Abstract
Many behaviors and physiological activities in living organisms display circadian rhythms, allowing them to anticipate and prepare for the diurnal changes in the living environment. In this way, metabolic processes are aligned with the periodic environmental changes and behavioral cycles, such as the sleep/wake and fasting/feeding cycles. Disturbances of this alignment significantly increase the risk of metabolic diseases. Meanwhile, the circadian clock receives signals from the environment and feedback from metabolic pathways, and adjusts its activity and function. Growing evidence connects the circadian clock with epigenomic regulators. Here we review the recent advances in understanding the crosstalk between the circadian clock and energy metabolism through epigenomic programming and transcriptional regulation.
Introduction
The approximately 24 hour cycling of light and dark drives the cyclic changes in the living environments for most organisms on Earth, from cyanobacteria to human beings. To adapt to changing environments, organisms anticipate the periodic changes and adjust their activities with the time of the day using an internal 24 hour clock system, known as the circadian clock. In human beings and other mammals, the clock governs many important behaviors and physiological processes including sleep/wake, feeding, body temperature, hormone secretion and metabolism.
Human beings are diurnal creatures. We conduct most of our activities during the day, including feeding, exercising and working, and rest at night. Circadian clocks in our bodies provide time cues for activities, and meanwhile synchronize the metabolic reactions with the anticipated activity cycles. The synchronization of behaviors and metabolism by the clock ensures the energy supply and maintains the internal homeostasis. However, this delicate system has been increasingly challenged in modern society. Modern life is characterized by increase in night activities, for instance, shift work, overtime work, night eating, sleep disruption and deprivation. Misalignment of activities with the internal clock and metabolic rhythms could disrupt the clock and energy homeostasis. Evidence suggests that shift workers have a higher risk of metabolic diseases, including obesity, diabetes, metabolic syndromes and cardiovascular diseases (Wang et al., 2011). Similar effects were also observed with sleep deprivation, sleep disruption and night eating (reviewed in Huang et al., 2011).
In recognition of these concerns, much recent research focuses on the crosstalk between the circadian clock and metabolism. The core mammalian biological clock consists of interlocked activators and repressors of transcription that function via epigenomic mechanisms, which can be tuned with metabolic signals, including hormones and metabolites, and also have direct effects on metabolic events. In this review, we will summarize recent advances in understanding how circadian clocks crosstalk with metabolic pathways through epigenomic mechanisms.
Environment, the epigenome, and metabolism
In addition to the linear genomic DNA sequences, information affecting the expression of individual genes can be encoded in the chromatin using mechanisms such as DNA methylation, histone modification and chromatin remodeling. This additional layer of gene regulation may be referred to as the epigenome. Epigenomic modification provides plasticity in gene expression and cellular functions in multicellular organisms, and allows reversible changes in response to changes in the their environment, including light, temperature, food availability and dietary composition which can affect many physiological processes, including development, aging, and metabolism (reviewed in Christensen and Marsit, 2011).
Metabolism is tightly regulated, and imbalance of energy intake and expenditure leads to accumulation of nutrients and metabolites and thus contributes to metabolic diseases, cardiovascular diseases, cancer and others. A common theme in metabolic control is transcriptional regulation of rate-limiting metabolic enzymes, usually involving epigenomic mechanisms. For instance, hepatic glucose production and secretion is regulated by phosphoenolpyruvate carboxykinase (PEPCK) and glucose-6-phosphatase (G6pase), respectively. PEPCK and G6pase are activated by glucagon and fasting through cAMP-responsive binding element protein (CREB), repressed by insulin through the forkhead O box protein 1 (FOXO1), and stimulated by glucocorticoids through glucocorticoid receptor (GR) (reviewed in Jitrapakdee, 2012). Environment factors such as nutrition, exercise, aging, and stress can signal through metabolic hormones, such as insulin and leptin, and metabolites, such as nicotinamide adenine dinucleotide (NAD+), ADP, acetyl-CoA, and S-adenosyl-methionine (SAM) (Christensen and Marsit, 2011). These signals regulate the epigenome by modulating the function of chromatin modifying enzymes as well as transcription factors that are responsible for recruiting these enzymes.
Take calorie restriction (CR) as an example, in which amount of daily calorie intake is reduced by 30% to 50% compared to ad libitum feeding. CR can increase lifespan and delay the onset of many aging related diseases, including cancer and diabetes. CR is known to decrease blood insulin and thyroid hormone levels in human (reviewed in Vaquero and Reinberg, 2009). Insulin signaling leads to phosphorylation and inhibition of FOXO1, whose targets include stress response genes and gluconeogenic genes (Dong et al., 2008).
CR response is also mediated by metabolites. Low energy intake in CR elevates the NAD+/NADH ratio and AMP/ATP ratio, which can be sensed by NAD+-dependent histone deacetylase (HDAC) sirtuins and AMP-activated protein kinase (AMPK), respectively. In mammals, sirtuin 1 (SIRT1), activated by NAD+, deacetylates histone marks H4K16Ac and H3K9Ac and histone methyltransferase (HMT) SUV39H1, thus promoting formation of facultative heterochromatin and represses transcription. SIRT1 also deacetylates and activates FOXO1 and PGC-1α, and promotes gluconeogenic gene expression (reviewed in Vaquero and Reinberg, 2009). This process might involve changes in the recruitment of histone acetyltransferases (HATs) by FOXO1 and PGC-1α. AMPK is activated by increase in AMP/ATP ratio, and directly phosphorylates PGC-1α, enabling SIRT1-mediated deacetylation and activation of PGC-1α. FOXOs might also be regulated similarly (reviewed in Cantó and Auwerx, 2011). Through these transcription factors and coregulators, AMPK can direct epigenomic remodeling and metabolic reprogramming. Sensing the nutritional/metabolic state through sirtuins and AMPK is a common theme, and also mediates responses to fasting and exercise (reviewed in Freyssenet, 2007; Cantó and Auwerx, 2011).
Environment and the clock
Metabolism is also controlled by the internal circadian rhythm. The cell-autonomous clock machinery consists of several transcriptional-translational feedback loops, which allows autonomous oscillation with a period of approximately 24 hours (Figure 1). In mammals, the first feedback loop in the basic clock machinery contains a heterodimer of transcription activators the brain and muscle ARNTL-like protein 1 (BMAL1) and the circadian locomoter output cycles kaput (CLOCK). BMAL1/CLOCK activates transcription of Crytochromes (CRYs) and Periods (PERs). CRY and PER proteins negatively regulates their own expression by binding BMAL1/CLOCK and inhibiting their transcriptional activity (reviewed in Bass and Takahashi, 2010). Another critical feedback loop drives the cyclic transcription of BMAL1 using nuclear receptor (NR) REV-ERBα/REV-ERBβ. BMAL1 activates Rev-erbα transcription, which then suppresses Bmal1 transcription (Preitner et al., 2002). This second loop, once considered auxiliary, was recently proved to be essential for the clock function (Bugge et al., 2012; Cho et al., 2012; Solt et al., 2012). The clock machinery is featured by the redundancy of their components: BMAL1/BMAL2, CLOCK/NPAS2 (neuronal PAS domain containing protein 2), CRY1/CRY2, PER1/PER2/PER3, REV-ERBα/REV-ERBβ (Bugge et al., 2012; Cho et al., 2012; reviewed in Bass and Takahashi, 2010) (Figure 1). Other factors that are important for clock function include the Retinoid-related orphan receptor RORs (RORα, RORβ, RORγ), which activate transcription of BMAL1 and REV-ERBα, and are repressed by REV-ERBα (reviewed in Jetten, 2009). Levels of the clock proteins are also subject to posttranslational regulation by casein kinases (CKIε and CKIδ), and ubiquitin E3 ligase β-TrCP, FBXL3 and ARF-BP1/PAM (Yin et al., 2010; reviewed in Bass and Takahashi, 2010). Although the transcriptional-translational clocks are ubiquitous in mammalian cells, non-transcriptional mechanisms are aslo sufficient to sustain circadian oscillation. In pico-eukaryotic alga Ostreococcus tauri, oxidation of peroxiredoxin proteins undergoes circadian cycles independent of its transcriptional circadian clock (O’Neill et al., 2011). Recently this mechanism was shown to be conserved in higher organisms including human, at least in human red blood cells (O’Neill and Reddy, 2011). The wide-spread presence of transcription-independent redox oscillations suggests that crosstalk between metabolic cycles and circadian clocks has long played a significant role in both physiology and evolution.
Figure 1. The basic clock machinery consists of negative transcriptional-translational feedback loops.
