Abstract
All cellular proteins undergo continuous synthesis and degradation. This permanent renewal is necessary to maintain a functional proteome and to allow for rapid changes in levels of specific proteins with regulatory purposes. Although for a long time lysosomes were considered unable to contribute to the selective degradation of individual proteins, the discovery of chaperone-mediated autophagy (CMA) changed this notion. Here, we review the characteristics that set CMA apart from other types of lysosomal degradation and the subset of molecules that confer cells the capability to identify individual cytosolic proteins and direct them across the lysosomal membrane for degradation.
Keywords: aging, cancer, chaperones, lysosomes, membrane proteins, neurodegeneration, protein degradation
Autophagy: more than the “cool” double membrane vesicles
Cells count on an organelle, the lysosome, fully dedicated to degradation of products coming from outside the cell and internalized by endocytosis or phagocytosis [1]. In addition, lysosomes can also degrade intracellular components (proteins and organelles) through a process generically known as autophagy (see Glossary) [2]. Most cells can perform autophagy through different mechanisms depending on the way that the products to be degraded (or cargo) are delivered to lysosomes. In addition, recent studies have subdivided the main autophagic pathways depending on the nature of the cargo or the stimuli that promote their degradation [3, 4]. In this review, we focus on a type of selective autophagy, known as chaperone-mediated autophagy (CMA), so far only identified in mammalian cells, that was set apart from other types of autophagy due to the unique way in which its substrates, mainly cytosolic proteins, enter the lysosome [5]. Proteins degraded by CMA are identified one-by-one by a cytosolic chaperone that delivers them to the surface of the lysosomes. Once there, the substrate proteins unfold and cross the lysosomal membrane. These three events i) individual recognition of single proteins, ii) unfolding before degradation and iii) translocation inside the lysosomes constitute the trademark signature of CMA and the basis for its cellular functions.
The degradation of cytosolic material by lysosomes was described almost in parallel to the identification of lysosomes as digestive organelles [6]. Shortly after the finding that cytosolic content could be found inside lysosomes, elegant morphological studies demonstrated two possible mechanisms for the arrival of this cytosolic cargo. The term macroautophagy was coined to describe the sequestration of regions of the cytosol inside double membrane vesicles that then fuse with lysosomes to transfer their luminal content for degradation [7]. The term microautophagy was reserved to describe the internalization of cytosolic content into lysosomes upon sequestration by invaginations of the lysosomal membrane that then pinch off in the shape of small vesicles into the lysosomal lumen for degradation [8]. When these two types of autophagy were described, the idea of selective degradation of single cytosolic proteins was not contemplated as both macroautophagy and microautophagy delivered to the lysosomes regions of the cytosol containing pools of soluble proteins and organelles. However, later analysis of the proteins internalized and degraded in lysosomes in cultured cells demonstrated that removal of nutrients from the culture media resulted in a preference for the lysosomal degradation of certain subsets of proteins [9]. These findings set the bases for the discovery and characterization of what is today known as CMA (Figure 1) [5].
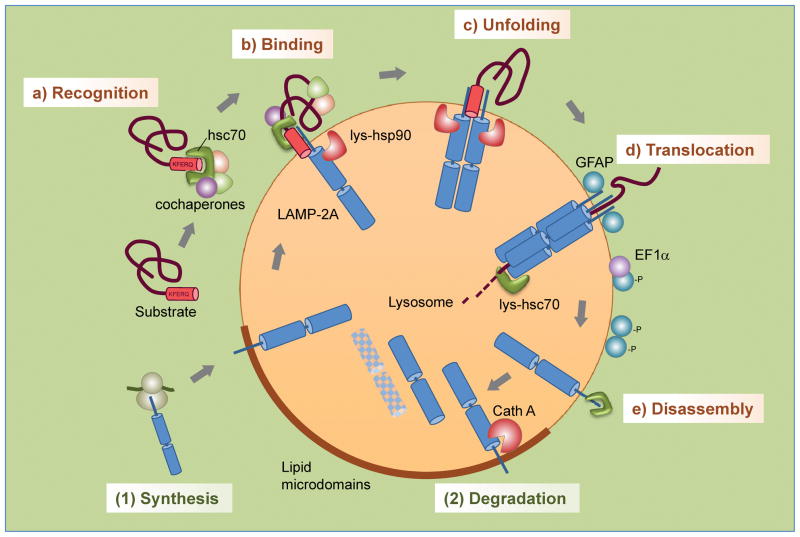
Figure 1.
Steps and regulation of CMA. Steps: (a) Recognition of substrate proteins by hsc70/cochaperones; (b) binding of substrate-chaperone complex to LAMP-2A; (c) unfolding of the substrate; (d) LAMP-2A multimerization, substrate translocation and subsequent degradation; (e) disassembly of LAMP-2A multimer/translocon. Regulation of levels of LAMP-2A at the lysosomal membrane is attained through (1) de novo synthesis and (2) degradation in specialized microdomains at the lysosomal membrane.
CMA: the basics
The discovery that individual cytosolic proteins could be rerouted for lysosomal degradation during nutrient deprivation, motivated attempts to reconstitute this process in vitro using isolated lysosomes [10]. Taking as a model the protease protection assays utilized to study the translocation of proteins inside other organelles, it was demonstrated that proteins presented to intact lysosomes in an assay tube could be detected inside these organelles after incubation at physiological temperatures [11].
Although in the beginning what was being reproduced in the test tube was thought to be some type of microautophagy, the unique characteristics of this transport provided support for a different mechanism of internalization. Lysosomal uptake occurred for some proteins but not for others, and this selectivity for individual proteins did not fit well with a model of internalization inside vesicles that more likely would contain a representative sample of the mixture of presented proteins [10, 11]. In addition, attempts to detect formation of vesicles on the surface of lysosomes during the internalization of proteins in the in vitro system were fruitless.
The studies with isolated lysosomes helped to further characterize the main properties of this new “direct” lysosomal uptake. Binding of substrates to the lysosomal membrane was saturable and addition of increasing concentrations of one substrate protein competed binding of others, supporting the existence of specific substrate-binding sites at the lysosomal membrane [12]. This saturability suggested the participation of a membrane receptor, also supported by the fact that substrate proteins could no longer bind to lysosomal membranes previously exposed to proteases (to eliminate proteins or protein regions exposed on the cytosolic side of the lysosomal membrane) [12].
The use of biochemical, and later on, genetic procedures has contributed to the current molecular characterization of this autophagic pathway, and the identification of a key role for chaperones in this direct lysosomal uptake motivated the renaming of this type of autophagy as chaperone-mediated autophagy or CMA.
Targeting cytosolic proteins one-by-one
All the proteins internalized in lysosomes through CMA contain in their amino acid sequence a pentapeptide motif that is necessary and sufficient for their targeting to lysosomes [13]. Studies with the first prototype substrate, ribonuclease A, demonstrated that removal of the pentapeptide KFERQ completely abolished its lysosomal degradation in response to nutrient deprivation [14]. Furthermore, attachment of this sequence to proteins that are not usually degraded by this lysosomal pathway is enough to induce their degradation through CMA [15]. In fact, this dependence on the KFERQ peptide constitutes the basis for the recent fluorescent reporters developed to monitor CMA in intact cells [15].
The consensus motif is based on the physical properties of the amino acid residues, rather than on the specific amino acids [13], and has been defined as the combination of a positively and a negatively charged residue, a hydrophobic residue and a forth amino acid that can be either positively charged or hydrophobic. These four residues are always flanked by a glutamine on either side (Box 1).
Box 1. Properties of the CMA-targeting motif.
