Summary
The field of epigenomics has been transformed by chromatin immunoprecipitation approaches that provide for the localization of a defined protein or posttranslationally-modified protein to specific chromosomal sites. While these approaches have helped us conceptualize epigenetic mechanisms, the field has been limited by the inability to define features like the proteome and histone modifications at a specific genomic locus in an unbiased manner. We developed an unbiased approach whereby a unique native genomic locus was isolated, which was followed by high resolution proteomic identification of specifically associated proteins and histone posttranslational modifications. This Chromatin Affinity Purification with Mass Spectrometry (ChAP-MS) technique was used to specifically enrich a ~1,000 base-pair section of GAL1 chromatin under transcriptionally active and repressive conditions, and to identify the specifically bound proteins and histone posttranslational modifications. ChAP-MS should yield unprecedented insight into the regulatory mechanisms of transcription and help identify factors that epigenetically control chromatin function.
Introduction
It has long been appreciated that chromatin-associated proteins and epigenetic factors play central roles in cell-fate reprogramming of genotypically identical stem cells thorough lineage-specific transcription or repression of precise genes and large chromosomal regions (Martin, 1981; Ho and Crabtree, 2010; Rossant, 2008). However, the hierarchy of chromatin-templated events orchestrating the formation and inheritance of different epigenetic states remains poorly understood at a molecular level. Since mis-regulation of chromatin structure and post-translational modification of histones (PTMs) is linked to cancer and other epigenetic diseases (Jones and Baylin, 2007; Chi et al., 2010), it is imperative to establish new methodologies that will allow comprehensive studies and unbiased screens for participants in epigenetic mechanisms. Unfortunately, defining how chromatin regulators collectively assemble and operate on a precise region of the genome is difficult to elucidate; there are no current methodologies that allow for determination of all proteins present at a defined, small region of chromatin.
Technical challenges have precluded the ability to determine positioning of chromatin factors along the chromosome. Chromatin immunoprecipitation (ChIP) assays have been used to better understand genome-wide distribution of proteins and histone modifications within a genome at the nucleosome level (Dedon et al., 1991; Ren et al., 2000; Pokholok et al., 2005; Robertson et al., 2007; Johnson et al., 2007; Barski et al., 2007; Mikkelsen et al., 2007). However, major drawbacks of ChIP-based chromatin enrichment methods are that experiments are largely confined to examining singular histone PTMs or proteins rather than simultaneous profiling of multiple targets, the inability to determine the co-occupancy of particular histone PTMs, and that ChIP is reliant on the previous identification of the molecular target. Affinity purification approaches have been devised for the isolation of a chromatin region using an engineered recombinase excision method (Griesenbeck et al., 2003); however, these approaches were not done at a level for proteomic analysis and they do not provide a mechanism for determining the specificity of protein interactions. More recently, groups biochemically enriching for intact chromatin have reported characterization of proteins associated with large chromatin structures such as telomeres (Dejardin and Kingston, 2009) and engineered plasmids (Akiyoshi et al., 2009; Unnikrishnan et al., 2010); however, these approaches do not enrich for a small integrated genomic locus, and do not employ specialized mass spectrometric techniques to detect protein contamination in purified material.
We sought to compare differences in chromatin between the transcriptionally active and silent states of a single genomic locus, and developed a technology, called Chromatin Affinity Purification with Mass Spectrometry (ChAP-MS). ChAP-MS provides for the site-specific enrichment of a given ~1,000 base-pair section of a chromosome followed by unambiguous identification of both proteins and histone PTMs associated with this chromosome section using highly selective mass spectrometry. Using ChAP-MS, we were able to purify chromatin at the S. cerevisiae GAL1 locus in transcriptionally silent and active states. We identified proteins and combinatorial histone PTMs unique to each of these functional states, and validated these findings with ChIP. The ChAP-MS technique will greatly improve the field of epigenomics as an unbiased approach to study regulatory mechanisms on chromatin.