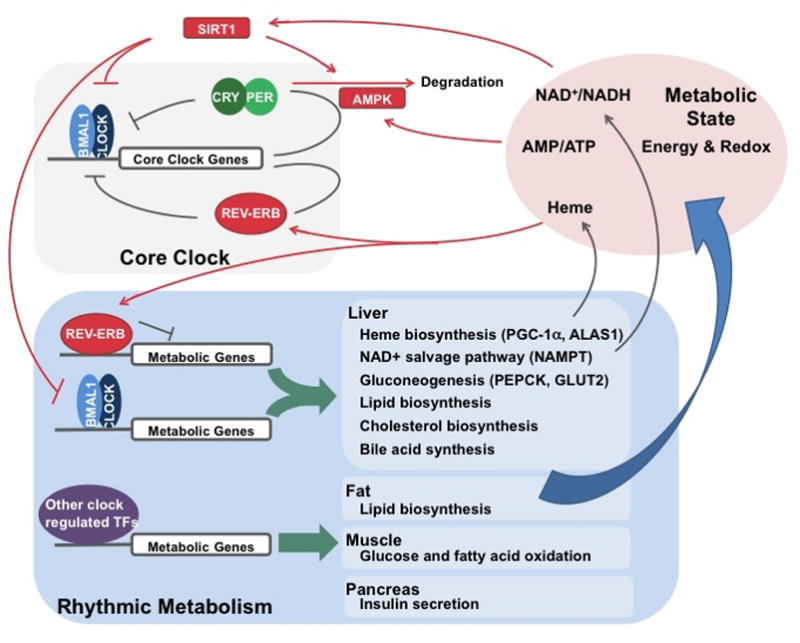
In the first loop, BMAL1/CLOCK drives Per/Cry transcription, while PER/CRY binds and inhibits transcriptional activity of BMAL1/CLOCK. In the second loop, BMAL1/CLOCK drives REV-ERB expression, which in turn represses Bmal1 transcription. Both loops are essential for maintaining circadian rhythm. In addition, post-translational modification, as shown for PER/CRY and REV-ERB, is also important in regulating clock activity. The core clock machinery can drive rhythmic behavioral and physiological activities, such as metabolism.
In mammals, the basic clock machinery is present in most organs, assembled in a hierarchical system in which the central clock can entrain peripheral clocks (Figure 2). The central clock is located in the suprachiasmatic nuclei (SCN) of the anterior hypothalamus, while peripheral clocks are present in most organs, including metabolic organs such as liver, adipose, heart, muscle and kidney (reviewed in Dibner et al., 2010). Both the central clock and peripheral clocks can be reset by environmental cues, also known as Zeitgebers (“time-giver”). The predominant Zeitgeber for the central clock is light, which is sensed by retina and signals directly to SCN. The central clock can entrain the peripheral clocks through neuronal and hormonal signals, as well as body temperature, aligning all clocks with the external light/dark cycle (Balsalobre et al., 2000; Brown et al., 2002; reviewed in Dibner et al., 2010). Peripheral clocks in metabolic tissues are also entrained by the central clock through feeding/fasting cycles (Figure 2). Fasting/feeding alters the levels of key endocrine hormones and the intracellular metabolic state, which can modulate the function of peripheral clocks. When restricted food availability shifts the fasting/feeding cycles, peripheral clocks can be reset separately from the central clock, suggesting that food availability is the predominant Zeitgeber for these clocks (Damiola et al., 2000; Stokkan et al., 2001).
Figure 2. Overview of the interplays between environment, circadian clocks and metabolism.
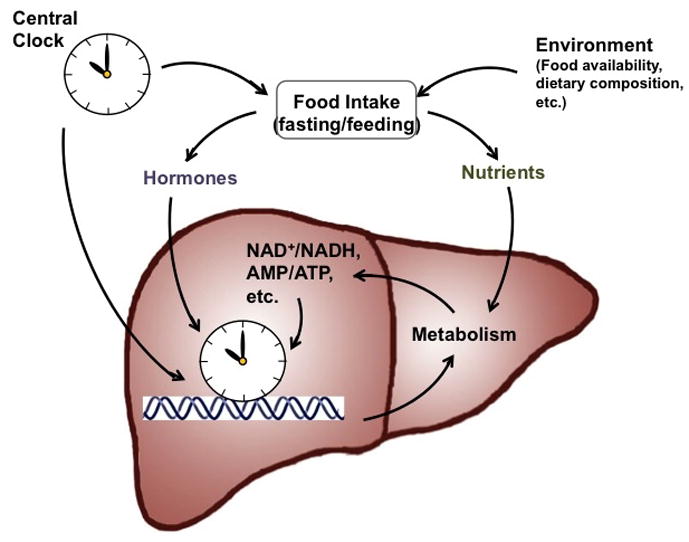
The circadian clock in the cells comprising metabolic organs, such as liver, functions as an epigenomic programmer and controls metabolic outputs. This autonomous apparatus is regulated by the central clock in the SCN and by food intake via hormones and nutrients/metabolites. Both the central clock and food availability contributes to the temporal regulation of food intake.
Rhythmic regulation of gene expression: The clock mechanism
How does the basic clock machinery coordinate metabolic activities in different metabolic organs, as well as other behaviors and physiology? Growing evidence suggests that circadian clocks control physiology through transcription. Transcriptomic studies in different mouse tissues revealed that a large proportion of the whole transcriptomes, from 3% to 20%, undergo circadian oscillation (reviewed in Green et al., 2008). The circadian transcriptomes are tissue-specific and tightly correlated with cellular functions. In SCN, it includes genes involved in protein-neuropeptide synthesis, secretion and degradation and regulation of mouse locomoter activity, while in liver, it includes genes involved in glucose, lipid and xenobiotic metabolism (Akhtar et al., 2002; Panda et al., 2002). Circadian control of activities can also be mediated through translational and post-translational regulation (Reddy et al., 2006). However, here we will focus on transcription regulation and epigenomic reprogramming.
In the core clock machinery, BMAL1/CLOCK, REV-ERBs, PERs, CRYs and RORs are all transcription regulators. Both BMAL1/CLOCK and REV-ERBs display rhythmic genome binding and can drive the rhythmic expression of their targets (Ripperger and Schibler, 2006; Feng et al., 2011; Rey et al., 2011; Bugge et al., 2012). For instance, REV-ERBα directly regulates gluconeogenic enzymes G6pase and PEPCK, and many lipid biosynthetic genes (Yin et al., 2007; Feng et al., 2011). More importantly, the core clock machinery can drive circadian expression of many transcription factors, thus extending and enhancing its regulatory function. Among these factors are nuclear receptors (NRs), such as retinoic acid receptors (RARs), TRs, peroxisome proliferator-activated receptors (PPARs), GR and short heterodimer partner (SHP) (Yang et al., 2006). Other transcription factors include the three members of the Prolin Acidic Amino acid Rich (PAR) domain basic leucine zipper proteins albumin D-site binding protein (DBP), thyrotroph embryonic factor (TEF), and hepatocyte leukemia factor (HLF), and the related protein E4BP4. All four of these proteins bind to D-boxes, but while DBP, TEF, and HLF are activators, E4BP4 is a repressor. Many of these factors are direct targets of the clock. BMAL1 controls PPARα and DBP, and REV-ERBα controls SHP and E4bp4 (Canaple et al., 2006; Duez et al., 2008). The circadian clock also regulates stability and activity of these factors. PER2 binding promotes ERα degradation, regulates PPARγ DNA binding, and co-activates PPARα-mediated transcription (Gery et al., 2007; Grimaldi et al., 2010; Schmutz et al., 2010), while CRY binding represses GR (Lamia et al., 2011). The clock can also regulate these factors indirectly, for instance, through their ligands such as glucocorticoid for GR (Reddy et al., 2007). The clock signal can also be mediated by transcription coactivators PGC-1α and BAF60a, as well as microRNAs (Wu et al., 2009; Tao et al., 2011; Gatfield et al., 2009).
Clock regulation of the epigenome
Transcription factors control gene transcription by facilitating recruitment and activation of the transcription machinery, or by altering epigenome to recreate a more favorable environment for transcription, or most of the time both (reviewed in Farnham, 2009). Accumulating evidence, summarized in this section, implicates the core clock genes as partners of chromatin modifying enzymes (Table 1).
Table 1.
