SUMMARY
Clofarabine (ClF) is a drug used in the treatment of leukemia. One of its primary targets is human ribonucleotide reductase (hRNR), a dual-subunit, (α2)m(β2)n, regulatory enzyme indispensable in de novo dNTP synthesis. We report that in live mammalian cells, ClF targets hRNR by converting its α-subunit into kinetically-stable hexamers. We established mammalian expression platforms that enabled isolation of functional α and characterization of its altered oligomeric associations in response to ClF treatment. Size exclusion chromatography and electron microscopy documented persistence of in-cell-assembled-α6. Our data validate hRNR as an important target of ClF, provide previously-unreported evidence that in vivo α’s quaternary structure can be perturbed by a non-natural ligand, and suggest small-molecule-promoted, persistent hexamerization as a new strategy to modulate hRNR activity. These studies lay foundations for documentation of RNR oligomeric state within a cell.
INTRODUCTION
Alterations of quaternary states of a protein can change its function dramatically (Nooren and Thornton, 2003; Arkin and Wells, 2004; Peihler, 2005). While many methods exist to establish protein oligomeric equilibria in vitro, validating the existence of these oligomers and their relevance in vivo is often challenging, despite its importance (Piehler, 2005). One specific case where these issues remain unresolved is ribonucleotide reductases (RNRs), highly regulated enzymes that catalyze the conversion of nucleoside 5´-diphosphates (NDPs) to deoxynucleotides (Stubbe and van der Donk, 1998; Stubbe et al., 2003; Nordlund and Reichard, 2006).
Human (h)RNRs belong to the class Ia family of RNRs that require two subunits (α2)m and (β2)n or p53(β2)n which form, as yet unresolved, active complex(es) proposed to be α2β2, α6β2 and/or α6β6 (Kashlan and Cooperman, 2003; Rofougaran et al., 2006; Fairman et al., 2011; Hofer et al., 2012). In addition to the NDP-binding, catalytic (C) site, (α2)m possesses two well-characterized allosteric sites. The activity (A) site, within the N-terminal ATP-cone-domain, binds either ATP, activating RNR activity, or dATP, inhibiting it. The specificity (S) site, at the α2 interface, binds dNTP/ATP, modulating the substrate preference at the C site (Fairman et al., 2011). In vitro studies on eukaryotic RNRs indicate that the dATP-inhibited complex is α6 (Kashlan and Cooperman, 2003; Rofougaran et al., 2006; Fairman et al., 2011; Aye and Stubbe, 2011), with its arrangement as a trimer-of-dimers demonstrated by electron microscopy (EM) (of particles preserved in stain) and 6.6 Å-resolution X-ray crystallography of the S. cerevisiae RNR (Fairman et al., 2011).
The importance of hRNR in dNTP pool homeostasis (Zhou and Elledge, 2000; Thelander, 2007; Bester et al., 2011) has rendered it a successful drug target (Shao et al., 2006), and drug-induced alterations in oligomeric equilibria have been demonstrated in vitro (Wang et al., 2007; Aye and Stubbe, 2011). We have recently shown that di- and triphosphates of the clinically-used leukemia-drug ClF (clofarabine, Clolar®, Figure 1) are both reversible inhibitors of hRNR, binding to the C and A-sites of α, respectively (Aye and Stubbe, 2011). Inactivation is accompanied by α-hexamerization that occurs independently of allosteric effectors and (β2)n. Size exclusion chromatography (SEC) studies further demonstrated that ClFD(T)P-induced hexamers are kinetically-stable in the absence of inhibitors in the SEC running buffer. These hexamers thus display fundamentally distinct kinetic properties from the dATP-induced α6 (Kashlan and Cooperman, 2003; Rofougaran et al., 2006; Fairman et al., 2011; Aye and Stubbe, 2011), which rapidly dissociate to an equilibrium mixture of lower order oligomers when the dATP is omitted from the elution buffer (Fairman et al., 2011; Aye and Stubbe, 2011).
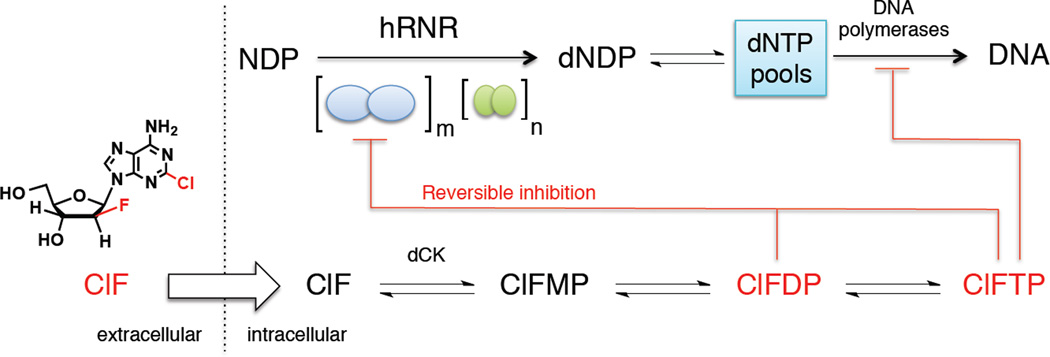
Figure 1. hRNR is a principal target of ClF.
