SUMMARY
Genome-wide studies have identified thousands of long noncoding RNAs (lncRNAs) lacking protein coding capacity. However, most lncRNAs are expressed at a very low level, and in most cases there is no genetic evidence to support their in vivo function. Malat1 (metastasis associated lung adenocarcinoma transcript 1) is among the most abundant and highly conserved lncRNAs, and it exhibits an uncommon 3′-end processing mechanism. In addition, its specific nuclear localization, developmental regulation, and dysregulation in cancer are suggestive of it having a critical biological function. We have characterized a Malat1 loss-of-function genetic model that indicates Malat1 is not essential for mouse pre- and post-natal development. Furthermore, depletion of Malat1 does not impact global gene expression, splicing factor level and phosphorylation status, or alternative pre-mRNA splicing. However, among a small number of genes that were dysregulated in adult Malat1 knockout mice, many were Malat1 neighboring genes, thus indicating a potential cis regulatory role of Malat1 gene transcription.
INTRODUCTION
Recent genome-wide studies have indicated that the majority of the human and mouse genomes are transcribed, yielding a complex network of transcripts that includes thousands of noncoding RNAs (ncRNAs) with no protein coding capacity (Chodroff et al., 2010; Guttman et al., 2009; reviewed in Kapranov et al., 2007; Orom et al., 2010). Long noncoding RNAs (lncRNAs), the largest and most complex class of ncRNAs, are mRNA-like RNA polymerase II transcripts ranging in size from 200nt to >100kb and many exhibit cell type specific expression (Chodroff et al., 2010; Guttman et al., 2009; Orom et al., 2010). The majority of lncRNAs are expressed at very low levels, some as low as one or less copy per cell (Mercer et al., 2011), and these RNAs generally exhibit poor primary sequence conservation over evolution. LncRNAs have been implicated in numerous molecular functions, including modulating transcriptional patterns, regulating protein activities, serving structural or organizational roles, altering RNA processing events, and serving as precursors to small RNAs (reviewed in Wilusz et al., 2009). However, most of these molecular functions were deduced from studies performed in cell lines upon lncRNA over-expression or knock-down. Except for a few examples, such as Xist involved in X-chromosome inactivation (Marahrens et al., 1997), Kcnq1ot1 (Mohammad et al., 2010) and Air (Sleutels et al., 2002) in genomic imprinting, genetic evidence supporting the in vivo function of most mammalian lncRNAs is lacking.
The lncRNA HOTAIR has recently been studied in knockdown cell lines and shown to regulate expression of HoxD genes in trans by associating with chromatin modification complexes such as PRC2, LSD1, and CoREST/REST (Khalil et al., 2009; Rinn et al., 2007; Tsai et al., 2010), and to be overexpressed in breast cancer and regulate metastasis by reprogramming chromatin via polycomb complexes (Gupta et al., 2010). Interestingly, genetic deletion of mouse HoxC cluster containing the Hotair gene did not result in misregulation of HoxD genes or any phenotype at the molecular or organismal level (Schorderet and Duboule, 2011). Noncoding nuclear-enriched abundant transcript 1 (Neat1, a.k.a. Men ε/β) lncRNA has been shown to be essential for the assembly and maintenance of nuclear paraspeckles (Clemson et al., 2009; Mao et al., 2011; Sasaki et al., 2009; Sunwoo et al., 2009) and nuclear retention of some hyperedited RNAs in hESCs and HeLa cells (Chen and Carmichael, 2009). Surprisingly, a study of Neat1 hypomorphic mouse mutants, although lacking paraspeckles, failed to exhibit any physiological defects (Nakagawa et al., 2011). While these lncRNAs do not appear to be essential during pre- or post-natal mouse development their regulatory function may be masked by redundant or compensatory mechanisms and may only be revealed upon specific stress conditions, which have thus far not been investigated. Interestingly, a recent study in zebrafish identified 29 evolutionarily conserved lncRNAs, and of these, two exhibited a significant regulatory function during zebrafish development as demonstrated by morpholino intervention (Ulitsky et al., 2011). Further in vivo analyses of lncRNAs are necessary to reveal their mechanisms of action.
Malat1 (metastasis associated lung adenocarcinoma transcript 1), also known as Neat2 (noncoding nuclear-enriched abundant transcript 2), is located on mouse chromosome 19qA (human chromosome 11q13.1). Malat1 is evolutionarily conserved among mammals in terms of its primary sequence, and is highly expressed in many tissues and regulated during tissue differentiation (Bernard et al., 2010; Hutchinson et al., 2007; Ji et al., 2003). MALAT1 is also upregulated in several human cancers, suggesting that it may have an important function during cancer progression (Guffanti et al., 2009; Ji et al., 2003). Knockdown of MALAT1 by ASOs (antisense oligonucleotides) has been shown to affect the recruitment of pre-mRNA splicing factors to a reporter locus in human U2OS cells, and to alter synapse number in cultured neurons (Bernard et al., 2010). Recently, MALAT1 depletion in HeLa cells was shown to perturb the protein level and phosphorylation status of two pre-mRNA splicing factors and the pre-mRNA splicing pattern of a set of transcripts (Tripathi et al., 2010). In addition, a large fraction of cells accumulated at the G2/M boundary and an increased apoptosis was observed (Tripathi et al., 2010). In contrast, a second study examining the effect of knock-down of MALAT1 in HeLa cells observed a loss of serum-induced cell proliferation and E2F1 target gene expression with profound G1/S arrest, but no apparent G2/M arrest or apoptosis (Yang et al., 2011). MALAT1 was shown to be part of a complex that binds to unmethylated Pc2 at nuclear speckles promoting E2F1 SUMOylation leading to activation of the growth-control gene program (Yang et al., 2011). However, the physiological function of Malat1 lncRNA at the tissue and organismal levels has not been investigated.
Here, we show that Malat1 is one of the most abundant lncRNAs in mouse liver and brain cortex. To assess the potential in vivo function of mouse Malat1 we first evaluated the consequences of its knock-down in adult mice using an antisense approach and found no significant morphologic change in tissue organization upon its transient depletion. The Malat1 locus is syntenically conserved from fish to human, and its high transcription rate is also maintained through evolution despite limited sequence conservation, suggesting that transcription of the Malat1 gene per se may carry a biological function. To examine the role of the Malat1 gene locus, we also established a mouse loss-of-function genetic model. Detailed whole body histopathologic characterization showed that Malat1 ncRNA and its transcription are dispensable for mouse pre- and post-natal development. Further cell biological and biochemical analyses indicated that Malat1 lncRNA is not essential for nuclear speckle assembly/maintenance, the level and phosphorylation status of SR splicing factors, or cell proliferation and apoptosis. Genome-wide expression and splicing profiling of mouse liver and brain cortex demonstrated that Malat1 loss results in minimal alterations in global gene expression and pre-mRNA splicing. However, inactivation of the Malat1 gene results in a nearly two-fold up-regulation of several genes that reside adjacent to the Malat1 locus, including the lncRNA Neat1. These results suggest a model whereby transcription by the highly active Malat1 promoter may be important for regulating the expression of nearby genes in cis.
RESULTS
Malat1 encodes a highly abundant lncRNA in mouse
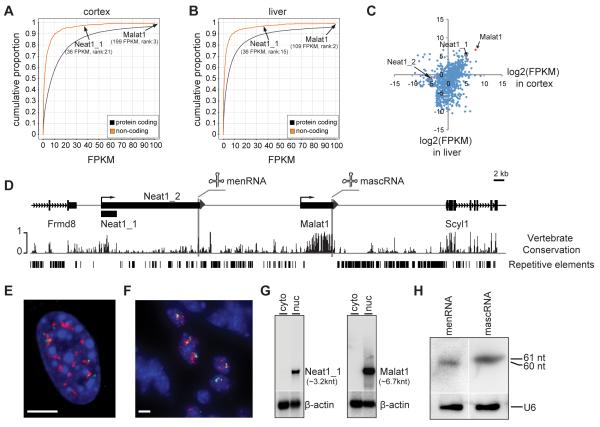
To assess the level of Malat1 lncRNA in mouse we performed RNA-seq profiling and obtained ~230 and ~220 million paired-end reads from three wild type mouse livers and three brain cortices, respectively. We first focused on profiles of ~1,700 lncRNA genes in RefSeq (see Methods). Consistent with previous findings, we found that most lncRNA genes are expressed at lower levels as compared to protein-coding genes (Fig. 1A, B). However, the fragments-per-kilobase-million-reads (FPKM) value for Malat1 is comparable or higher than several house-keeping protein-coding genes (in liver: Malat1, 109 FPKM; Actb, 177 FPKM; Gapdh, 21 FPKM; in brain cortex: Malat1, 199 FPKM; Actb, 484 FPKM; Gapdh, 16 FPKM). Malat1 is highly expressed in both brain cortex and liver (Fig. 1A-C and Table S1) and comprised ~15.2% of total lncRNA sequence reads in liver and ~5.7% in brain cortex.
