SUMMARY
The stromal interaction molecule, STIM1 regulates Ca2+ entry via Orai1 channels in response to decreased concentration of ER luminal Ca2+. In search of a mechanism that switches the cytoplasmic aspect of STIM1 from an inactive to an active state, we identified an acidic motif within the STIM1 coiled-coil region that keeps the Ca2+ activation domain (CAD/SOAR) inactive. The STIM1 acidic motif shows significant homology to the C-terminal coiled-coil segment of Orai1, the postulated site of interaction with STIM1. Mutations within the acidic region make STIM1 constitutively active while those within a short basic segment of CAD/SOAR prevent Orai1 activation. We propose that during STIM1 activation, the CAD/SOAR domain is released from an intramolecular clamp allowing the basic segment to activate Orai1 channels. This evolutionary conserved activation mechanism of STIM1 resembles the regulation of protein kinases by intramolecular silencing with pseudosubstrate binding.
INTRODUCTION
Store-operated Ca2+ entry (SOCE) was postulated as a Ca2+ entry pathway regulated by the ER luminal Ca2+ concentration more than 20 years ago (1). Although a current, named ICRAC, corresponding to SOCE in T-cells and mast cells have long been identified by patch clamp studies (2,3), the molecular mechanism(s) underlying ICRAC or SOCE has eluded identification until quite recently. Stromal interaction molecules (STIM1 and-2) were found in small interfering RNA screens as the ER Ca2+ sensors (4,5) and the Orai/CRACM proteins as the plasma membrane (PM) Ca2+ channels, working together to constitute ICRAC (6–8). STIM1 and Orai1 have most prominent functions in T cell activation, their defects causing severe immunodeficiences (6,9), but they are also important for other cells such as platelets (10) and for skeletal muscle development (11).
Since their identification, rapid progress has been made in analyzing the molecular details of the STIM1 and Orai1 activation process (recently reviewed in (12). The N-terminal luminal segment of the ER-localized, single membrane spanning STIM1 protein contains EF hand and SAM domains that are responsible for luminal Ca2+ sensing and oligomerization, respectively (13). The C-terminal segment of STIM1 (STIM-ct) facing the cytosol contains coiled-coil domains as well as several acidic, serine-proline-rich and basic segments (recently reviewed in (14) (Fig. 1A). The STIM-ct is capable of activating Orai1 channels (15–17) and it was shown in elegant experiments that oligomerization alone is sufficient to activate STIM1 (18). Several studies identified small ~100 amino acid partly overlapping segments within the cytosolic aspect of STIM1 as the minimal activating domain variably termed CAD, SOAR and OASF (19–21). At the same time, the C-terminal putative coiled-coiled domain of Orai1 was shown to be critical for the activation by STIM1 proteins (22–24). In spite of these impressive advances, it is still not understood how oligomerization and subsequent clustering of STIM1 facilitates the interaction of the STIM1 CAD/SOAR domain with Orai1 channels.
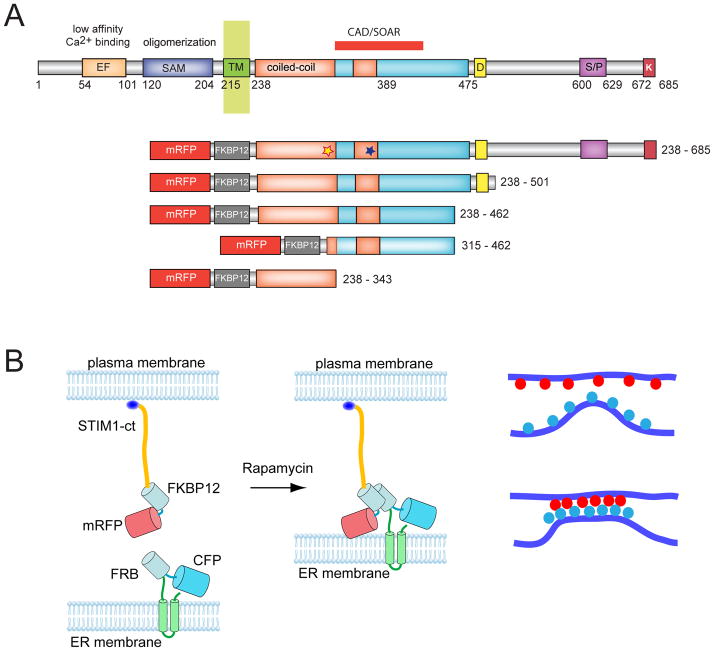
Figure 1.
Clustering of the cytosolic domain of STIM1 activates Orai1 channels. (A) Schematics of STIM1 structure and the constructs used in the present study. The numbering corresponds to the human STIM1 protein. EF, Ca2+ binding EF-hand motif; SAM, sterile alpha motif; CAD/SOAR minimal Orai1 activation domain; D, acidic region; S/P, proline-, serine/threonine-rich segment, K, polybasic domain (after(39). The blue and yellow asterisks indicate the positions of the basic and acidic regions, respectively, identified in the present study. (B) Schematics of clustering by rapamycin-induced heterodimerization of the FKBP12 fused cytosolic STIM1 (FK-STIM1-ct) and the FRB targeted to the ER surface. The blue oval represents the C-terminal polybasic domain of STIM1 that is important for the PM localization of the protein (C) Localization of the indicated proteins expressed in COS-7 cells before (top row) and 5 min after (bottom row) rapamycin addition. Confocal images were taken in live cells 1 day after transfection. The area in the white box is shown enlarged. Note the significant membrane localization of the cytoplasmic STIM1 segment and its co-clustering with the Orai1 at the ER-PM contact zones after addition of rapamycin (100 nM). (D) Rapamycin-induced clustering increases FRET between YFP- and mRFP-tagged recruitable STIM1-ct indicating that clustering forces them to get within FRET distance. (E) Cytosolic Ca2+ increases evoked by rapamycin-induced clustering of FK-STIM1-ct. COS-7 cells were transfected with the ER-targeted CFP-FRB, the mRFP-FKBP12-STIM1-ct and untagged Orai1. Cytosolic Ca2+ changes were followed by Fura2. Blue represent cells with no visible transfections. Means ± S.E.M. are shown (n=51 and 58 cells for red and blue, respectively, form two separate experiments).
In the present study we used a unique oligomerization strategy to show that the full cytoplasmic segment of STIM1 is a poor activator of Orai1, but it becomes highly active upon clustering. In contrast, the CAD/SOAR domain is very active to open Orai1 channels without oligomerization, suggesting that it must be kept inactive in the context of the whole cytosolic segment of STIM1. In search of an intramolecular silencing mechanism, we identified a short acidic segment within the first coiled-coiled domain of STIM1 that could form an intramolecular interaction with a basic sequence within CAD/SOAR recently identified as key for Orai1 activation (25). We show that mutations within the acidic stretch make STIM1 constitutively active and confirm that the basic sequence within CAD/SOAR is essential for STIM1-mediated Orai1 activation. Intriguingly, the acidic stretch within STIM1 shows significant sequence homology with the C-terminus of Orai1 and could be used as a decoy to interfere with the ability of STIM1 to activate Orai1 channels but only when expressed as part of the juxtamembrane first coiled-coil domain of STIM1. We suggest that the initial oligomerization of STIM1 unmasks the CAD/SOAR domain by breaking the intramolecular interaction that keeps the CAD/SOAR domain inactive in the quiescent STIM1 molecule.
