULT1A1, TCF23, TMEM204, TMEM45A, WFDC2 | |
| Cell cycle, Reproductive system development and function, cellular development | AGR2, COL1A1, CWH43, GGH, HTRA3, ITGA8, Prl2a1, Prl2c2, RNASE1, ROBO2, SPACA7 | |
| Cellular movement, Hematological system development and function | ACTG2, CXCL14, DIO2, EDNRB, MYLK, NUPR1, PAX8, PCP4, PENK, WNT5A | |
| E6.5 FVB/N Prnp KO Sprn KD versus Prnp KO | Tissue Morphology. Cellular Growth and Differentiation. Hematological System | ACTA2, ACTG2, AGR2, CNN1, CTSK, IL15, MIXL1, MYH11, MYLK, NUPR1, SAA3P, TAGLN |
| Genetic Disorder. Hematological Disease | PCP4, PFAS, SYNPO2, TMEM102 | |
| Developmental and Genetic Disorder | RMRP, RPPH1 | |
| E7.5 FVB/N Sprn KD versus WT | Cell Cycle. Cardiovascular System Development and Function, Organismal Development | A2M, ANGPT2, ARG1, DCN, DIO3, GJA1, LYVE1, MMP7, PRRX2, PTN, S100A4, SLC2A12, SLPI, TDGF1, TNFRSF11B |
| Cellular Development. Cellular Growth and Proliferation. Hair and Skin Development | ANXA8, COL5A2, CYP11B1, FBLN2, HAVCR2, MFAP5, NAPSA, PDZK1IP1, PRAP1, SMOC2 | |
| Lipid Metabolism. Small Molecule Biochemistry. Carbohydrate Metabolism | CRIP1, EMCN, HSPB7, PTRF, TDO2 | |
| E7.5 FVB/N Prnp KO Sprn KD versus WT | Connective tissue development and function, skeletal and muscular system development | ANGPT2, CRYAA, CYP286, DES, DIO3, FBLN2, FBN1, H5D11B1, LBP, LYVE1, MFAP5, PTGS1, PTN, SRD5A1, TDGF1, TNFRSF11B |
| Cellular development, Embryonic development | ANXA8, COL5A2, CRIP1, CTSO, CYP286, FBLN2, HOXA11, IGSF11, Ly6a, NDUFAF1, OLFML3, PTRF, SDPR, TDO2, ZNF503 | |
| E7.5 FVB/N Prnp KO Sprn KD versus Prnp KO | Embryonic Development | ALDH1A2, ARG1, CDX1 |
Bold faced genes: upregulated genes in Sprn-knockdown embryos. Un-bolded genes: downregulated genes in Sprn-knockdown embryos.
We also performed similar in silico analysis on genes that were differentially expressed between Sprn KD Prnp KO embryos and Prnp KO or Sprn KD embryos at both developmental stages (Table 3 and Table S1). At E6.5, three biological pathways were identified for genes differentially expressed between Sprn KD Prnp KO and Prnp KO embryos, cellular growth and differentiation, hematological system development and disease, developmental and genetic disorder. It corresponded to top functions already documented. In E7.5 FVB/N Prnp KO Sprn KD embryos, the Ingenuity-detected pathway was suggestive of a general distress (Table 3), in agreement with the embryonic lethality that is observed in such animals [25]. Only few genes were found to be differentially expressed between Sprn KD Prnp KO and Sprn KD embryos, 8 at E6.5 and 14 at E7.5 (Table S1). Five out of the 8 identified genes at E6.5 (Actg2, Myh11, Cnn1, Acta2 and Pep4) are involved in cellular movement and hematological system development and differentiation functions (Table 3). However, none of these genes were share between the two developmental stages and none were specifically deregulated in the Sprn KD Prnp KO embryos. At E7.5, 12 out of the 14 genes that were identified comparing Sprn KD Prnp KO and Sprn KD embryos were inversely differentially expressed by LS1 or LS2 It probably reflects the overall more pronounced phenotype observed with LS2-treated embryos.