Figure 1. Malat1 encodes a highly abundant lncRNA in vertebrates.
A-B. RNA-seq transcriptome analyses of mouse brain cortex and liver. Note that Malat1 and Neat1_1 are among the most abundant lncRNAs and that expression of Malat1 in both cortex and liver is higher than most protein-coding genes. C. Tissue-specific expression of lncRNA genes. x, y axes, log2(FPKM) values of lncRNAs in cortex and liver, respectively. D. The reverse strand of the genomic locus of mouse Chr19:5,760,586-5,860,585. Malat1 is ~40 kb downstream of Neat1. Frmd8 and Scyl1 are adjacent protein coding genes. E-F. RNA FISH shows that both Malat1 (red) and Neat1 (green) occupy distinct subnuclear domains in MEFs (E) and interstitial cells of testis (F). Blue, DAPI staining; scale bars = 5 μm. G. Northern blot analysis shows that Neat1_1 and Malat1 are enriched in the nuclear fraction and that β-actin is distributed in both cytosolic and nuclear fractions. H. small RNA Northern blot analysis shows that menRNA and mascRNA are detected in mouse liver. U6 is the loading control. Note that the blot in H is the same as that used in lanes 3 and 4 in Fig. 4E to avoid cross-hybridization of the menRNA probe to mascRNA.
Interestingly, the Malat1 gene resides adjacent to Neat1, another lncRNA gene on mouse Chr.19 (Fig. 1D). The nuclear-retained Malat1 ncRNA is ~6.7 knt (kilonucleotide) and the nuclear retained Neat1 RNA exists as two isoforms of 3.2 knt (Neat1_1) and ~20 knt (Neat1_2)(Sunwoo et al., 2009)(Fig. 1D). Conservation analysis showed that both Malat1 and Neat1_1 are among the most conserved lncRNAs during vertebrate evolution (data not shown) with minimal repetitive elements (Fig. 1D). Similar to Malat1, Neat1 is also highly and widely expressed in adult tissues (Fig. 1A-C)(Hutchinson et al., 2007). A recent study has shown that both RNA molecules have a unique 3′ end processing module, which is conserved from fish to human (Fig. 1D)(Sunwoo et al., 2009; Wilusz et al., 2008). Malat1 and Neat1 nascent RNAs are each processed by RNases P and Z to produce nuclear-retained ncRNAs and small cytoplasmic RNAs (mascRNA from Malat1, menRNA from Neat1) that exhibit a tRNA-like structure (Sunwoo et al., 2009; Wilusz et al., 2008)(Fig. 1D). RNA fluorescence in situ hybridization (FISH) demonstrated that Neat1 RNA localizes to nuclear regions in close proximity to Malat1 RNA in mouse embryo fibroblasts (MEFs) as well as interstitial cells of testis (Fig. 1E, F), consistent with the previously reported localization of these RNAs in nuclear speckles (Malat1) and paraspeckles (Neat1) of cultured cells (Clemson et al., 2009; Hutchinson et al., 2007; Mao et al., 2011; Sasaki et al., 2009; Sunwoo et al., 2009). Northern blot analyses showed that both Malat1 and Neat1 are nuclear retained in MEFs (Fig. 1G), consistent with previous findings (Hutchinson et al., 2007), and that both mascRNA and menRNA are detected in mouse liver (Fig. 1H) and kidney (data not shown). In contrast to a previous finding indicating that menRNA is unstable in cultured cells due to a noncanonical CCACCA addition at its 3′ end (Wilusz et al., 2011), our data suggest that menRNA generated in liver and kidney is a stable small RNA exhibiting the classical CCA addition (Fig. S1). However, in wild type mouse brain, while we saw relatively high level of mascRNA, menRNA is not detectable (data not shown). This is likely due to the lower expression of the long isoform of Neat1 (Neat1_2) in brain, as observed by Northern blot (data not shown) and RNA-seq analyses (Neat1_2: 0.44 FPKM in liver; 0.04 FPKM in cortex, Fig. 1C). Collectively, our results demonstrated that Malat1 lncRNA is expressed at a very high level, processed at its 3′ end to generate mascRNA, and localized in distinct subnuclear domains in mouse tissues as well as cultured cells.
Antisense knockdown of Malat1 ncRNA in adult mice does not alter organ organization
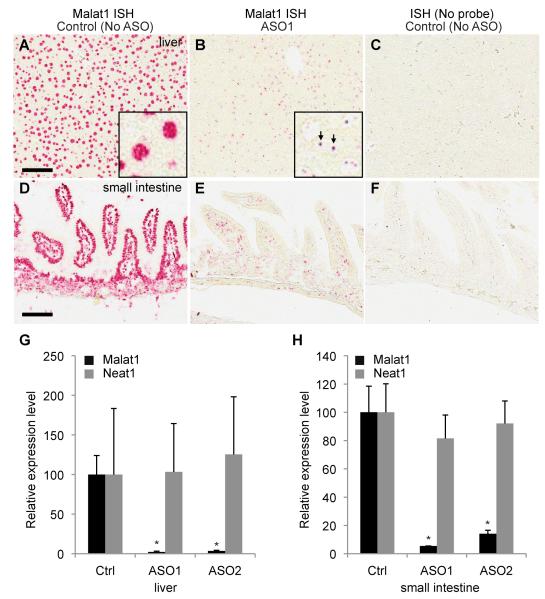
The high abundance, strong evolutionary conservation, specific subcellular localization and developmental regulation of Malat1 suggest significant in vivo biological functions. However, such function could be derived from either the Malat1 RNA transcript itself or the act of transcriptional activity of the Malat1 gene. To distinguish between these two possibilities we first examined the functional consequence of in vivo depletion of Malat1 lncRNA. Malat1 ASOs were administered into adult mice subcutaneously and the knockdown efficiency was examined by qRT-PCR and RNA in situ hybridization (ISH) (Fig 2A-H). As compared with saline treated animals, significant Malat1 knockdown was observed in both liver (97-98% knockdown, Fig. 2G) and small intestine (86-95% knockdown, Fig. 2H) from animals treated with Malat1 ASO1 or ASO2. Knockdown of Malat1 with either of these ASOs does not alter the overall expression level of Neat1 in liver or small intestine (Fig. 2G, H). ASO1 was slightly more effective than ASO2 in both liver and small intestine. In liver, Malat1 depletion in hepatocytes was more dramatic than in bile duct epithelial cells (data not shown). In small intestine, Malat1 knockdown in villi epithelial cells was more effective than in crypts and lamina propria (Fig. 2D-F). No detectable Malat1 signal was observed in saline treated tissues incubated without Malat1 probes (Fig. 2C, F). Although significant knockdown was achieved, no noticeable abnormality was seen in serum chemistry and tissue morphology between Malat1 ASO and control, PBS, treated mice.
Figure 2. Antisense knockdown of Malat1 lncRNA in adult mice does not alter organ organization.