RESULTS
The STIM1 cytosolic segment requires clustering to become active
It has been shown that the STIM1-ct is capable of activating Orai1 channels (15) a finding confirmed by several subsequent studies (see (14) for a recent Review). However, when STIM1-ct is expressed in COS-7 cells in the form of an mRFP fusion protein (together with Orai1) we and others ((19–21) found only a moderate increase in basal cytosolic Ca2+ with only very few cells showing substantially increased basal Ca2+ (above a Fura2 ratio of 0.3) in spite of prominent PM localization of the protein (Fig. 1C). We assumed that this was because the expressed protein was mostly in an inactive conformation. In order to cluster the STIM1-ct protein we devised a strategy whereby we placed an FKBP12 module between the mRFP and the cytosolic STIM fragment (mRFP-FKBP12-STIM1-ct or FK-STIM1-ct for short) and expressed this protein together with the ER-targeted FRB-CFP recruiter construct used in our previous study (26) (Fig. 1B). Since the STIM1-ct protein showed substantial PM localization, we reasoned that addition of rapamycin would connect these molecules to the ER-bound FRB, thereby enriching them in the ER-PM contact zones (Fig. 1B). This indeed was the case, as rapamycin induced the formation of such contact zones enriched in both binding partners and also bringing Orai1 proteins to these sites (Fig. 1C). Importantly, this manipulation led to a FRET increase between the STIM1-ct molecules (Fig. 1D) and evoked a substantial increase in cytoplasmic Ca2+ indicating that the so oligomerized STIM1-ct construct was now able to activate Orai1 channels (Fig. 1E).
This was an important observation because it suggested that the cytosolic segment of STIM1 contained all the information necessary for turning from an inactive to an active state. Although previous studies showed that STIM1-ct is sufficient to activate Orai1 channels and that artificial clustering of STIM1 molecules via a rapamycin-inducible multimerization system placed in the luminal side of STIM1 was able to evoke Orai1 activation (reviewed in (14), our studies show for the first time that an isolated soluble STIM1-ct can be switched from an inactive to an active state. In order to rule out that activation of STIM1-ct by this manipulation was due to bringing a yet unidentified ER-localized protein to PM proximity, we performed similar experiments using an FRB domain targeted to the surface of mitochondria. Here, rapamycin caused the clustering of the STIM1-ct protein at the contact zones formed between the PM and adjacent mitochondria (Fig. S1A). Again, this manipulation was associated with significant Ca2+ increases indicating the activation of the STIM1 protein by clustering (Fig. S1B). No visible clustering or activation of Ca2+ influx was observed when STIM1-ct molecules were dimerized with AP1510. This compound binds and dimerizes FKBP12 molecules, which then can be reversed by FK506 (27). FRET analysis showed that AP1510 did induce dimerization of STIM1-ct that was reversible by FK506 (Fig. S1C). Similarly, no activation of Ca2+ influx was demonstrable when the Orai1 channel was directly tagged with a C-terminal FKBP12 module and clustered with the ER-targeted FRB by rapamycin treatment (Fig. S2AB). Since the FKBP12-tagged Orai1 construct is functional as it still can be activated via STIM1 by ER-Ca2+ depletion (Fig. S2C), this result showed that it is not the clustering of Orai1 per se but clustering of STIM1-ct that activated the Orai1 channels. It is important to note that the large Ca2+ responses shown here were dependent on the presence of expressed Orai1 (the responses were substantially smaller in cells expressing only the recruitable STIM1 with endogenous Orai1) and that 2-APB (50 μM) prevented or rapidly reversed the Ca2+ increase, indicating that the responses shown here were due to STIM1-Orai1 activation and not an ER Ca2+ release or other Ca2+ signaling mechanism (Fig. S3).
An inhibitory domain within the coiled-coil domain keeps STIM1 inactive
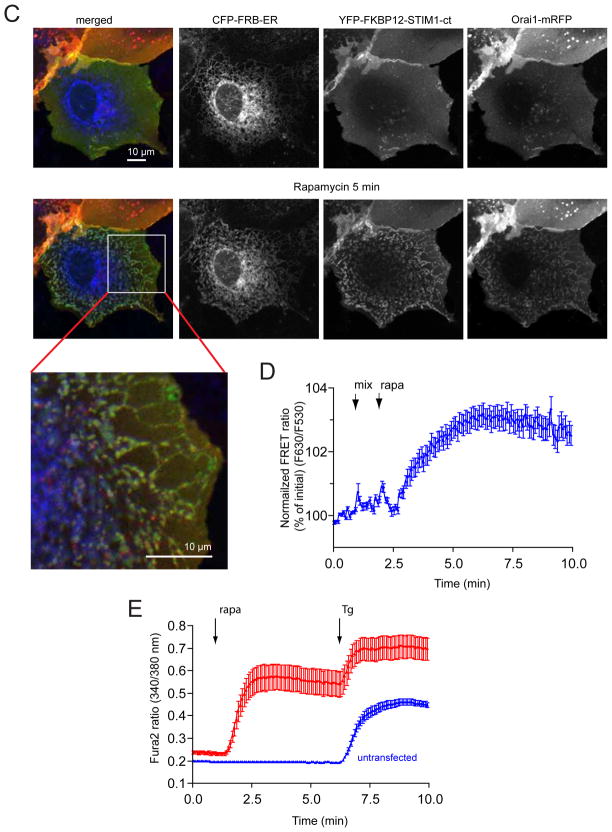
Increased truncations from the C-terminus in the full-STIM1 molecule up to residue 463 yielded proteins that were able to activate Orai1 by ER-Ca2+ depletion, as shown previously by others (reviewed in (12) and (14) (Fig. 2A). The same truncations on the recruitable FK-STIM1-ct proteins resulted notable differences in their abilities to increase Ca2+ influx after rapamycin-induced clustering (Fig. 2B). Although these results pointed to important regions in the STIM1 molecule for activation and inactivation that were consistent with published reports (and not detailed further in this study), these were only modulatory, since truncations down to 462 yielded a protein that was capable of Orai1 activation upon clustering (Fig. 2B). When this molecule was now truncated from the N-terminal direction, a significant change occurred in that the fragment became constitutively active no more requiring clustering (Fig. 2C). This activating fragment (315–462) closely corresponded to the CAD (342–448) and SOAR (344–442) domains described in recent studies (19,20) and its activity was indistinguishable from that of CAD (Fig 2C). Addition of rapamycin to the recruitable versions of these activating pieces elicited a notable decrease in cytoplasmic Ca2+ concentration in a fraction of cells (not plotted separately) that was not unexpected as the free concentration of the domains in the cytosol decreased after their recruitment to the ER surface and these domains do not efficiently bridge the ER and PM after rapamycin addition. It is important to note that due to the Ca2+ toxicity of the constructs causing constitutive activation, cells were kept in a low Ca2+ (0.2 mM) medium during transfection (see Methods), which greatly increased the number of cells showing the high basal Ca2+ during the experiments. Even with these modifications in culturing, CAD was more toxic to the cells than the 315–462 STIM1 piece.