Results
ChAP-MS technology
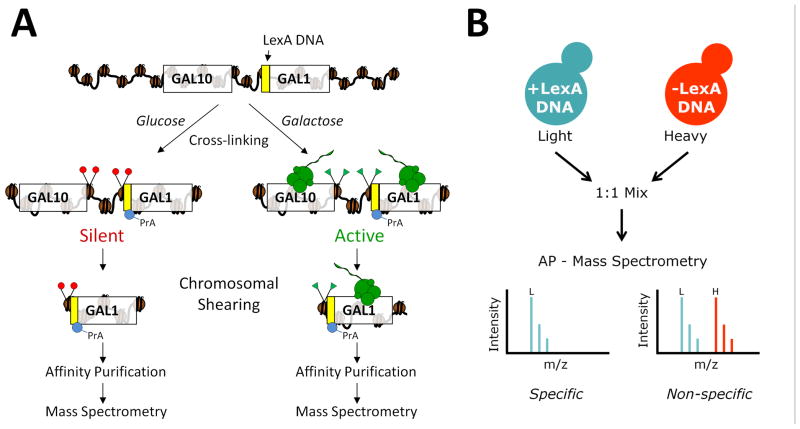
Figure 1A provides an overview of the ChAP-MS approach that was used to screen for proteins and histone PTMs associated with a specific genomic locus in transcriptionally active or repressive states. A LexA DNA binding site was engineered immediately upstream of the GAL1 start codon in a S. cerevisiae strain constitutively expressing a LexA-Protein A (LexA-PrA) fusion protein. The LexA DNA binding site directs the localization of the LexA-PrA protein affinity “handle” to the GAL1 promoter in vivo. The positioning of the LexA DNA was designed so we could specifically enrich for chromatin-associated proteins and histone PTMs regulating gene expression near the transcriptional start site of GAL1. This strain was cultured in glucose to repress gene transcription, or galactose to activate gene transcription. Following in vivo chemical cross-linking to preserve the native protein-protein interactions at GAL1 promoter chromatin, the chromatin was sheared to ~1,000 base-pair sections. The PrA moiety of the LexA-PrA fusion protein was then used to affinity purify the ~1,000 base-pair section of chromatin at the 5’-end of the GAL1 gene for high resolution mass spectrometric identification of proteins and histone PTMs. We anticipated culturing these cells in glucose would result in the isolation of proteins and PTMs correlated to silent chromatin, while culturing cells in the presence of galactose would purify histone PTMs and proteins, like RNA polymerase, that are involved with active gene transcription.
Figure 1. Chromatin affinity purification with mass spectrometry (ChAP-MS).
(A) The ChAP-MS approach provides for the specific enrichment of a given chromosome section and identification of specifically associated proteins and posttranslational modifications. A LexA DNA affinity handle was engineered just upstream of the GAL1 start codon in S. cerevisiae. Strains containing the LexA DNA binding site and a plasmid expressing LexA-PrA protein affinity handle were cultured in glucose or galactose to provide transcriptional repression or activation, respectively, and subjected to in vivo chemical cross-linking to trap protein interactions. Following shearing of the chromatin to ~1,000 bp, LexA-PrA was affinity purified on IgG-coated Dynabeads and co-enriched proteins/posttranslational modifications were identified by high resolution mass spectrometry. (B) To control for non-specifically enriched proteins, a strain lacking the LexA DNA binding site, but containing the LexA-PrA plasmid, was cultured isotopically heavy (13C615N2-lysine) in glucose or galactose and mixed equally with the corresponding isotopically light culture containing the LexA DNA binding site prior to cell lysis. Following affinity purification (AP) and mass spectrometric analysis, non-specifically enriched proteins were identified as a 1:1 ratio of light to heavy lysine-containing peptides, while proteins specifically enriched with the chromosome section were identified with a higher level of isotopically light lysine-containing peptides.