A-B. ISH analysis of Malat1 on saline-treated or Malat1 ASO1-treated livers. Malat1 is highly expressed in most liver cells (A) with its signal enriched in nuclei (inset of A). ASO1 efficiently knocks down Malat1 expression in most liver cells (B) and the remaining signals form a few distinctive dots in nuclei (arrows, inset of B), which are likely the nascent transcripts at the transcription sites of the Malat1 gene locus. C. As a negative control, probe-free ISH on the saline-treated liver shows no hybridization signals. D-E. ISH analysis of Malat1 on saline-treated or Malat1 ASO1-treated small intestine. Malat1 is highly expressed in most small intestine cells (D) with its signal enriched in nuclei. ASO1 efficiently knocks down Malat1 expression in most cell types (E). F. As a negative control, probe-free ISH on the saline-treated small intestine shows no hybridization signal. Scale bars = 100 μm. G-H. qRT-PCR analyses show that Malat1 ncRNA is significantly knocked down in ASO-treated liver (G) and small intestine (H), while Neat1 ncRNA exhibits no significant change upon Malat1 depletion in both liver (G) and small intestine (H). ISH, in situ hybridization. ASO, antisense oligonucleotide. Ctrl, control. Error bars represent S.D. (standard deviation). *, p < 0.05, Student’s unpaired t-test.
Generation and characterization of a Malat1−/− mouse model
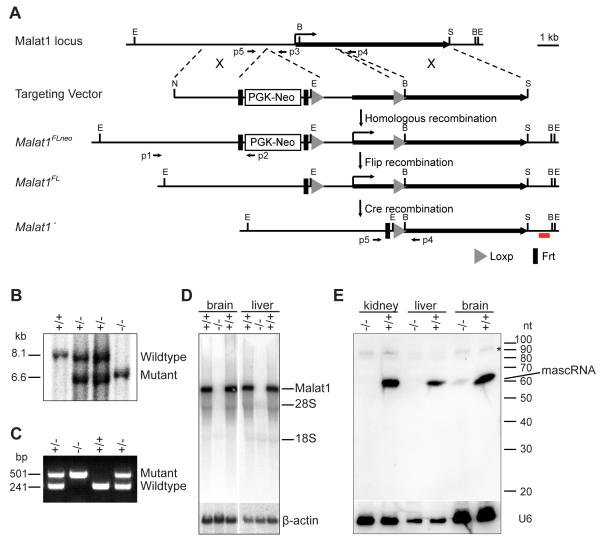
As in vivo knockdown studies did not reveal a function for Malat1 lncRNA in adult mice, we developed a knockout mouse to delineate its potential role during early development and to assess whether the direct function of the Malat1 gene is the act of its transcription. Towards this end we generated Malat1 mutant mice using homologous recombination in ES cells (Fig. 3A). We found the recombination rate at the Malat1 locus to be ~36% (24 recombination positives from 67 clones), higher than other loci in the genome. This is consistent with previous reports of the high susceptibility for translocation and DNA damage, and an exceptionally high transcriptional activity at the Malat1 locus (Davis et al., 2003; Ferris et al., 2010; Kato et al., 2012; Rajaram et al., 2007).
Figure 3. Generation and characterization of a Malat1−/− knockout mouse.
A. The strategy for Malat1 targeting using homologous recombination. E, EcoRI; S, SalI; N, NotI; B, BglII; X, homologous recombination; p1, p2, p3, p4, p5 PCR primers; red line, 3′ external probe for Southern blot analysis. B, C. Southern blot (B) and PCR analyses (C) show detection of wild type and Malat1 mutant alleles. D. Northern blot analyses show that Malat1 lncRNA is depleted in homozygous mutant brain and liver. 28S and 18S indicate the positions of their size. E. Small RNA Northern blot analyses using oligo probe shows that mascRNA is depleted in Malat1 mutant liver and kidney and that a small amount of mascRNA is detected in the mutant brain. *, non-specific bands. β-actin and U6 are the loading controls.
The mutant mice, lacking a ~3 kb genomic region containing the 5′ end of the Malat1 gene and its promoter, were genotyped by Southern blotting and tail DNA PCR analyses (Fig. 3B, C). Northern blotting analysis using a ~2 kb partial cDNA probe showed that the Malat1 transcript is completely depleted in Malat1−/− tissues (Fig. 3D). In addition, small RNA Northern blotting analysis using an oligonucleotide probe demonstrated that mascRNA is absent in Malat1−/− kidney and liver, but is detectable in Malat1−/− brain at a significantly lower level as compared to that in wild type brain (Fig. 3E). The minimal amount of mascRNA in mutant brain may be the result of a transcript derived from a brain-specific promoter upstream of the 3′ tRNA-like structure. However, we did not see any other RNA bands of a different size recognized by a Malat1 partial cDNA probe in both wild type and mutant brains (Fig. 3D).
Since knockdown of Malat1 in human HeLa cells was previously shown to result in alterations in alternative pre-mRNA splicing and apoptosis (Tripathi et al., 2010), we expected that homozygous deletion of Malat1 would result in aberrations in pre- and/or post-natal development or tissue maturation. However, Malat1−/− mice are grossly normal and fertile. The offspring of heterozygous breedings follow Mendelian segregation (Fig. S2A), suggesting that deletion of Malat1 does not affect mouse pre- and post-natal viability. Breedings between Malat1 homozygotes and wild type produce normal-sized litters (Fig. S2A), indicating Malat1−/− mice are fertile. A large number of tisssues from wild type and mutant mice were assessed for potential histo-pathological alterations upon Malat1 deletion. No obvious gross phenotypes were observed in mutants (Fig. S2B-G). Blood cell counts and chemistry did not show any significant defects in Malat1 mutant mice (data not shown). We also performed immunohistochemical labeling of Ki67 and Cleaved Caspase3 and did not see any significant difference in terms of the number of proliferating or apoptotic cells between wild type and mutant tissue sections (data not shown). This is in stark contrast to apoptosis and growth arrest induced by Malat1 knockdown using ASOs or RNAi in human and mouse cell lines (Tripathi et al., 2010; Yang et al., 2011). Together, these data suggest that neither Malat1 nor its transcription is essential for mouse development, viability, and fertility, under normal physiological and environmental conditions. However, it remains to be determined whether Malat1 and/or its transcriptional activity are required under specific stress conditions, or if functional redundancy exists in the mouse.
Malat1 lncRNA is not necessary for the establishment or maintenance of nuclear speckles
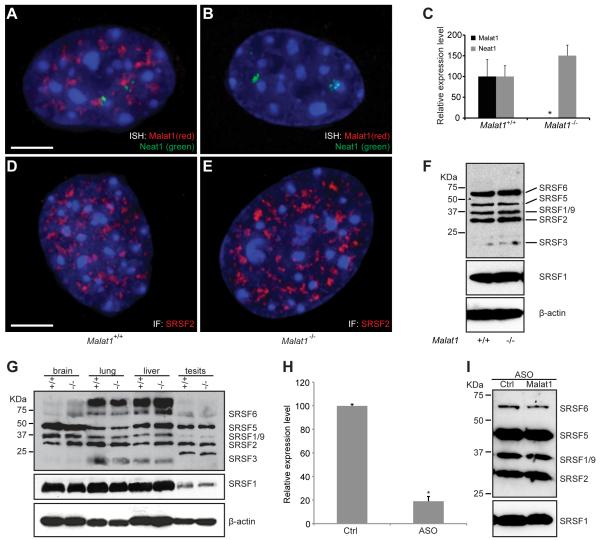
Malat1 lncRNA localizes to nuclear speckles and interacts with nuclear speckle associated proteins such as SRSF1 (Hutchinson et al., 2007; Tripathi et al., 2010), suggesting a potential function for Malat1 in nuclear speckle assembly and/or maintenance. To investigate the effects of loss of Malat1 lncRNA on nuclear speckles, we isolated MEFs. No proliferation or apoptotic defects were seen in mutant MEFs as compared to wild type (data not shown). RNA FISH and qRT-PCR analyses indicated that Malat1 lncRNA is completely depleted from the mutant MEFs while its neighboring lncRNA Neat1 is slightly upregulated and forms two paraspeckle clusters, presumably around its transcription sites (Fig. 4A-C).
Figure 4. Depletion of Malat1 lncRNA does not alter Neat1 localization, nuclear speckle morphology, or the phosphorylation status of SR proteins.