Composition: All CMA-targeting motifs contain: i) one or two of the following positively charged residues: K, R; ii) one or two of the following hydrophobic residues: I, L, V, F; iii) one of the following negatively charged residues: D, E; and iv) one glutamine (Q) on either side of the pentapeptide [13].
Location: The CMA-targeting motif can be located in the C-terminus, N-terminus or in central regions of the protein, the only requirement being that it becomes exposed or accessible for chaperone binding. Scenarios that could promote exposure of the CMA-targeting motif include partial unfolding of the protein, dissociation of protein complexes in which interacting proteins mask the targeting motif, or protein release from intracellular membranes.
Number: Although several cytosolic proteins contain more than one targeting motif, experimental addition of tandem motifs does not make proteins better CMA substrates (one motif suffices for lysosomal targeting) [13]. In proteins with multiple targeting motifs, it is plausible that different motifs become available for chaperone recognition depending on the stimuli that induce the degradation of the protein (i.e. partial unfolding versus dissociation from a protein complex when still fully folded). In proteins that undergo physiological cleavage to release functional peptides, the presence of multiple motifs could guarantee selective removal of the resulting peptides by CMA.
Abundance: Sequence analysis reveals that about 30% of soluble cytosolic proteins contain a putative CMA-targeting motif [16]. However, the fact that posttranslational modifications may generate additional motifs increases the number of possible substrates [18, 19]. Motifs are only detected in soluble cytosolic proteins, or proteins inside organelles that may end in the cytosolic compartment under specific conditions. Membrane proteins cannot be translocated into lysosomes and subsequently this pool of proteins lacks CMA-targeting motifs. The only known exceptions are specific membrane proteins in which the targeting motif is present in regions of the membrane protein that can be released in the cytosol upon cleavage.
Selectivity: Although the presence of a CMA-targeting motif is a sin equanum requirement for all CMA substrates, classification of a protein as CMA substrate cannot be done solely based on this criterion. This motif has recently been shown to be also utilized for targeting of proteins to late endosomes in order to undergo microautophagy [17]. In addition, it is possible that some KFERQ-like motifs are utilized for targeting of proteins that exert a function at the lysosomal membrane and do not necessarily undergo degradation in this compartment [28].
These pentapeptide motifs are recognized specifically by the cytosolic protein hsc70 (heat shock cognate protein of 70KDa), the constitutively expressed member of the hsp70 family of cytosolic chaperones (Figure 1a) [16]. Hsc70 is a multifunctional chaperone that participates in many other cellular functions (i.e. protein refolding, disassembly of multiprotein complexes such as clathrin, protein targeting into other organelles, etc.). For many of these functions, hsc70 interacts with proteins on the basis of their hydrophobicity or other amino acid motifs; however, mutagenesis of the KFERQ-like motif in substrate proteins has demonstrated that this is the only binding region used by hsc70 for their delivery to lysosomes [16].
A very recent development has been the discovery that binding of hsc70 to the KFERQ region of cytosolic proteins can also mediate their delivery to late endosomes where they undergo degradation through endosomal microautophagy (e-MI) [17] (the characteristics of this pathway and the differences between CMA and e-MI are summarized in Box 2).
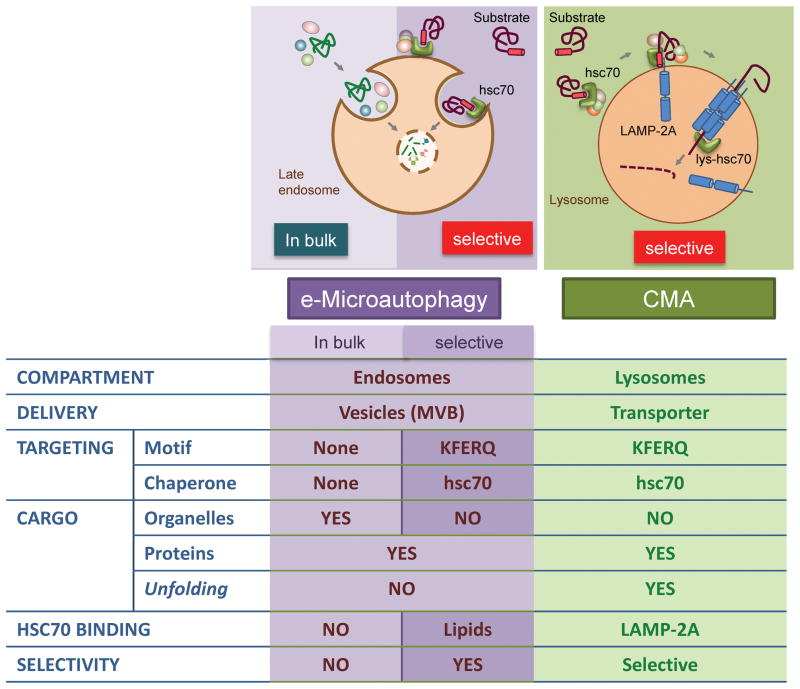
Box 2. CMA and microautophagy – how are they different?
Out of the three autophagic pathways, microautophagy has been the least studied, with mainly morphological evidence demonstrating the entrapment of cytosolic components via the invaginations of the lysosomal membrane, which subsequently pinch off and get degraded [8]. Recently, a similar internalization of cytosolic components has been described to occur in late endosomes [17]. Interestingly, in this process, now termed endosomal microautophagy (e-MI), apart from the in-bulk entrapment of cargo, selective internalization of cytosolic proteins also occurs. Selective e-MI and CMA share the requirement for the KFERQ-like motif in their substrates, which are recognized by the cytosolic chaperone hsc70 [17]. However, e-MI does not require LAMP-2A for binding of the hsc70/substrate complex, which instead directly binds the phospholipid phosphatidylserine at the endosomal membrane through specific residues in hsc70. Also, unfolding of substrate proteins is not a requirement for e-MI [17] (similarities and differences between e-MI and CMA are summarized in Figure I).
Contrary to other degradation tags, such as ubiquitin, that are added to a protein only when it needs to be degraded, the KFERQ-motif is always present in the substrate sequence; how then is this degradation regulated? Regulation may depend, for the most part, on whether or not this motif is accessible to the chaperone. Partial unfolding may favor recognition of motifs buried in the core of the folded proteins, and the motif could also be masked by interacting proteins or by binding to intracellular membranes. Subcompartmentalization may also make targeting possible only when the proteins reach the cytosol. Recent studies have also revealed regulation by posttranslational modifications in proteins missing only one of the residues necessary to complete a CMA-targeting motif. For example, phosphorylation can contribute a missing negatively charged residue, or acetylation of a lysine can provide a missing glutamine (as acetylated lysine is comparable to glutamine). In fact, acetylation of the glycolytic enzyme embryonic M2 isoform of pyruvate kinase (PKM2) initiates its lysosomal degradation by CMA [18]. Likewise, degradation of the N-terminal region of huntingtin is initiated by phosphorylation followed by acetylation that completes a targeting motif [19]. Generation of CMA-targeting motifs through posttranslational modifications expands the subset of cellular proteins that can undergo degradation by this pathway and provides an additional level of regulation for this type of degradation. A list of proteins carrying the KFERQ-like motif confirmed experimentally to be CMA substrates can be found in [20].
Cytosolic hsc70: the decision maker
Hsc70 is the only chaperone demonstrated to mediate substrate targeting for CMA [16]. The chaperone CHIP has been shown to mediate lysosomal degradation of proteins such as alpha-synuclein, previously identified as a CMA substrate [21], however, whether CHIP-mediated degradation occurs by CMA or other forms of autophagy requires further investigation.