Clock-associated histone modifying activities.
| Clock Component | Associated Chromatin Modifier | Targets | References |
|---|---|---|---|
| Bmal1/Clock | Clock | H3K9, H3K14 Bmal1(K537), GR | (Doi et al., 2006; Nader et al., 2009; Charmandari et al., 2011) |
| p300 | H3 | (Takahata et al., 2000; Etchegaray et al., 2002) | |
| PCAF | (Curtis et al., 2004) | ||
| CBP | (Takahata et al., 2000; Curtis et al., 2004) | ||
| Sirt1 | H3K9, H3K14, Bmal1(K537), Per2 | (Asher et al., 2008; Nakahata et al., 2008) | |
| MLL1 | H3K4 | (Katada and Sassone-Corsi, 2010) | |
| EZH2 | H3K27 | (Etchegaray et al., 2006) | |
| JARID1a | (DiTacchio et al., 2011) | ||
| Bmal1/Npas2 | p300 | H3, H4 | (Curtis et al., 2004) |
| PCAF | H3, H4 | (Curtis et al., 2004) | |
| ACTR | (Curtis et al., 2004) | ||
| Per | PSF-Sin3A-HDAC1 | H3, H3K9, H4K5 | (Duong et al., 2011) |
| WDR5 | H3K4 | (Brown et al., 2005) | |
| Cry | Sin3B-HDAC1/2 | Histone H3, H4 | (Naruse et al., 2004) |
| Rev-erbα,β | NCoR-HDAC3 | H3, H4, H3K9 | (Ishizuka and Lazar, 2003; Yin and Lazar, 2005; Feng et al, 2011; Bugge et al, 2012) |
Histone acetylation
Histone acetylation undergoes cyclic oscillation with the clock, as evident in mouse liver, not only at the promoters of the clock genes and but also in a genome-wide scale (Etchegaray et al., 2003; Curtis et al., 2004; Naruse et al., 2004; Ripperger and Schibler, 2006; Feng et al., 2011). Histone acetylation is controlled by a battle between HATs and HDACs, both of which participate in clock-regulated rhythmic gene transcription.
The BMAL1/CLOCK or BMAL1/NPAS2 heterodimer recruits both HATs and HDACs. BMAL1 binds transcription coactivator p300 and possibly the CREB binding protein (CBP), while CLOCK and NPAS2 bind p300 and the CBP-associated factor (PCAF) (Takahata et al., 2000; Etchegaray et al., 2003; Curtis et al., 2004). All three coactivators have intrinsic HAT activity. In liver and heart, p300 exhibits a circadian association with CLOCK or NPAS2, correlating with increase in Per1 mRNA and histone H3 acetylation on the Per1 promoter (Etchegaray et al., 2003; Curtis et al., 2004). BMAL1/CLOCK also directly interacts with SIRT1, recruits SIRT1 to the Dbp gene in a timely manner, and drives rhythmic histone acetylation and gene transcription (Asher et al., 2008; Nakahata et al., 2008) (Figure 3). Moreover, CLOCK itself is a HAT, and its HAT activity is essential for the rhythmic acetylation at the Per1 promoter and the circadian expression of Per1 and Dbp (Doi et al., 2006).
Figure 3. The core clock machinery drives rhythmic metabolic activities and receives feedback from intracellular metabolites.
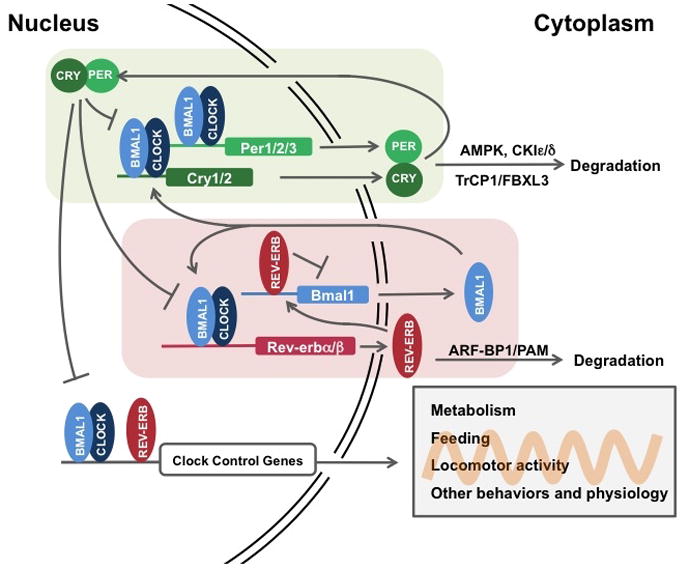
BMAL1/CLOCK and REV-ERB drive rhythmic metabolic outputs, including NAD+ and heme biosynthesis, while intracellular NAD+ and heme feedback on the clock through their sensors, SIRT1 and REV-ERB, respectively. Intracelluar AMP levels regulates the circadian clock through activation of AMPK and degradation of PER and CRY. The core clock also drives many metabolic pathways in different tissues, which also contribute to the intracellular metabolite pool and metabolic state.
PER and CRY direct the negative feedback signal to BMAL1/CLOCK probably through attenuating their affinity for DNA (Figure 1) (Ripperger and Schibler, 2006). In addition, through interaction with BMAL1/CLOCK, CRY1 can recruit Sin3B corepressor and HDAC1/2 to the Per1 promoter, and repress Per1 transcription through histone deacetylation (Naruse et al., 2004). Similarly, PER2 recruit Sin3A corepressor and HDAC1 through the polypyrimidine tract–binding protein–associated splicing factor (PSF) to the Per1 promoter (Duong et al., 2011).
REV-ERBs and RORs are members of the NR family, a family known to employ multiple coactivator and corepressor complexes for chromatin modification. REV-ERBα recruits nuclear receptor corepressor (NCoR) and HDAC3 to deacetylate histones and repress transcription (Yin and Lazar, 2005). RORs interact with both corepressors and coactivators (reviewed in Jetten, 2009), and the rhythmic binding of RORs to the Npas2 promoter are positively correlated with DNA accessibility and histone acetylation (Takeda et al., 2011).
Histone methylation
Whereas histone acetylation is usually associated with transcriptional activation, methylation has a mixed effect on transcription, depending on the modification sites. CLOCK interacts with and recruits the mixed lineage leukemia 1 (MLL1), a HMT that specifically promotes H3K4me3, an activation mark. MLL1 is required for circadian H3K4 methylation and H3 acetylation and for circadian gene expression (Katada and Sassone-Corsi, 2010). PER1 associates with WD repeat-containing protein 5 (WDR5), and loss of WDR5 abolishes the circadian H3K4 and H3K9 methylation at the Rev-erbα promoter (Brown et al., 2005). Interestingly, MLL1 and WDR5 can be present in the same methyltransferase complex. BMAL1/CLOCK binds polycomb protein EZH2 and methylates H3K27 at the Per1 and Per2 promoters, which is essential for CRY-mediated transcriptional repression (Etchegaray et al., 2006). BMAL1/CLOCK also recruits JumonjiC and ARID domain-containing histone lysine demethylase 1a (JARID1a) to the Per2 promoter, though it appears that the circadian functions of JARID1a may be independent of its histone modifying activity (DiTacchio et al., 2011).
Chromatin Remodeling
Chromatin remodeling often accompanies histone modification and transcriptional regulation (reviewed in Strahl and Allis, 2000). One study showed that RORs recruit the SWItch/Sucrose Non-Fermentable (SWI/SNF) chromatin remodeling complex subunit BAF60a to drive rhythmic Bmal1 and G6pase expression (Tao et al., 2011). However, this aspect of clock function is understudied at the present time.
Rhythmic chromatin modification is not limited to the promoter of clock genes that have been studied, as ChIP-seq revealed that BMAL1 and REV-ERBs bind thousands of sites throughout the genome (Dufour et al., 2011; Feng et al., 2011; Rey et al., 2011; Bugge et al., 2012). In liver, REV-ERBs drive genome-wide circadian recruitment of HDAC3. As a consequence, although HDAC3 expression is constant, it cyclically deacetylates histone and represses transcription at regions targeted by REV-ERBα and REV-ERBβ (Feng et al., 2011; Bugge et al., 2012). In addition, the clock can drive rhythmic epigenomic programming indirectly through clock-regulated transcription factors, such as NRs, which then recruit HATs and HDACs.
Non-histone targets of clock-associated histone modifiers
Histone modifying enzymes also target transcription factors, cofactors and enzymes. CLOCK acetylates its partner BMAL1, and facilitates CRY1 binding and CRY1-mediated transcriptional repression (Hirayama et al., 2007). CLOCK also acetylates GR, attenuates its DNA binding, and regulates glucocorticoid response (Nader et al., 2009; Charmandari et al., 2011). SIRT1 counteracts CLOCK-mediated BMAL1 acetylation, deacetylates and degrades PER2 thus derepressing BMAL1/CLOCK (Nakahata et al., 2008; Asher et al., 2008) (Figure 3). Activation or inhibition of transcription activity will also alter the epigenomic state of the target genes. Also clock-associated histone modifiers can be recruited by other transcription factors and function independent of the clock machinery.
Circadian regulation of metabolism
Metabolic activities undergo diurnal changes, as reflected by the oscillating hormone levels in blood and rhythmic activities of metabolic pathways. Hormones mediate the crosstalk between the central nervous system and major metabolic organs, and are essential to metabolic homeostasis. Some of them are secreted in a circadian manner, including insulin and glucagon from pancreas, adiponectin and leptin from adipose, and ghrelin from stomach (reviewed in Froy, 2007). Many metabolic pathways display rhythmic activities, consistent with levels and activities of the rate-limiting enzymes. For instance, gluconeogenesis peaks in the daytime with PEPCK activity (Kida et al., 1980). Rhythmic activities were also observed in xenobiotic metabolism, glycogen metabolism and amino acid metabolism, and in other metabolic tissues, such as fat (reviewed in Davidson et al., 2004; Gimble and Floyd, 2009).