The GAL1 gene is present at one copy per haploid cell; due to the relative low abundance of the targeted chromatin region in cellular lysates, we fully anticipated that proteins non-specifically associating with GAL1 chromatin would complicate our analysis of the resulting purified material. Co-purification of non-specifically associating proteins is one of the major complications of affinity purifications, however, isotopic labeling of media provides a means to gauge in vivo protein-protein interactions and quantitate differences in peptide abundance (Smart et al., 2009; Tackett et al., 2005a). We previously developed a variation of this labeling technique called iDIRT (isotopic differentiation of interactions as random or targeted) that provides a solution for determining which co-enriched proteins are specifically or non-specifically associated with a complex of proteins (Smart et al., 2009; Tackett et al., 2005a). The iDIRT technique was adapted (as described in Figure 1B) to control for proteins non-specifically enriching with LexA-PrA and the resin. By using this adaptation of iDIRT on chromatin enriched from active and repressed chromatin states, the proteins non-specifically enriching with the isolated GAL1 chromatin section were identified. The strain containing the LexA DNA binding site and LexA-PrA fusion protein was cultured in isotopically light media, while a strain lacking the LexA DNA binding site (but still containing the LexA-PrA fusion protein) was cultured in isotopically heavy media (13C615N2-lysine). Following isolation of the cells, the light and heavy strains were mixed and co-lysed. The growth and mixing of light/heavy strains was performed separately under glucose and galactose growth conditions. The affinity purification of the GAL1 chromatin was performed from this mixture of light/heavy lysates. Proteins and histone PTMs specifically associated with the GAL1 chromatin containing the LexA DNA binding site were isotopically light as they arose from the cells grown in light media. Proteins that were non-specific to the purification were a 1:1 mix of light and heavy as they were derived equally from the light and heavy lysates. Analysis of peptides from the enriched proteins with high resolution mass spectrometry was used to determine the level of isotopically light and heavy proteins – thereby determining whether the detected protein was either a specific in vivo constituent of GAL1 chromatin or a non-specific contaminant.
Affinity purification of a specific chromosome section
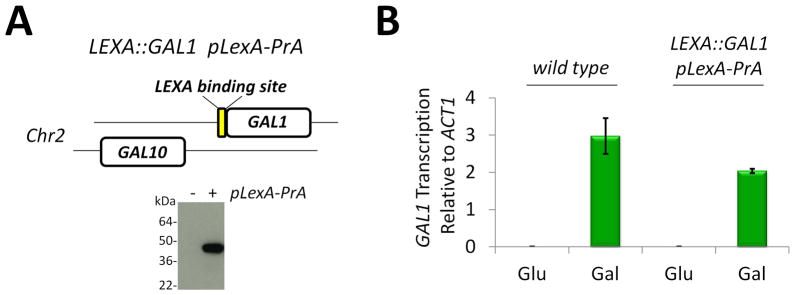
To provide for enrichment of a specific chromosome section, a DNA affinity handle was engineered at the GAL1 gene in S. cerevisiae (Fig. 2A). A LexA DNA binding site was inserted via homologous recombination just upstream of the GAL1 start codon to create strain LEXA::GAL1. To create this strain, GAL1 was genomically deleted with URA3 in the W303a background, and then the GAL1 gene was re-inserted with the upstream LEXA DNA sequence by homologous recombination. A plasmid constitutively expressing LexA-PrA was introduced into the strain to create LEXA::GAL1 pLexA-PrA (Fig. 2A). This strain provides a DNA affinity handle at the GAL1 gene and a protein affinity handle for specific enrichment. To determine if insertion of this LexA DNA binding site at GAL1 affected gene transcription, we cultured LEXA::GAL1 pLexA-PrA in glucose to repress gene transcription at GAL1, and separately in galactose to activate transcription. From these growths, we prepared cDNA and used real-time PCR to measure the activation of GAL1 transcription in the presence of galactose (Fig. 2B). Insertion of the LexA DNA binding site just upstream of the GAL1 start codon did not drastically affect the activation of gene transcription; thus, this strain was used for ChAP-MS purification of GAL1 chromatin in the transcriptionally active and silent states.