Hsc70 also targets KFERQ-containing proteins to late endosomes for e-MI (Box 2) [17], and has been shown to interact with protein aggregates and mediate their degradation by macroautophagy through a process named chaperone-assisted selective autophagy (CASA) [22]. Whether or not CASA requires the KFERQ-motif is still a pending question. Of interest now, is to identify the factors that determine targeting through one type of autophagy or another once hsc70 interacts with a KFERQ-bearing protein. It is possible that different subsets of cochaperones, when associated to hsc70, modulate targeting to one particular pathway. The fact that the KFERQ-motif is necessary and sufficient for CMA, and only necessary but not sufficient for e-MI supports the possible participation of additional proteins/factors in e-MI targeting [17]. Alternatively, properties of the substrate protein such as its ability to undergo complete unfolding, an absolute requirement for CMA [23] but not for e-MI [17], may also determine its degradative fate.
Cytosolic hsc70 and its associated cochaperones may participate in steps of CMA beyond substrate targeting (Figure 1). For example, substrate unfolding could be the responsibility of the same hsc70 and cochaperones involved in the targeting of substrate to lysosomes. However, it is also possible that chaperones resident at the cytosolic side of the lysosomal membrane are the main unfoldases of the CMA substrates, because unfolding is not required for membrane binding, but only for translocation [23].
LAMP-2A at lysosomal entry
Binding of substrate proteins to the lysosomal membrane occurs at the cytosolic tail of the single span membrane protein LAMP-2A (lysosome-associated membrane protein type 2A) (Figure 1b) [24]. This protein is one of the three splice variants of the gene lamp2 that share identical lumenal regions and different transmembrane and cytosolic tails [25]. Substrate binding to LAMP-2A is limiting for CMA and, in fact, cells use changes in the levels of this membrane protein to rapidly upregulate or downregulate CMA [26]. Changes in the rates of LAMP-2A synthesis, its regulated degradation at the lysosomal membrane and its subcompartmentalization in this organelle all contribute to modulate CMA activity (Figure 1).
Binding of substrate proteins to monomeric forms of LAMP-2A promotes its assembly into a high molecular weight complex of about 700KDa at the lysosomal membrane (Figure 1c,d) [27]. Multimerization is required for substrate translocation as LAMP-2A mutants unable to associate into this complex still bind substrate proteins but prevent translocation. Unexpectedly, the translocation complex is not stable at the lysosomal membrane and once the substrate has been released into the lysosomal lumen, LAMP-2A dissociates into monomers (Figure 1e) [27]. Disassembly is an active process mediated by hsc70 that generates monomeric forms of LAMP-2A required for substrate binding to sustain cycles of binding and uptake.
Different subsets of proteins have been described to modulate the stability of the CMA translocation complex. In addition of the disassembling function of hsc70, the pair of proteins glial fibrillary acidic protein (GFAP) and elongation factor 1 alpha (EF1α) work in a coordinated manner to regulate the amount of translocation complex at the lysosomal membrane [27]. GFAP associates to the lysosomal membrane in two forms, a non-phosphorylated variant that binds and stabilizes LAMP-2A in the multimeric complex, and phosphorylated GFAP (pGFAP) that binds the lysosomal membrane outside this complex, usually associated to EF1α (Figure 1d). Binding to this GTP-binding protein prevents pGFAP from dimerizing with GFAP. However, in the presence of GTP, EF1α is released from the membrane and GFAP abandons the translocation complex to associate with pGFAP, resulting in the disassembly of the translocation complex and the subsequent decrease in CMA [28]. Oscillatory changes in GTP concentration in the proximity of the lysosome may contribute to regulate CMA activity under physiological conditions.
Membrane lipids also exert a modulatory effect on CMA by affecting the stability of LAMP-2A at the lysosomal membrane. Animals subjected to diets with high lipid content show reduced CMA activity due to the instability of LAMP-2A in their lysosomes [29]. Comparative lipidomic analysis revealed that these diets induce qualitative and quantitative changes in the lipid compositions of the lysosomal membrane that alter LAMP-2A multimerization and favor its recruitment to specific membrane microdomains where this protein is normally degraded [30] (Figure 1). Interestingly, the changes in the lipid profile resemble those observed during physiological aging, a condition also associated with reduced CMA activity.
Lysosomal chaperones in CMA
Besides the cytosolic form of hsc70, variants of this protein associated with lysosomes also contribute to CMA. Levels of lysosome-associated hsc70 (lys-hsc70) increase proportionally to the increase in CMA activity and become limiting when LAMP-2A is in excess. Part of lys-hsc70 is localized in the lumen of lysosomes and blockage of this intralysosomal hsc70, with antibodies delivered by endocytosis, does not affect membrane binding of substrate proteins but abolishes their internalization [31]. The presence or not of luminal lys-hsc70 has helped to identify lysosomal populations with different competence for CMA [32]. Although LAMP-2A is present in all types of lysosomes, only the subset of lysosomes that contain lys-hsc70 is competent for substrate uptake. The percentage of CMA-active lysosomes (containing both hsc70 and CMA substrates) increases in response to the stressors that upregulate this pathway (i.e. starvation, oxidative stress) and can change from 20%, in resting conditions, to up to 80% of the total cellular lysosomes when CMA is maximally active [32].
The electrophoretic pattern of cytosolic and lys-hsc70 is markedly different (9 cytosolic isoforms and only 2 in lysosomes) [31], but the fact that cytosolic hsc70 substitutes for lys-hsc70 when artificially introduced inside lysosomes incompetent for CMA [32], suggests that both forms of hsc70 likely originate from the same gene, and that only modified forms of hsc70 can reach the lysosomal lumen through yet unknown mechanisms. Further studies are needed to identify the basis for the resistance of hsc70 to degradation in this compartment. Other lysosome-resident proteins avoid digestion through assembly into semi-aggregated complexes from which they are released in a pH-dependent manner. It is interesting that higher acidification is a distinctive characteristic of CMA-competent lysosomes and that small increase in their pH is enough to destabilize and rapidly degrade lys-hsc70 [32].
Hsc70 also associates with the cytosolic side of the lysosomal membrane where it may contribute to unfolding of the substrate proteins [33] and active disassembly of the LAMP-2A translocation complex [27]. Hsp90 localizes on both sides of the lysosomal membrane, but the luminal form is tightly bound to the lysosomal membrane [27] (Figure 1d). This luminal variant stabilizes LAMP-2A when transitioning from a monomeric to a multimeric state. To date, the specific cochaperones modulating the activity of the membrane-associated hsc70 and hsp90 have not been identified. In addition, it is not known if there are luminal lysosome-specific cochaperones or how the intralysosomal chaperones obtain the ATP required for their function.
Physiology of CMA
Independent of the cell type, CMA contributes to two critical functions in cellular physiology – maintenance of energy homeostasis and protein quality control. In addition, CMA also takes part in cell type-specific functions that depend on the substrate protein being degraded by this pathway (Figure 2).