Figure 2.
Cytoplasmic Ca2+ responses evoked by various STIM1 constructs truncated from the C-terminus. (A) C-terminal truncations on the full-STIM1 (with intact N-terminus) protein up to residue 463 have only small effects on the thapsigargin (Tg, 200 nM) –induced Ca2+ elevations. Cells were co-transfected with untagged Orai1 and the indicated mRFP-tagged (in the luminal side) STIM1 constructs driven by the thimidine kinase promoter (TK-STIM1). Means ± S.E.M. are shown (n = 89, 37, 32 and 198 cells for black, green, red and blue traces, respectively obtained in 2–5 separate experiments). (B) The same truncations have significant impact on the abilities of the cytoplasmic STIM1 (STIM1-ct) pieces to activate Orai1 channels after rapamycin-induced clustering. Here the indicated STIM1-ct fragments were fused with the mRFP-FKBP12 module (FK-STIM-ct) and co-expressed with untagged Orai1 and the ER-targeted CFP-FRB. Means ± S.E.M. are shown (n = 356, 254, 259 and 639 cells for black, green, red and blue traces, respectively obtained in 9–10 separate experiments). (C) Truncation of the cytoplasmic STIM1 fragments from the N-terminal direction makes them constitutively active not requiring clustering. Cells were transfected with the indicated constructs as described in panel B. Means ± S.E.M. are shown (n = 208, 82, 124 and 305 cells for black, green, red and blue traces, respectively obtained in 4–6 separate experiments).
These results together suggested that the juxtamembrane coiled-coiled segment of STIM1 (within the sequence of 238–343) has the ability to keep the CAD/SOAR domain inactive and that clustering of STIM1 was required for unmasking the latter. We hypothesized that the “inhibitory“ coiled-coil segment makes an intramolecular contact with part(s) of the CAD/SOAR domain that is important for Orai1 activation. Therefore, we speculated that the interaction site within the STIM1 coiled-coil domain that keeps CAD/SOAR in an inactive state might share some similarity with the Orai1 C-terminal tail. The coiled-coiled domain of STIM1 has several acidic residues with spacing that suggests that these form a highy acidic surface and coincidentally the C-terminal putative coiled-coiled domain of Orai1 has similar features (Fig. 3A). Indeed, an aligment between the latter and the STIM1 coiled-coil segment revealed a sequence similarity that was remarkable with identical spacing of the acidic residues that is best demonstrated in a wheel diagram (Fig. 3A). This segment also had a highly acidic cluster a few amino acids downstream.
Figure 3.
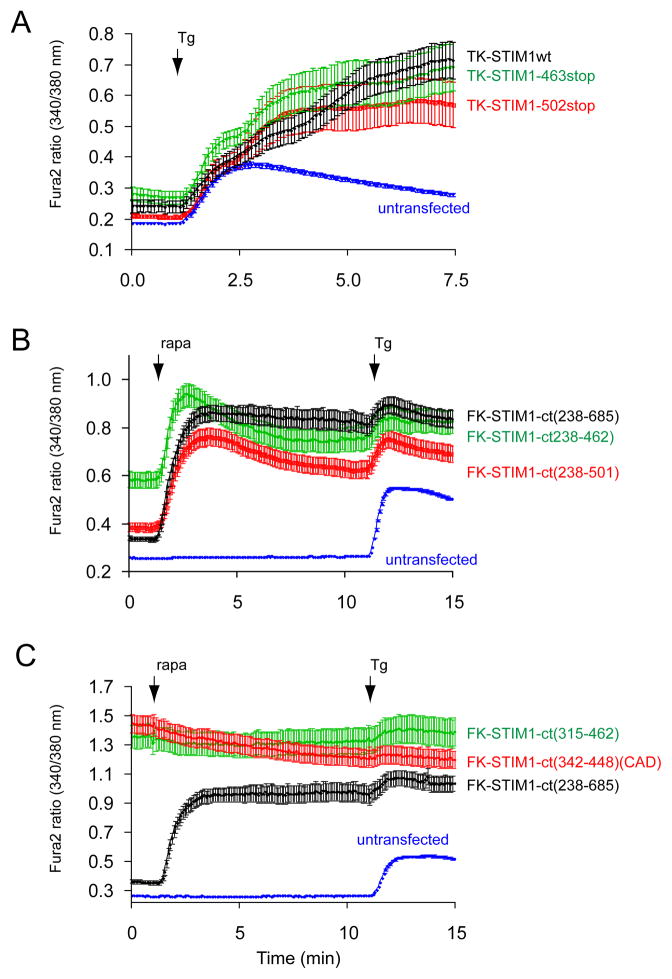
An acidic segment within the coiled-coil domain of STIM1 is homologous with the Orai1 C-terminal helix and is essential to keep STIM1 in an inactive state. (A) Sequence alignment between STIM1 orthologues and the Orai1 C-terminus identifies a homologous region within the coiled-coil domain. Helical wheel diagram shows the high degree of positional conservation between the Orai1 helix and the proximal part of the acidic STIM1 segment. A Leu to Ser substitution of the residue shown by the red arrow in Orai1 has been shown to prevent its activation by STIM1 (22). Mutations within this segment of STIM1 are labeled by an asterisk (AGAA, or 3EA). Changing only the EL residues in STIM1 (to AG) had negligible effects (not shown). (B) Mutations within this segment (3EA) make the cytoplasmic STIM1 construct partially active without clustering (red). However, mutation of the four E residues in the distal part of the acidic motif (4EA) renders the cytoplasmic STIM1 piece fully constitutively fully active. Means ± S.E.M. are shown (n = 220, 345, 143 and 802 cells for black, green, red and blue traces, respectively obtained in 6–38 separate dishes from 3–9 independent experiment). (C) The same mutations within the full-length STIM1 protein show similar partial or full constitutive activity. For comparison, the effects of the known constitutively active D76A mutant (4) STIM1 is shown. For these experiments the thymidine kinase (TK) driven mRFP-STIM1 constructs were expressed together with untagged Orai1. Means ± S.E.M. are shown (n = 143, 109, 80, 111 and 785 cells for black, green, pink, red and blue traces, respectively, obtained in 3–36 dishes from 3–8 separate experiments). (D) Unlike wild-type STIM1 (upper two rows), the 4EA mutant mRFP-STIM1 forms constitutive clusters together with Orai1-YFP without Ca2+ release from the ER (lower row). This analysis was performed at room temperature. (E) Plasma membrane recruitment of the STIM1 coiled-coil domain containing the inhibitory acidic segment inhibits Orai1-mediated Ca2+ influx activated by the constitutively active STIM1 fragment (315–462). For this experiments COS-7 cells were transfected with the mRFP-fused STIM1(315–462) (non-recruitable!), untagged Orai1 and the ER-recruiter FRB together with either the mRFP-FKBP12-fused STIM1-ct(238–343) [FK-STIM1-ct (238–443), red] or mRFP-FKBPonly (FK-only, black). Rapamycin addition decreased the high basal Ca2+ in a significant fraction of cells (~16 %) in the STIM1-ct(238–343) expressing group while had no effect in any cells in the FKBPonly group. Means ± S.E.M. are shown (n = 119, 80 and 247 cells for black, red and blue traces, respectively, obtained in 5–10 separate experiments).