ClF enters the cell by passive diffusion and/or via nucleoside transporters, and is rapidly phosphorylated (Bonate et al., 2006; Zhenchuk et al., 2009). ClFDP and ClFTP both target hRNR (α2)m, resulting in the depletion of dNTP pools including dCTP. Reduction of the latter potentiates production of ClF metabolites by removing negative feedback inhibition of deoxycytidine kinase, dCK, an enzyme that catalyzes monophosphorylation of ClF. Diminution of overall dNTP pool sizes amplifies misincorporation of ClFTP into DNA by DNA polymerase-α and –ε where it functions as a chain terminator. Induction of apoptosis is thought to be ultimately responsible for anticancer activity. ClFTP is a rapid reversible inhibitor of α via binding to the activity (A) site (Ki = 40 nM) and ClFDP is a slow-release reversible inhibitor (Ki* = 17 nM, t1/2= 23 min) of α via binding to the catalytic (C) site, and both cause α hexamerization (Aye and Stubbe, 2011).  and
and  ellipses represent (α2) and (β2 or p53 β2) respectively.
ellipses represent (α2) and (β2 or p53 β2) respectively.
We now report biochemical and structural evidence that hexamers generated subsequent to RNR inhibition by ClFD(T)P in vitro (Aye and Stubbe, 2011), are also assembled in ClF-treated cultured live cells. Initially, crosslinking studies on homogeneous recombinant α at concentrations measured in vivo (Åkerblom et al., 1981; Håkansson et al., 2006; this study) showed, by gel analysis, that in the presence of ClFD(T)P, α was almost exclusively hexameric. However, similar studies to demonstrate hexamerization of the endogenous mammalian α in highly dilute lysates resulting from ClF-treated COS-1 cells, proved to be challenging due to very low endogenous α expression. Thus in an effort to assess the oligomeric state of α in vivo, the protein was first over-expressed minimally (3.5-fold above the endogenous level as judged by western blot analysis). Under these conditions, α from ClF treated live COS-1 cells, subsequent to crosslinking in the resulting lysates, underwent a gel-shift to a molecular weight consistent with α hexamers and similar to that characterized in vitro. To obtain biochemical and structural data in support of α-hexamerization, we then constructed a bicistronic reporter cassette that simultaneously overexpressed a fluorescent protein, Discosoma Red (DsRed), and His6-α (Wang et al., 2007; Aye and Stubbe, 2011). This platform enabled assessment of the oligomeric state of native α by SEC and single particle EM, subsequent to cell-lysis and affinity purification. Our data together validate the current model that hRNR is a target of ClF metabolites prior to the onset of cytotoxicity (Bonate et al., 2006). Remarkably, targeting hRNR results in the in-cell assembly of persistent hexamers that are also shown in vitro to remain stable subsequent to inhibitor dissociation. We provide the first evidence that the oligomeric state of (α2)m in live cells can be perturbed by an external ligand.
RESULTS
Crosslinking Studies on (α2)m Expressed at Near-Endogenous Levels Provide Preliminary Evidence for Drug-Induced-Hexamerization in Cultured Cells
The α subunit of hRNR has a half-life of 15–24 h and is present throughout the cell cycle in proliferating cells at very low levels (Åkerblom et al., 1981, Engström et al., 1984, 1985; Håkansson et al., 2006; Thelander et al., 2007). The amount of α in non-synchronized cells has been estimated to be ~0.07% wt/wt of total proteins in 3T6 cells (Åkerblom et al., 1981) and 0.04% in COS-1 cells (this study, Figure S1A). These low expression levels technically limit the isolation and the biochemical and structural characterization of the oligomeric state of α in the cell.
As a prelude to this goal, we initially attempted native gel electrophoresis on hexameric His6-α generated in vitro as previously described (Aye and Stubbe, 2011). However, α6 failed to penetrate a 4% polyacrylamide gel. We then tested chemical crosslinking in an effort to covalently stabilize the hexameric state for analysis by denaturing gel electrophoresis. BS-3 [bis (sulfosuccinimidyl) suberate, Fig S1B–A] was chosen as the crosslinker and the optimized conditions resulted in a shift of α from 92 kDa (monomer of α) to > 500 kDa (consistent with hexamer of α) (Figure S1B–B). Hexamerization was detected over a range of α concentrations (5 nM – 5 µM) in the presence of BS3 (1 – 3 mM), subsequent to an incubation period of as early as 1 min at 37 °C. Only when the samples had been treated with ClFD(T)P and BS3, were the shifts observed. In a control with no inhibitor, or with inhibitor and no BS3, the large molecular weight species was absent. In addition, crosslinked complexes with an approximate molecular weight of α dimers were also detected (to < 20% of total α) in the presence or absence of inhibitor (Figure S1B–B). The observed gel shift thus serves as a diagnostic marker for α hexamerization and provided the impetus to use crosslinking as an initial strategy to examine the ClF-induced quaternary state perturbation of (α2)m in mammalian cells.