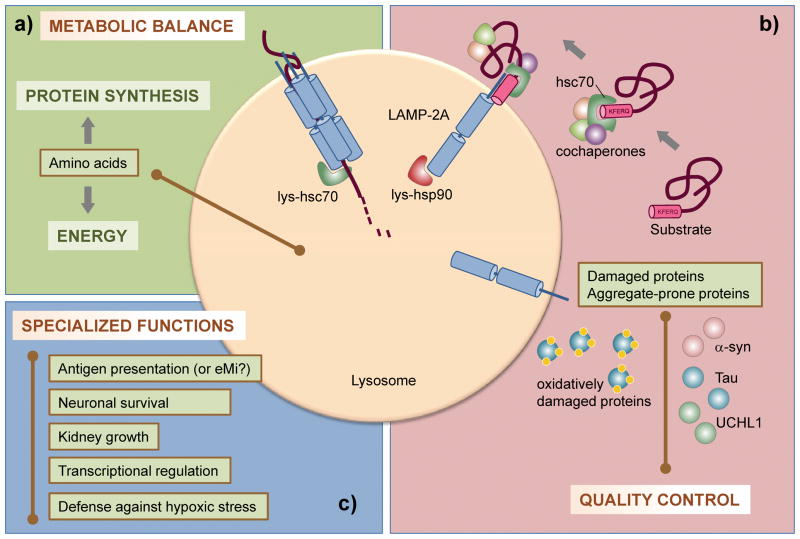
Figure 2.
Functions of CMA. a) By degrading no longer needed proteins during nutrient deprivation, CMA provides amino acids for the synthesis of needed proteins and generates energy. b) By degrading damaged and prone-to-aggregate proteins, CMA is an essential component of the cellular quality control system. c) CMA also has specialized functions, depending on the substrate protein and cells/tissues involved. Some examples discussed in the text are listed.
General functions
In almost all cell types, nutrient scarcity maximally activates CMA [34, 35]. Degradation of proteins through this autophagic pathway provides free amino acids to sustain synthesis of other proteins (Figure 2a). Activation of CMA is initiated after ~10 hours of starvation and follows the upregulation of macroautophagy that typically occurs ~30 minutes into the starvation and does not last more than 8–10 hours. The selectivity of CMA for single soluble cytosolic proteins, might allow degradation of proteins no longer required (such as glycolytic enzymes [11] or catalytic subunits of the 20S proteasome [36]) and channel their amino acids for the synthesis of proteins needed [37]. In tissues such as liver, CMA activity plateaus at ~36 hours of starvation and remains active until 3 days [34]. In addition to assuring protein synthesis, part of the amino acids provided through CMA can be utilized by the cells to generate energy, as CMA-deficient cells have reduced levels of ATP when deprived of nutrients [38].
Another important function of CMA is protein quality control via the selective degradation of damaged or altered proteins (Figure 2b). CMA is upregulated in response to stressors that cause protein damage, such as mild oxidative stress [39, 40], or protein denaturing compounds [41]. Partial unfolding or formation of protein adducts can upregulate degradation of certain proteins via CMA, preventing their aggregation into insoluble protein inclusions [42, 43]. In fact, oxidized and aggregated proteins accumulate in CMA-incompetent cells, rendering them more susceptible to a variety of stressors and further supporting the contribution of CMA to maintain the stability of the proteome [38].
Recent studies have reported CMA activation in response to hypoxic stress and the need for functional CMA under these conditions to guarantee neuronal survival [44]. Future studies are needed to clarify whether CMA is required to maintain the energetic cellular balance or to facilitate removal of altered components during hypoxia. However, the fact that CMA activation occurs 7 days after ischemic injury is more supportive of the latter possibility.
Specific functions
CMA participates in specialized functions, in a cell-type specific context, depending on the substrate proteins being degraded by this pathway (Figure 2c).
Several well-characterized CMA substrates are transcription factors or inhibitors of transcription factors, making transcriptional regulation an important function of CMA. For example, during nutrient deprivation, CMA degrades a fraction of cellular IκBα, the endogenous inhibitor of the transcription factor NFκB, allowing NFκB to activate the transcriptional program required for adaptation to prolonged starvation [45]. Changes in the oxidative status of IκBα during starvation are the trigger for its degradation by CMA. In other instances, CMA controls changes in transcription factors only in specific tissues. For example, malfunctioning CMA in kidney epithelial cells has been shown to compromise the degradation of the paired box gene 2 (Pax2), a protein important for cell proliferation and differentiation [46]. Failure to degrade Pax2 in these cells results in abnormal growth of kidney cells and kidney hypertrophy. Similarly, functional compromise of CMA in neurons favors accumulation of inactive molecules of the myocyte-specific enhancer factor 2D (MEF2D), a transcription factor required for neuronal survival, rendering cells more susceptible to stressors [47].
CMA also contributes to MHC class II antigen presentation in professional antigen-presenting cells (macrophages) (Figure 2c). Manipulations in hsc70 and hsp90 in these cells reduce their CMA rates and antigen presentation [48, 49]. In light of the recently described participation of hsc70 in the delivery of cytosolic proteins to late endosomes, a compartment classically associated with antigen presentation, it now becomes of great interest to elucidate the relative contribution of CMA and e-MI to this immune function. More CMA functions are bound to be added to the already growing list, as new substrates for CMA are identified.
CMA in pathology and aging
Neurodegeneration
Over the past years, defects in CMA have been linked to the pathogenesis of some severe neurodegenerative disorders. Accumulation of pathogenic proteins, in the form of insoluble inclusions, is a common factor underlying all these diseases. Many of these pathogenic proteins bear CMA-targeting motifs making them potential CMA substrates. For example, KFERQ-like sequences have been identified in Parkinson’s disease (PD)-associated proteins alpha-synuclein, parkin, UCHL1 and pink-1; the Alzheimer’s disease (AD)-associated proteins APP and tau; and the protein mutated in Huntington’s disease (HD), huntingtin. These proteins can be degraded by CMA only when they are in soluble forms; once the insoluble inclusions are formed, they can only be degraded via other proteolytic pathways [50].
Interestingly, in many instances not only is CMA unable to degrade the pathogenic variants of these proteins, but they also exert an inhibitory effect on CMA function. Studies with isolated lysosomes [42], neuronal cultured cells [42, 43, 51], and mouse models of PD [52] demonstrated that wild-type alpha-synuclein is a CMA substrate, but its pathogenic variants are less amenable to this autophagic degradation. Pathogenic alpha-synucleins are properly recognized by cytosolic hsc70 and targeted to lysosomes, but they fail to cross the lysosomal membrane due to their organization into irreversible oligomeric complexes, which at the same time, block the degradation of other CMA substrates via sheer steric hindrance (Figure 3a) [42, 43]. Similar abnormal interaction with CMA components has been shown for the mutant forms of UCHL1 [53].
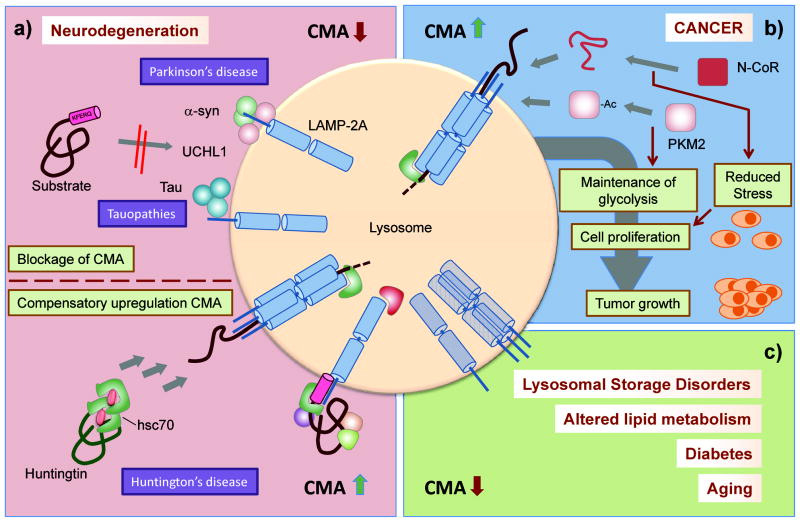
Figure 3.