An acidic segment within the STIM1 coiled-coil domain similar to that in the Orai1 C-terminal helix is an autoinhibitory domain
To determine whether the acidic segment within the coiled-coil domain is responsible for keeping the basic domain from activating Orai1 we mutated the acidic residues. This segment contains three Glu residues that show a good alignment with the Orai1 C-terminal helix, and an additional acidic stretch with four Glu residues in an adjacent region (Fig. 3A). First we mutated three of the acidic residues into alanines (3EA) both in the recruitable full cytoplasmic STIM1 and in the full-length STIM1 molecule (these constructs also had a mutation in the Leu residue that corresponded to the Leu residue of Orai1 necessary for activation (22) but mutation of the Leu alone in STIM1 was without effect (not shown)). Intriguingly, the 3EA mutation turned the cytoplasmic STIM1 partially active without the need for rapamycin-induced clustering although clustering still caused further activation (Fig. 3B). However, a stronger constitutive activation was observed when the distal four Glu residues were mutated to alanines (4EA) and this mutant showed no response to rapamycin indicating that the CAD/SOAR domain was fully exposed (Fig. 3B). Similarly, the full-length STIM1 molecules carrying the 3EA mutation showed a partial constitutive activity when expressed with Orai1, while the 4EA mutant was fully active (Fig. 3C). Combination of the two mutations (7EA) did not significantly enhance the activity of the constructs above that of the 4EA mutants. Moreover, all of the constitutively active mutant full-length STIM1 molecules showed clustering and co-localization with expressed Orai1 without emptying the Ca2+ stores (Fig. 3D shows the 4EA mutant). These results supported the assumption that the acidic segment acts as a negative regulator of the STIM1 molecule and the negative cluster of Glu residues had a critical role in this respect.
Intriguingly, the 4EA mutant STIM1 molecules did not show the massive clustering characteristic of the D76A mutant when expressed without the Orai1 molecules. This construct showed microtubule association, yet it was significantly different from the wild-type in that it had a rosary-like appearance along the microtubules. These structures rapidly turned to clusters after store depletion (Fig. S4). This suggested that these mutations did not cause a gross alteration in the STIM1 molecules that could lead to artificial aggregation and activation. However, the subtle conformational change is sufficient to promote activation of the endogenous Orai1 molecules and to create visible clusters when Orai1 molecules are also expressed (Fig 3D).
The acidic segment within the STIM1 coiled-coil domain can be used as a decoy to inhibit Orai1 activation
If the acidic segment of STIM1 indeed is similar to the C-terminal Orai1 activation domain, then it is expected to act as a competitor when brought to the plasma membrane. For this, we co-expressed Orai1with the isolated acidic segment of STIM1 (238–343) with the constitutively active CAD/SOAR domain (315–462) to see if this segment is capable of inhibiting CAD/SOAR. Since we assumed that the affinity of the two interacting STIM1 fragments is relatively low, we used the rapamycin-induced recruitment of the acidic segment to the PM to increase its concentration at the proximity of Orai1. Using this manipulation we found that a significant fraction of cells showed a decrease in cytoplasmic Ca2+ concentration after rapamycin addition (not all cells express all of the constructs in the right proportion). No such inhibition was found in any cells when the mRFP-FKBP12only construct was used instead of the acidic domain in similar experiments (Fig. 3E). These data suggested that the acidic segment within the coiled-coil domain could keep the CAD/SOAR domain of STIM1 in an inactive state and it is able to act as a decoy in the context of Orai1 activation.
A polybasic domain within CAD/SOAR is required for Orai1 activation
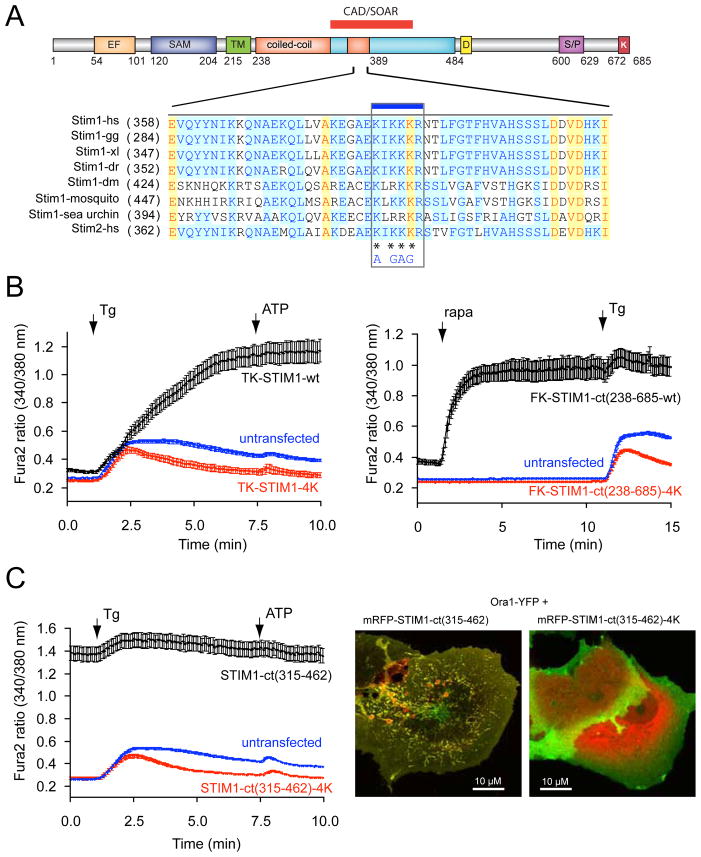
The importance of the acidic stretch in keeping STIM1 in an inactive state presumably by masking the CAD/SOAR domain, made it logical to assume that the segment within the CAD/SOAR domain important for both the intramolecular interaction and Orai1 activation has a basic character. We identified such a sequence in the middle of the CAD/SOAR domain (382–386) that is highly conserved in the STIM orthologues (Fig. 4A). When four of the basic residues within the CAD/SOAR were mutated to AGAG within either our recruitable STIM1-ct or in the full-length STIM1 molecule, they were found unable to activate Ca2+ influx via the Orai1 proteins (Fig. 4B). Also, the constitutively active wild-type 315–462 fragment that causes a strong constitutive activation of the expressed Orai1 protein (19,21) was unable to do so with the 4K (AGAG) mutation (Fig. 4C). These experiments showed that indeed the basic region within the CAD/SOAR domain is necessary for Orai1 activation. While this manuscript was in preparation, the same basic residues were reported as critical for Orai1 activation by Calloway et al. (25).