Discussion
While the inactivation of the sole Prnp gene did not affect the survival rate of mammals despite early embryonic expression [13]–[16], it induced embryonic lethality in Zebrafish [36], [37] arguing for the involvement of the Prion protein family in embryonic development. Such a lethal phenotype was also described in early FVB/N Prnp KO Sprn KD mouse embryos [25] suggesting that in mammals, the absence of Prnp was compensated by the expression of the PrP-related protein, Shadoo. Here we provide further evidence of the specificity of this compensation by showing that such a knockdown approach performed on a transgenic line recently established that expresses physiological levels of ovine PrP on an FVB/N Prnp KO genetic background, led to results similar to those obtained on FVB/N control mice.
The surviving FVB/N Prnp KO Sprn KD embryos suffer from a neural defect with a failure of closure of the cranial tube [25]. However, it is commonly accepted that such a default could not explain the observed lethality. As a major finding of this study, we show that trophoblastic-targeted Sprn-down-regulation led to developmental defects of the trophoblastic lineage in an FVB/N Prnp KO genetic background that were sufficient to explain the observed lethal phenotype. It does not preclude that other embryonic detrimental biological functions might be also affected. The above-mentioned absence of neural tube closure for example might be a direct consequence of a placental developmental defect [38], or directly result from embryonic transcriptomic alterations. It was indeed reported that alterations of trypsin-like serine protease activities, such as granzymes and PRSS found to be differentially expressed in Prnp KO Sprn KD embryos, could alter both the placental development and neurogenesis [39]. Although Sprn transcriptional regulation appears to be more tightly controlled than that of Prnp in terms of tissue-specificity [8], [21], [24], [40], we could detect Sprn expression in E10.5 extra-embryonic annexes, supporting the suggested role of these genes in the trophoblastic lineage (Figure S3 and [24]).
Comparative analyses of E6.5 and E7.5 FVB/N Prnp KO or Prnp WT Sprn KD embryos with their wild-type FVB/N counterparts, performed by RNASeq, revealed that relatively few genes were differentially expressed following Sprn downregulation. Overall, these transcriptomic data suggested that Shadoo and PrP have complementary, not necessary overlapping, functions associated with cellular movement and hematological system development and differentiation. Such biological roles were already highlighted in Prnp-invalidated Zebrafish [37], [41] and mouse [30] embryos. It also emphasized Shadoo potential synergetic involvement in the development of extra-embryonic lineages with the differential expression of specific ectoplacental genes (Table 3). Strikingly few differentially genes were consistently found at both E6.5 and E7.5, suggesting a fast evolution/adaptation of the embryo biology at these early developmental stages. Furthermore, the histological and the pathways’ analyses at E6.5 and E7.5 suggest that the lack of expression of these two genes synergically affects the establishment of the chorioallantoic placenta (Figure 3). The trophectoderm-derived compartment is a differentiated tissue that forms with invading cells needing complex adhesive structures. Differential expression of several genes involved in the establishment, modeling and maintenance of the extracellular matrix as well as that of genes specifically expressed in the ectoplacental cone were observed only when both Prnp and Sprn genes were invalidated. It is likely to impair placental formation, expansion and maturation, thus reducing its invasive capacity and depriving the embryos of their vital resources. Consequently, it may result in the activation of macrophagic and pro-apoptotic reactions leading to embryonic resorption [36], [42]. Overall, our results suggest an as yet unknown function of the prion protein family in controlling the trophoblastic cell lineage maintenance and differentiation, potentially expending the involvement of these proteins in stem cell biology. It re-enforces the interest in looking at the prion protein family involvement in normal and pathological human placenta biology [43], [44].
Figure 3. Schematic representation of the embryonic phenotype induced by the lack of PrP and Shadoo.