Pathology of CMA. a) Top: CMA is primarily compromised in some neurodegenerative diseases due to direct blockage of the translocation machinery by the pathogenic proteins. Bottom: Upregulation of CMA activity in Huntington’s disease compensates for the blockage in other degradative pathways and has been utilized for artificial targeting of the toxic protein. b) Cancer cells depend on CMA to sustain cellular proliferation and tumor growth. Continuous CMA activation maintains glycolytic activity and reduces stress in cancer cells. c) CMA activity is compromised in disorders that compromise lysosomal function and in metabolic diseases such as those related to alterations in lipid metabolism and diabetes. CMA activity also decreases with age.
Two proteins related with AD, tau [54] and regulator of calcineurin 1 (RCAN1) [55] have been confirmed as CMA substrates. Mutant variants of tau demonstrate impaired CMA activity, in this case due to their partial translocation across the membrane, that results in their cleavage while still bound to the membrane and generation of toxic amyloidogenic peptides that disrupt lysosomes [54]. Blocking CMA of this Tau mutant by preventing its binding to hsc70 improves cell viability as it reduces lysosomal toxicity [54].
Interestingly, in the case of HD, CMA acts more as a rescue compensatory pathway for the primary defects in the ubiquitin-proteasome system and in macroautophagy that occur in the affected cells [56–58]. HD models and cells from patients display constitutive activation of CMA [59], and experimental upregulation of CMA decreases toxicity in HD brain slices [19]. This compensatory activation is possible because of the close interrelation of CMA with the other proteolytic systems (Box 3). Interestingly, despite the presence of multiple KFERQ-like motifs in huntingtin, the protein mutated in HD patients, its degradation via CMA under normal conditions is negligible [59]. However, recent studies have succeeded in artificially enhancing targeting of mutant huntingtin for CMA degradation [60], supporting that the constitutive upregulation of CMA in this disorder could be exploited for therapeutic purposes (Figure 3a).
Box 3. CMA in the context of other proteolytic systems.
Autophagy and the ubiquitin-proteasome system coexist in all mammalian cells and growing evidence supports some level of crosstalk between them. As part of the autophagic system, CMA actively interacts with other autophagic pathways and with the ubiquitin-proteasome system.
Crosstalk with macroautophagy
Cultured cells knocked down for the CMA receptor, LAMP-2A, show compensatory upregulation of macroautophagy [38]. Even though macroautophagy and CMA are distinct and not redundant in terms of specific functions, they can compensate for each other. Cells respond to blockage of one of these pathways by upregulating the other, and in the absence of any added stress, this compensation is enough to maintain normal cell viability. However, when the cells are exposed to stressors (i.e. mild oxidation, ultraviolet light, toxic compounds), which increase abundance of damaged proteins normally degraded by CMA, the cells with compromised CMA become significantly less viable, despite macroautophagic activation [38]. Conversely, cells with abrogated macroautophagic activity demonstrate constitutive upregulation of CMA [73], highlighting the bidirectional nature of the crosstalk between these two pathways. Compensatory activation of CMA in response to macroautophagic compromise has been confirmed in vivo in an HD mouse model, highlighting the importance of the autophagic crosstalk in the prevention of pathological manifestations [59]. Although the mechanisms that modulate the intercommunication between macroautophagy and CMA remain, for the most part unknown, in the HD setting, the increase in CMA was attained through both transcriptional upregulation and downregulation of degradation of LAMP-2A to increase the levels of this receptor [59]. Increased CMA in HD seems to be initially sufficient to maintain proteomic stability, but the functional decline in this pathway with age accelerates pathological manifestations of this neurodegenerative disorder.
Crosstalk with the ubiquitin-proteasome system
The crosstalk between the autophagic pathways is also extended to the ubiquitin-proteasome system. Acute blockage of the proteasome results in both activation of macroautophagy [74] and of CMA [15], whereas, chronic blockage leads to macroautophagic deregulation [75]. In contrast, inhibition of macroautophagy seems to exert a negative effect on the proteasome, in part through competition for the enzymes that remove the ubiquitin tag from the substrates [76]. The blockage of these two major proteolytic pathways under these conditions makes even more relevant the compensatory activation of CMA in response to blockage of either of them. Specific proteasome activities are also compromised in situations with chronic blockage of CMA [38]. It is possible that the decrease in the proteolytic activities of the proteasome observed in cells with compromised CMA is a consequence of their poor turnover, as some of the subunits of the proteasome have been previously identified as bona fide CMA substrates [36].
Cancer
Growing numbers of connections have been recently established between CMA and cancer biology. CMA has been shown to be upregulated in a wide collection of cancer cell lines and levels of LAMP-2A, a surrogate for CMA activation, are also consistently higher in human tumor samples than in the corresponding healthy tissues [61]. Genetic manipulations have revealed that CMA is essential for cancer cell proliferation, tumor growth and generation of metastasis and that blockage of CMA in already-established tumors induces their shrinkage [61]. The favorable effect of CMA on cancer cell proliferation likely results from the combination of several mechanisms (Figure 3b). So far, the strongest candidate is the connection between CMA and glucose metabolism. Cancer cells are highly dependent on glycolysis for the generation of ATP and intermediate metabolites that promote cell proliferation. CMA activation in cancer cells is required to maintain upregulated transcriptional expression of rate-limiting glycolytic enzymes contributing to increased flux through this pathway [61]. In addition, CMA specifically regulates the balance between the active and inactive form of PKM2, an enzyme highly expressed in tumor cells, recently identified to be a CMA substrate [18]. Interestingly, CMA reduced the levels of the inactive enzyme upon its tagging by acetylation and thus maintains a positive balance for the active form [18].
Beyond the glycolytic control, in specific types of cancer CMA reduces the cellular stress of the rapidly growing tumor. For example, degradation by CMA of the unfolded protein nuclear receptor co-repressor (N-CoR) highly expressed in some types of lung cancer ameliorates ER stress and promotes the activation of survival pathways in these cells (Figure 3b) [62].
Despite this apparent favorable impact of CMA in cancer progression, CMA could exert an anti-oncogenic effect in normal cells by maintaining the cellular response to stressors and reducing cellular damage. In addition, CMA contributes to the degradation of at least one pro-oncogenic protein, the epidermal growth factor receptor pathway substrate 8 (Eps8), associated with promotion of numerous solid malignancies [63]. Future studies are needed to better characterize the contribution of CMA to the prevention of cancer.
Other pathologies
Changes in CMA have been described in different toxic-induced kidney pathologies. CMA is initially upregulated to eliminate the altered proteins generated by the toxic insult, but eventually becomes insufficient to fight against toxicity [41]. The kidney is also the organ predominantly affected by the decrease in CMA observed in diabetic mouse models, where compromised CMA contributes to the diabetes-induced nephropathy [64]. Diseases affecting lysosomes (i.e. lysosomal storage disorders) also impact CMA indirectly because the undegraded material that accumulates in lysosomes in these disorders interferes with normal lysosomal functioning. In addition, CMA is primarily affected in lysosomal disorders such as galactosialidosis or mucolipidosis, because the protein mutated in these conditions also has a function in CMA [65, 66].