Experiments on COS-1 cells treated with ClF to detect hexamerization of the endogenous protein proved considerably more challenging. Ultimately success was achieved using a mammalian expression plasmid (monocistronic cassette, Figure 2A and Table S1A) transfected into COS-1 cells that allowed ectopic expression of untagged hRNR α at levels 3.5-fold above the endogenous level (Figure S1B–C). As ClFD(T)P-induced oligomerization is (β2-independent (Aye and Stubbe, 2011), only α was part of the construct. The expressed α in the transfected samples had activity ~10-fold higher than the non-transfected samples (Figure 2B, Figure S1A–A). The basis behind the 2–3-fold higher lysate activities, relative to the fold-increment in expression level assessed by western blot (Figure S1B–C), is not well understood.
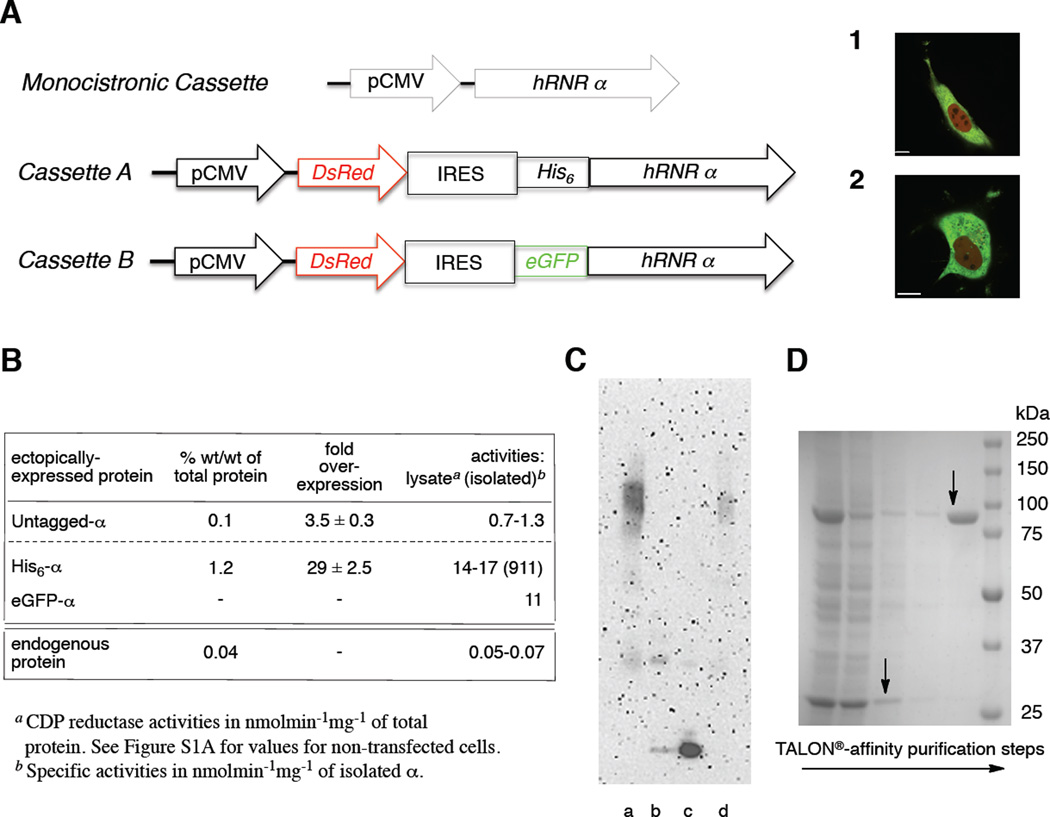
Figure 2. Initial crosslinking studies on in-cell relevance of α-hexamerization and a bicistronic reporter cassette for expression/rapid purification of hRNR (α2)m from mammalian cells.
(A) hRNR (α2)m expression platforms (Table S1A and S1B): Monocistronic cassette results in expression of untagged protein; Cassette A of DsRed and His6-α; Cassette B of DsRed and eGFP-α. Merged confocal fluorescence images of live HeLa (1) and COS-1 (2) cells transfected with Cassette B. Scale bar in (1) and (2) each corresponds to 10 µm. (B) Protein content, fold-overexpression and activities in lysate (and of isolated, where applicable) of untagged-, His6-and eGFP-α ectopically expressed in COS-1 cells, subsequent to transient transfection with either monocistronic cassette (Table S1A) or bicistronic reporter cassettes A or B (Table S1B). Corresponding values for the endogenous α were shown for comparison. Data, where applicable, are represented as mean +/− SD. Also see Figure S1A and S1B–C. (C) α-Hexamerization, subsequent to crosslinking in lysates, in response to ClF (50 nM) treatment of live COS-1 cells expressing untagged-α at 3.5 fold above the endogenous levels. Lanes a and b, results from BS3 (1 mM) treatment of the lysates (0.1 mg/ml) from ClF-treated (a) and untreated (b) cells. Lane c, recombinant His6-α (0.15 µg/ml) treated in vitro with BS3 and subjected to identical sample preparation procedures as for lysates. Lane d, identical to lane c except additional presence of ClFTP (effectively, crosslinked α6 standard - see Figure S1B–B for in vitro characterization). (D) SDS-PAGE of steps involved in His6-α purification from COS-1 cells transfected with Cassette A. Lanes (left to right): lysate; supernatant after TALON® incubation; wash 1; wash 2; elution; MW ladder. Arrows indicate His6-α (92 kDa) and DsRed (27 kDa). Also see Figure S1D.