Figure 4.
A polybasic segment within the CAD/SOAR domain is responsible for Orai1 activation. (A) sequence alignment between various STIM1 orthologues and STIM2 within the CAD/SOAR domain (gg, Gallus gallus; xl, Xenopus laevis; dr, danio rero; dm, Drosophila melanogaster. The mutated Lys residues are labeled with an asterisk. (B) The 4K mutation renders full-length STIM1 proteins inactive. COS-7 cells were transfected with the thimidine-kinase (TK) driven STIM1 (black) or its 4K mutant (red) together with untagged Orai1 and their cytoplasmic Ca2+ monitored by Fura2. Thapsigargin (Tg, 200 nM) was added to empty the Ca2+ stores. Means ± S.E.M. are shown (n = 135, 132 and 160 cells for black, red and blue traces, respectively, obtained in 4–5 separate experiments). (C) The same 4K mutation makes the STIM1-ct fragment inactive not responding to clustering with Ca2+ increase (red). Means ± S.E.M. are shown (n = 170, 189 and 239 cells for black, red and blue traces, respectively obtained in 6–7 separate experiments). (D) The 4K(AGAG) mutation inactivates the otherwise constitutively active STIM1-ct(315–462) segment. Means ± S.E.M. are shown (n = 188, 163 and 244 cells for black, red and blue traces, respectively, obtained in 5–7 separate experiments). (E) The constitutively active STIM1(315–462) piece causes clustering and co-localization with Orai1 in COS-7 cells. However, the 4K mutation prevents clustering of Orai1 and the STIM1 construct is largely remains in the cytosol. Confocal images of live cells are shown one day after transfection.
DISCUSSION
In search of an autoinhibitory domain that keeps the STIM1-ct in a quiescent state, we identified an inhibitory acidic segment within the first coiled-coil domain of the STIM1 molecule. Mutation of the acidic residues within this domain rendered STIM1 fully constitutively active both in its native full-length form and as a STIM1-ct tail. To our knowledge, this is the first fully constitutively active STIM1 molecule other than the D76A mutant. Importantly, while the D76A mutation triggers the natural STIM1 activation process by decreasing the Ca2+ affinity of the luminal EF-hand domain (4), the 4EA mutant bypasses this step and becomes active even when its luminal domain is Ca2+ bound.
Our attention to this region of STIM1 was drawn by its significant homology with the C-terminal helical tail of the Orai1. This raised the possibility that this segment may mediate an intramolecular interaction with the CAD/SOAR domain of STIM1 as the latter was shown to interact and activate the Orai1 channels (19,20). We hypothesize that STIM1 molecules bearing neutralizing mutations within this region (4EA) are unable to keep the STIM1-ct in an inactive state allowing unimpeded access of the CAD/SOAR domain to the Orai1 channels. Together with the importance of the basic stretch within the CAD/SOAR domain in Orai1 activation, shown here and also reported recently (25), these findings would be consistent with a mechanistic model in which the positive residues of CAD/SOAR form an intramolecular interaction with the acidic segment in the quiescent state of STIM1 molecules. This intramolecular silencing would then be interrupted during normal STIM1 activation and is disrupted in the 4EA mutant form. The high degree of similarity between the acidic inhibitory segment of STIM1 and the C-terminal tail of Orai1, gives support for this model, which is reminiscent of how certain protein kinses are kept inactive by their own pseudosubstrate sequences (28). Direct evidence to support this model, however, would require demonstration of an interaction between the acidic and basic segments of STIM1, which has so far been unsuccessful in our hand when using separately expressed domains. Nevertheless it is likely that this interaction has significant structural constraints as it depends on a conformation that is regulated by luminal Ca2+ binding of STIM1 (likely being transduced by oligomerizaion). Also, the isolated CAD domain already forms a dimer (19), which may hampers its interaction with the STIM1 acidic region. More studies are in progress to address this question with recombinant STIM1-ct molecules.
An alternative explanation for the constitutive activity of the 4EA STIM1 mutants could be a simple disruption of the structure of the first coiled-coil domain thereby causing the unmasking of CAD/SOAR domain. Theoretically, the 4EA mutation could even cause the clustering of STIM1 and trigger the activation process that way. However, unlike the D76A mutant the 4EA or 7EA mutant STIM1 molecules did not show massive clustering without the expression of Orai1 molecules and still underwent clustering after store depletion arguing against clustering or even a major structural distortion within the coiled-coil region. Moreover, a major structural defect would more likely render STIM1 inactive then making it constitutively active.
An important question that remains to be answered is how the luminal ER Ca2+ decrease leads to the unmasking of the CAD domain. A major conformational change within the isolated luminal EF hand and SAM domains have been demonstrated upon Ca2+ unbinding (13), and oligomerization of full-length STIM1 in response to Ca2+ depletion has been well documented by FRET analysis (29). The role of the various STIM domains in this process was thoroughly studied in a recent study by Covington et al.(30). These authors showed that overexpressed STIM1 molecules could form oligomers even in the resting state but only when they contain the STIM-ct. A STIM1 mutant lacking the entire cytoplasmic domain showed no basal oligomers but still responded to ER Ca2+ depletion with oligomerization. Most importantly, Covington et al. also found that STIM1 truncated after the CC1 domain promoted a constitutive oligomerization that was unresponsive to Ca2+ depletion unless the CAD domain was added. This study clearly established an important role of the CAD domain in regulating oligomerization with data to suggest that a communication exists between the CC1 and CAD domains. In this context it is noteworthy that the 4K mutant STIM1 failed to show clustering after Ca2+ store depletion in our experiments (data not shown). Our rapamycin-inducible conformational change in STIM1-ct suggests that forcing STIM1 molecules to tightly line up with an orderly fashion (their N-termini being fixed) is sufficient to mimic their clustered state. The possibility was considered that a mechanical stretching of STIM1-ct during the ER-PM bridge formation contributes to its activation. While such a mechanism cannot be ruled out (even for the natural activation process), the fact that we observed similar activation with the mitochondrial anchor, which probably does not represent the same pulling force, and with a significantly longer [9x-helical linker, see (26)] ER-targeted FRB construct (not shown), makes the mechanical pulling a less likely explanation.