Aging
Decline in CMA activity with age has been described in almost all cell types and tissues, both in rodents and in cells derived from human subjects [67, 68]. This decrease is primarily due to a reduction in the levels of LAMP-2A at the lysosomal membrane [67]. Transcription, synthesis and targeting of LAMP-2A to lysosomes once synthesized, occur at similar rates in young and old individuals. Instead, the stability of LAMP-2A at the lysosomal membrane is markedly reduced with age resulting in a decrease in the net content of this protein in lysosomes [69]. Changes in the lipid composition of the lysosomal membrane with age are one of the primary reasons for the reduced stability of LAMP-2A [29, 30]. Genetic manipulations aimed at preventing the decrease of LAMP-2A with age by expressing an exogenous copy of this transgene in mouse liver, have proven sufficient to restore CMA activity, improve cellular homeostasis and preserve the function of this organ in aged rodents [70]. These studies support the notion of an active anti-aging function for CMA and the possible therapeutic value of modulating levels of LAMP-2A in aged organisms to extend their health-span.
Concluding remarks
In summary, the identification of selective degradation of proteins by CMA has expanded the number of regulatory functions attributable to the breakdown of proteins in lysosomes. Despite the number of outstanding questions about this autophagic pathway (Box 4), recent studies have stressed that cytosolic hsc70 determines which proteins are degraded by CMA and when, but the rate of degradation depends on the unique characteristics of the transient translocation system. The methods currently available to study CMA allow for measurement of both the overall activity of this pathway as well as changes in its individual steps [71, 72]. Combination of both types of approaches should be useful to identify additional pathological conditions in which CMA is altered and to dissect the steps of CMA responsible for its failure.
Box 4. Outstanding questions.
Despite recent advances in our understanding of the molecular mechanisms that govern CMA and its contribution to cellular physiology, there are still important pending questions about this autophagic pathway:
What are the groups of CMA substrates that reach this pathway upon posttranslational modifications to complete a CMA-targeting motif? Are there dedicated kinases and acetylases to modulate their CMA targeting?
How does hsc70 discriminate between targeting for CMA and e-MI? What are the subsets of cytosolic and lysosomal cochaperones that modulate hsc70 function in CMA?
What are the changes in the lipidome and proteome of the lysosomal membrane that physiologically contribute to regulation of CMA?
What are the energetic requirements and driving forces of substrate translocation through the LAMP-2A-enriched multimeric complex?
What are the molecular regulators of the crosstalk of CMA with other proteolytic pathways? Does this crosstalk expand to e-MI?
Is the age-related defect of CMA universal, or are there cell-type specific differences in the steps and components of this pathway responsible for its functional decline in aging?
Does CMA contribute to cellular functions common for other autophagic pathways such as host defense against pathogens, cellular remodeling, protein secretion, programmed cell death, etc.?
All together, the array of cellular functions in which CMA participates highlights the importance of this autophagic pathway in the forefront of response to cellular stress. The ability of CMA to identify and degrade single proteins, a characteristic shared with the proteasome, makes it a preferred route when removal of specific proteins is needed (selective degradation of proteins has been described for macroautophagy but only when they form part of large protein complexes). However, as for other cellular processes, the strength of CMA (the one-by-one translocation of single proteins to allow for selective removal) also becomes its weakness under pathological conditions, due to the ease of clogging of this translocation system by abnormal proteins.
Interventions to prevent CMA blockage and to improve its functioning in old organisms may have therapeutic value in age-related diseases. In contrast, the recently described dependence of cancer cells on CMA suggests that inhibition of this pathway may be of use as anti-oncogenic therapy. In this respect, it is possible that neutralizers of lysosomal pH such as cloroquine, currently in use in anti-cancer clinical trials, exert their effect in part by compromising CMA, because the lysosome is shared by the different autophagic pathways. However, the challenge remains in designing more selective ways to compromise CMA activity without affecting the other types of autophagy.
Figure I.
Comparison of endosomal-microautophagy (e-MI) and chaperone-mediated autophagy (CMA)
Acknowledgments
Work in our laboratory is supported by NIH grants from NIA, NINDS, the Beatrice and Roy Backus Foundation, the Rainwaters Foundation, a Hirsch/Weill-Caulier Career Scientist Award to AMC and the generous support of Robert and Renee Belfer. SK is supported by a NIH/NIA training grant.
Glossary
- Autophagy
degradation of intracellular proteins and organelles in lysosomes
- Cathepsins
proteases located inside lysosomes that reach maximal activity at acidic pH
- Chaperone
protein that assists other proteins in their folding and unfolding and intracellular trafficking by preventing non-specific interactions with other surrounding proteins
- Chaperone-mediated autophagy
autophagic pathway in which cytosolic proteins are targeted one-by-one to the surface of the lysosome from where they reach the lumen by crossing the lysosomal membrane
- Endosomal microautophagy
degradation of cytosolic proteins in late endosomes after internalization by mechanisms that resemble those of microautophagy
- Lysosome
single-membrane bound vesicles fully dedicated to breakdown of cellular structures for their recycling. They are characterized by their luminal acidic pH and their high abundance of hydrolases
- Lysosome-associated membrane proteins
family of single-span membrane proteins preferentially located in lysosomes that display short C-terminus (10–12 amino acids) at the cytosolic side of the lysosomal membrane. The luminal part of these proteins is heavily glycosylated to protect them from degradation
- Macroautophagy
autophagic pathway in which cytosolic proteins and organelles are sequestered into a double-membrane vesicle and then fuse with lysosomes to assure their degradation
- Microautophagy
internalization of cytosolic proteins and organelles into lysosomes through invaginations of the lysosomal membrane that then pinch off as single-membrane vesicles into the lysosomal lumen
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Saftig P, Klumperman J. Lysosome biogenesis and lysosomal membrane proteins: trafficking meets function. Nat Rev Mol Cell Biol. 2009;10:623–635. doi: 10.1038/nrm2745. [DOI] [PubMed] [Google Scholar]
- 2.Mizushima N, et al. Autophagy fights disease through cellular self-digestion. Nature. 2008;451:1069–1075. doi: 10.1038/nature06639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dikic I, et al. Selective autophagy in cancer development and therapy. Cancer Res. 2010;70:3431–3434. doi: 10.1158/0008-5472.CAN-09-4027. [DOI] [PubMed] [Google Scholar]