The role of circadian clocks in metabolic regulation is well supported by genetic evidence that mutation in clock genes disturbs rhythmic expression of key metabolic genes and causes metabolic disorders. Key metabolic pathways under clock control will be discussed in this section. In addition, other metabolic or related pathways are also regulated by the circadian clock, including cardiovascular function and inflammation (reviewed in Bass and Takahashi, 2010).
Glucose homeostasis
Loss of BMAL1 attenuates the diurnal variation in glucose and triglyceride levels, impairs gluconeogenesis and shows glucose intolerance, while the Clock mutant mice (ClockΔ19) develop hyperglycemia and hypoinsulinemia (Rudic et al., 2004; Lamia et al., 2008; Turek et al., 2005). Both genetic models suggest that BMAL1/CLOCK plays a critical role in glucose homeostasis. The study was followed by tissue-specific deletion of Bmal1 in liver and pancreatic β cells, both of which support the critical role of peripheral clocks in metabolic tissues. Loss of BMAL1 in liver abolishes rhythmic expression of glucose metabolic genes, e.g. Pepck and Glut2, and leads to hypoglycemia only in the fasting phase of the day, while loss of BMAL1 in pancreatic β cells impairs insulin secretion (Figure 3) (Lamia et al., 2008; Marcheva et al., 2010).
Other clock components also play a role. CRYs inhibit gluconeogenic gene expression, probably through regulating CREB activity. Therefore, hepatic depletion of CRY1/2 increases circulating glucose, while CRY1 overexpression reduces fasting blood glucose and improves whole body insulin sensitivity in db/db mice (Zhang et al., 2010). CRYs also transrepress glucocorticoid-induced Pepck transcription, and loss of CRYs results in glucose intolerance (Lamia et al., 2011). Similarly depletion of both REV-ERBs increases fasting blood glucose (Cho et al., 2012). And RORα is known to activate G6pase through SRC-2 and BAF60a (Chopra et al., 2008; Tao et al., 2011).
Lipid metabolism
Clock mutation and Bmal1 deficiency also impairs lipid metabolism, as shown by hyperleptinemia, hyperlipidemia and hepatic steatosis (Turek et al., 2005; Shimba et al., 2011). The mechanism behind this remains unclear. Ablation of REV-ERBs also causes hepatic steatosis, in part via derepression of lipogenesis (Feng et al., 2011; Bugge et al., 2012). Indeed evidence suggests that REV-ERBs might be responsible for circadian lipid biosynthesis in liver, which has been recognized for over 3 decades (Hems et al., 1975; Alenghat et al., 2008; Feng et al., 2011). Moreover, by inhibiting the lipid biosynthesis and driving lipid storage, it reroutes gluconeogenic metabolites and indirectly promotes gluconeogenesis (Sun et al., 2012). REV-ERB agonists inhibit lipid and cholesterol synthesis in liver and fat, and promote fatty acid and glucose oxidation in muscle, resulting in increased energy expenditure (Figure 3). These agonists significantly improve dyslipidemia and hyperglycaemia in a diet-induced obesity model (Solt et al., 2012). In addition, BMAL1 and REV-ERBα both regulates adipocyte differentiation (Laitinen et al., 2005; Shimba et al., 2005).
Bile acid metabolism
REV-ERBα is an important regulator of bile acid synthesis. Genetic ablation of REV-ERBα lowers bile acid synthesis and decreases bile acid accumulation in the liver, correlated with altered phase and total decrease of Cyp7a1 expression (Duez et al., 2008; Le Martelot et al., 2009). REV-ERBα may indirectly activates Cyp7a1 via E4bp4 and SHP, or via LXR (Duez et al., 2008; Le Martelot et al., 2009). REV-ERBα also regulates SREBP function through its regulator INSIG2 and thus the SREBP targets involved in cholesterol and lipid metabolism (Le Martelot et al., 2009). Loss of BMAL1 increases circulating cholesterol, and loss of PER1/2 up-regulates bile acid synthesis and results in hepatic cholestasis (Ma et al., 2009; Shimba et al., 2011).
Rhythmic regulation of metabolism by fasting/feeding
A key unanswered question is whether the rhythmic feature of metabolism is an passive response to the fasting/feeding cycles in metabolic tissues or an active anticipatory mechanism directly governed by the circadian clock. Recent transcriptomic studies in mouse liver provide some insight. Liver-specific overexpression of REV-ERBα abolishes oscillation of almost all the rhythmic transcripts found in wildtype mice, suggesting that they are controlled by the liver clock (Kornmann et al., 2007). Meanwhile, only a small subset of these transcripts, including the clock genes, maintain their oscillation under fasting, suggesting most of the rhythmic transcripts are also regulated by fasting/feeding (Vollmers et al., 2009). Interestingly, in clock deficient mice restricted feeding can resume oscillation of about 60% of the rhythmic transcripts and even increase their amplitude, along with many other non-rhythmic genes (Vollmers et al., 2009). These observations suggest that under normal conditions, peripheral clocks and fasting/feeding cycles work together to drive rhythmic gene expression and metabolic activities.
Fasting/feeding activities are usually aligned with sleep/wake cycles, and are timed by the central clock. Therefore, normally both the central clock and peripheral clocks synergize in rhythmic metabolic regulation. In addition to feeding/fasting, the central clock can also drive metabolic activities through hormones and body temperature (Brown et al., 2002; Reddy et al., 2007). For instance, blood glucocorticoid levels exhibits circadian oscillation (Oster et al., 2006). In the absence of the central clock, daily glucocorticoid injection can restore about 60% of the liver circadian transcriptome, probably through hepatocyte nuclear factor 4α (HNF4α) (Maywood et al., 2007; Reddy et al., 2007). Therefore, under normal conditions, the central clock synchronize peripheral clocks, fasting/feeding cycles, hormone secretion and body temperature changes, and all of these contribute to the rhythmic metabolic activities throughout the body (Figure 2).
On the other hand, fasting/feeding is also subject to food availability in the environment, and changes in metabolic profiles as induced by fasting/feeding can impact and even resept the peripheral clocks in metabolic tissues. Thus, in addition to the clock regulating metabolism, metabolism can regulate the clock.
Metabolic regulation of the clock
The circadian clock is regulated by metabolic signals such as fasting/feeding and dietary factors. Fasting and refeeding regulates the clock gene expression in peripheral tissues, particularly liver (Kawamoto et al., 2006; Tahara et al., 2011). High fat diet (HFD) lengthens the circadian period and attenuates clock oscillation (Kohsaka et al., 2007). Clock-associated histone modifiers such as p300 and SIRT1 are also regulated. Fasting decreases p300 phosphorylation and activates p300, and induces Sirt1 expression, while high fat diet (HFD) inhibits p300 phosphorylation, lengthens the circadian period and attenuates clock oscillation (Rodgers et al., 2005; Liu et al., 2008). The impact of metabolism might be mediated through hormones, such as glucagon and insulin (Liu et al., 2008; Tahara et al., 2011), as well as metabolites. The circadian clock can sense the intracellular metabolite levels, adjust its own rhythm and functions and allows a fine and precise temporal regulation of metabolic pathways within individual cells. We will focus on the key metabolites such as NAD+, AMP and heme, though other metabolites may also play a role.
NAD+/NADH
NAD+/NADH redox equilibrium indicates the metabolic state of the cell. NADH and NADPH directly binds CLOCK or NPAS2 and enhances BMAL1/CLOCK (or NPAS2) DNA binding, while the reduced form NAD+ binds and inhibits (Rutter et al., 2001). NAD+/NADH ratio also regulate SIRT1, which deacetylates PER2, BMAL1 and histones (Asher et al., 2008; Nakahata et al., 2008) (Figure 3). Intracelluar NAD+ levels are circadian, and are critical for circadian clock functions (Sahar et al., 2011).
NAD+ levels are determined both by the fasting/feeding cycles and circadian rhythms. Fasting induces NAD+ levels in liver, as well as SIRT1 protein levels (Rodgers et al., 2005). Circadian clocks regulates NAD+ mainly through the NAD+ salvage pathway, in which NAD+ is synthesized from nicotinamide (NAM), the byproduct of sirtuins, through the rate-limiting enzyme nicotinamide phosphoribosyltransferase (NAMPT). BMAL1/CLOCK drives the circadian expression of NAMPT and contributes to the rhythmic NAD+ levels. NAD+ activates while NAM represses SIRT1, which interacts with BMAL1/CLOCK and inhibits NAMPT transcription. Therefore, NAD+ drives a negative feedback on its own synthesis through SIRT1 and the circadian clock, and through NAD+, the intracellular metabolic state can regulate the clock (Figure 3) (Nakahata et al., 2009; Ramsey et al., 2009).