Using the crosslinking method described above, the ectopic expression of hRNR α at 3.5 fold above the endogenous levels enabled, as shown below, the demonstration of α-hexamerization within the cell in response to ClF treatment. Throughout this work, all studies were carried out in a time frame where essentially all cells are viable (Figure S1C and S1D–A), to minimize the possibility that results are affected by ClF-induced apoptosis (Carson et al., 1992; Bonate et al., 2006; Zhenchuk et al., 2009). The results obtained are therefore most likely a consequence of the direct interaction between (α and ClFD(T)P (Aye and Stubbe, 2011). Transfected COS-1 cells were treated with ClF (50 nM) and incubated for 3 h and compared with control experiments under identical conditions. Choice of nucleoside concentration and incubation time were guided by the viability data of ClF-treated cells relative to untreated controls (Figure S1C) and by previous pharmacological studies (Parker et al., 1991; Carson et al., 1992; Xie et al., 1995 and 1996; Lofti et al., 1999; Bonate et al., 2006; Zhenchuk et al., 2009). Subsequently, lysates from treated and untreated cells were diluted (25× – 250×) prior to treatment with the crosslinker. Initial tests were performed under different dilutions since crosslinking of pre-existing α6 (an intramolecular reaction) should not be influenced by dilution, while undesired crosslinking to other proteins in the crude extract would be minimized. A lysate concentration of ~0.1 mg/ml was found to be optimum. The high dilution also required that lysate solutions be lyophilized and ethanol-precipitated prior to western blot analysis. Under these conditions, incubation with BS3 (1 mM) over 1 min at 37 °C showed that > 95 ± 1 % of α is present in hexameric form in the treated cells, whereas < 1 % of α is hexameric in the control (Figure 2C). These data provide preliminary evidence for the formation of hexameric α within a cell upon ClF treatment.
Reporter Assay Advances Optimization of hRNR Expression in Live Mammalian Cells
To further characterize ClF-induced hexamerization of α in vivo, expression of sufficient amounts of protein and rapid affinity isolation were required. Since N-terminal His6-tagging of α has been previously shown not to affect α’s activity or ClFD(T)P-promoted hexamerizations (Aye and Stubbe, 2011), ectopic expression of His6-α in mammalian cells was pursued. An internal ribosomal entry site (IRES)-based cassette encoding both DsRed and His6-α was designedw(Cassette A, Figure 2A and Table S1B). As both DsRed and His6-α are derived from a single mRNA transcript downstream of the CMV promoter, the presence of one mandates that of the other (Yen et al., 2008). The intrinsic co-expression of the fluorescent marker, DsRed, allowed rapid optimization of the transfection efficiency. This strategy enabled overexpression of α at the levels sufficient for biochemical analysis [29-fold above the endogenous in COS-1 cells, as judged by western blot (Figure S1B–C)]. We reasoned that the shift to the hexamer would not be negatively impacted by high overexpression of α, because inhibitor-induced oligomerization is (β2-independent (Aye and Stubbe, 2011). Furthermore, since mammalian RNR activity in vivo is limited by (β2 because of the constitutive expression of (αm throughout the cell cycle (Thelander, 2007), minimum perturbation to the native system was expected, despite the high level of α expression. His6-α was additionally shown to be ectopically expressed in HeLa and HEK-293 cell lines while maintaining good cell viability (Figure S1D–A). We chose COS-1 as our representative cell line. The α expressed was shown to be functional by lysate activity assays (Figure 2B and Figure S1A). As previously observed for COS-1 cells transfected with the monocistronic plasmid (Figure 2A and Table S1A), lysate activities from cells expressing DsRed and His6-α also are higher than what was expected from the corresponding level of protein overexpression (Figure 2B and Figure S1B–C).
As an additional proof-of-principle for a functional IRES-mediated hRNR expression platform, an alternative dual-reporter cassette B (Figure 2A and Table S1B) was constructed that directly reports on α expression levels relative to the DsRed using FACS. Lysates of ectopically expressed enhanced green fluorescent protein-fused α (eGFP-α) from COS-1 cells have reductase activity similar to lysates containing His6-α (Figure 2B), as do recombinantly expressed and purified His6-α and His6-eGFP-α from E. coli (Table S1C and Figure S1E). Preferential nuclear exclusion of GFP-fused α observed in live COS-1 and HeLa cells (Figure 2A) is consistent with previously reported data using cell fractionation and immunofluorescence (Engström et al., 1984; Pontarin et al., 2008).
Rapid Affinity Purification Affords Isolation of Ectopically Expressed Active hRNR α
E. coli expressed recombinant (r)-His6-α was previously purified to homogeneity using a cobalt-affinity chromatography, TALON® (Aye and Stubbe, 2011); thus a similar strategy was pursued to isolate His6-α from mammalian cell lines subsequent to transfection (Cassette A, Figure 2A and Table S1B). The optimized strategy led to the isolation of α (>95% pure as judged by SDS-PAGE) with activity similar to that of recombinant enzyme (Aye and Stubbe, 2011), and its identity was further confirmed by western blot (Figure 2B, 2D, and Figure S1D–B and S1D–C). Typical yields from COS-1 and HEK-293 cell lines ranged from 2–4 µg α from a 75 cm2 of confluent monolayer culture.