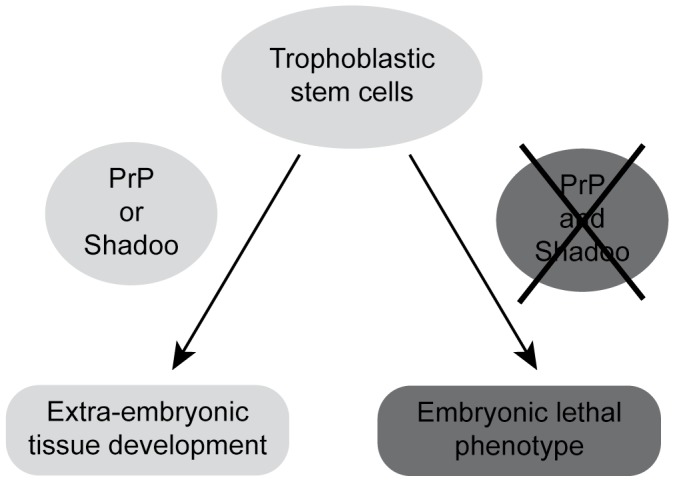
The lethal consequence of the absence of PrP and Shadoo during early mouse embryogenesis is indicated.
During the revision process of this article, Daude et al. reported the creation and analysis of Sprn-knockout mice that did not produce embryonic lethality in combination with Prnp-invalidation, although the output of crosses between Sprn KO Prnp KO x Sprn KO Prnp KO mice was not apparently assessed (TableS1 in [45]). Three hypotheses were proposed by the authors that could conciliate their data with that of the previously published Sprn-knockdown ones [25] and of the present article; i) the use of similar but not identical genetic backgrounds, ii) an adaptation of the Sprn-knockout embryos to the lack of this protein and iii) an alteration of the Sprn-overlapping Mtg1 transcript expression level by the shRNA. This latter hypothesis can be ruled out by i) the location of the targeting sequences of LS1 and LS2 that are located outside the overlapping region between Mtg1 and Sprn [25] and ii) the present analysis that indicates no alteration of the Mtg1 gene expression by either LS1 or LS2 (Table S1 and data not shown). We are currently testing the former hypotheses by invalidation of Sprn in the FVB/N Prnp-knockout genetic background by the use of Zing Finger Nucleases.
Materials and Methods
Animal experiments were carried out in strict accordance with the recommendations in the guidelines of the Code for Methods and Welfare Considerations in Behavioural Research with Animals (Directive 86/609EC). And all efforts were made to minimize suffering. Experiments were approved by the Local ethics committee of Jouy-en-Josas on the Ethics of Animal Experiments of the author’s institution, INRA (Permit Number RTA06-091). All animal manipulations were done according to the recommendations of the French Commission de Génie Génétique (Permit Number N°12931 (01.16.2003)).
Generation of the P10 Transgenic Line
The ovine 136A154R171R PrP cDNA was inserted into the phgPrP half-genomic vector as previously described [46], leading to the P10 construct. The NotI-SalI gel-purified, plasmid-free, insert was micro-injected into FVB/N Prnp-knockout (Prnp KO) oocytes [13], [47]. A transgenic line, that expressed the transgene in its brain at physiological levels, could be derived and bred to a homozygous status.
Lentiviral Injection in Mouse Embryos and Blastocysts
Intra-perivitellin space injections of lentiviral solutions were done according to [48]. Trophoblast-specific lentiviral infections were done as previously described [28], [29]. The sh-RNA lentiviral solutions used that target either mouse Sprn or mouse FoxL2 were purchased from Sigma (LSI : TRCN0000179960, LSII : TRCN0000184740 and Lfox : TRCN0000086505) [25]. Chi2 statistical analyses of the differences observed for survival rates were performed.
Histological Analysis
Collected embryos alongside their deciduas and uterine tissue were fixed in 4% PFA, dehydrated in ethanol before being embedded in paraffin and 5 µm sections cut on a microtome. Sections were stained by hematoxylin, eosin, and saffron then photographed using the Nanozoomer (Hamamatsu). On average, 50 sections per embryos were made and analyzed.
RNAseq Analysis
Total RNA was isolated from pools of E6.5 and E7.5 mouse embryos. RNA extractions were performed using the RNeasy Lipid Tissue Mini kit (Qiagen cat # 75842). RNA concentration was calculated by electro-spectrophotometry and the RNA integrity checked with the Agilent Bioanalyser (Waldbroom, Germany).