A, B. RNA FISH shows the subnuclear distribution of Malat1 (red) and Neat1 (green) in wild type (A) and Malat1−/− (B) MEFs. Note that Neat1 ncRNAs form two nuclear clusters adjacent to Malat1 ncRNAs. C. qRT-PCR analysis shows that Malat1 ncRNA is depleted in Malat1−/− MEFs, while Neat1 ncRNA exhibits no significant change in the mutants as compared to wild type. Error bars represent S.D.. *, p < 0.05, Student’s unpaired t-test. D, E. Immunofluorescence labeling of SRSF2(SC35) for nuclear speckles shows no significant changes of nuclear speckle morphology, distribution, and number in the mutant (E) as compared to wild type (D) MEFs. F. Western blot analyses of phospho SR proteins labeled by 3C5 antibody and total SRSF1 labeled by anti-SRSF1 show no change of SR protein level or phosphorylation status in the mutant as compared to wild type MEFs. β-actin is the loading control. Scale bars = 5 μm. G. Western blot analyses of phospho SR proteins show no changes of SR protein phosphorylation status or total SRSF1 level in brain, lung, liver, and testis from the mutant as compared to wild type. β-actin is the loading control. H. qRT-PCR analysis shows that free-uptake ASO against MALAT1 knocks down MALAT1 lncRNA by ~80% compared to the control ASO (Ctrl) in MCF7 cells. *, p < 0.05, Student’s unpaired t-test. I. Western blot analyses of phospho SR proteins show no changes of SR protein phosphorylation status or protein level in MCF7 cells treated with the MALAT1 ASO compared to the control ASO (Ctrl).
To examine whether loss of Malat1 lncRNA affects nuclear speckle or paraspeckle morphology, we performed immunofluorescence (IF) labeling for these nuclear domains using antibodies against SRSF2 and PSP1α in MEFs. We found that Malat1 deletion has no overall effect on the number, size, and distribution of either nuclear speckles (Fig. 4D,E) or paraspeckles (Fig. S3).
Loss of Malat1 does not alter the level and/or phosphorylation status of SR proteins
Nuclear speckles are characterized by the enrichment of pre-mRNA splicing factors including those of the SR family and small nuclear ribonucleoprotein complexes (reviewed in Lamond and Spector, 2003). It has been previously reported that Malat1 knock-down in HeLa cells alters the level and phosphorylation status of two SR proteins, resulting in changes in the alternative splicing pattern of certain pre-mRNAs, and apoptosis. Since Malat1 mutant mice are fertile and do not exhibit any obvious cellular or tissue defects, we examined the phosphorylation status of SR family members in wild type versus mutant mice. Initially, the status of SR protein phosphorylation in MEFs was evaluated by immunoblot and probing against members of the SR family, including SRSF1, using a phospho-epitope specific antibody (3C5) that recognizes the conserved serine arginine residues of SR family members of splicing factors (Turner and Franchi, 1987). We did not observe any alteration in the level or phosphorylation status of SR proteins, including SRSF1, in Malat1−/− MEFs as compared to wild type (Fig. 4F).
Next, we examined the phosphorylation status of these SR proteins in adult tissues of wild type and mutant mice, and found that there is no change in the total pool of phosphorylated splicing factors and the level of SRSF1 remained the same in each of the tissues (adult brain, lung, liver, and testis) of wild type and Malat1 mutant samples (Fig. 4G). Immunofluorescence of SRSF2 in wild type and mutant MEFs also did not show any change in nuclear speckle number or morphology (Fig. 4D, E). The speckled pattern was also studied in adult mouse tissue sections, by immunofluroscence using SRSF2 antibody, and cells exhibited no change in the number or pattern of nuclear speckles in Malat1−/− tissues as compared to wild type (data not shown). These observations clearly demonstrate that Malat1 loss does not result in a change in the level or phosphorylation status of SR splicing factors or the association of SRSF2 with nuclear speckles in vivo.
This was very surprising given the fact that in HeLa cells knock-down of Malat1 had a profound effect on splicing factor phosphorylation status (Tripathi et al., 2010). As Malat1 is among the most conserved lncRNAs in mammals, it is highly unlikely that it will have a different function between mouse and human. To address this discrepancy, we repeated the Malat1 knock-down experiment in human HeLa cells using lipofectamine 2000-mediated transfection of ASOs. While knock-down of Malat1 at 48 and 72 hrs after transfection resulted in dephosphorylation of SRSF1 (Fig. S4) accompanied by significant apoptosis, SRSF1 phosphorylation changes and apoptosis were also observed at some level in control ASO transfected cells 72 hrs post treatment. However, similar knockdown experiments using Fugene, a less toxic lipid reagent, resulted in no significant cell death after 48 hrs and minimal apoptosis after 72 hrs in both control ASO and MALAT1 ASO treated human MCF7 cells (data not shown). To further confirm that knock-down of MALAT1 in cultured cells does not cause apoptosis and alter SR phosphorylation status, we used ASOs with a MOE gapmer structure, which can be taken up effectively by some cultured cell lines freely without transfection reagents. The MALAT1 knock-down using ASOs at 75 nM concentration in MCF7 cells reached a level of 80-90% RNA reduction after 48 hrs (Fig. 4H), which is comparable to that of ASO knock-down using lipid transfection reagents. The MALAT1 knock-down by free-uptake did not alter the phosphorylation status of SR proteins and the SRSF1 level at the 48hr time point in MCF7 cells (Fig. 4I) and did not result in apoptosis at the 72 hr time point (data not shown). We also see no effects of Malat1 knock-down on SR phosphorylation in human SW480 (Fig. S4) and MCF10A cells, and mouse mammary tumor 4T1, 4T07, 67NR, and 168FARN cells (data not shown). Taken together, MALAT1 loss in and of itself does not result in changes in the level of SRSF1, the phosphorylation status of SR proteins, or in apoptosis in many human and mouse cell lines as well as in mouse tissues.
Malat1 lncRNA does not regulate global pre-mRNA splicing
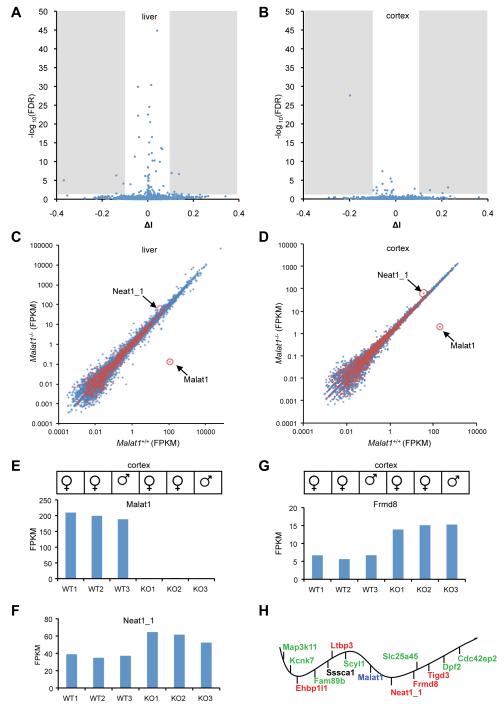
While depletion of Malat1 RNA in mouse tissues does not alter nuclear speckle morphology nor cause any significant changes in SR protein level and phosphorylation, we were interested in examining the effect of loss of Malat1 on global gene expression and alternative pre-mRNA splicing. To probe molecular changes in Malat1 mutant tissues, we performed RNA-seq profiling and obtained ~70 million paired-end reads for each sample (liver: 3 wild types and 3 mutants; and brain cortex: 3 wild types and 3 mutants; data from wild type were used for the lncRNA abundance quantification in Fig. 1). We examined all (~13,000) cassette exons annotated in the mouse genome, which represent the most predominant pattern of alternative splicing, and did not see global changes in the average inclusion level between wild-type and Malat1 mutant livers (Fig. S5A and Fig. 5A) or cortices (Fig. S5B and Fig. 5B). Only 10 exons in liver and 5 exons in brain cortex show statistically significant changes (FDR < 0.05 and |ΔI| > 0.1, see Methods; Fig. 5A, B), of moderate magnitude, and sometimes with substantial variation between biological replicates. Therefore, Malat1 does not appear to regulate global pre-mRNA splicing in adult mouse liver and brain cortex.
Figure 5. Malat1 RNA does not regulate global pre-mRNA splicing, but its transcription inactivation alters gene expression in cis.