α Inhibited by ClFD(T)P or dATP in Lysates, but Only Drug-InducedIα6 Survives
We initially examined whether preassembled-α6 [formed by treatment of r-His6-α with ClFD(T)P (Aye and Stubbe, 2011)] could be added to COS-1 lysates and isolated intact using TALON®. SEC of isolated protein and western blot analysis of eluted fractions revealed that hexameric states were observed (Figure S2A). We then showed that ectopically-expressed functional His6-α in mammalian cells is also responsive to ClF-nucleotides as expected from in vitro studies with r-His6-α (Aye and Stubbe, 2011). Treatment of COS-1 cell lysates with ClFD(T)P (2 µM) depleted lysate activities (Figure S2B) and the α6 state(s) assembled in lysates were retained throughout the purification (Figure S2C). These outcomes contrast with controls without ClFD(T)P or when saturating amounts of dATP (20 µM) (Kashlan and Cooperman, 2003; Rofougaran et al., 2006; Fairman et al., 2011; Aye and Stubbe, 2011), were added to the lysates under identical conditions.
ClF Treatment Drives Persistent Hexamerization in Live Mammalian Cells
Our subsequent studies employed the bicistronic plasmid (Cassette A, Figure 2A and Table S1B) as it gave the highest total protein yield and the native His6-tagged-α could be isolated from the lysate. As in the crosslinking experiments above that used the monocistronic plasmid, COS-1 cells were incubated with ClF for 3 h where high cell viability was still maintained (Figure S1C and S1D–A). However, since α expression was now higher (Figure S1B–C), 5 µM ClF was used as opposed to the 0.05 µM in crosslinking experiments. After cell lysis and TALON® purification of His6-α, the oligomeric state of eluted protein, from both treated and untreated cells, was determined by SEC. The SEC profile of the isolated protein from drug-treated cells closely resembled α6 characterized in vitro (Aye and Stubbe, 2011) and was markedly different from the isolated α from untreated cells or the recombinant protein in the absence of ClFD(T)P. Proteins in SEC fractions were ethanol-precipitated and analyzed by gel electrophoresis and silver-staining (Chevallet et al., 2006), confirming the presence of a homogeneous hexameric α in the experiment and not in the control (Figure 3A–D).
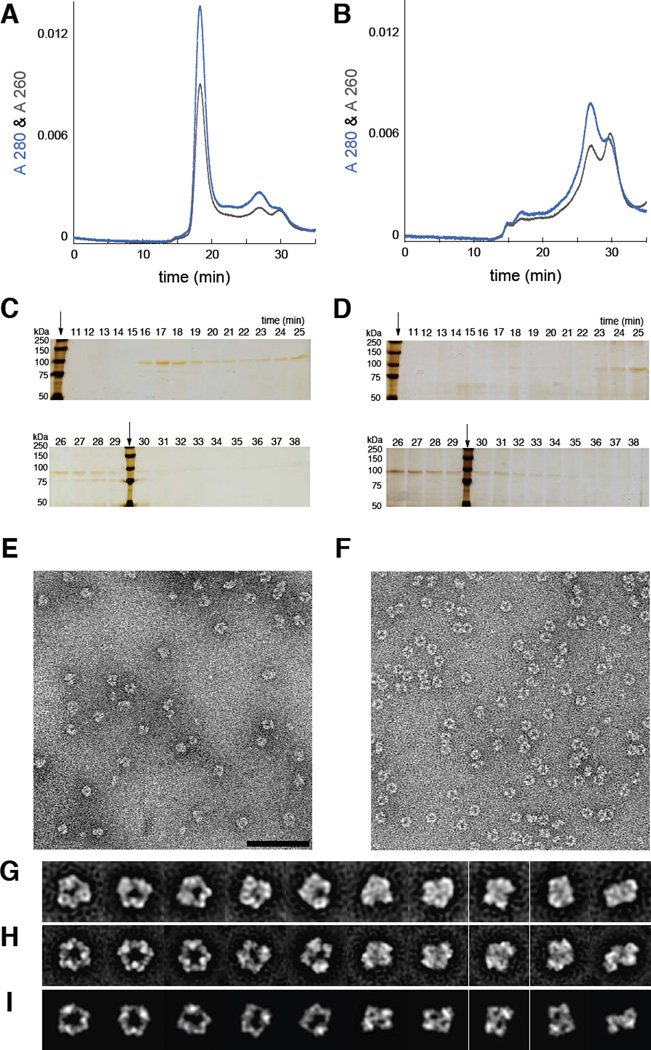
Figure 3. ClF treatment results in persistent α6 in live cells which are analyzed by size exclusion chromatography (SEC) (A–D) and single particle electron microscopy (EM) (E–J).
SEC elution profiles of TALON®-purified (α2)m,,monitored by A280nm and A260nm, from (A) ClF-treated COS-1 cells and (B) untreated COS-1 cells. The major peak in (A) elutes at 17 min and is proposed to be α6 by comparison with in vitro- ClFTP and ClFDP-treated r-His6-α [inset in (A)] and standard proteins of known molecular weight (Figure S2C- Panel F). Note the absence of this peak in (B). Silver-stained-SDS-PAGE of SEC fractions, subsequent to ethanol precipitation, from ClF-treated (C) and untreated cells (D). Arrows indicate MW-standards. (E) EM image of negatively stained complexes from the sample eluted at 17 min in (A). (F) EM image of negatively stained complexes of recombinant (r)-His6-α incubated with ClFDP in vitro. Similar results were obtained with ClFTP. (G) ISAC averages of α6 isolated from ClF-treated cells. (H) ISAC averages derived from (F) were matched with the averages in (G). (I) Averages in (G and H) correspond with 2D projections of an α6 model derived by superimposing 3 copies of a human r-α2 X-ray structure onto the 6.6 Å-resolution X-ray structure of yeast α6 (Fairman et al., 2011). Scale bar corresponds to 100 nm in (E and F), or 314 Å in (G, H and I).