RNA samples of 5 µg, obtained from around 30 embryos each collected from 3 to 4 females, were sent to GATC Biotech SARL for RNAseq analysis. A standard cDNA library was derived from each sample. These cDNAs were analyzed on an Illumina Genome Analyzer II with raw data output of up to 350 Mb and 42,000,000 reads per sample and a read length of 36 bases (single read). Sequence cleaning was done using Seqclean (http://compbio.dfci.harvard.edu/tgi/sofware/seqclean_README). Cleaned reads were mapped to the NCBI mouse transcript database (ftp://ftp.ncbi.nih.gov/genomed/M_musculus/RNA/) using BWA software [49].
Differentially expressed genes between FVB/N and FVB/N Prnp KO embryos were identified at 5% FDR using the DESeq package of the R software [50]. They were clustered using the software DAVID [51], then classified in pathways and networks by using Ingenuity (http://www.ingenuity.com/) and the GEPS application of Genomatix (http://www.genomatix.de).
Supporting Information
Trophoblastic-specific GFP expression pattern. Trophoblastic-specific expression pattern of an ubiquitin-EGFP lentiviral-expressing vector (FG12, Addgene) was achieved in E10 mouse embryos following infection as described in Okada et al., 2007. No GFP signal was detected in non-infected control embryos of similar age. Images were merged using the AxioVision 4.8 software.
(TIF)
Evidence for Sprn downregulation. RT-PCR was performed on total RNA isolated from pooled E7.5 embryos. The used oligonucleotides and PCR conditions were as previously described (25). Actb: actin control RT-PCR. M: 1 kb ladder molecular weight marker (InVitrogen). 1: FVB/N embryos. 2: LS2-injected FVB/N embryos. 3: FVB/N PrnpKO embryos. 4: LS1-injected FVB/N Prnp KO embryos.
(TIF)
Evidence for Sprn expression in mouse placenta. RT-PCRs were performed on total RNA isolated from E12 mouse placenta embryos (P1 and P2) and adult brain (Br). The used oligonucleotides and PCR conditions were as previously described (25). Actb: actin control RT-PCR. M: 1 kb ladder molecular weight marker (InVitrogen). + with reverse transcriptase. – without reverse transcriptase.
(TIF)
Complete list of genes differentially expressed in mouse embryos of various genotypes at E6.5 and E7.5. The genotypes of the compared embryos are indicated in the top line. Upregulated genes are in green. Downregulated genes are in red. Indicated numbers in each case refer to the observed fold ratio (p Value). For data involving Sprn knockdown experiments, in each case the first set of numbers is that observed with LS1, the second set is that observed with LS2. ??: upregulation of the gene with no observed expression in the control genotype.
(DOC)
Acknowledgments
FVB/N Prnp-knockout mice were kindly provided by S. Prusiner (San Francisco, USA).
Funding Statement
RY is a post-doctoral scientist supported by the ANR-09-BLAN-0015-01. SH is a post-doctoral scientist supported by an Institut National de la Recherche Agronomique-Transfert fellowship and by the ANR-09-BLAN-0015-01.This work was supported by the ANR-09-BLAN-0015-01 and by the Institut National de la Recherche Agronomique AIP BioRessources PRIFASTEM. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Prusiner S B (1982) Novel proteinaceous infectious particles cause scrapie. Science 216: 136–44. [DOI] [PubMed] [Google Scholar]
- 2. Aguzzi A, Heikenwalder M, Polymenidou M (2007) Insights into prion strains and neurotoxicity. Nat Rev Mol Cell Biol 8: 552–61. [DOI] [PubMed] [Google Scholar]
- 3. Kovacs G, Budka H (2009) Molecular pathology of human prion diseases. Int J Mol Sci 10: 976–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Zomosa-Signoret V, Arnaud JD, Fontes P, Alvarez-Martinez MT, Liautard J P (2008) Physiological role of the cellular prion protein. Vet Res 39: 9–25. [DOI] [PubMed] [Google Scholar]