Lastly, the similarity between the STIM1 acidic domain and the Orai1 C-tail drew attention to the importance of the acidic residues in the latter during Orai1 activation. We mutated the two DH residues located at the distal end of this acidic cassette in Orai1 and saw no major difference in its activation (not shown). However, Calloway et al. showed that removal of all Glu and Asp residues within this cassette was needed to eliminate Tg-induced Ca2+ influx (24). Their recent FRET studies with the polybasic domain mutant STIM1 and acidic mutant Orai1 molecules indicated that the putative electrostatic interaction was important for Orai1 activation but the two molecules were still able to show FRET when these charges were neutralized (25). Clearly, more studies are needed to fully understand the molecular sequences that transmit the ER luminal Ca2+ change to the Orai1 channels.
Autoinhibitory segments in the cytosolic STIM1 different from the one identified in this study have already been described in three separate studies (19,21,31). One such region is located between residues 470–490 of STIM1 in the two former studies, and between 445–475 in the latter. In our present studies, deletion of this region in the STIM1-ct constructs (as in the 238–463 STIM1 piece) also yielded higher constitutive activity in a number of cells as already described in (19,21). A recent elegant study, analyzing the direct interaction between recombinant cytosolic STIM1 pieces and Orai1 channels expressed in yeast, found that the 233–463 piece of STIM1 was a very poor interactor and activator of Orai1 (32), which is consistent with the transient activation by this fragment in our studies and suggests that sequences between the 463 and 502 contain sites that stabilize STIM1 Orai1 interactions. Recently, this acidic segment encoded by STIM1-(475–483) was found responsible for the fast Ca2+ dependent inactivation of Orai1 (33,34). These data together suggest that the 475–483 segment is involved in Ca2+-dependent inactivation rather than keeping the full-STIM1 molecule inactive in quiescent cells.
Several modifiers are superimposed on the basic activation mechanism of Orai1 by STIM1 suggested in this study. Drosophila STIM roughly corresponds to the minimal segment (1–499) (35) that still shows almost full Orai1 activation. Nevertheless, STIM1 truncations in the cytoplasmic region that extends beyond residue 502 have identified several other regions within STIM1 that modify the activation and inactivation pattern of ICRAC. These regions have been reviewed in detail recently (12). Also, as shown in a recent study, phosphorylation of STIM1 in regions beyond residue 482 can prevent STIM1 activation and being responsible for the inability of Tg to induce Ca2+ influx during the cell cycle (36). Therefore, more studies are needed to fully understand the complex activation mechanism of STIM1.
In summary, the present studies revealed important regions within the STIM1 molecule suggesting an autoinhibitory intramolecular interaction within the cytoplasmic segment of STIM1 molecules. Identification of an acidic region within the STIM1 coiled-coil domain that keeps STIM1 in an inactive state and a short basic region within the activation domain of STIM1 are suggestive of an intramolecular silencing mechanism. The significant similarity between the acidic inhibitory STIM1 segment and the C-terminal helical segment of Orai1 presumed to be the site of activating interaction with the STIM1 molecule gives support to this model. This mechanism of intramolecular silencing resembles the regulation of kinases by intramolecular pseudosubstrate binding and may reveal a new paradigm in Ca2+ influx regulation.
MATERIALS AND METHODS
Materials
Rapamycin and thapsigargin were purchased from Calbiochem. ATP was obtained from Sigma. Fura2/AM was from Molecular Probes (Invitrogen). All other chemicals were of the highest analytical grade.
DNA Constructs
The list of constructs and the primers used to generate them are listed in supplementary Table 1. All STIM1 constructs were based on the CMV- and TK- driven mRFP- or YFP-tagged (luminally) human STIM1 protein described in (26). In all experiments using STIM1 molecules with intact ER luminal domains the TK-driven versions were used to keep expression levels low. For the recruitable cytosolic fragments, the various STIM1 pieces obtained by PCR (using Pfu polymerase) were inserted in place of the polyphosphoinositide 5-phosphatase enzyme between PvuI and KpnI restriction sites in the mRFP-FKBP12-5-ptase construct described in (37). Mutations were introduced with the Quik Change mutagenesis Kit (Statagene). The Orai1 constructs used in this study have been described previously (26) as were the PM and ER targeted FRB constructs. The mitochondrial recruiter contained the small sequence from AKAP1 (34–63) (MAIQLRSLFPLALPGLLALLGWWWFFSRKK) described in (38). Orai1-mRFP-FKBP12 was obtained by cloning a PCR product of FKBP12 flanked with XbaI and SgrAI sites. PCR product for this cloning was generated with mRFP-FKBP12-5′phosphatase as a template [previously described in (26)]. To cut the Orai1-mRFP constructs with XbaI, plasmids were amplified and purified from a Dam- bacterial strain. All constructs were verified by dideoxy sequencing.
Cytoplasmic Ca2+ Measurements and confocal microscopy
COS-7 cells were cultured on glass coverslips (3 × 105 cells/35-mm dish) and transfected with the indicated constructs (0.5μg of DNA/dish, each) using Lipofectamine 2000 for 24 h as described previously (26). For calcium experiments cells were loaded with Fura2/AM (3 μM) for 45 min in HEPES-buffered M199, Hanks’ salt solution containing 0.1% bovine serum albumin, 0.06% Pluronic acid and 200 μM sulfinpyrazone, at room temperature. Due to the Ca2+ toxicity caused by expression of the constitutively active constructs, cells were kept in a low Ca2+ (0.2 mM) medium during transfection, which greatly increased the number of cells showing the high basal Ca2+ during the experiments. Calcium measurements with Fura2 were performed in modified Krebs-Ringer solution containing 120mM NaCl, 4.7mM KCl, 1.2mM CaCl2, 0.7mM MgSO4, 10mM glucose, 10mM Na-Hepes, pH 7.4 at room temperature using an Olympus IX70 inverted microscope equipped with a Lamda-DG4 illuminator and a MicroMAX-1024BFT digital camera and the appropriate filter sets. The MetaFluor (Molecular Devices) software was used for data acquisition and analysis. For confocal analysis, the coverslips with the transfected cells were mounted on the heated (35 °C) stage of a Zeiss 510Meta scanning confocal microscope and images were taken in at selected times before and after stimulation with the appropriate chemicals. Some experiments were performed at room temperature that allows a better assessment of the clusterization process of full-length STIM1. Images were analyzed post-acquisition with Adobe Photoshop but only linear transformations were allowed to use the full dynamic range of the 12 bit images.
FRET Measurements
COS-7 cells were co-transfected with the YFP- and mRFP-tagged versions of the various STIM1 constructs. Cells were examined in the same microscope used for Ca2+ measurements but a different dual dichroic mirror was used in the microscope and a Dual-View beam splitter (565D, 535/30 and 630/75, Photometrics) to separate the YFP-and mRFP signals. The ratio of the signals obtained in the respective channels (red/green) using 488 excitation was used as an indication of a FRET change after stimulation. Some FRET experiments were done in cell suspension using a PTI fluorescence Spectrophotometer.