NAD+ is also a substrate of poly(ADP-ribose) polymerase 1 (PARP-1), an NAD+-dependent ADP-ribosyltransferase. PARP-1 activity oscillates in a daily manner, and is regulated by feeding. PARP-1 binds and poly(ADP-ribosyl)ates clock at the onset of the light phase, regulates BMAL1/CLOCK interaction with PER and CRY and BMAL1/CLOCK DNA binding, and mediates food entrainment of the liver clock. NAD+ might play a role in regulating PARP-1 activity by feeding, but other mechanisms are also involved (Asher et al., 2010).
AMP/ADP
AMP and ADP bind AMPK, inhibit AMPK dephosphorylation and activate its kinase activity. Therefore, AMPK functions as a sensor of energy state in the cell, which is reflected by AMP and ADP levels (Cantó and Auwerx, 2011). Several studies place AMPK as a circadian clock regulator (Um et al., 2007, 2011; Vieira et al., 2008; Lamia et al., 2009) with two different mechanisms: AMPK act ivates CKIε and promotes PER2 degradation(Um et al., 2007); AMPK phosphorylates Cry1 and destabilizes it (Lamia et al., 2009) (Figure 3). AMPK also regulates the NAD+/NADH ratio, through which it crosstalks with SIRT1 (Cantó et al., 2009).
Heme
Heme is a ligand of REV-ERBα and REV-ERBβ, and enhances corepressor recruitment and transcriptional repression (Raghuram et al., 2007; Yin et al., 2007; Pardee et al., 2009; Phelan et al., 2010) (Figure 3). Through heme, REV-ERBs can sense the redox state and gas such as O2, NO and CO, as they regulate REV-ERB structure and corepressor recruitment (Pardee et al., 2009). Also heme binding can be regulated by the redox state of the REV-ERBβ protein (Gupta and Ragsdale, 2011). Heme is also a component of NPAS2, whose heterodimeric DNA binding with Bmal1 is inhibited by CO (Dioum et al., 2002; Gilles-Gonzalez and Gonzalez, 2004).
Like NAD+, heme can also negatively regulate its own synthesis through the circadian clock. The control is exerted on the expression of the rate-limiting enzyme in heme biosynthesis, δ-aminolevulinate synthase (ALAS1), whose expression is circadian. Heme inhibits BMAL1/NPAS2-activated ALAS1 expression and heme biosynthesis (Kaasik and Lee, 2004). In parallel, through REV-ERBα, heme represses PGC-1α transcription, a potent inducer of heme biosynthesis (Wu et al., 2009) (Figure 3). Both NAD+ and heme are critical metabolites and indicators/sensors of the metabolic state in the cells. The negative metabolite feedback loops connect metabolism with the circadian clock, and are critical for metabolic homeostasis and fine tuning of the clock function.
Conclusions and Perspective
In this review, we have discussed the functions of the circadian clock as a rhythmic epigenomic programmer and transcriptional regulator of gene expression and metabolism, and the interplays between the circadian clock and metabolism on epigenome. Crosstalk between the circadian rhythm and metabolism is essential for maintaining metabolic homeostasis and prevent metabolic disorders. In addition, the environment, particularly food availability and energy intake, participates in the crosstalk, regulating the circadian clock as well as metabolic pathways. However, there are still gaps in our understanding. First, while much of our current knowledge focuses on the gene expression of clock components as well as metabolic factors, their effects of cellular and organismal phenotype depend upon protein levels and activity, which are highly regulated as the translational and post-translational levels, respectively. Moreover, metabolic pathways are interdependent with multiple control points, such that the concept of a single rate-limiting step is likely simplistic. In addition, it remains to be determined whether transcriptional regulation can be predicted from knowledge of the modification of the epigenome, or if this is an epiphenomenon of other cellular determinants. Last, but not least, to what extent rhythmic metabolism is a passive response to fasting/feeding or an anticipatory effect of the clock remains to be further elucidated. Future studies will advance our understanding, and it is hoped that this knowledge will lead to alleviation of the adverse effects of circadian misalignment and metabolic diseases in modern society.
Acknowledgments
We thank Anne Bugge and Zheng Sun for critical comments on the manuscript. Work on circadian regulation of the epigenome and metabolism in the authors’ lab is supported by NIH grants DK45586 and DK43806 (to MAL), and the JPB Foundation.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Akhtar RA, Reddy AB, Maywood ES, Clayton JD, King VM, Smith AG, Gant TW, Hastings MH, Kyriacou CP. Circadian cycling of the mouse liver transcriptome, as revealed by cDNA microarray, is driven by the suprachiasmatic nucleus. Current Biology. 2002;12:540–550. doi: 10.1016/s0960-9822(02)00759-5. [DOI] [PubMed] [Google Scholar]
- Alenghat T, Meyers K, Mullican SE, Leitner K, Adeniji-Adele A, Avila J, Bućan M, Ahima RS, Kaestner KH, Lazar MA. Nuclear receptor corepressor-histone deacetylase 3 governs circadian metabolic physiology. Nature. 2008;456:997–1000. doi: 10.1038/nature07541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asher G, Gatfield D, Stratmann M, Reinke H, Dibner C, Kreppel F, Mostoslavsky R, Alt FW, Schibler U. SIRT1 regulates circadian clock gene expression through PER2 deacetylation. Cell. 2008;134:317–328. doi: 10.1016/j.cell.2008.06.050. [DOI] [PubMed] [Google Scholar]
- Asher G, Reinke H, Altmeyer M, Gutierrez-Arcelus M, Hottiger MO, Schibler U. Poly(ADP-Ribose) Polymerase 1 participates in the phase entrainment of circadian clocks to feeding. Cell. 2010;142:943–953. doi: 10.1016/j.cell.2010.08.016. [DOI] [PubMed] [Google Scholar]
- Balsalobre A, Brown SA, Marcacci L, Tronche F, Kellendonk C, Reichardt HM, Schütz G, Schibler U. Resetting of circadian time in peripheral tissues by glucocorticoid signaling. Science. 2000;289:2344–2347. doi: 10.1126/science.289.5488.2344. [DOI] [PubMed] [Google Scholar]
- Bass J, Takahashi JS. Circadian integration of metabolism and energetics. Science. 2010;330:1349–1354. doi: 10.1126/science.1195027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown SA, Ripperger J, Kadener S, Fleury-Olela F, Vilbois F, Rosbash M, Schibler U. PERIOD1-associated proteins modulate the negative limb of the mammalian circadian oscillator. Science. 2005;308:693–696. doi: 10.1126/science.1107373. [DOI] [PubMed] [Google Scholar]
- Brown SA, Zumbrunn G, Fleury-Olela F, Preitner N, Schibler U. Rhythms of mammalian body temperature can sustain peripheral circadian clocks. Current Biology. 2002;12:1574–1583. doi: 10.1016/s0960-9822(02)01145-4. [DOI] [PubMed] [Google Scholar]
- Bugge A, Feng D, Everett LJ, Briggs ER, Mullican SE, Wang F, Jager J, Lazar MA. Rev-Erbα and Rev-Erbβ coordinately protect the circadian clock and normal metabolic function. Genes Dev. 2012;26:657–667. doi: 10.1101/gad.186858.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canaple L, Rambaud J, Dkhissi-Benyahya O, Rayet B, Tan NS, Michalik L, Delaunay F, Wahli W, Laudet V. Reciprocal regulation of brain and muscle Arnt-like protein 1 and peroxisome proliferator-activated receptor alpha defines a novel positive feedback loop in the rodent liver circadian clock. Mol Endocrinol. 2006;20:1715–1727. doi: 10.1210/me.2006-0052. [DOI] [PubMed] [Google Scholar]