- 5. Linden R, Martins VR, Prado MA, Cammarota M, Izquierdo I, Brentani RR (2008) Physiology of the prion protein. Physiol Rev 88: 673–728. [DOI] [PubMed] [Google Scholar]
- 6. Martins VR, Beraldo FH, Hajj GN, Lopes MH, Lee KS, et al. (2009) Prion Protein: Orchestrating Neurotrophic Activities. Curr Issues Mol Biol 12: 63–86. [PubMed] [Google Scholar]
- 7. Schneider B, Pietri M, Pradines E, Loubet D, Launay JM, et al. (2011) Understanding the neurospecificity of Prion protein signaling. Front Biosci 16: 169–86. [DOI] [PubMed] [Google Scholar]
- 8. Watts JC, Drisaldi B, Ng V, Yang J, Strome B, et al. (2007) The CNS glycoprotein Shadoo has PrP(C)-like protective properties and displays reduced levels in prion infections. EMBO J 26: 4038–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Sakthivelu V, Seidel RP, Winklhofer KF, Tatzelt J (2011) Conserved stress-protective activity between prion protein and Shadoo. J Biol Chem 286: 8901–8908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Daude N, Westaway D (2011) Biological properties of the PrP-like Shadoo protein. Front Biosci 16, 1505–1516. [DOI] [PubMed] [Google Scholar]
- 11. Behrens A, Genoud N, Naumann H, Rulicke T, Janett F, et al. (2002) Absence of the prion protein homologue Doppel causes male sterility. EMBO J 21: 3652–3658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Paisley D, Banks S, Selfridge J, McLennan NF, Ritchie AM, et al. (2004) Male infertility and DNA damage in Doppel knockout and prion protein/Doppel double-knockout mice. Am J Pathol 164: 2279–2288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Bueler H, Fischer M, Lang Y, Bluethmann H, Lipp HP, et al. (1992) Normal development and behaviour of mice lacking the neuronal cell-surface PrP protein. Nature 356: 577–582. [DOI] [PubMed] [Google Scholar]
- 14. Manson JC, Clarke AR, Hooper ML, Aitchison L, McConnell I, et al. (1994) 129/Ola mice carrying a null mutation in PrP that abolishes mRNA production are developmentally normal. Mol Neurobiol 8: 121–127. [DOI] [PubMed] [Google Scholar]
- 15. Richt JA, Kasinathan P, Hamir AN, Castilla J, Sathiyaseelan T, et al. (2007) Production of cattle lacking prion protein. Nat Biotechnol 25: 132–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Yu G, Chen J, Xu Y, Zhu C, Yu H, et al. (2009) Generation of goats lacking prion protein. Mol Reprod Dev 76: 3–12. [DOI] [PubMed] [Google Scholar]
- 17. Shmerling D, Hegyi I, Fischer M, Blattler T, Brandner S, et al. (1998) Expression of amino-terminally truncated PrP in the mouse leading to ataxia and specific cerebellar lesions. Cell 93: 203–214. [DOI] [PubMed] [Google Scholar]
- 18. Watts JC, Westaway D (2007) The prion protein family: diversity, rivalry, and dysfunction. Biochim Biophys Acta 1772: 654–672. [DOI] [PubMed] [Google Scholar]
- 19. Manson J, West JD, Thomson V, McBride P, Kaufman MH, et al. (1992) The prion protein gene: a role in mouse embryogenesis? Development 115: 117–122. [DOI] [PubMed] [Google Scholar]
- 20. Miele G, Alejo Blanco AR, Baybutt H, Horvat S, et al. (2003) Embryonic activation and developmental expression of the murine prion protein gene. Gene Expr 11: 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Tremblay P, Bouzamondo-Bernstein E, Heinrich C, Prusiner SB, DeArmond SJ (2007) Developmental expression of PrP in the post-implantation embryo. Brain Res 1139: 60–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hajj GN, Santos TG, Cook ZS, Martins VR (2009) Developmental expression of prion protein and its ligands stress-inducible protein 1 and vitronectin. J Comp Neurol 517: 371–384. [DOI] [PubMed] [Google Scholar]
- 23. Peralta OA, Huckle WR, Eyestone WH (2011) Expression and knockdown of cellular prion protein (PrPC) in differentiating mouse embryonic stem cells. Differentiation 81: 68–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Young R, Bouet S, Polyte J, Le Guillou S, Passet B, et al. (2011) Expression of the prion-like Shadoo protein in the developing embryo. Biochem Biophys Res Commun. 416: 184–187. [DOI] [PubMed] [Google Scholar]