Figure 2. Introduction of a DNA affinity handle for purification of a specific chromosome section.
(A) S. cerevisiae strain LEXA::GAL1 pLexA-PrA was created by insertion of a LEXA DNA binding site upstream of the GAL1 start codon via homologous recombination. The pLexA-PrA plasmid was introduced into this strain and the constitutive expression of the LexA-PrA fusion protein was confirmed by western blotting for PrA. (B) Introduction of the LEXA DNA binding site does not impede GAL1 transcription. cDNA from wild type or LEXA::GAL1 pLexA-PrA strains grown in glucose or galactose was used as a template for real time PCR analysis of GAL1 vs ACT1 gene transcription. Error bars are the standard deviation of triplicate analyses.
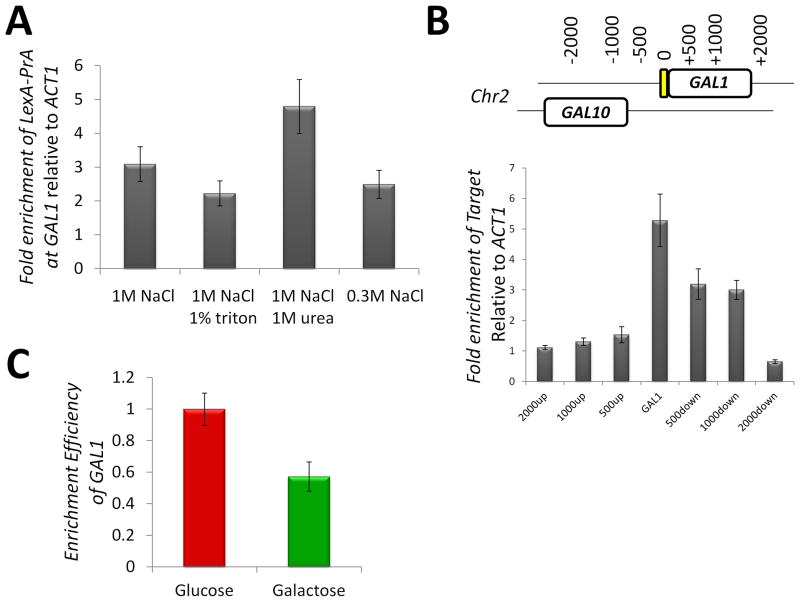
To determine the effectiveness of isolation of GAL1 chromatin, we analyzed the stringency and specificity of different purification conditions. Purification of protein complexes under increasing stringencies such as high salt levels provides for the isolation of fewer non-specifically interacting proteins (Smart et al., 2009; Taverna et al., 2006). Since the proteins purified with GAL1 chromatin will be chemically cross-linked, the stringency of the purification can potentially be quite high. Indeed, ChIP-qPCR against GAL1 showed that the PrA-based purification can survive relatively stringent conditions (Fig. 3A). From these studies, 1M NaCl and 1M urea were selected for future purifications, as these conditions are quite stringent and provide for enrichment of the GAL1 chromatin. Using an identical ChIP approach, the specificity of the GAL1 chromatin enrichment was determined (Fig. 3B). Using primers targeted to the indicated regions of chromatin surrounding GAL1, we detected that the first 1,000 base-pair section of the GAL1 gene was indeed enriched. Enrichment of GAL1 chromatin was observed at a similar level under glucose and galactose growth conditions (Fig. 3C). The slightly less efficient isolation under galactose growth conditions may reflect availability of the DNA affinity site due to alterations in chromatin structure.
Figure 3. Efficiency of GAL1 chromatin purification.
(A) The effect of buffer stringency on purification of LexA-PrA with associated chromatin was evaluated with ChIP. Strain LEXA::GAL1 pLexA-PrA was subjected to ChIP using the following buffer with the reagents indicated on the graph: 20 mM hepes, pH 7.4, 0.1% tween-20, and 2 mM MgCl2. Enrichment of GAL1 DNA relative to ACT1 DNA was monitored by real-time PCR. (B) ChIP was used to measure the specificity of enrichment of LexA-PrA bound chromatin. Enrichment was monitored by real-time PCR with primer sets at the indicated chromosomal locations. (C) GAL1 chromatin is enriched in both glucose and galactose growth conditions. The relative efficiency of GAL1 enrichment was monitored by real-time PCR with primers targeted to the “0” position in panel B and to ACT1. The standard error is indicated.