A,B. The volcano plot shows proportional change of inclusion level (x axis, ΔI) of each exon and their statistical significance (y axis, -log10(FDR)) upon Malat1 depletion. Shaded regions represent statistical significant changes. C,D. The scatter plot shows the average expression of protein-coding (blue) and lncRNA (red) genes in wild type and Malat1 mutant livers and brain cortices. Malat1 and Neat1_1 are highlighted. E. Malat1 is significantly depleted in Malat1 mutant brain cortexes. F, G, Neat1_1 and Frmd8 show consistent overexpression in mutant cortexes. WT, wild type (Malat1+/+); KO, knockout (Malat1−/−). H. The ~240 kb genomic locus of Malat1 with adjacent genes distributed in order but not at the exact scale. Upregulated genes with statistical significance after multiple testing correction (red), and with statistical significance without multiple testing correction (green). Sssca1 expression is not significantly altered. Vertical bars under the wavy line represent transcription from the negative strand of the chromosome; vertical bars above the line represent transcription from the positive strand.
Inactivation of Malat1 transcription alters local but not global gene expression
It has been recently reported that Malat1 interacts with unmethylated Pc2 to promote E2F1 SUMOylation, resulting in relocation of its downstream growth-control genes to nuclear speckles for expression, and that knockdown of Malat1 inhibits cell growth with a failure of these genes to reposition in the nucleus (Yang et al., 2011). Malat1 knockdown has also been shown to alter several synaptogenesis-associated genes in cultured Neuro2A neuroblastoma cells (Bernard et al., 2010). To address whether Malat1 regulates global gene expression, we compared expression levels of ~1,700 lncRNAs and ~19,000 protein-coding genes in wild type and mutant livers and brain cortices (see Methods). Interestingly, Malat1 lncRNA is absent in mutant liver (wild type, 109 FPKM; mutant, 0.1 FPKM) (Table S2), but still detectable at a very low level in mutant brain cortex (wild type, 199 FPKM; mutant, 2 FPKM) (Table S3). RNA-seq reads of Malat1 in mutant brain cortex were mapped to a ~3kb region upstream of mascRNA (data not shown), consistent with the presence of a low level of mascRNA in mutant brain cortex (Fig. 3E). It remains to be determined whether this 3 kb transcript is expressed by an alternative promoter to compensate for the loss of full-length Malat1 in mutant brain.
In both liver and cortex, no significant global changes in steady state mRNA level were detected (Fig. 5C, D). After multiple testing correction, only one gene (Saa3, serum amyloid A 3) in liver and 12 genes in cortex showed statistically significant changes between wild type and Malat1 mutants (Tables S2 and S3). However, we observed a bigger sex difference of gene expression in liver than in cortex (data not shown) which might have masked some changes. For example, Neat1 shows higher expression in males, and consistent ~1.5-2 fold increases in Malat1 mutant livers in both sexes (Fig. S6A, B, and Table S4A). When we compared gene expression in livers only from males, 22 genes, including Saa3 and Upp2 (uridine phosphorylase 2), have statistically significant changes between wild type and Malat1 mutant liver samples (Table S2).
Interestingly, among the 12 genes with significant expression changes in brain cortex, five are adjacent to Malat1, including Neat1, Frmd8, Tigd3, Ehbp1l1, and Ltbp3 (Fig. 5E-G; Table S4B). All of these genes are upregulated 1.5 to 2.3 fold upon inactivation of the Malat1 gene (Table S3). Upon further examination of other genes adjacent to Malat1 (~240 kb region centered around Malat1), we found seven additional genes, Map3k11, Kcnk7, Fam89b, Scyl1, Slc25a45, Dpf2 and Cdc42ep2, except for Sssca1, whose up-regulation reaches statistical significance without genome-wide multiple testing correction (1.15-1.3 fold; Fig. 5G; Table S4B). Collectively, our data demonstrate that inactivation of the Malat1 gene up-regulates gene expression in cis, but does not alter global gene expression patterns in mouse liver and cortex.
DISCUSSION
Genome-wide approaches have been successfully applied to identify thousands of lncRNAs some of which are transcribed spatiotemporally. A complete understanding of their in vivo function(s) requires molecular and cell biological characterization as well as the development of genetic models to assess their role(s) at the more complex organismal level. Here, we have developed a loss-of-function genetic model for Malat1, one of the most well conserved and highly expressed lncRNAs. While Malat1 does not have a direct role in mouse pre- and post-natal development, inactivation of its transcription results in an up-regulation of a small number of adjacent genes.
Malat1 nascent transcripts are processed by RNases P and Z to produce a 6.7 knt long nuclear-retained ncRNA with a genomically encoded short polyA-like moiety at its 3′ end, and a tRNA-like small RNA, mascRNA (Wilusz et al., 2008) that is transported to the cytoplasm. Although Malat1 is highly abundant in polyA(+) samples, given its lack of a traditional poly(A) tail it is very likely that our RNA-seq analysis of such samples underestimates the abundance of Malat1. Consistent with this, it has been reported that ~0.4-2.0% total sequencing reads from the transcriptome profiling for ribo (-) RNA species in a number of mouse tissues are mapped to the Malat1 gene (Castle et al., 2010), indicating the high abundance and wide expression pattern of this lncRNA.
The Malat1 gene is located on mouse Chr.19qA (human Chr.11q13.1). Interestingly, ~40 kb upstream of Malat1 in the mouse genome, another lncRNA gene, Neat1, produces two isoforms with different 3′ ends. The short Neat1 transcript (Neat1_1) is 3.2 knt, while the nascent long Neat1 transcript (Neat1_2) is also processed by the tRNA processing machinery to produce a nuclear retained ~20 knt long Neat1_2 transcript and menRNA, a small tRNA-like RNA (Sunwoo et al., 2009). In mammalian genomes, the primary sequences of both Malat1 and Neat1_1 are highly conserved and are nearly devoid of annotated repeat-derived sequences, implying biological functions of these RNAs. Neat1 is not computationally detected outside of mammals; however, the Malat1 gene is syntenically conserved in most sequenced gnathostome genomes (Stadler, 2010). Except for the conserved 3′ end processing module, the rest of Malat1 lncRNA is surprisingly divergent without homologous fragments detected between fishes and mammals. Interestingly, similar to that observed in human and mouse, the transcriptional activity of the Malat1 gene in lizard, xenopus, and zebrafish is exceptionally high (Zhang B and Spector DL, unpublished data)(Ulitsky et al., 2011), suggesting that maintaining the high transcriptional activity of the Malat1 locus during evolution may be important. The strong constraint on the 3′ end processing module during the evolution of gnathostomes implies that the processing mechanism and/or its products may carry a biological role. However, the functional carrier for the Malat1 locus during evolution is not necessarily limited to only one component. Instead, it is very likely to be combinations of the lncRNA transcript, the transcription of the Malat1 gene, and/or the small mascRNA.
The chromatin structure at the Malat1-Neat1 genomic locus exhibits several interesting features. 1) Transcription of these two lncRNA genes is very active (Castle et al., 2010), and in some cell types, RNA Pol II binding at their promoters is extremely high as compared to the entire genome (Martianov et al., 2010). 2) The human 11q13 region, where human MALAT1 is located, is a ‘RIDGE’ chromosome domain of very high gene-density and gene expression that supports strong transcription activity of inserted transgenes (Gierman et al., 2007) and has a very decompact higher-order chromatin structure (Gilbert et al., 2004). 3) The homologous recombination frequency is exceptionally high for both Malat1 (this study) and Neat1 (Zhang B and Spector DL, unpublished data). 4) Malat1 is frequently translocated in renal carcinoma and embryonic sarcoma (Davis et al., 2003; Rajaram et al., 2007). 5) Malat1 is the most favored hotspot for HIV integration mediated by the ING2-IBD fusion (Ferris et al., 2010) and is among the top targets of activation-induced cytidine deaminase (AID) with accumulated mutations as frequent as the Ig locus after AID activation in B cells (Kato et al., 2012). Taken together, the high susceptibility for recombination, translocation, and DNA breaks, is likely associated with a distinct chromatin structure and/or histone modifications of the locus, which is responsible for its high transcriptional activity. Given these characteristics this genomic locus could be utilized to efficiently knock-in any transgene of interest to achieve stable and high expression in most tissues and cell types.