Single Particle Electron Microscopy Directly Identifies Drug-Induced Hexameric Assemblies
To gain further insight into the subunit organization of α assembled in live cells, we turned to electron microscopy (EM) analysis of complexes isolated from ClF-treated cells subsequent to both affinity purification and SEC characterization (Figure 3E). For comparison, we also collected EM images of r-His6-α incubated with ClFDP in vitro (Aye and Stubbe, 2011) (Figure 3F). Images were analyzed using a recently described iterative stable alignment and clustering (ISAC) algorithm (Yang et al., 2012). ISAC averages of the α complexes from ClF-treated cells reveal a subunit arrangement that strikingly resembles the complex formed in vitro (Figures 3G and H, respectively, and Figure S2D). Furthermore, ISAC averages from both preparations closely resemble projections of an atomic model of human r-α6 generated by arranging six copies of the human r-α X-ray structure with dATP/TTP bound according to the 6.6 Å-resolution X-ray structure of α6 from yeast (Fairman et al., 2011) (Figure 3I). These observations establish that the drug-induced complexes formed in cells or assembled in vitro adopt similar trimer-of-dimers architectures.
ClFD(T)P Induces Kinetically-Stable Hexamers Persisting Beyond Inhibitor Dissociation
Given the isolation procedure, the α-hexamers isolated from live cells are likely unliganded. To support this proposal, [8-3H]-ClFD(T)P were synthesized (see Supplemental Experimental Procedures) and SEC experiments were replicated using r-His6-α in vitro (Figure S3A). No observable radioactivity was associated with the ClFTP-induced hexameric protein fractions. In the case of [8-3H]-ClFDP, 1.0 ± 0.2 ClFDP/α6 was detected at 1.5 or 15 µM α, consistent with its slow release from E•ClFDP* (Aye and Stubbe, 2011). We additionally showed that in the case with ClFTP, the SEC-eluted unliganded-α6 persists in this state, even subsequent to overnight standing at 4 °C in 5 mM DTT (Figure S3B).
DISCUSSION
Our data establish that alterations of (α2)m quaternary equilibria observed in vitro with ClFD(T)P can be recapitulated in live cells. They demonstrate ClF-nucleotide(s)-driven in-cell assembly of persistent hexamers, even in the presence of dNTP/ATP pools and limited amounts of (β2)n/p53(β2)n (Thelander, 2007). Remarkably the hexameric states remain kinetically-stable subsequent to cell-lysis and affinity isolation. The markedly different results observed with the non-treated lysates and dATP-treated controls emphasize that hexamerization is ClFD(T)P-mediated and most likely cell-type-independent. Our results further suggest that small-molecule-promoted persistent oligomerization is a new avenue to target hRNR. Once the molecular basis for these inhibitory states are understood, additional small-molecules might be discovered that stabilize these states and function as new types of hRNR inhibitors. Finally, the IRES-mediated dual expression system that we have built here (Figure 2A) provides a platform to study further aspects of hRNR in live mammalian cells.
EXPERIMENTAL PROCEDURES
See Supplemental Information for experimental procedures and additional supporting figures. Supplemental Information can be found with this article online at
Supplementary Material
SIGNIFICANCE.
Human ribonucleotide reductase (hRNR), an essential enzyme in DNA synthesis, is a proven target of the leukemia drug, Clofarabine (ClF). Two distinct subunits, (α2)m and (β2)n, complete the active unit required for the catalysis of NDP (N=A,C,G,U) reduction to dNDPs. Oligomeric equilibria of (α2)m governed by binding of various NDP substrate/(d)NTP allosteric effector pairs remain a poorly understood area of mammalian RNR, even in vitro, and the relevance of these states [(α2)m(β2)n; m, n = 1–3] is yet to be established in the cell. Our previous biochemical studies have demonstrated that hRNR inhibition by ClFD(T)P is coupled to α-hexamerization in vitro. We now report that this mechanism is operative in mammalian cells: α-targeted inhibition by phosphorylated ClF involves in-cell assembly of kinetically-stable hexamers that persist beyond inhibitor departure. The oligomeric state of α isolated from live cells exposed to ClF has been analyzed by chemical crosslinking, size exclusion chromatography and single-particle electron microscopy. Together these studies reveal that α-hexamerization in vivo occurs independent of (β2)n, and even in the presence of various pools of NDP substrates and (d)NTP allosteric effectors. This is in stark contrast to previously characterized nucleotide analog inhibitors of RNR where (α2)m(β2)n holocomplex formation is a prerequisite for the initiation of mechanism-based inactivation. Given the constitutive expression of α in mammalian cells and the contrasting expression of β which is primarily S-phase specific, our studies unveil a new avenue to down-regulate activity of this key enzyme, solely by targetingt(α2)m through persistent oligomerization. The approach we have established here also represents a potentially useful strategy to characterize modulations of RNR quaternary structure in live mammalian cells in response to external stimuli. In a broader context, these findings present an interesting example of non-natural small-molecule(s)-induced perturbation of protein quaternary structures in cells, demonstrated using the clinically-important drug, ClF.