- Cantó C, Auwerx J. Calorie restriction: is AMPK a key sensor and effector? Physiology. 2011;26:214–224. doi: 10.1152/physiol.00010.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantó C, Gerhart-Hines Z, Feige JN, Lagouge M, Noriega L, Milne JC, Elliott PJ, Puigserver P, Auwerx J. AMPK regulates energy expenditure by modulating NAD+ metabolism and SIRT1 activity. Nature. 2009;458:1056–1060. doi: 10.1038/nature07813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charmandari E, Chrousos GP, Lambrou GI, Pavlaki A, Koide H, Ng SSM, Kino T. Peripheral CLOCK regulates target-tissue glucocorticoid receptor transcriptional activity in a circadian fashion in man. PLoS One. 2011;6:25612. doi: 10.1371/journal.pone.0025612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho H, Zhao X, Hatori M, Yu RT, Barish GD, Lam MT, Chong LW, DiTacchio L, Atkins AR, Glass CK, et al. Regulation of circadian behaviour and metabolism by REV-EPBα and REV-ERBβ. Nature. 2012;485:123–127. doi: 10.1038/nature11048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chopra AR, Louet JF, Saha P, An J, DeMayo F, Xu J, York B, Karpen S, Finegold M, Moore D, et al. Absence of the SRC-2 coactivator results in a glycogenopathy resembling Von Gierke’s Disease. Science. 2008;322:1395–1399. doi: 10.1126/science.1164847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen BC, Marsit CJ. Epigenomics in environmental health. Front Genet. 2011;2:84. doi: 10.3389/fgene.2011.00084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtis AM, Seo S, Westgate EJ, Rudic RD, Smyth EM, Chakravarti D, FitzGerald GA, McNamara P. Histone acetyltransferase-dependent chromatin remodeling and the vascular clock. J Biol Chem. 2004;279:7091–7097. doi: 10.1074/jbc.M311973200. [DOI] [PubMed] [Google Scholar]
- Damiola F, Le Minh N, Preitner N, Kornmann B, Fleury-Olela F, Schibler U. Restricted feeding uncouples circadian oscillators in peripheral tissues from the central pacemaker in the suprachiasmatic nucleus. Genes Dev. 2000;14:2950–2961. doi: 10.1101/gad.183500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dibner C, Schibler U, Albrecht U. The mammalian circadian timing system: organization and coordination of central and peripheral clocks. Annual Review of Physiology. 2010;72:517–549. doi: 10.1146/annurev-physiol-021909-135821. [DOI] [PubMed] [Google Scholar]
- Dioum EM, Rutter J, Tuckerman JR, Gonzalez G, Gilles-Gonzalez MA, McKnight SL. NPAS2: A gas-responsive transcription factor. Science. 2002;298:2385–2387. doi: 10.1126/science.1078456. [DOI] [PubMed] [Google Scholar]
- DiTacchio L, Le HD, Vollmers C, Hatori M, Witcher M, Secombe J, Panda S. Histone lysine demethylase JARID1a activates CLOCK-BMAL1 and dinfluences the circadian clock. Science. 2011;333:1881–1885. doi: 10.1126/science.1206022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doi M, Hirayama J, Sassone-Corsi P. Circadian regulator CLOCK is a histone acetyltransferase. Cell. 2006;125:497–508. doi: 10.1016/j.cell.2006.03.033. [DOI] [PubMed] [Google Scholar]
- Dong XC, Copps KD, Guo S, Li Y, Kollipara R, DePinho RA, White MF. Inactivation of hepatic Foxo1 by insulin signaling is required for adaptive nutrient homeostasis and endocrine growth regulation. Cell Metab. 2008;8:65–76. doi: 10.1016/j.cmet.2008.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duez H, van der Veen JN, Duhem C, Pourcet B, Touvier T, Fontaine C, Derudas B, Baugé E, Havinga R, Bloks VW, et al. Regulation of bile acid synthesis by the nuclear receptor Rev-erbalpha. Gastroenterology. 2008;135:689–698. doi: 10.1053/j.gastro.2008.05.035. [DOI] [PubMed] [Google Scholar]
- Dufour CR, Levasseur MP, Pham NHH, Eichner LJ, Wilson BJ, Charest-Marcotte A, Duguay D, Poirier-Héon JF, Cermakian N, Giguère V. Genomic convergence among ERRα, PROX1, and BMAL1 in the control of metabolic clock outputs. PLoS Genet. 2011;7:e1002143. doi: 10.1371/journal.pgen.1002143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duong HA, Robles MS, Knutti D, Weitz CJ. A molecular mechanism for circadian clock negative feedback. Science. 2011;332:1436–1439. doi: 10.1126/science.1196766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etchegaray JP, Lee C, Wade PA, Reppert SM. Rhythmic histone acetylation underlies transcription in the mammalian circadian clock. Nature. 2003;421:177–182. doi: 10.1038/nature01314. [DOI] [PubMed] [Google Scholar]
- Etchegaray J-P, Yang X, DeBruyne JP, Peters AHFM, Weaver DR, Jenuwein T, Reppert SM. The Polycomb group protein EZH2 is required for mammalian circadian clock function. J Biol Chem. 2006;281:21209–21215. doi: 10.1074/jbc.M603722200. [DOI] [PubMed] [Google Scholar]
- Farnham PJ. Insights from genomic profiling of transcription factors. Nature Reviews Genetics. 2009;10:605–616. doi: 10.1038/nrg2636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng D, Liu T, Sun Z, Bugge A, Mullican SE, Alenghat T, Liu XS, Lazar MA. A circadian rhythm orchestrated by histone deacetylase 3 controls hepatic lipid metabolism. Science. 2011;331:1315–1319. doi: 10.1126/science.1198125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freyssenet D. Energy sensing and regulation of gene expression in skeletal muscle. J Appl Physiol. 2007;102:529–540. doi: 10.1152/japplphysiol.01126.2005. [DOI] [PubMed] [Google Scholar]
- Froy O. The relationship between nutrition and circadian rhythms in mammals. Front Neuroendocrinol. 2007;28:61–71. doi: 10.1016/j.yfrne.2007.03.001. [DOI] [PubMed] [Google Scholar]
- Gatfield D, Le Martelot G, Vejnar CE, Gerlach D, Schaad O, Fleury-Olela F, Ruskeepää AL, Oresic M, Esau CC, Zdobnov EM, et al. Integration of microRNA miR-122 in hepatic circadian gene expression. Genes Dev. 2009;23:1313–1326. doi: 10.1101/gad.1781009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gery S, Virk RK, Chumakov K, Yu A, Koeffler HP. The clock gene Per2 links the circadian system to the estrogen receptor. Oncogene. 2007;26:7916–7920. doi: 10.1038/sj.onc.1210585. [DOI] [PubMed] [Google Scholar]
- Gilles-Gonzalez M-A, Gonzalez G. Signal transduction by heme-containing PAS-domain proteins. J Appl Physiol. 2004;96:774–783. doi: 10.1152/japplphysiol.00941.2003. [DOI] [PubMed] [Google Scholar]
- Gimble JM, Floyd ZE. Fat circadian biology. J Appl Physiol. 2009;107:1629–1637. doi: 10.1152/japplphysiol.00090.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green CB, Takahashi JS, Bass J. The meter of metabolism. Cell. 2008;134:728–742. doi: 10.1016/j.cell.2008.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimaldi B, Bellet MM, Katada S, Astarita G, Hirayama J, Amin RH, Granneman JG, Piomelli D, Leff T, Sassone-Corsi P. PER2 controls lipid metabolism by direct regulation of PPARγ. Cell Metab. 2010;12:509–520. doi: 10.1016/j.cmet.2010.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta N, Ragsdale SW. Thiol-disulfide redox dependence of heme binding and heme ligand switching in nuclear hormone receptor rev-erb{beta} J Biol Chem. 2011;286:4392–4403. doi: 10.1074/jbc.M110.193466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hems DA, Rath EA, Verrinder TR. Fatty acid synthesis in liver and adipose tissue of normal and genetically obese (ob/ob) mice during the 24-hour cycle. Biochem J. 1975;150:167–173. doi: 10.1042/bj1500167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirayama J, Sahar S, Grimaldi B, Tamaru T, Takamatsu K, Nakahata Y, Sassone-Corsi P. CLOCK-mediated acetylation of BMAL1 controls circadian function. Nature. 2007;450:1086–1090. doi: 10.1038/nature06394. [DOI] [PubMed] [Google Scholar]
- Huang W, Ramsey KM, Marcheva B, Bass J. Circadian rhythms, sleep, and metabolism. J Clin Invest. 2011;121:2133–2141. doi: 10.1172/JCI46043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jetten AM. Retinoid-related orphan receptors (RORs): critical roles in development, immunity, circadian rhythm, and cellular metabolism. Nucl Recept Signal. 2009;7:e003. doi: 10.1621/nrs.07003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jitrapakdee S. Transcription factors and coactivators controlling nutrient and hormonal regulation of hepatic gluconeogenesis. The International Journal of Biochemistry & Cell Biology. 2012;44:33–45. doi: 10.1016/j.biocel.2011.10.001. [DOI] [PubMed] [Google Scholar]
- Kaasik K, Lee CC. Reciprocal regulation of haem biosynthesis and the circadian clock in mammals. Nature. 2004;430:467–471. doi: 10.1038/nature02724. [DOI] [PubMed] [Google Scholar]