Western Analysis
To determine the expression level of the individual constructs, cells were transfected as described above and the next day they were washed with PBS and lysed in Laemmli buffer. After boiling and a brief sonication, lysates were subjected to SDS PAGE using 8–12% precast gradient gels (Novex) and transferred to Nitrocellulose membranes. A rat monoclonal anti mRFP antibody (5F8) (ChromoTek GmbH, Martinsried, Germany) was used to visualize the expressed proteins using the Odyssey infrared detection system. These results are shown in Fig. S5.
Supplementary Material
Acknowledgments
Confocal imaging was performed at the Microscopy & Imaging Core of the National Institute of Child Health and Human Development, NIH with the kind assistance of Drs. Vincent Schram and James T. Russell. This research was supported in part by the Intramural Research Program of the National Institute of Child Health and Human Development of the National Institutes of Health. P.V. was also supported by the Hungarian Scientific Research fund (OTKA NF-68563) and the Medical Research Council (ETT 440/2006). IMM was supported by a Fellowship from the Spanish Personnel Research Training Program from the Ministry of Science and Education of Spain.
ABBREVIATIONS
- ER
endoplasmic reticulum
- FRB
fragment of mTOR that binds FKBP12
- PM
plasma membrane
- rapa
rapamycin
- STIM1
stromal interaction molecule 1
- Tg
thapsigargin
Footnotes
This manuscript has been accepted for publication in Science Signaling. This version has not undergone final editing. Please refer to the complete version of record at http://www.sciencesignaling.org/. The manuscript may not be reproduced or used in any manner that does not fall within the fair use provisions of the Copyright Act without the prior, written permission of AAAS.
REFRENCES
- 1.Putney JW., Jr A model for receptor-regulated calcium entry. Cell Calcium. 1986;7:1–12. doi: 10.1016/0143-4160(86)90026-6. [DOI] [PubMed] [Google Scholar]
- 2.Hoth M, Penner R. Depletion of intracellular calcium stores activates a calcium current in mast cells. Nature. 1992;355:353–356. doi: 10.1038/355353a0. [DOI] [PubMed] [Google Scholar]
- 3.Zweifach A, Lewis RS. Mitogen-regulated Ca2+ current of T-lymphocytes is activated by depletion of intracellular Ca2+ stores. Proc Natl Acad Sci U S A. 1993;90:6295–6299. doi: 10.1073/pnas.90.13.6295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liou J, Kim ML, Heo WD, Jones JT, Myers JW, Ferrell JE, Jr, Meyer T. STIM is a Ca2+ sensor essential for Ca2+-store-depletion-triggered Ca2+ influx. Curr Biol. 2005;15:1235–1241. doi: 10.1016/j.cub.2005.05.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Roos J, DiGregorio PJ, Yeromin AV, Ohlsen K, Lioudyno M, Zhang S, Safrina O, Kozak JA, Wagner SL, Cahalan MD, Velicelebi G, Stauderman KA. STIM1, an essential and conserved component of store-operated Ca2+ channel function. J Cell Biol. 2005;169:435–445. doi: 10.1083/jcb.200502019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Feske S, Gwack Y, Prakriya M, Srikanth S, Puppel SH, Tanasa B, Hogan PG, Lewis RS, Daly M, Rao A. A mutation in Orai1 causes immune deficiency by abrogating CRAC channel function. Nature. 2006;441:179–185. doi: 10.1038/nature04702. [DOI] [PubMed] [Google Scholar]
- 7.Vig M, Beck A, Billingsley JM, Lis A, Parvez S, Peinelt C, Koomoa DL, Soboloff J, Gill DL, Fleig A, Kinet JP, Penner R. CRACM1 Multimers Form the Ion-Selective Pore of the CRAC Channel. Curr Biol. 2006;16:2073–2079. doi: 10.1016/j.cub.2006.08.085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Prakriya M, Feske S, Gwack Y, Srikanth S, Rao A, Hogan PG. Orai1 is an essential pore subunit of the CRAC channel. Nature. 2006;443:230–233. doi: 10.1038/nature05122. [DOI] [PubMed] [Google Scholar]
- 9.Gwack Y, Srikanth S, Oh-Hora M, Hogan PG, Lamperti ED, Yamashita M, Gelinas C, Neems DS, Sasaki Y, Feske S, Prakriya M, Rajewsky K, Rao A. Hair loss and defective T and B cell function in mice lacking ORAI1. Mol Cell Biol. 2008;28:5209–5222. doi: 10.1128/MCB.00360-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Varga-Szabo D, Braun A, Kleinschnitz C, Bender M, Pleines I, Pham M, Renne T, Stoll G, Nieswandt B. The calcium sensor STIM1 is an essential mediator of arterial thrombosis and ischemic brain infarction. J Exp Med. 2008;205:1583–1591. doi: 10.1084/jem.20080302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stiber J, Hawkins A, Zhang ZS, Wang S, Burch J, Graham V, Ward CC, Seth M, Finch E, Malouf N, Williams RS, Eu JP, Rosenberg P. STIM1 signalling controls store-operated calcium entry required for development and contractile function in skeletal muscle. Nat Cell Biol. 2008;10:688–697. doi: 10.1038/ncb1731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schindl R, Muik M, Fahrner M, Derler I, Fritsch R, Bergsmann J, Romanin C. Recent progress on STIM1 domains controlling Orai activation. Cell Calcium. 2009;46:227–232. doi: 10.1016/j.ceca.2009.08.003. [DOI] [PubMed] [Google Scholar]
- 13.Stathopulos PB, Zheng L, Li GY, Plevin MJ, Ikura M. Structural and mechanistic insights into STIM1-mediated initiation of store-operated calcium entry. Cell. 2008;135:110–122. doi: 10.1016/j.cell.2008.08.006. [DOI] [PubMed] [Google Scholar]
- 14.Deng X, Wang Y, Zhou Y, Soboloff J, Gill DL. STIM and Orai: dynamic intermembrane coupling to control cellular calcium signals. J Biol Chem. 2009;284:22501–22505. doi: 10.1074/jbc.R109.018655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huang GN, Zeng W, Kim JY, Yuan JP, Han L, Muallem S, Worley PF. STIM1 carboxyl-terminus activates native SOC, I(crac) and TRPC1 channels. Nat Cell Biol. 2006;8:1003–1010. doi: 10.1038/ncb1454. [DOI] [PubMed] [Google Scholar]
- 16.Baba Y, Hayashi K, Fujii Y, Mizushima A, Watarai H, Wakamori M, Numaga T, Mori Y, Iino M, Hikida M, Kurosaki T. Coupling of STIM1 to store-operated Ca2+ entry through its constitutive and inducible movement in the endoplasmic reticulum. Proc Natl Acad Sci U S A. 2006;103:16704–16709. doi: 10.1073/pnas.0608358103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li Z, Lu J, Xu P, Xie X, Chen L, Xu T. Mapping the interacting domains of STIM1 and Orai1 in Ca2+ release-activated Ca2+ channel activation. J Biol Chem. 2007;282:29448–29456. doi: 10.1074/jbc.M703573200. [DOI] [PubMed] [Google Scholar]