The specific subcellular localization of Malat1 has led some to suggest that this RNA could function along with speckle-associated protein factors (i.e., SRSF1 and SRSF2) and other nuclear proteins to regulate cell and tissue-specific gene expression and/or pre-mRNA splicing. In this regard, two recent studies have revealed different phenotypes upon Malat1 knockdown in HeLa cells. Tripathi et al. (2010) demonstrated that knockdown of Malat1 resulted in a G2/M phase arrest and significant levels of apoptosis in both human HeLa cells and mouse EpH4 cells (Tripathi et al., 2010). Contrary to this finding, Yang et al. (2011) showed that Malat1 depletion resulted in a G1/S phase arrest (Yang et al., 2011). However, we have found that Malat1 knockout mice are viable and fertile with no obvious gross abnormalities and cells do not exhibit apoptosis or defects in cell cycle progression. This discrepancy of Malat1 function between in vitro and in vivo analyses could be due to redundant compensatory mechanisms during development, however, the transient knockdown of Malat1 in adult mouse tissues and cultured cells by free-uptake of ASOs in the absence of any transfection reagent, did not induce either apoptosis or cell cycle arrest.
The different cellular phenotypes observed in Malat1-depleted HeLa cells, G2/M arrest/increased apoptosis versus G1/S arrest, were explained by distinct molecular mechanisms of Malat1 in splicing versus gene expression, respectively (Tripathi et al., 2010; Yang et al., 2011). Tripathi et al. (2010) demonstrated that Malat1 regulates pre-mRNA splicing through modulating pre-mRNA splicing factor level and the phosphorylation status of one or more SR-family splicing factors (Tripathi et al., 2010). In contrast, Yang et al. (2011) showed that Malat1 interacts with unmethylated Pc2 to promote E2F1 target gene expression during cell cycle progression (Yang et al., 2011). However, our cellular, biochemical, and molecular analyses showed no significant changes in the level and/or phosphorylation status of SR proteins and global splicing patterns in Malat1 depleted human and mouse cells and tissues. We also did not see global changes of steady state mRNA levels nor E2F1 target gene mis-regulation upon Malat1 depletion in mouse liver and cortex. Furthermore, neither apoptosis nor alterations in proliferation were observed when Malat1 was knocked down using free-uptake ASOs. Taken together, our data on mouse tissues and mouse and human cells suggest that loss of Malat1 does not alter global pre-mRNA splicing and/or gene expression.
Although we did not observe changes in either global gene expression or alternative pre-mRNA splicing in the Malat1 knockout mutants, we did identify a small number of genes with statistically significant changes in expression in liver and brain cortex. For example, Neat1 is up-regulated by ~1.5-2 fold in both liver and cortex, suggesting a potential compensation between Malat1 and Neat1. Interestingly, among the 12 genes with statistically significant changes in expression level upon inactivation of the Malat1 gene in brain cortex, five of these genes reside adjacent to Malat1 and are up-regulated by ~1.5-2.3 fold. Furthermore, several additional genes in the proximity of Malat1 also showed significant up-regulation when performing statistical analysis without using a multiple testing correction against the entire genome. This cis effect on gene expression is similar to the effect observed upon loss of imprinting lncRNAs, such as Air (Sleutels et al., 2002). However, so far there is no evidence in support of the role of the Malat1 locus in imprinting. Since we did not see Neat1 expression changes upon depletion of Malat1 RNA by ASO treatment in liver and small intestine, this cis effect is probably not mediated by Malat1 RNA itself, implying a different mechanism than imprinting. The syntenic conservation and strong transcription of the Malat1 locus from fish to human suggests that transcription of the Malat1 gene may carry a function that is under selective constraints during evolution. This may also explain why the Malat1 primary sequence is very divergent between fish and human except for the 3′ end tRNA-like processing module. Several mechanisms underlying ncRNA transcription-mediated cis gene regulation have been recently identified. It has been demonstrated that transcription of ncRNA genes can directly interfere with downstream/overlapping gene transcription by altering transcription factor binding at the yeast SER3, FLO11, and IME4 loci (Bumgarner et al., 2012; Hongay et al., 2006; Martens et al., 2004), and the Drosophila Ubx locus (Petruk et al., 2006). This mechanism is called transcriptional inference, the direct negative influence of one transcriptional process on a second transcriptional process in cis (reviewed in Shearwin et al., 2005). Moreover, the act of ncRNA transcription can also induce histone modifications and thus indirectly alter the downstream/overlapping or neighboring gene transcription as demonstrated at the yeast PHO84 locus (Camblong et al., 2007), GAL1-10 gene clusters (Houseley et al., 2008), and the fbp1+ locus in S. pombe (Hirota et al., 2008). Given that a number of Malat1 neighboring genes showed significant upregulation upon Malat1 inactivation and that Malat1 transcription does not overlap with any nearby protein-coding genes, the cis effect may be attributed to altered histone modification, which could have a broad impact on local chromatin organization. Alternatively, the strong Malat1 promoter activity may sequester RNA pol II from its adjacent genes, thereby modulating their expression in wild type cells. However, we cannot rule out the possibility that the deleted 3 kb region of the Malat1 gene contains cis DNA elements that negatively regulate expression of genes adjacent to Malat1. In addition, given the distinct subnuclear localization and high abundance of the Malat1 lncRNA and its conservation in mammals, it is highly likely that the Malat1 transcripts will also exhibit a trans-effect in mouse tissues under specific physiological conditions. Examining these possibilities merits future investigation.
Taken together, our in vivo and in vitro studies demonstrated that Malat1 is not essential for mouse pre- and post-natal development and its deletion does not affect global gene expression and pre-mRNA splicing under the conditions examined. Among a small number of genes with significant changes in expression in mouse liver and cortex, a significant number of these genes are Malat1 neighboring genes. Despite being dispensable for development, the unique features of Malat1, syntenic conservation, high abundance, conserved 3′ end processing, specific nuclear localization, and developmental regulation, argue for a broader functional role for this lncRNA. A lack of phenotype upon the loss of Malat1 lncRNA transcripts could be attributed to functional redundancy with other RNA transcripts (e.g. Neat1 lncRNA) or to compensatory mechanisms during development, as occurs with respect to many protein-coding genes. Alternatively, some lncRNAs, including Malat1, could have a subtle role and regulate cellular processes via a fine-tuning mechanism. Given the initial identification of Malat1 as a gene whose up-regulation was correlated with tumors that have the propensity to metastasize (Ji et al., 2003) it will be important to investigate the potential role of Malat1 in cancer progression and upon other pathological stresses.
Material and Methods
In vivo knockdown of Malat1
Male Balb/c mice (n=4) at the age of 7 week old were subcutaneously dosed with saline, Malat1 ASO1 or ASO2 (sequence information in Table S5) at 100mg/kg/week for 4 weeks. At 24 hours after the last dose, the animals were sacrificed. The liver and small intestine were harvested and homogenized in RLT buffer (Qiagen, CA) containing 1% 2-mercaptoethanol. Total mRNA was prepared with the PureLink™ Total RNA Purification Kit (Invitrogen, CA) according to the manufacturer’s instruction. A second set of tissues were fixed in 10% buffered formalin for 72 hours and further paraffin embedded and sectioned at 4 μm for histology and RNA in situ hybridization analysis using Affymetrix QuantiGene ViewRNA Assay (Affymetrix, CA).
Generation of Malat1 knockout mice
Malat1-deficient mice were generated by homologous recombination and genotypes were determined by Southern blot and PCR analyses (sequence information in Table S5; see details in Supplementary Material and Methods).
RNA-seq expression and splicing analyses
Raw reads obtained from the Illumina pipeline were mapped back to the mouse reference genome (NCBI37/mm9) together with a comprehensive database of annotated exon junctions using the program OLego (Wu J. and Zhang C., unpublished data). Only those reads that were mapped to unique loci with ≤ 4 mismatches (substitutions, insertions or deletions) were used for further analysis. For junction reads, we required ≥ 8 nt matches on each side for novel junctions, and ≥ 5 nt matches on each side for known junctions. Transcript structure of each cDNA fragment unobserved between the paired ends due to alternative splicing was then inferred using a Bayes method, followed by analysis of alternative splicing in ~13,000 annotated cassette exons in the mouse genome (Charizanis K. and Swanson, M.S., unpublished data). Briefly, fragments of the biological replicates were first pooled together, and cassette exons were then filtered by junction fragment coverage to reduce multiple testing (junction_in + junction_skip ≥ 20, junction_WT + junction_KO ≥ 20). A Fisher’s exact test was performed to evaluate the statistical significance of splicing change using both exon or exon-junction fragments, followed by Benjamini multiple testing correction to estimate the false discovery rate (FDR). In addition, proportional change of exon inclusion (ΔI) was calculated using inclusion or skipping junction fragments.