- Katada S, Sassone-Corsi P. The histone methyltransferase MLL1 permits the oscillation of circadian gene expression. Nature Structural & Molecular Biology. 2010;17:1414–1421. doi: 10.1038/nsmb.1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawamoto T, Noshiro M, Furukawa M, Honda KK, Nakashima A, Ueshima T, Usui E, Katsura Y, Fujimoto K, Honma S, et al. Effects of fasting and re-feeding on the expression of Dec1, Per1, and other clock-related genes. J Biochem. 2006;140:401–408. doi: 10.1093/jb/mvj165. [DOI] [PubMed] [Google Scholar]
- Kida K, Nishio T, Yokozawa T, Nagai K, Matsuda H, Nakagawa H. The circadian change of gluconeogenesis in the liver in vivo in fed rats. J Biochem. 1980;88:1009–1013. doi: 10.1093/oxfordjournals.jbchem.a133051. [DOI] [PubMed] [Google Scholar]
- Kohsaka A, Laposky AD, Ramsey KM, Estrada C, Joshu C, Kobayashi Y, Turek FW, Bass J. High-fat diet disrupts behavioral and molecular circadian rhythms in mice. Cell Metabolism. 2007;6:414–421. doi: 10.1016/j.cmet.2007.09.006. [DOI] [PubMed] [Google Scholar]
- Kornmann B, Schaad O, Bujard H, Takahashi JS, Schibler U. System-driven and oscillator-dependent circadian transcription in mice with a conditionally active liver clock. PLoS Biol. 2007;5:e34. doi: 10.1371/journal.pbio.0050034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laitinen S, Fontaine C, Fruchart JC, Staels B. The role of the orphan nuclear receptor Rev-Erb alpha in adipocyte differentiation and function. Biochimie. 2005;87:21–25. doi: 10.1016/j.biochi.2004.12.006. [DOI] [PubMed] [Google Scholar]
- Lamia KA, Papp SJ, Ruth TY, Barish GD, Uhlenhaut NH, Jonker JW, Downes M, Evans RM. Cryptochromes mediate rhythmic repression of the glucocorticoid receptor. Nature. 2011;480:552–556. doi: 10.1038/nature10700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamia KA, Sachdeva UM, DiTacchio L, Williams EC, Alvarez JG, Egan DF, Vasquez DS, Juguilon H, Panda S, Shaw RJ, et al. AMPK regulates the circadian clock by cryptochrome phosphorylation and degradation. Science. 2009;326:437–440. doi: 10.1126/science.1172156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamia KA, Storch K-F, Weitz CJ. Physiological significance of a peripheral tissue circadian clock. Proc Natl Acad Sci USA. 2008;105:15172–15177. doi: 10.1073/pnas.0806717105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Dentin R, Chen D, Hedrick S, Ravnskjaer K, Schenk S, Milne J, Meyers DJ, Cole P, Yates J, et al. A fasting inducible switch modulates gluconeogenesis via activator-coactivator exchange. Nature. 2008;456:269–273. doi: 10.1038/nature07349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma K, Xiao R, Tseng HT, Shan L, Fu L, Moore DD. Circadian dysregulation disrupts bile acid homeostasis. PLoS ONE. 2009;4:e6843. doi: 10.1371/journal.pone.0006843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcheva B, Ramsey KM, Buhr ED, Kobayashi Y, Su H, Ko CH, Ivanova G, Omura C, Mo S, Vitaterna MH, et al. Disruption of the clock components CLOCK and BMAL1 leads to hypoinsulinaemia and diabetes. Nature. 2010;466:627–631. doi: 10.1038/nature09253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Martelot G, Claudel T, Gatfield D, Schaad O, Kornmann B, Sasso GL, Moschetta A, Schibler U. REV-ERBα participates in circadian SREBP signaling and bile acid homeostasis. PLoS Biol. 2009;7:e1000181. doi: 10.1371/journal.pbio.1000181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maywood ES, O’Neill JS, Reddy AB, Chesham JE, Prosser HM, Kyriacou CP, Godinho SIH, Nolan PM, Hastings MH. Genetic and molecular analysis of the central and peripheral circadian clockwork of mice. Cold Spring Harb Symp Quant Biol. 2007;72:85–94. doi: 10.1101/sqb.2007.72.005. [DOI] [PubMed] [Google Scholar]
- Nader N, Chrousos GP, Kino T. Circadian rhythm transcription factor CLOCK regulates the transcriptional activity of the glucocorticoid receptor by acetylating its hinge region lysine cluster: potential physiological implications. Faseb J. 2009;23:1572–1583. doi: 10.1096/fj.08-117697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakahata Y, Kaluzova M, Grimaldi B, Sahar S, Hirayama J, Chen D, Guarente LP, Sassone-Corsi P. The NAD+-dependent deacetylase SIRT1 modulates CLOCK-mediated chromatin remodeling and circadian control. Cell. 2008;134:329–340. doi: 10.1016/j.cell.2008.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakahata Y, Sahar S, Astarita G, Kaluzova M, Sassone-Corsi P. Circadian control of the NAD+ salvage pathway by CLOCK-SIRT1. Science. 2009;324:654–657. doi: 10.1126/science.1170803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naruse Y, Oh-hashi K, Iijima N, Naruse M, Yoshioka H, Tanaka M. Circadian and light-induced transcription of clock gene Per1 depends on histone acetylation and deacetylation. Mol Cell Biol. 2004;24:6278–6287. doi: 10.1128/MCB.24.14.6278-6287.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Neill JS, Ooijen G van, Dixon LE, Troein C, Corellou F, Bouget FY, Reddy AB, Millar AJ. Circadian rhythms persist without transcription in a eukaryote. Nature. 2011;469:554–558. doi: 10.1038/nature09654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Neill JS, Reddy AB. Circadian clocks in human red blood cells. Nature. 2011;469:498–503. doi: 10.1038/nature09702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oster H, Damerow S, Kiessling S, Jakubcakova V, Abraham D, Tian J, Hoffmann MW, Eichele G. The circadian rhythm of glucocorticoids is regulated by a gating mechanism residing in the adrenal cortical clock. Cell Metabolism. 2006;4:163–173. doi: 10.1016/j.cmet.2006.07.002. [DOI] [PubMed] [Google Scholar]
- Panda S, Antoch MP, Miller BH, Su AI, Schook AB, Straume M, Schultz PG, Kay SA, Takahashi JS, Hogenesch JB. Coordinated transcription of key pathways in the mouse by the circadian clock. Cell. 2002;109:307–320. doi: 10.1016/s0092-8674(02)00722-5. [DOI] [PubMed] [Google Scholar]
- Pardee KI, Xu X, Reinking J, Schuetz A, Dong A, Liu S, Zhang R, Tiefenbach J, Lajoie G, Plotnikov AN, et al. The structural basis of gas-responsive transcription by the human nuclear hormone receptor REV-ERBbeta. PLoS Biol. 2009;7:e43. doi: 10.1371/journal.pbio.1000043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phelan CA, Gampe RT, Jr, Lambert MH, Parks DJ, Montana V, Bynum J, Broderick TM, Hu X, Williams SP, Nolte RT, et al. Structure of Rev-erbalpha bound to N-CoR reveals a unique mechanism of nuclear receptor-co-repressor interaction. Nat Struct Mol Biol. 2010;17:808–814. doi: 10.1038/nsmb.1860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Portela A, Esteller M. Epigenetic modifications and human disease. Nature Biotechnology. 2010;28:1057–1068. doi: 10.1038/nbt.1685. [DOI] [PubMed] [Google Scholar]
- Preitner N, Damiola F, Lopez-Molina L, Zakany J, Duboule D, Albrecht U, Schibler U. The orphan nuclear receptor REV-ERBalpha controls circadian transcription within the positive limb of the mammalian circadian oscillator. Cell. 2002;110:251–260. doi: 10.1016/s0092-8674(02)00825-5. [DOI] [PubMed] [Google Scholar]
- Raghuram S, Stayrook KR, Huang P, Rogers PM, Nosie AK, McClure DB, Burris LL, Khorasanizadeh S, Burris TP, Rastinejad F. Identification of heme as the ligand for the orphan nuclear receptors REV-ERBalpha and REV-ERBbeta. Nat Struct Mol Biol. 2007;14:1207–1213. doi: 10.1038/nsmb1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsey KM, Yoshino J, Brace CS, Abrassart D, Kobayashi Y, Marcheva B, Hong HK, Chong JL, Buhr ED, Lee C, et al. Circadian clock feedback cycle through NAMPT-mediated NAD+ biosynthesis. Science. 2009;324:651–654. doi: 10.1126/science.1171641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy AB, Karp NA, Maywood ES, Sage EA, Deery M, O’Neill JS, Wong GKY, Chesham J, Odell M, Lilley KS, et al. Circadian orchestration of the hepatic proteome. Current Biology. 2006;16:1107–1115. doi: 10.1016/j.cub.2006.04.026. [DOI] [PubMed] [Google Scholar]