- 4.Yang Z, Klionsky DJ. Mammalian autophagy: core molecular machinery and signaling regulation. Curr Opin Cell Biol. 2010;22:124–131. doi: 10.1016/j.ceb.2009.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cuervo AM. Chaperone-mediated autophagy: Dice’s ‘wild’ idea about lysosomal selectivity. Nat Rev Mol Cell Biol. 2011;12:535–541. doi: 10.1038/nrm3150. [DOI] [PubMed] [Google Scholar]
- 6.De Duve C, Wattiaux R. Functions of lysosomes. [Review] Ann Rev Physiol. 1966;28:435–492. doi: 10.1146/annurev.ph.28.030166.002251. [DOI] [PubMed] [Google Scholar]
- 7.Mortimore GE, Pösö AR. The lysosomal pathway of intracellular proteolysis in liver: regulation by amino acids. Adv Enzyme Regul. 1986;25:257–276. doi: 10.1016/0065-2571(86)90018-x. [DOI] [PubMed] [Google Scholar]
- 8.Mortimore GE, et al. Regulation of microautophagy and basal protein turnover in rat liver. Effects of short-term starvation. Journal of Biological Chemistry. 1988;263:2506–2512. [PubMed] [Google Scholar]
- 9.Neff N, et al. Degradation of proteins microinjected into IMR-90 human diploid fibroblasts. J Cell Biol. 1981;91:184–194. doi: 10.1083/jcb.91.1.184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Terlecky S, Dice J. Polypeptide import and degradation by isolated lysosomes. J Biol Chem. 1993;268:23490–23495. [PubMed] [Google Scholar]
- 11.Aniento F, et al. Uptake and degradation of glyceraldehyde-3- phosphate dehydrogenase by rat liver lysosomes. J Biol Chem. 1993;268:10463–10470. [PubMed] [Google Scholar]
- 12.Cuervo AM, et al. Selective binding and uptake of ribonuclease A and glyceraldehyde-3-phosphate dehydrogenase by isolated rat liver lysosomes. J Biol Chem. 1994;269:26374–26380. [PubMed] [Google Scholar]
- 13.Dice JF. Peptide sequences that target cytosolic proteins for lysosomal proteolysis. Trends Biochem Sci. 1990;15:305–309. doi: 10.1016/0968-0004(90)90019-8. [DOI] [PubMed] [Google Scholar]
- 14.Dice JF, et al. Regulation of catabolism of microinjected ribonuclease A: Identification of residues 7–11 as the essential pentapeptide. J Biol Chem. 1986;262:6853–6859. [PubMed] [Google Scholar]
- 15.Koga H, et al. A photoconvertible fluorescent reporter to track chaperone-mediated autophagy. Nat Commun. 2011;2:386. doi: 10.1038/ncomms1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chiang H, et al. A role for a 70-kilodalton heat shock protein in lysosomal degradation of intracellular proteins. Science. 1989;246:382–385. doi: 10.1126/science.2799391. [DOI] [PubMed] [Google Scholar]
- 17.Sahu R, et al. Microautophagy of cytosolic proteins by late endosomes. Develop Cell. 2011;20:131–139. doi: 10.1016/j.devcel.2010.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lv L, et al. Acetylation targets the M2 isoform of pyruvate kinase for degradation through chaperone-mediated autophagy and promotes tumor growth. Mol Cell. 2011;42:719–730. doi: 10.1016/j.molcel.2011.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Thompson LM, et al. IKK phosphorylates Huntingtin and targets it for degradation by the proteasome and lysosome. J Cell Biol. 2009;187:1083–1099. doi: 10.1083/jcb.200909067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kaushik S, et al. Chaperone-mediated autophagy at a glance. J Cell Sci. 2011;124:495–499. doi: 10.1242/jcs.073874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shin Y, et al. The co-chaperone carboxyl terminus of Hsp70-interacting protein (CHIP) mediates alpha-synuclein degradation decisions between proteasomal and lysosomal pathways. J Biol Chem. 2005;280:23727–23734. doi: 10.1074/jbc.M503326200. [DOI] [PubMed] [Google Scholar]
- 22.Arndt V, et al. Chaperone-assisted selective autophagy is essential for muscle maintenance. Curr Biol. 2010;20:143–148. doi: 10.1016/j.cub.2009.11.022. [DOI] [PubMed] [Google Scholar]
- 23.Salvador N, et al. Import of a cytosolic protein into lysosomes by chaperone-mediated autophagy depends on its folding state. Journal of Biological Chemistry. 2000;275:27447–27456. doi: 10.1074/jbc.M001394200. [DOI] [PubMed] [Google Scholar]
- 24.Cuervo AM, Dice JF. A receptor for the selective uptake and degradation of proteins by lysosomes. Science. 1996;273:501–503. doi: 10.1126/science.273.5274.501. [DOI] [PubMed] [Google Scholar]
- 25.Gough NR, et al. The family of LAMP-2 proteins arises by alternative splicing from a single gene: characterization of the avian LAMP-2 gene and identification of mammalian homologs of LAMP-2b and LAMP-2c. DNA Cell Biol. 1995;14:863–867. doi: 10.1089/dna.1995.14.863. [DOI] [PubMed] [Google Scholar]
- 26.Cuervo AM, Dice JF. Regulation of lamp2a levels in the lysosomal membrane. Traffic. 2000;1:570–583. doi: 10.1034/j.1600-0854.2000.010707.x. [DOI] [PubMed] [Google Scholar]
- 27.Bandyopadhyay U, et al. The chaperone-mediated autophagy receptor organizes in dynamic protein complexes at the lysosomal membrane. Mol Cell Biol. 2008;28:5747–5763. doi: 10.1128/MCB.02070-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bandyopadhyay U, et al. Identification of regulators of chaperone-nediated autophagy. Mol Cell. 2010;39:535–547. doi: 10.1016/j.molcel.2010.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rodriguez-Navarro JA, et al. Inhibitory effect of dietary lipids on chaperone-mediated autophagy. Proc Natl Acad Sci U S A. 2012 doi: 10.1073/pnas.1113036109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kaushik S, et al. Lysosome membrane lipid microdomains: novel regulators of chaperone-mediated autophagy. EMBO J. 2006;25:3921–3933. doi: 10.1038/sj.emboj.7601283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Agarraberes F, et al. An intralysosomal hsp70 is required for a selective pathway of lysosomal protein degradation. J Cell Biol. 1997;137:825–834. doi: 10.1083/jcb.137.4.825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cuervo AM, et al. A population of rat liver lysosomes responsible for the selective uptake and degradation of cytosolic proteins. J Biol Chem. 1997;272:5606–5615. doi: 10.1074/jbc.272.9.5606. [DOI] [PubMed] [Google Scholar]
- 33.Agarraberes F, Dice JF. A molecular chaperone complex at the lysosomal membrane is required for protein translocation. J Cell Sci. 2001;114:2491–2499. doi: 10.1242/jcs.114.13.2491. [DOI] [PubMed] [Google Scholar]
- 34.Cuervo AM, et al. Activation of a selective pathway of lysosomal proteolysis in rat liver by prolonged starvation. Am J Physiol. 1995;269:C1200–C1208. doi: 10.1152/ajpcell.1995.269.5.C1200. [DOI] [PubMed] [Google Scholar]
- 35.Wing S, et al. Proteins containing peptide sequences related to KFERQ are selectively depleted in liver and heart, but not skeletal muscle, of fasted rats. Biochem J. 1991;275:165–169. doi: 10.1042/bj2750165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cuervo AM, et al. Degradation of proteasomes by lysosomes in rat liver. Eur J Biochem. 1995;227:792–800. doi: 10.1111/j.1432-1033.1995.tb20203.x. [DOI] [PubMed] [Google Scholar]
- 37.Backer J, Dice J. Covalent linkage of ribonuclease S-peptide to microinjected proteins causes their intracellular degradation to be enhanced by serum withdrawal. Proc Nat Acad Sci USA. 1986;83:5830–5834. doi: 10.1073/pnas.83.16.5830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Massey AC, et al. Consequences of the selective blockage of chaperone-mediated autophagy. Proc Nat Acad Sci USA. 2006;103:5905–5910. doi: 10.1073/pnas.0507436103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Finn PF, Dice JF. Ketone bodies stimulate chaperone-mediated autophagy. J Biol Chem. 2005;280:25864–25870. doi: 10.1074/jbc.M502456200. [DOI] [PubMed] [Google Scholar]