- 25. Young R, Passet B, Vilotte M, Cribiu EP, Beringue V, et al. (2009) The prion or the related Shadoo protein is required for early mouse embryogenesis. FEBS Lett 583: 3296–3300. [DOI] [PubMed] [Google Scholar]
- 26. Chadi S, Young R, Le Guillou S, Tilly G, Bitton F, et al. (2010) Brain transcriptional stability upon prion protein-encoding gene invalidation in zygotic or adult mouse. BMC Genomics 11: 448–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Sigoillot FD, King RW (2011) Vigilance and validation: Keys to success in RNAi screening. ACS Chem Biol 6: 47–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Okada Y, Ueshin Y, Isotani A, Saito-Fujita T, Nakashima H, et al. (2007) Complementation of placental defects and embryonic lethality by trophoblast-specific lentiviral gene transfer. Nat Biotechnol 25: 233–237. [DOI] [PubMed] [Google Scholar]
- 29. Georgiades P, Cox B, Gertsenstein M, Chawengsaksophak K, Rossant J (2007) Trophoblast-specific gene manipulation using lentivirus-based vectors. Biotechniques 42: 322–325. [DOI] [PubMed] [Google Scholar]
- 30. Khalifé M, Rachel Y, Passet B, Haillez S, Vilotte M, et al. (2011) Transcriptomic analysis brings new insight into the biological role of the prion protein during mouse embryogenesis. PloS One 6(8): e23253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Simmons DG, Rawn S, Davies A, Hughes M, Cross JC (2008) Spatial and temporal expression of 23 murine prolactin/Placental lactogen-related genes is not associated with their position in the locus. BMC Genomics 9: 352–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Hamlin GP, Lu X J, Roby KF, Soares M J (1994) Recapitulation of the pathway for trophoblast giant cell differentiation in vitro: stage-specific expression of members of the prolactin gene family. Endocrinology 134: 2390–2396. [DOI] [PubMed] [Google Scholar]
- 33. Muller H, Liu B, Croy BA, Head JR, Hunt JS, et al. (1999) Uterine natural killer cells are targets for a trophoblast cell-specific cytokine, prolactin-like protein A. Endocrinology. 140: 2711–2720. [DOI] [PubMed] [Google Scholar]
- 34. Liu Y, Zhou J, Chen J, Gao W, Le Y, et al. (2009) PRL-3 promotes epithelial mesenchymal transition by regulating cadherin directly. Cancer Biol Ther 8: 1352–1359. [DOI] [PubMed] [Google Scholar]
- 35. Stevens RL, Adachi R (2007) Protease-proteoglycan complexes of mouse and human mast cells and importance of their beta-tryptase-heparin complexes in inflammation and innate immunity. Immunol. Rev. 217: 155–167. [DOI] [PubMed] [Google Scholar]
- 36. Caucheteux SM, Kanellopoulos-Langevin C, Ojcius DM (2003) At the innate frontiers between mother and fetus: linking abortion with complement activation. Immunity 18: 169–172. [DOI] [PubMed] [Google Scholar]
- 37. Malaga-Trillo E, Solis GP, Schrock Y, Geiss C, Luncz L, et al. (2009) Regulation of embryonic cell adhesion by the prion protein. PLoS Biol 7: e55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Zeltser LM, Leibel RL (2011) Roles of the placenta in fetal brain development. Proc. Natl. Acad. Sci. USA 108: 15667–15668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Szabo R, Hobson JP, Christoph K, Kosa P, List K, et al. (2009) Regulation of cell surface protease matriptase by HAI2 is essential for placental development, neural tube closure and embryonic survival in mice. Development 136: 2653–2663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Young R, Le Guillou S, Tilly G, Passet B, Vilotte M, et al. (2011) Generation of Sprn-regulated reporter mice reveals gonadic spatial expression of the prion-like protein Shadoo in mice Biochem Biophys Res Commun. 412: 752–756. [DOI] [PubMed] [Google Scholar]
- 41. Malaga-Trillo E, Salta E, Figueras A, Panagiotidis C, Sklaviadis T (2011) Fish models in prion biology: underwater issues Biochim Biophys Acta. 1812: 402–414. [DOI] [PubMed] [Google Scholar]