ChAP-MS analysis of transcriptionally active and silent GAL1 chromatin
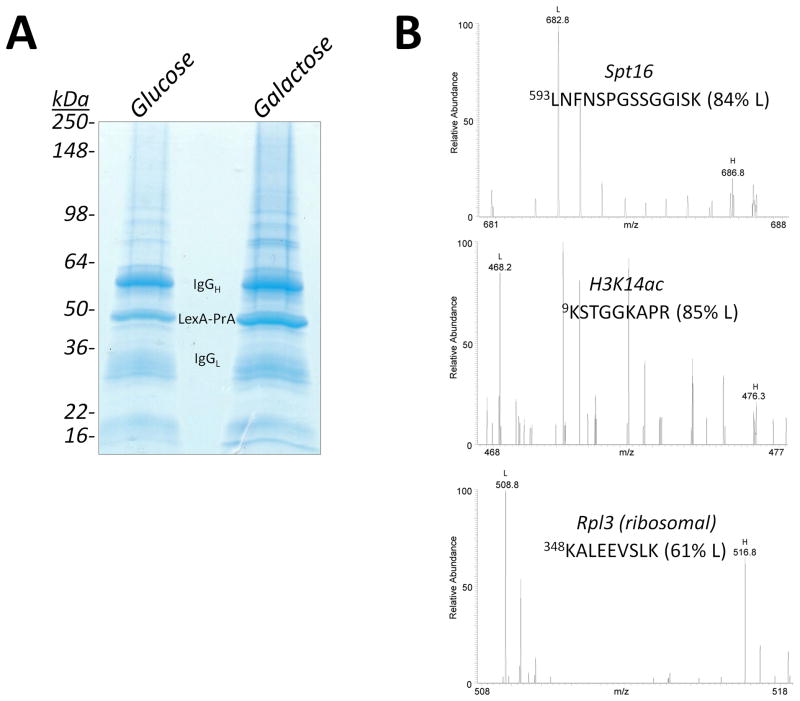
Strain LEXA::GAL1 pLexA-PrA was subjected to the ChAP-MS procedure as outlined in Figure 1. Strain LEXA::GAL1 pLexA-PrA was grown in isotopically light media, while strain pLexA-PrA was grown in isotopically heavy media. Following growth of each strain to mid-log phase, the cells were treated with 1.25% formaldehyde to trap protein interactions on the chromosomes. We have recently published a detailed analysis of the amount of formaldehyde cross-linking required to preserve the in vivo state of chromatin during affinity purifications (Byrum et al., 2011a; Byrum et al., 2011b). Approximately 2.5 × 1011 LEXA::GAL1 pLexA-PrA cells were mixed with an equivalent amount of isotopically heavy pLexA-PrA cells (separately for media containing glucose and galactose) and then subjected to lysis under cryogenic conditions with a Retch MM301 ball mill (Tackett et al., 2005a). Lysates were suspended in 20 mM hepes, pH 7.4, 0.1% tween-20, 1 M NaCl, 1 M urea, and 2 mM MgCl2. Lysates were then subjected to sonication and chromatin was sheared to sections of ~1,000 base-pairs. LexA-PrA was collected on IgG-coated Dynabeads and proteins co-enriching were resolved by SDS-PAGE and visualized by Coomassie-staining (Fig. 4A). Gel lanes were sliced into 2 mm sections and subjected to in-gel trypsin digestion (Smart et al., 2009; Tackett et al., 2005a; Tackett et al., 2005b). Peptides from proteins were identified by high resolution mass spectrometry with a Thermo Velos Orbitrap mass spectrometer equipped with a Waters nanoACQUITY UPLC system. Proteins and PTM-containing peptides were identified and the level of isotopically light to heavy peptide was calculated with Mascot Distiller (Smart et al., 2009). Representative spectra are shown in Figure 4B. Major bands observed in the gel lanes correspond to the affinity purification protein LexA-PrA and IgG chains as anticipated. Other proteins identified correspond to specifically and non-specifically enriched proteins. Supplemental Tables S1–4 list the proteins identified and percent isotopically light peptides (352 proteins from the glucose ChAP-MS and 399 proteins from the galactose ChAP-MS).