- 40.Kiffin R, et al. Activation of chaperone-mediated autophagy during oxidative stress. Mol Biol Cell. 2004;15:4829–4840. doi: 10.1091/mbc.E04-06-0477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cuervo AM, et al. Direct lysosomal uptake of alpha2-microglobulin contributes to chemically induced nephropathy. Kidney Int. 1999;55:529–545. doi: 10.1046/j.1523-1755.1999.00268.x. [DOI] [PubMed] [Google Scholar]
- 42.Cuervo AM, et al. Impaired degradation of mutant alpha-synuclein by chaperone-mediated autophagy. Science. 2004;305:1292–1295. doi: 10.1126/science.1101738. [DOI] [PubMed] [Google Scholar]
- 43.Martinez-Vicente M, et al. Dopamine-modified alpha-synuclein blocks chaperone-mediated autophagy. J Clin Invest. 2008;118:777–788. doi: 10.1172/JCI32806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dohi E, et al. Hypoxic stress activates chaperone-mediated autophagy and modulates neuronal cell survival. Neurochem Int. 2012;60:431–442. doi: 10.1016/j.neuint.2012.01.020. [DOI] [PubMed] [Google Scholar]
- 45.Cuervo AM, et al. IkB is a substrate for a selective pathway of lysosomal proteolysis. Mol Biol Cell. 1998;9:1995–2010. doi: 10.1091/mbc.9.8.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Franch H, et al. A mechanism regulating proteolysis of specific proteins during renal tubular cell growth. J Biol Chem. 2001;276:19126–19131. doi: 10.1074/jbc.M101777200. [DOI] [PubMed] [Google Scholar]
- 47.Yang Q, et al. Regulation of neuronal survival factor MEF2D by chaperone-mediated autophagy. Science. 2009;323:124–127. doi: 10.1126/science.1166088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhou D, et al. Lamp-2a facilitates MHC class II presentation of cytoplasmic antigens. Immunity. 2005;22:571–581. doi: 10.1016/j.immuni.2005.03.009. [DOI] [PubMed] [Google Scholar]
- 49.Rajagopal D, et al. A role for the Hsp90 molecular chaperone family in antigen presentation to T lymphocytes via major histocompatibility complex class II molecules. Eur J Immunol. 2006;36:828–841. doi: 10.1002/eji.200535326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wong E, Cuervo AM. Autophagy gone awry in neurodegenerative diseases. Nat Neurosci. 2010;13:806–811. doi: 10.1038/nn.2575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vogiatzi T, et al. Wild type alpha-synuclein is degraded by chaperone-mediated autophagy and macroautophagy in neuronal cells. J Biol Chem. 2008;283:23542–23556. doi: 10.1074/jbc.M801992200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mak SK, et al. Lysosomal degradation of alpha-synuclein in vivo. J Biol Chem. 2010;285:13621–13629. doi: 10.1074/jbc.M109.074617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kabuta T, Wada K. Aberrant interaction between Parkinson disease-associated mutant UCH-L1 and the lysosomal receptor for chaperone-mediated autophagy. J Biol Chem. 2008;283:23731–22373. doi: 10.1074/jbc.M801918200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang Y, et al. Tau fragmentation, aggregation and clearance: the dual role of lysosomal processing. Hum Mol Genet. 2009;18:4153–4170. doi: 10.1093/hmg/ddp367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Liu H, et al. Degradation of regulator of calcineurin 1 (RCAN1) is mediated by both chaperone-mediated autophagy and ubiquitin proteasome pathways. FASEB J. 2009;23:3383–3392. doi: 10.1096/fj.09-134296. [DOI] [PubMed] [Google Scholar]
- 56.Ciechanover A, Brundin P. The ubiquitin proteasome system in neurodegenerative diseases: sometimes the chicken, sometimes the egg. Neuron. 2003;40:427–446. doi: 10.1016/s0896-6273(03)00606-8. [DOI] [PubMed] [Google Scholar]
- 57.Ravikumar B, et al. Aggregate-prone proteins with polyglutamine and polyalanine expansions are degraded by autophagy. Hum Mol Genet. 2002;11:1107–1117. doi: 10.1093/hmg/11.9.1107. [DOI] [PubMed] [Google Scholar]
- 58.Martinez-Vicente M, et al. Cargo recognition failure is responsible for inefficient autophagy in Huntington’s Disease. Nat Neurosci. 2010 doi: 10.1038/nn.2528. E-pub ahead of printing. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Koga H, et al. Constitutive Upregulation of Chaperone-Mediated Autophagy in Huntington’s Disease. J Neurosci. 2011;31:18492–18505. doi: 10.1523/JNEUROSCI.3219-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bauer PO, et al. Harnessing chaperone-mediated autophagy for the selective degradation of mutant huntingtin protein. Nat Biotechnol. 2010;28:256–263. doi: 10.1038/nbt.1608. [DOI] [PubMed] [Google Scholar]
- 61.Kon M, et al. Chaperone-mediated autophagy is required for turmor growth. Sci Trans Med. 2011;3:109ra117. doi: 10.1126/scitranslmed.3003182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ali AB, et al. Role of chaperone mediated autophagy (CMA) in the degradation of misfolded N-CoR protein in non-small cell lung cancer (NSCLC) cells. PLoS One. 2011;6:e25268. doi: 10.1371/journal.pone.0025268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Welsch T, et al. Eps8 is recruited to lysosomes and subjected to chaperone-mediated autophagy in cancer cells. Exp Cell Res. 2010;316:1914–1924. doi: 10.1016/j.yexcr.2010.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sooparb S, et al. Suppression of chaperone-mediated autophagy in the renal cortex during acute diabetes mellitus. Kidney Int. 2004;65:2135–2144. doi: 10.1111/j.1523-1755.2004.00639.x. [DOI] [PubMed] [Google Scholar]
- 65.Cuervo AM, et al. Cathepsin A regulates chaperone-mediated autophagy through cleavage of the lysosomal receptor. EMBO J. 2003;22:12–19. doi: 10.1093/emboj/cdg002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Venugopal B, et al. Chaperone-mediated autophagy is defective in mucolipidosis type IV. J Cell Physiol. 2009;219:344–353. doi: 10.1002/jcp.21676. [DOI] [PubMed] [Google Scholar]
- 67.Cuervo AM, Dice JF. Age-related decline in chaperone-mediated autophagy. J Biol Chem. 2000;275:31505–31513. doi: 10.1074/jbc.M002102200. [DOI] [PubMed] [Google Scholar]
- 68.Dice JF. Altered degradation of proteins microinjected into senescent human fibroblasts. J Biol Chem. 1982;257:14624–14627. [PubMed] [Google Scholar]
- 69.Kiffin R, et al. Altered dynamics of the lysosomal receptor for chaperone-mediated autophagy with age. J Cell Sci. 2007;120:782–791. doi: 10.1242/jcs.001073. [DOI] [PubMed] [Google Scholar]
- 70.Zhang C, Cuervo AM. Restoration of chaperone-mediated autophagy in aging liver improves cellular maintenance and hepatic function. Nat Med. 2008;14:959–965. doi: 10.1038/nm.1851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kaushik S, Cuervo AM. Chaperone-mediated autophagy. Methods Mol Biol. 2008;445:227–244. doi: 10.1007/978-1-59745-157-4_15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kaushik S, Cuervo AM. Methods to monitor chaperone-mediated autophagy. Methods Enzymol. 2009;452:297–324. doi: 10.1016/S0076-6879(08)03619-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kaushik S, et al. Constitutive Activation of Chaperone-mediated Autophagy in Cells with Impaired Macroautophagy. Mol Biol Cell. 2008;19:2179–2192. doi: 10.1091/mbc.E07-11-1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Iwata A, et al. Increased susceptibility of cytoplasmic over nuclear polyglutamine aggregates to autophagic degradation. Proc Natl Acad Sci. 2005;102:13135–13140. doi: 10.1073/pnas.0505801102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ding Q, et al. Characterization of chronic low-level proteasome inhibition on neural homeostasis. J Neurochem. 2003;86:489–497. doi: 10.1046/j.1471-4159.2003.01885.x. [DOI] [PubMed] [Google Scholar]
- 76.Korolchuk VI, et al. Autophagy inhibition compromises degradation of ubiquitin-proteasome pathway substrates. Mol Cell. 2009;33:517–527. doi: 10.1016/j.molcel.2009.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]