- 42. Girardi G (2011) Role of tissue factor in pregnancy complications: cross talks between coagulation and inflammation. Thrombosis Res. 127: S43–46. [DOI] [PubMed] [Google Scholar]
- 43. Donadio S, Alfaidy N, De Keukeleire B, Micoud J, Feige JJ, et al. (2007) Expression and localization of cellular prion and COMMD1 proteins in human placenta throughout pregnancy. Placenta 28: 907–911. [DOI] [PubMed] [Google Scholar]
- 44. Hwang HS, Park SH, Park YW, Kwon HS, Sohn IS (2010) Expression of cellular prion protein in the placentas of women with normal and preeclamptic pregnancies. Acta Obstet Gynecol Scand 89: 1155–1161. [DOI] [PubMed] [Google Scholar]
- 45. Daude N, Wohlgemuth S, Brown R, Pitstick R, Giapeshina H, et al. (2012) Knockout of the prion protein (PrP)-like Sprn gene does not produce embryonic lethality in combination with PrPC-deficiency. Proc. Natl. Acad. Sci. USA 109: 9035–9040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Vilotte JL, Soulier S, Essalmani R, Stinnakre MG, Vaiman D, et al. (2001) Markedly increased susceptibility to natural sheep scrapie of transgenic mice expressing ovine PrP. J Virol 75: 5977–5984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Giri RK, Young R, Pitstick R, DeArmond SJ, Prusiner SB, et al. (2006) Prion infection of mouse neurospheres. Proc Natl Acad Sci U S A 103: 3875–3880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Lois C, Hong EJ, Pease S, Brown EJ, Baltimore D (2002) Germline transmission and tissue-specific expression of transgenes delivered by lentiviral vectors. Science 295: 868–872. [DOI] [PubMed] [Google Scholar]
- 49. Li H, Durbin R (2010) Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics 26: 589–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Anders S, Huber W (2010) Differential expression analysis for sequence count data Genome Biol. 11: R106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Dennis G Jr, Sherman BT, Hosack DA, Yang J, Gao W, et al. (2003) DAVID: Database for Annotation, Visualization, and Integrated Discovery Genome Biol. 4: P3. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Trophoblastic-specific GFP expression pattern. Trophoblastic-specific expression pattern of an ubiquitin-EGFP lentiviral-expressing vector (FG12, Addgene) was achieved in E10 mouse embryos following infection as described in Okada et al., 2007. No GFP signal was detected in non-infected control embryos of similar age. Images were merged using the AxioVision 4.8 software.
(TIF)
Evidence for Sprn downregulation. RT-PCR was performed on total RNA isolated from pooled E7.5 embryos. The used oligonucleotides and PCR conditions were as previously described (25). Actb: actin control RT-PCR. M: 1 kb ladder molecular weight marker (InVitrogen). 1: FVB/N embryos. 2: LS2-injected FVB/N embryos. 3: FVB/N PrnpKO embryos. 4: LS1-injected FVB/N Prnp KO embryos.
(TIF)
Evidence for Sprn expression in mouse placenta. RT-PCRs were performed on total RNA isolated from E12 mouse placenta embryos (P1 and P2) and adult brain (Br). The used oligonucleotides and PCR conditions were as previously described (25). Actb: actin control RT-PCR. M: 1 kb ladder molecular weight marker (InVitrogen). + with reverse transcriptase. – without reverse transcriptase.
(TIF)
Complete list of genes differentially expressed in mouse embryos of various genotypes at E6.5 and E7.5. The genotypes of the compared embryos are indicated in the top line. Upregulated genes are in green. Downregulated genes are in red. Indicated numbers in each case refer to the observed fold ratio (p Value). For data involving Sprn knockdown experiments, in each case the first set of numbers is that observed with LS1, the second set is that observed with LS2. ??: upregulation of the gene with no observed expression in the control genotype.
(DOC)