Figure 4. ChAP-MS analysis of GAL1 chromatin.
(A) Enrichment of GAL1 chromatin under transcriptionally repressive glucose and active galactose growth conditions. Strain LEXA::GAL1 pLexA-PrA was grown in either glucose or galactose and subjected to affinity purification of GAL1 chromatin via LexA-PrA as detailed in Figure 1. Addition of an equivalent amount of isotopically heavy (13C615N2-lysine) cells lacking the LexA DNA binding site provided for the identification of proteins specifically enriched with GAL1 chromatin. Proteins co-enriching with LexA-PrA were resolved by SDS-PAGE and visualized by Coomassie-staining. Each gel lane was sliced into 2 mm sections. Gel slices were treated with trypsin and resulting peptides were analyzed by high resolution mass spectrometry. (B) Representative high resolution mass spectra from proteins and histone posttranslational modifications identified from the purification of transcriptionally active GAL1 chromatin.
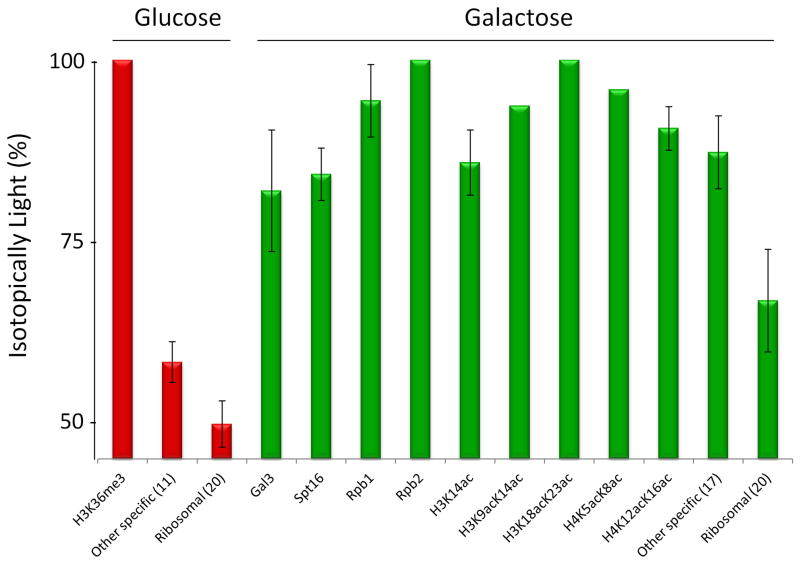
Once proteins were identified, a baseline was established for non-specifically associated proteins in accordance to our previously published iDIRT approach (Smart et al., 2009). Non-specifically enriching ribosomal proteins were used to establish the non-specifically associating baseline (Smart et al., 2009). The average percent isotopically light peptides from 20 ribosomal proteins from the glucose and galactose growth conditions were used to establish this non-specifically associating baseline (Supplemental Table S5). This resulted in a non-specifically associating baseline of 49.93 ± 2.12 % light for the glucose ChAP-MS and 66.8 ± 7.1 % light for the galactose ChAP-MS (Fig. 5). Proteins were categorized as specifically associating with GAL1 chromatin if the percent light was greater than two standard deviations above the ribosomal level (Smart et al., 2009). Figure 5 shows the proteins and histone PTMs specifically enriched with GAL1 chromatin under glucose and galactose growth conditions. Supplemental Tables S6 & S7 list proteins that were identified as specifically enriched in both the glucose and galactose ChAP-MS analyses. Specifically enriched proteins or histone PTMs known to be involved in transcriptional regulation are listed in Figure 5. For the glucose and galactose ChAP-MS analyses, 11 and 17 (respectively) additional proteins were detected as specifically enriched (Fig. 5, Supplemental Table S6 & S7). These additional proteins are abundant metabolic and heat shock proteins that are typical contaminants and false positives for this study. However, narrowing down 352 proteins identified from the glucose ChAP-MS and 399 proteins from the galactose ChAP-MS to 12 proteins and 27 proteins/PTMs specifically enriched produced a short list of candidates that was easily validated.