HIGHLIGHTS.
New system allows expression/isolation of active human RNR-α from mammalian cells
Strategy unveils Clofarabine mechanism and in-cell-relevance of α-oligomerization
Independent biochemical & structural methods document in-cell-assembled α-hexamers
Drug-induced hexameric states have remarkable kinetic stability
ACKNOWLEDGEMENTS
This research was supported by the National Institutes of Health Grants (GM29595 to J.S.), GM67167 (to F.J.A.), and the Damon Runyon Cancer Research postdoctoral fellowship (DRG2015-09) to Y.A. M.J.C.L acknowledges a Howard Hughes Medical Institute international student fellowship. C.L.D. is a Howard Hughes Medical Institute Investigator. Electron microscopy was conducted at the National Resource for Automated Molecular Microscopy, which is supported by the National Institutes of Health though the National Center for Research Resources’ P41 program (RR017573). Author contributions: Y.A. and J.S. conceived of and designed the experiments and assisted in the design of the EM experiments carried out by E.J.B., J.C. and F.J.A. M.J.C.L assisted Y.A. in the design and construction of the reporter cassettes and the confocal imaging microscopy. Y.A. and J.S. wrote the paper with the description of the EM experiments and results provided by E.J.B. and F.J.A. All authors participated in editing the manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Authors declare no conflict of interest.
REFERENCES
- Åkerblom L, Ehrenberg A, Gräslund A, Lankinen H, Reichard P, Thelander L. Overproduction of the free radical of ribonucleotide reductase in hydroxyurea-resistant mouse fibroblast 3T6 cells. Proc. Natl. Acad. Sci. USA. 1981;78:2159–2163. doi: 10.1073/pnas.78.4.2159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arkin MR, Wells JA. Small-molecule inhibitions of protein-protein interactions: progressing towards the dream. Nat. Rev. Drug Discov. 2004;3:301–317. doi: 10.1038/nrd1343. [DOI] [PubMed] [Google Scholar]
- Aye Y, Stubbe J. Clofarabine 5´-di and –triphosphates inhibit human ribonucleotide reductase by altering the quaternary structure of its large subunit. Proc. Natl. Acad. Sci. USA. 2011;108:9815–9820. doi: 10.1073/pnas.1013274108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bester AC, Roniger M, Oren YS, Im MM, Sarni D, Chaoat M, Bensimon A, Zamir G, Shewach DS, Kerem B. Nucleotide deficiency promotes genomic instability in early stages of cancer development. Cell. 2011;145:435–446. doi: 10.1016/j.cell.2011.03.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonate PL, Arthaud L, Cantrell WR, Stephenson K, Secrist JA, Weitman S. Discovery and development of clofarabine: a nucleoside analogue for treating cancer. Nat. Rev. Drug Discovery. 2006;5:855–240. doi: 10.1038/nrd2055. [DOI] [PubMed] [Google Scholar]
- Carson DA, Wasson DB, Esparza LM, Carrera CJ, Kipps TJ, Cottam HB. Oral antilymphocyte activity and induction of apoptosis by 2-chloro-2´-arabino-fluoro-2´-deoxyadenosine. Proc. Natl. Acad. Sci. USA. 1992;89 doi: 10.1073/pnas.89.7.2970. 2970– (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chevallet M, Luche S, Rabilloud T. Silver-staining of proteins in polyacrylamide gels. Nat. Protoc. 2006;1:1852–1858. doi: 10.1038/nprot.2006.288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engström Y, Rozell B, Hansson H-A, Stemme S, Thelander L. Localization of ribonucleotide reductase in mammalian cells. EMBO J. 1984;3:863–867. doi: 10.1002/j.1460-2075.1984.tb01897.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engström Y, Eriksson S, Jildevik I, Skog S, Thelander L, Tribukait B. Cell cycle-dependent expression of mammalian ribonucleotide reductase. J. Biol. Chem. 1985;260:9114–9116. [PubMed] [Google Scholar]
- Fairman JW, Wijerathna SR, Ahmad MF, Xu H, Nakano R, Jha S, Prendergast J, Welin RM, Flodin S, Roos A, et al. Structural basis for allosteric regulation of human ribonucleotide reductase by nucleotide-induced oligomerization. Nat. Struct. Mol. Biol. 2011;3:316–322. doi: 10.1038/nsmb.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Håkansson P, Hofer A, Thelander L. Regulation of mammalian ribonucleotide reduction and dNTP pools after DNA damage and in resting cells. J. Biol. Chem. 2006;281:7834–7841. doi: 10.1074/jbc.M512894200. [DOI] [PubMed] [Google Scholar]