- 18.Luik RM, Wang B, Prakriya M, Wu MM, Lewis RS. Oligomerization of STIM1 couples ER calcium depletion to CRAC channel activation. Nature. 2008;454:538–542. doi: 10.1038/nature07065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Park CY, Hoover PJ, Mullins FM, Bachhawat P, Covington ED, Raunser S, Walz T, Garcia KC, Dolmetsch RE, Lewis RS. STIM1 clusters and activates CRAC channels via direct binding of a cytosolic domain to Orai1. Cell. 2009;136:876–890. doi: 10.1016/j.cell.2009.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yuan JP, Zeng W, Dorwart MR, Choi YJ, Worley PF, Muallem S. SOAR and the polybasic STIM1 domains gate and regulate Orai channels. Nat Cell Biol. 2009;11:337–343. doi: 10.1038/ncb1842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Muik M, Fahrner M, Derler I, Schindl R, Bergsmann J, Frischauf I, Groschner K, Romanin C. A Cytosolic Homomerization and a Modulatory Domain within STIM1 C Terminus Determine Coupling to ORAI1 Channels. J Biol Chem. 2009;284:8421–8426. doi: 10.1074/jbc.C800229200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Muik M, Frischauf I, Derler I, Fahrner M, Bergsmann J, Eder P, Schindl R, Hesch C, Polzinger B, Fritsch R, Kahr H, Madl J, Gruber H, Groschner K, Romanin C. Dynamic coupling of the putative coiled-coil domain of ORAI1 with STIM1 mediates ORAI1 channel activation. J Biol Chem. 2008;283:8014–8022. doi: 10.1074/jbc.M708898200. [DOI] [PubMed] [Google Scholar]
- 23.Frischauf I, Muik M, Derler I, Bergsmann J, Fahrner M, Schindl R, Groschner K, Romanin C. Molecular determinants of the coupling between STIM1 and Orai channels: differential activation of Orai1-3 channels by a STIM1 coiled-coil mutant. J Biol Chem. 2009;284:21696–21706. doi: 10.1074/jbc.M109.018408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Calloway N, Vig M, Kinet JP, Holowka D, Baird B. Molecular clustering of STIM1 with Orai1/CRACM1 at the plasma membrane depends dynamically on depletion of Ca2+ stores and on electrostatic interactions. Mol Biol Cell. 2009;20:389–399. doi: 10.1091/mbc.E07-11-1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Calloway N, Holowka D, Baird B. A Basic Sequence in STIM1 Promotes Ca(2+) Influx by Interacting with the C-Terminal Acidic Coiled Coil of Orai1. Biochemistry. 49:1067–1071. doi: 10.1021/bi901936q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Varnai P, Toth B, Toth DJ, Hunyady L, Balla T. Visualization and manipulation of plasma membrane-endoplasmic reticulum contact sites indicates the presence of additional molecular components within the STIM1-Orai1 Complex. J Biol Chem. 2007;282:29678–29690. doi: 10.1074/jbc.M704339200. [DOI] [PubMed] [Google Scholar]
- 27.Amara JF, Clackson T, Rivera VM, Guo T, Keenan T, Natesan S, Pollock R, Yang W, Courage NL, Holt DA, Gilman M. A versatile synthetic dimerizer for the regulation of protein-protein interactions. Proc Natl Acad Sci U S A. 1997;94:10618–10623. doi: 10.1073/pnas.94.20.10618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kemp BE, Parker MW, Hu S, Tiganis T, House C. Substrate and pseudosubstrate interactions with protein kinases: determinants of specificity. Trends Biochem Sci. 1994;19:440–444. doi: 10.1016/0968-0004(94)90126-0. [DOI] [PubMed] [Google Scholar]
- 29.Liou J, Fivaz M, Inoue T, Meyer T. Live-cell imaging reveals sequential oligomerization and local plasma membrane targeting of stromal interaction molecule 1 after Ca2+ store depletion. Proc Natl Acad Sci U S A. 2007;104:9301–9306. doi: 10.1073/pnas.0702866104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Covington ED, Wu MM, Lewis RS. Essential role for the CRAC activation domain in store-dependent oligomerization of STIM1. Mol Biol Cell. 2010;21:1897–1907. doi: 10.1091/mbc.E10-02-0145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kawasaki T, Lange I, Feske S. A minimal regulatory domain in the C terminus of STIM1 binds to and activates ORAI1 CRAC channels. Biochem Biophys Res Commun. 2009;385:49–54. doi: 10.1016/j.bbrc.2009.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhou Y, Meraner P, Kwon HT, Machnes D, Masatsugu OH, Zimmer J, Huang Y, Stura A, Rao A, Hogan PG. STIM1 gates the store-operated calcium channel ORAI1 in vitro. Nat Struct Mol Biol. 17:112–116. doi: 10.1038/nsmb.1724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mullins FM, Park CY, Dolmetsch RE, Lewis RS. STIM1 and calmodulin interact with Orai1 to induce Ca2+-dependent inactivation of CRAC channels. Proc Natl Acad Sci U S A. 2009;106:15495–15500. doi: 10.1073/pnas.0906781106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lee KP, Yuan JP, Zeng W, So I, Worley PF, Muallem S. Molecular determinants of fast Ca2+-dependent inactivation and gating of the Orai channels. Proc Natl Acad Sci U S A. 2009;106:14687–14692. doi: 10.1073/pnas.0904664106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Williams RT, Manji SS, Parker NJ, Hancock MS, Van Stekelenburg L, Eid JP, Senior PV, Kazenwadel JS, Shandala T, Saint R, Smith PJ, Dziadek MA. Identification and characterization of the STIM (stromal interaction molecule) gene family: coding for a novel class of transmembrane proteins. Biochem J. 2001;357:673–685. doi: 10.1042/0264-6021:3570673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Smyth JT, Petranka JG, Boyles RR, DeHaven WI, Fukushima M, Johnson KL, Williams JG, Putney JW., Jr Phosphorylation of STIM1 underlies suppression of store-operated calcium entry during mitosis. Nat Cell Biol. 2009;11:1465–1472. doi: 10.1038/ncb1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Varnai P, Thyagarajan B, Rohacs T, Balla T. Rapidly inducible changes in phosphatidylinositol 4,5-bisphosphate levels influence multiple regulatory functions of the lipid in intact living cells. J Cell Biol. 2006;175:377–382. doi: 10.1083/jcb.200607116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Csordas G, Varnai P, Golenar T, Roy S, Purkins G, Schneider TG, Balla T, Hajnoczky G. Imaging interorganelle contacts and local calcium dynamics at the ER-mitochondrial interface. Mol Cell. 2010;39:121–132. doi: 10.1016/j.molcel.2010.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Soboloff J, Spassova MA, Dziadek MA, Gill DL. Calcium signals mediated by STIM and Orai proteins--a new paradigm in inter-organelle communication. Biochim Biophys Acta. 2006;1763:1161–1168. doi: 10.1016/j.bbamcr.2006.09.023. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.