Figure 5. Proteins and histone posttranslational modifications enriched with GAL1 chromatin.
Proteins and histone posttranslational modifications identified from the ChAP-MS analysis of GAL1 chromatin in the transcriptionally active galactose and repressive glucose growth conditions are listed in accordance to their percent isotopically light. Proteins or posttranslational modifications were considered specifically enriched with GAL1 chromatin if the percent isotopically light was 2 standard deviations from the non-specific baseline established by the average of contaminant ribosomal proteins. Other proteins shown to be specifically enriched, but not correlated to gene transcription, are averaged together and listed individually in Supplemental Tables S6 & S7. The number of proteins averaged is shown in parentheses. The standard deviation is indicated.
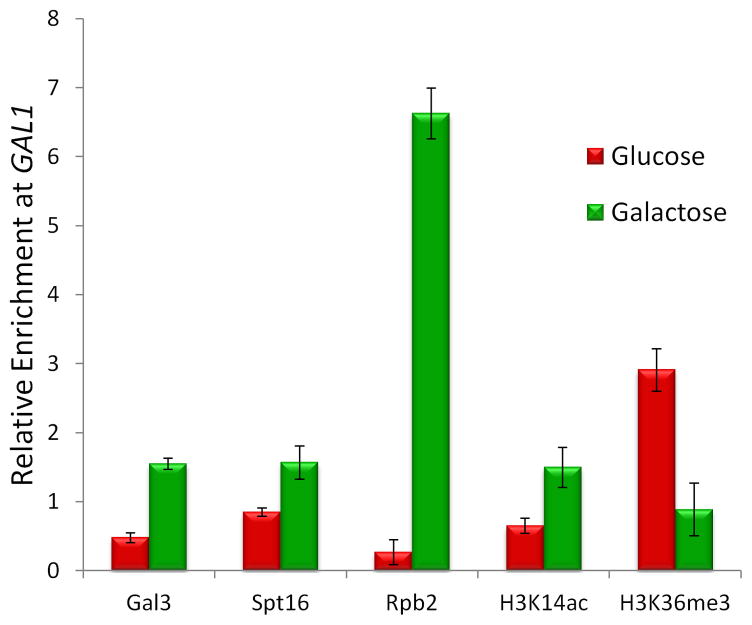
The ChAP-MS analyses of GAL1 chromatin revealed association of Gal3, Spt16, Rpb1, Rpb2, H3K14ac, H3K9acK14ac, H3K18acK23ac, H4K5acK8ac and H4K12acK16ac under transcriptionally active conditions; while transcriptionally repressive conditions showed the enrichment of H3K36me3. In order to validate our approach, standard ChIP was performed to specific interactions detected in the transcriptionally active and silent chromatin state at GAL1 (Fig. 6). These ChIP experiments validated the proteins and PTMs found associated with the transcriptionally active and repressed states of GAL1 chromatin determined from the ChAP-MS approach.
Figure 6. Validation of proteins and histone posttranslational modifications on GAL1 chromatin.
ChIP was targeted to Gal3-TAP, Spt16-TAP, Rpb2-TAP, H3K14ac and H3K36me3 under transcriptionally active galactose and repressive glucose growth conditions. ChIP to general H3 was used as a nucleosome occupancy control for each histone posttranslational modification ChIP. Enrichment of the 5’-end of GAL1 DNA relative to ACT1 DNA was monitored by real-time PCR. The standard error is indicated.