- Hofer A, Crona M, Logan DT, Sjoberg BM. DNA building blocks: keeping control of manufacture. Crit. Rev. Biochem. Mol. Biol. 2012;47:50–63. doi: 10.3109/10409238.2011.630372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashlan OB, Cooperman BS. Comprehensive model for allosteric regulation of mammalian ribonucleotide reductase: refinements and consequences. Biochemistry. 2003;42:1696–1706. doi: 10.1021/bi020634d. [DOI] [PubMed] [Google Scholar]
- Lofti K, Mansson E, Spasokoukotskaja T, Pettersson B, Liliemark J, Peterson C, Eriksson S, Albertioni F. Biochemical pharmacology and resistance to 2-chloro-2´-arabino-fluoro-2´-deoxyadenosine, a novel analogue of cladribine in human leukemic cells. Clin. Cancer Res. 1999;5:2438–2444. [PubMed] [Google Scholar]
- Nooren IMA, Thornton JM. Diversity of protein–protein interactions. EMBO J. 2003;22:3486–3492. doi: 10.1093/emboj/cdg359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordlund N, Reichard P. Ribonucleotide reductases. Annu. Rev. Biochem. 2006;75:681–706. doi: 10.1146/annurev.biochem.75.103004.142443. [DOI] [PubMed] [Google Scholar]
- Parker WB, Shaddix SC, Chang CH, White EL, Rose LM, Brockman RW, Shortnacy AT, Montgomery JA, Secrist JA, III, Bennett LL., Jr Effects of 2-chloro-9-(2´-deoxy-2´-fluoro-β-D-arabinofuranosyl)adenine on K562 cellular metabolism and the inhibition of human ribonucleotide reductase and DNA polymerases by its 5´-triphosphate. Cancer Res. 1991;51:2386–2394. [PubMed] [Google Scholar]
- Piehler J. New methodologies for measuring protein interactions in vivo and in vitro. Curr. Opin. Struct. Biol. 2005;15:4–14. doi: 10.1016/j.sbi.2005.01.008. [DOI] [PubMed] [Google Scholar]
- Pontarin G, Fijolek A, Pizzo P, Ferraro P, Rampazzo C, Pozzan T, Thelander L, Reichard PA, Bianchi V. Ribonucleotide reduction is a cytosolic process in mammalian cells independently of DNA damage. Proc. Natl. Acad. Sci. USA. 2008;105:17801–17806. doi: 10.1073/pnas.0808198105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rofougaran R, Vodnala M, Hofer A. Enzymatically active mammalian ribonucleotide reductase exists primarily as an α6β6 octamer. J. Biol. Chem. 2006;281:27705–27711. doi: 10.1074/jbc.M605573200. [DOI] [PubMed] [Google Scholar]
- Shao J, Zhou B, Chu B, Yen Y. Ribonucleotide reductase inhibitors and future drug design. Curr. Cancer Drug Targets. 2006;6:409–431. doi: 10.2174/156800906777723949. [DOI] [PubMed] [Google Scholar]
- Stubbe J, van der Donk WA. Protein radicals in enzyme catalysis. Chem. Rev. 1998;98:705–762. doi: 10.1021/cr9400875. [DOI] [PubMed] [Google Scholar]
- Stubbe J, Nocera DG, Yee CS, Change MCY. Radical initiation in the class I ribonucleotide reductase: long-range proton-coupled electron transfer. Chem. Rev. 2003;103:2167–2202. doi: 10.1021/cr020421u. [DOI] [PubMed] [Google Scholar]
- Thelander L. Ribonucleotide reductase and mitochondrial DNA synthesis. Nat. Genet. 2007;39:703–704. doi: 10.1038/ng0607-703. and references cited therein. [DOI] [PubMed] [Google Scholar]
- Wang J, Lohman GJS, Stubbe J. Enhanced subunit interactions with gemcitabine-5´-diphosphate inhibit ribonucleotide reductases. Proc. Natl. Acad. Sci. USA. 2007;104:14324–14329. doi: 10.1073/pnas.0706803104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie KC, Plunkett W. Metabolism and actions of 2-chloro-9-(2´-deoxy-2´-fluoro-D-arabinofuranosyl)adenine in human lymphoblastoid cells. Cancer Res. 1995;55:2847–2852. [PubMed] [Google Scholar]
- Xie KC, Plunkett W. Deoxynucleotide pool depletion and sustained inhibition of ribonucleotide reductase and DNA synthesis after treatment of human lymphoblastoidcells with 2-chloro-(2´-deoxy-fluoro-D-arabinofuranosyl) adenine. Cancer Res. 1996;56:3030–3037. [PubMed] [Google Scholar]
- Yang Z, Fang J, Chittuluru J, Asturias FJ, Penczek PA. Iterative stable alignment and clustering of 2D transmission electron microscope images. Structure. 20:237–247. doi: 10.1016/j.str.2011.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yen H-CS, Xu Q, Chou DM, Zhao Z, Elledge SJ. Global protein stability profiling in mammalian cells. Science. 2008;322:918–923. doi: 10.1126/science.1160489. [DOI] [PubMed] [Google Scholar]
- Zhenchuk A, Lotfi K, Juliusson G, Albertioni F. Mechanisms of anti-cancer action and pharmacology of clofarabine. Biochem. Pharmacol. 2009;78:1351–1359. doi: 10.1016/j.bcp.2009.06.094. [DOI] [PubMed] [Google Scholar]
- Zhou BB, Elledge SJ. The DNA damage response: Putting checkpoints in perspective. Nature. 2000;408:433–439. doi: 10.1038/35044005. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.