We analyzed the mRNA steady state level of 18,974 protein-coding genes, and 1,722 lncRNAs. For protein-coding genes, we defined a set of non-overlapping “core” exons, which consists of a comprehensive collection of annotated exons in RefSeq and UCSC known genes, as well as in mRNA and EST sequences; exons showing inclusion level < 0.5, as estimated from mRNA/EST data, were excluded. LncRNA genes were obtained from RefSeq based on the annotated gene type “miscRNA”. A vast majority of these ncRNAs in this collection are long and have multiple exons, but also included miRNAs and scaRNAs, which were excluded for this study. The remaining transcripts are from 1,722 unique loci, and all exons were used to estimate gene expression level. To estimate FPKM (fragments per kilobase per million) values, the number of fragments overlapping with exons in each gene was counted, normalized by the total length of core exons and the total number of exonic reads in all genes. Statistical significance of differential expression was assessed by the edgeR program (Robinson et al., 2010) using fragment counts in each sample.
Supplementary Material
Malat1, a highly abundant lncRNA, is dispensable for mouse development.
Loss of Malat1 does not alter the level and phosphorylation status of SR proteins.
Malat1 does not regulate global splicing and gene expression in mouse liver and cortex.
Transcription of Malat1 regulates brain cortex gene expression in cis.
Acknowledgement
We thank Carmen Berasain, Jan Bergmann, Megan Bodnar, Melanie Eckersley-Maslin, Stephen Hearn, Michael Hübner, Ileng Kumaran, Jingjing Li, Cinthya Zepeda-Mendoza, and Rui Zhao of the Spector laboratory and C. Frank Bennett at Isis Pharmaceuticals for helpful discussions and comments. We thank Sang Yong Kim and Ying Lin for technical assistance. We are grateful to Angus Lamond for PSP1α antibody and Adrian Krainer for SRSF1 antibody (AK96). This work was supported by grants from NCI 5P01CA013106-40, NCI 2P30CA45508-24, and NIGMS 42694 (to D.L.S.), K99GM95713 (to C.Z.). B.Z. is supported by a Department of Defense Prostate Cancer Research Program Postdoctoral Fellowship (W81XWH-10-1-0190). Y.S.M. is supported by a National Cancer Center Postdoctoral Fellowship.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Competing Interests
DLS is a consultant to Isis Pharmaceuticals. The other authors have declared that no competing interest exists.
REFERENCES
- Bernard D, Prasanth KV, Tripathi V, Colasse S, Nakamura T, Xuan Z, Zhang MQ, Sedel F, Jourdren L, Coulpier F, et al. A long nuclear-retained non-coding RNA regulates synaptogenesis by modulating gene expression. Embo J. 2010;29:3082–3093. doi: 10.1038/emboj.2010.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bumgarner SL, Neuert G, Voight BF, Symbor-Nagrabska A, Grisafi P, van Oudenaarden A, Fink GR. Single-cell analysis reveals that noncoding RNAs contribute to clonal heterogeneity by modulating transcription factor recruitment. Molecular cell. 2012;45:470–482. doi: 10.1016/j.molcel.2011.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camblong J, Iglesias N, Fickentscher C, Dieppois G, Stutz F. Antisense RNA stabilization induces transcriptional gene silencing via histone deacetylation in S. cerevisiae. Cell. 2007;131:706–717. doi: 10.1016/j.cell.2007.09.014. [DOI] [PubMed] [Google Scholar]
- Castle JC, Armour CD, Lower M, Haynor D, Biery M, Bouzek H, Chen R, Jackson S, Johnson JM, Rohl CA, et al. Digital genome-wide ncRNA expression, including SnoRNAs, across 11 human tissues using polyA-neutral amplification. PLoS One. 2010;5:e11779. doi: 10.1371/journal.pone.0011779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen LL, Carmichael GG. Altered nuclear retention of mRNAs containing inverted repeats in human embryonic stem cells: functional role of a nuclear noncoding RNA. Molecular cell. 2009;35:467–478. doi: 10.1016/j.molcel.2009.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chodroff RA, Goodstadt L, Sirey TM, Oliver PL, Davies KE, Green ED, Molnar Z, Ponting CP. Long noncoding RNA genes: conservation of sequence and brain expression among diverse amniotes. Genome Biol. 2010;11:R72. doi: 10.1186/gb-2010-11-7-r72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clemson CM, Hutchinson JN, Sara SA, Ensminger AW, Fox AH, Chess A, Lawrence JB. An architectural role for a nuclear noncoding RNA: NEAT1 RNA is essential for the structure of paraspeckles. Molecular cell. 2009;33:717–726. doi: 10.1016/j.molcel.2009.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis IJ, Hsi BL, Arroyo JD, Vargas SO, Yeh YA, Motyckova G, Valencia P, Perez-Atayde AR, Argani P, Ladanyi M, et al. Cloning of an Alpha-TFEB fusion in renal tumors harboring the t(6;11)(p21;q13) chromosome translocation. Proc Natl Acad Sci U S A. 2003;100:6051–6056. doi: 10.1073/pnas.0931430100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferris AL, Wu X, Hughes CM, Stewart C, Smith SJ, Milne TA, Wang GG, Shun MC, Allis CD, Engelman A, et al. Lens epithelium-derived growth factor fusion proteins redirect HIV-1 DNA integration. Proc Natl Acad Sci U S A. 2010;107:3135–3140. doi: 10.1073/pnas.0914142107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gierman HJ, Indemans MH, Koster J, Goetze S, Seppen J, Geerts D, van Driel R, Versteeg R. Domain-wide regulation of gene expression in the human genome. Genome Res. 2007;17:1286–1295. doi: 10.1101/gr.6276007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert N, Boyle S, Fiegler H, Woodfine K, Carter NP, Bickmore WA. Chromatin architecture of the human genome: gene-rich domains are enriched in open chromatin fibers. Cell. 2004;118:555–566. doi: 10.1016/j.cell.2004.08.011. [DOI] [PubMed] [Google Scholar]
- Guffanti A, Iacono M, Pelucchi P, Kim N, Solda G, Croft LJ, Taft RJ, Rizzi E, Askarian-Amiri M, Bonnal RJ, et al. A transcriptional sketch of a primary human breast cancer by 454 deep sequencing. BMC genomics. 2009;10:163. doi: 10.1186/1471-2164-10-163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta RA, Shah N, Wang KC, Kim J, Horlings HM, Wong DJ, Tsai MC, Hung T, Argani P, Rinn JL, et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464:1071–1076. doi: 10.1038/nature08975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttman M, Amit I, Garber M, French C, Lin MF, Feldser D, Huarte M, Zuk O, Carey BW, Cassady JP, et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature. 2009;458:223–227. doi: 10.1038/nature07672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirota K, Miyoshi T, Kugou K, Hoffman CS, Shibata T, Ohta K. Stepwise chromatin remodelling by a cascade of transcription initiation of non-coding RNAs. Nature. 2008;456:130–134. doi: 10.1038/nature07348. [DOI] [PubMed] [Google Scholar]
- Hongay CF, Grisafi PL, Galitski T, Fink GR. Antisense transcription controls cell fate in Saccharomyces cerevisiae. Cell. 2006;127:735–745. doi: 10.1016/j.cell.2006.09.038. [DOI] [PubMed] [Google Scholar]
- Houseley J, Rubbi L, Grunstein M, Tollervey D, Vogelauer M. A ncRNA modulates histone modification and mRNA induction in the yeast GAL gene cluster. Molecular cell. 2008;32:685–695. doi: 10.1016/j.molcel.2008.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchinson JN, Ensminger AW, Clemson CM, Lynch CR, Lawrence JB, Chess A. A screen for nuclear transcripts identifies two linked noncoding RNAs associated with SC35 splicing domains. BMC genomics. 2007;8:39. doi: 10.1186/1471-2164-8-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji P, Diederichs S, Wang W, Boing S, Metzger R, Schneider PM, Tidow N, Brandt B, Buerger H, Bulk E, et al. MALAT-1, a novel noncoding RNA, and thymosin beta4 predict metastasis and survival in early-stage non-small cell lung cancer. Oncogene. 2003;22:8031–8041. doi: 10.1038/sj.onc.1206928. [DOI] [PubMed] [Google Scholar]