- Reddy AB, Maywood ES, Karp NA, King VM, Inoue Y, Gonzalez FJ, Lilley KS, Kyriacou CP, Hastings MH. Glucocorticoid signaling synchronizes the liver circadian transcriptome. Hepatology. 2007;45:1478–1488. doi: 10.1002/hep.21571. [DOI] [PubMed] [Google Scholar]
- Rey G, Cesbron F, Rougemont J, Reinke H, Brunner M, Naef F. Genome-wide and phase-specific DNA-binding rhythms of BMAL1 control circadian output functions in mouse liver. PLoS Biol. 2011;9:e1000595. doi: 10.1371/journal.pbio.1000595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ripperger JA, Schibler U. Rhythmic CLOCK-BMAL1 binding to multiple E-box motifs drives circadian Dbp transcription and chromatin transitions. Nature Genetics. 2006;38:369–374. doi: 10.1038/ng1738. [DOI] [PubMed] [Google Scholar]
- Rodgers JT, Lerin C, Haas W, Gygi SP, Spiegelman BM, Puigserver P. Nutrient control of glucose homeostasis through a complex of PGC-1α and SIRT1. Nature. 2005;434:113–118. doi: 10.1038/nature03354. [DOI] [PubMed] [Google Scholar]
- Rutter J, Reick M, Wu LC, McKnight SL. Regulation of Clock and NPAS2 DNA binding by the redox state of NAD cofactors. Science. 2001;293:510–514. doi: 10.1126/science.1060698. [DOI] [PubMed] [Google Scholar]
- Sahar S, Nin V, Barbosa MT, Chini EN, Sassone-Corsi P. Altered behavioral and metabolic circadian rhythms in mice with disrupted NAD+ oscillation. Aging (Albany NY) 2011;3:794–802. doi: 10.18632/aging.100368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmutz I, Ripperger JA, Baeriswyl-Aebischer S, Albrecht U. The mammalian clock component PERIOD2 coordinates circadian output by interaction with nuclear receptors. Genes Dev. 2010;24:345–357. doi: 10.1101/gad.564110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimba S, Ishii N, Ohta Y, Ohno T, Watabe Y, Hayashi M, Wada T, Aoyagi T, Tezuka M. Brain and muscle Arnt-like protein-1 (BMAL1), a component of the molecular clock, regulates adipogenesis. Proc Natl Acad Sci USA. 2005;102:12071–12076. doi: 10.1073/pnas.0502383102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimba S, Ogawa T, Hitosugi S, Ichihashi Y, Nakadaira Y, Kobayashi M, Tezuka M, Kosuge Y, Ishige K, Ito Y, et al. Deficient of a clock gene, brain and muscle Arnt-like protein-1 (BMAL1), induces dyslipidemia and ectopic fat formation. PLoS ONE. 2011;6:e25231. doi: 10.1371/journal.pone.0025231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solt LA, Wang Y, Banerjee S, Hughes T, Kojetin DJ, Lundasen T, Shin Y, Liu J, Cameron MD, Noel R, et al. Regulation of circadian behaviour and metabolism by synthetic REV-ERB agonists. Nature. 2012;485:62–68. doi: 10.1038/nature11030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stokkan KA, Yamazaki S, Tei H, Sakaki Y, Menaker M. Entrainment of the circadian clock in the liver by feeding. Science. 2001;291:490–493. doi: 10.1126/science.291.5503.490. [DOI] [PubMed] [Google Scholar]
- Sun Z, Miller RA, Patel RT, Cheng J, Dhir RD, Wang H, Zhang D, Graham MJ, Unterman TG, Shulman GI, Sztalryd C, Bennett MJ, Ahima RS, Birnbaum MJ, Lazar MA. Hepatic HDAC3 promotes gluconeogenesis by repressing lipid synthesis and sequestration. Nat Medicine. 2012 doi: 10.1038/nm.2744. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tahara Y, Otsuka M, Fuse Y, Hirao A, Shibata S. Refeeding after fasting elicits insulin-dependent regulation of Per2 and Rev-Erbα with shifts in the liver clock. J Biol Rhythms. 2011;26:230–240. doi: 10.1177/0748730411405958. [DOI] [PubMed] [Google Scholar]
- Takahata S, Ozaki T, Mimura J, Kikuchi Y, Sogawa K, Fujii-Kuriyama Y. Transactivation mechanisms of mouse clock transcription factors, mClock and mArnt3. Genes to Cells. 2000;5:739–747. doi: 10.1046/j.1365-2443.2000.00363.x. [DOI] [PubMed] [Google Scholar]
- Takeda Y, Kang HS, Angers M, Jetten AM. Retinoic acid-related orphan receptor γ directly regulates neuronal PAS domain protein 2 transcription in vivo. Nucleic Acids Res. 2011;39:4769–4782. doi: 10.1093/nar/gkq1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao W, Chen S, Shi G, Guo J, Xu Y, Liu C. SWItch/sucrose nonfermentable (SWI/SNF) complex subunit BAF60a integrates hepatic circadian clock and energy metabolism. Hepatology. 2011;54:1410–1420. doi: 10.1002/hep.24514. [DOI] [PubMed] [Google Scholar]
- Turek FW, Joshu C, Kohsaka A, Lin E, Ivanova G, McDearmon E, Laposky A, Losee-Olson S, Easton A, Jensen DR, et al. Obesity and metabolic syndrome in circadian clock mutant mice. Science. 2005;308:1043–1045. doi: 10.1126/science.1108750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Um JH, Pendergast JS, Springer DA, Foretz M, Viollet B, Brown A, Kim MK, Yamazaki S, Chung JH. AMPK regulates circadian rhythms in a tissue- and isoform-specific manner. PLoS ONE. 2011;6:e18450. doi: 10.1371/journal.pone.0018450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Um JH, Yang S, Yamazaki S, Kang H, Viollet B, Foretz M, Chung JH. Activation of 5’-AMP-activated kinase with diabetes drug metformin induces casein kinase Iepsilon (CKIepsilon)-dependent degradation of clock protein mPer2. J Biol Chem. 2007;282:20794–20798. doi: 10.1074/jbc.C700070200. [DOI] [PubMed] [Google Scholar]
- Vaquero A, Reinberg D. Calorie restriction and the exercise of chromatin. Genes Dev. 2009;23:1849–1869. doi: 10.1101/gad.1807009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieira E, Nilsson EC, Nerstedt A, Ormestad M, Long YC, Garcia-Roves PM, Zierath JR, Mahlapuu M. Relationship between AMPK and the transcriptional balance of clock-related genes in skeletal muscle. Am J Physiol Endocrinol Metab. 2008;295:E1032–1037. doi: 10.1152/ajpendo.90510.2008. [DOI] [PubMed] [Google Scholar]
- Vollmers C, Gill S, DiTacchio L, Pulivarthy SR, Le HD, Panda S. Time of feeding and the intrinsic circadian clock drive rhythms in hepatic gene expression. Proc Natl Acad Sci U S A. 2009;106:21453–21458. doi: 10.1073/pnas.0909591106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang XS, Armstrong MEG, Cairns BJ, Key TJ, Travis RC. Shift work and chronic disease: the epidemiological evidence. Occup Med (Lond) 2011;61:78–89. doi: 10.1093/occmed/kqr001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu N, Yin L, Hanniman EA, Joshi S, Lazar MA. Negative feedback maintenance of heme homeostasis by its receptor, Rev-erbα. Genes Dev. 2009;23:2201–2209. doi: 10.1101/gad.1825809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Downes M, Yu RT, Bookout AL, He W, Straume M, Mangelsdorf DJ, Evans RM. Nuclear receptor expression links the circadian clock to metabolism. Cell. 2006;126:801–810. doi: 10.1016/j.cell.2006.06.050. [DOI] [PubMed] [Google Scholar]
- Yin L, Joshi S, Wu N, Tong X, Lazar MA. E3 ligases Arf-bp1 and Pam mediate lithium-stimulated degradation of the circadian heme receptor Rev-erbα. Proc Natl Acad Sci U S A. 2010;107:11614–11619. doi: 10.1073/pnas.1000438107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin L, Lazar MA. The orphan nuclear receptor Rev-erbα recruits the N-CoR/Histone Deacetylase 3 corepressor to regulate the circadian Bmal1 gene. Molecular Endocrinology. 2005;19:1452–1459. doi: 10.1210/me.2005-0057. [DOI] [PubMed] [Google Scholar]
- Yin L, Wu N, Curtin JC, Qatanani M, Szwergold NR, Reid RA, Waitt GM, Parks DJ, Pearce KH, Wisely GB, et al. Rev-erbα, a heme sensor that coordinates metabolic and circadian pathways. Science. 2007;318:1786–1789. doi: 10.1126/science.1150179. [DOI] [PubMed] [Google Scholar]
- Zhang EE, Liu Y, Dentin R, Pongsawakul PY, Liu AC, Hirota T, Nusinow DA, Sun X, Landais S, Kodama Y, et al. Cryptochrome mediates circadian regulation of cAMP signaling and hepatic gluconeogenesis. Nature Medicine. 2010;16:1152–1156. doi: 10.1038/nm.2214. [DOI] [PMC free article] [PubMed] [Google Scholar]