- Kapranov P, Willingham AT, Gingeras TR. Genome-wide transcription and the implications for genomic organization. Nature reviews. 2007;8:413–423. doi: 10.1038/nrg2083. [DOI] [PubMed] [Google Scholar]
- Kato L, Begum NA, Burroughs AM, Doi T, Kawai J, Daub CO, Kawaguchi T, Matsuda F, Hayashizaki Y, Honjo T. Nonimmunoglobulin target loci of activation-induced cytidine deaminase (AID) share unique features with immunoglobulin genes. Proc Natl Acad Sci U S A. 2012;109:2479–2484. doi: 10.1073/pnas.1120791109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khalil AM, Guttman M, Huarte M, Garber M, Raj A, Rivea Morales D, Thomas K, Presser A, Bernstein BE, van Oudenaarden A, et al. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc Natl Acad Sci U S A. 2009;106:11667–11672. doi: 10.1073/pnas.0904715106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamond AI, Spector DL. Nuclear speckles: a model for nuclear organelles. Nat Rev Mol Cell Biol. 2003;4:605–612. doi: 10.1038/nrm1172. [DOI] [PubMed] [Google Scholar]
- Mao YS, Sunwoo H, Zhang B, Spector DL. Direct visualization of the co-transcriptional assembly of a nuclear body by noncoding RNAs. Nat Cell Biol. 2011;13:95–101. doi: 10.1038/ncb2140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marahrens Y, Panning B, Dausman J, Strauss W, Jaenisch R. Xist-deficient mice are defective in dosage compensation but not spermatogenesis. Genes Dev. 1997;11:156–166. doi: 10.1101/gad.11.2.156. [DOI] [PubMed] [Google Scholar]
- Martens JA, Laprade L, Winston F. Intergenic transcription is required to repress the Saccharomyces cerevisiae SER3 gene. Nature. 2004;429:571–574. doi: 10.1038/nature02538. [DOI] [PubMed] [Google Scholar]
- Martianov I, Choukrallah MA, Krebs A, Ye T, Legras S, Rijkers E, Van Ijcken W, Jost B, Sassone-Corsi P, Davidson I. Cell-specific occupancy of an extended repertoire of CREM and CREB binding loci in male germ cells. BMC genomics. 2010;11:530. doi: 10.1186/1471-2164-11-530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercer TR, Gerhardt DJ, Dinger ME, Crawford J, Trapnell C, Jeddeloh JA, Mattick JS, Rinn JL. Targeted RNA sequencing reveals the deep complexity of the human transcriptome. Nat Biotechnol. 2011;30:99–104. doi: 10.1038/nbt.2024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohammad F, Mondal T, Guseva N, Pandey GK, Kanduri C. Kcnq1ot1 noncoding RNA mediates transcriptional gene silencing by interacting with Dnmt1. Development. 2010;137:2493–2499. doi: 10.1242/dev.048181. [DOI] [PubMed] [Google Scholar]
- Nakagawa S, Naganuma T, Shioi G, Hirose T. Paraspeckles are subpopulation-specific nuclear bodies that are not essential in mice. J Cell Biol. 2011;193:31–39. doi: 10.1083/jcb.201011110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orom UA, Derrien T, Beringer M, Gumireddy K, Gardini A, Bussotti G, Lai F, Zytnicki M, Notredame C, Huang Q, et al. Long noncoding RNAs with enhancer-like function in human cells. Cell. 2010;143:46–58. doi: 10.1016/j.cell.2010.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petruk S, Sedkov Y, Riley KM, Hodgson J, Schweisguth F, Hirose S, Jaynes JB, Brock HW, Mazo A. Transcription of bxd noncoding RNAs promoted by trithorax represses Ubx in cis by transcriptional interference. Cell. 2006;127:1209–1221. doi: 10.1016/j.cell.2006.10.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajaram V, Knezevich S, Bove KE, Perry A, Pfeifer JD. DNA sequence of the translocation breakpoints in undifferentiated embryonal sarcoma arising in mesenchymal hamartoma of the liver harboring the t(11;19)(q11;q13.4) translocation. Genes, chromosomes & cancer. 2007;46:508–513. doi: 10.1002/gcc.20437. [DOI] [PubMed] [Google Scholar]
- Rinn JL, Kertesz M, Wang JK, Squazzo SL, Xu X, Brugmann SA, Goodnough LH, Helms JA, Farnham PJ, Segal E, et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129:1311–1323. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki YT, Ideue T, Sano M, Mituyama T, Hirose T. MENepsilon/beta noncoding RNAs are essential for structural integrity of nuclear paraspeckles. Proc Natl Acad Sci U S A. 2009;106:2525–2530. doi: 10.1073/pnas.0807899106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schorderet P, Duboule D. Structural and functional differences in the long non-coding RNA hotair in mouse and human. PLoS Genet. 2011;7:e1002071. doi: 10.1371/journal.pgen.1002071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shearwin KE, Callen BP, Egan JB. Transcriptional interference--a crash course. Trends Genet. 2005;21:339–345. doi: 10.1016/j.tig.2005.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sleutels F, Zwart R, Barlow DP. The non-coding Air RNA is required for silencing autosomal imprinted genes. Nature. 2002;415:810–813. doi: 10.1038/415810a. [DOI] [PubMed] [Google Scholar]
- Stadler P. Evolution of the Long Non-coding RNAs MALAT1 and MENβ/ε. Advances in Bioinformatics and Computational Biology. 2010;6268:1–12. [Google Scholar]
- Sunwoo H, Dinger ME, Wilusz JE, Amaral PP, Mattick JS, Spector DL. MEN epsilon/beta nuclear-retained non-coding RNAs are up-regulated upon muscle differentiation and are essential components of paraspeckles. Genome Res. 2009;19:347–359. doi: 10.1101/gr.087775.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tripathi V, Ellis JD, Shen Z, Song DY, Pan Q, Watt AT, Freier SM, Bennett CF, Sharma A, Bubulya PA, et al. The nuclear-retained noncoding RNA MALAT1 regulates alternative splicing by modulating SR splicing factor phosphorylation. Molecular cell. 2010;39:925–938. doi: 10.1016/j.molcel.2010.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai MC, Manor O, Wan Y, Mosammaparast N, Wang JK, Lan F, Shi Y, Segal E, Chang HY. Long noncoding RNA as modular scaffold of histone modification complexes. Science. 2010;329:689–693. doi: 10.1126/science.1192002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner BM, Franchi L. Identification of protein antigens associated with the nuclear matrix and with clusters of interchromatin granules in both interphase and mitotic cells. J Cell Sci. 1987;87(Pt 2):269–282. doi: 10.1242/jcs.87.2.269. [DOI] [PubMed] [Google Scholar]
- Ulitsky I, Shkumatava A, Jan CH, Sive H, Bartel DP. Conserved function of lincRNAs in vertebrate embryonic development despite rapid sequence evolution. Cell. 2011;147:1537–1550. doi: 10.1016/j.cell.2011.11.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilusz JE, Freier SM, Spector DL. 3′ end processing of a long nuclear-retained noncoding RNA yields a tRNA-like cytoplasmic RNA. Cell. 2008;135:919–932. doi: 10.1016/j.cell.2008.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilusz JE, Sunwoo H, Spector DL. Long noncoding RNAs: functional surprises from the RNA world. Genes Dev. 2009;23:1494–1504. doi: 10.1101/gad.1800909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilusz JE, Whipple JM, Phizicky EM, Sharp PA. tRNAs marked with CCACCA are targeted for degradation. Science. 2011;334:817–821. doi: 10.1126/science.1213671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Lin C, Liu W, Zhang J, Ohgi KA, Grinstein JD, Dorrestein PC, Rosenfeld MG. ncRNA- and Pc2 methylation-dependent gene relocation between nuclear structures mediates gene activation programs. Cell. 2011;147:773–788. doi: 10.1016/j.cell.2011.08.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.