Discussion
The chromatin biology and epigenomics research communities have been limited to biased technologies that restrict targeted genome localization studies to previously identified proteins or histone PTMs. Here we have described a novel technology called Chromatin Affinity Purification with Mass Spectrometry (ChAP-MS) that circumvents this limitation by providing for isolation of a ~1,000 base-pair section of a chromosome for proteomic identification of specifically bound proteins and PTMs. In essence, the ChAP-MS approach allows one to take a “molecular snapshot” of chromatin dynamics at a specific genomic locus. Furthermore, employing this approach to target other chromatin regions will likely provide unprecedented insight on a variety of epigenetic regulatory mechanisms, chromatin structure, and genome metabolism.
Validation of the ChAP-MS approach
The ChAP-MS approach was validated on the well-studied GAL1 locus in S. cerevisiae. The GAL1 gene is activated for gene transcription in the presence of galactose, while glucose represses transcription. Accordingly, we rationalized that a purified ~1,000 base-pair section of chromatin at the 5’-end of the GAL1 gene from cells grown in galactose would contain histone PTMs correlated with active transcription and cellular machinery necessary for transcription, while the same chromatin section from cells grown in glucose would be enriched with histone PTMs associated with transcriptional repression. Prior publications have documented that H3 acetylation is enriched on the 5’-end of the active galactose-induced GAL1 gene, while in the presence of glucose it contains H3K36me3 (Shukla et al., 2006; Houseley et al., 2008). Our results with ChAP-MS support each of these prior findings (Fig. 5). Furthermore, we identified the presence of doubly acetylated histones (H3K9acK14ac, H3K18acK23ac, H4K5acK8ac, H4K12acK16ac) during transcriptional activation. This demonstrates how ChAP-MS can be used to study the combinatorial “code” of histone modifications at given chromosome regions without the need for prior identification of PTMs, PTM-specific antibodies, or sequential chromatin pull-downs. Considering H4K12acK16ac for example, the identification of the double acetylation is unique to the ChAP-MS approach as antibodies to this double acetylation do not exist, thus one could not have performed a biased ChIP analysis. Additionally, it has been reported that commercially available antibodies to single acetylation at H4K12 or H4K16 are cross-reactive with other H4 acetylations and that double acetylation of H4K12K16 significantly alters the specificity of the antibody to the singly acetylated sites (Bock et al., 2011) – a limitation specific to antibodies used in biased ChIP studies and not to the unbiased ChAP-MS approach that uses quantitative mass spectrometric readout. The ChAP-MS approach simultaneously identified the presence of RNA polymerase (Rpb1, Rpb2) and FACT component Spt16 (which aids in re-organizing chromatin for RNA pol activity) under these transcriptionally active conditions. Also of interest was the identification of Gal3 at actively transcribing GAL1, which has previously been shown to inhibit the repressive activity of Gal80 at the GAL10/GAL1 locus (Platt and Reece, 1998). We have demonstrated how the ChAP-MS approach can be utilized to study chromatin dynamics at GAL1 under different states of gene transcription. Of particular interest for future functional studies will be the upstream activating sequence which binds the Gal4 activator and Gal80 repressor which will allow us to better understand the events surrounding the switch from repression to activation at GAL1 and GAL10, as well as the middle and 3’ end of GAL1 to understand the processes of elongation and termination, respectively.
Utility of ChAP-MS as a general tool to study chromatin biology
The ChAP-MS technology presented here demonstrates the ability to purify a unique chromosome section on the order of 4 to 5 nucleosomes in length from an in vivo source that can subsequently be subjected to sensitive proteomic studies. ChAP-MS has numerous advantages relative to traditional ChIP – including the ability to unbiasedly detect proteins/PTMs at a specific genomic locus and the identification of combinatorial histone modifications on a single histone molecule. Furthermore, ChAP-MS only requires approximately an order of magnitude more cells relative to biased ChIP studies, which is a huge advantage if you factor in doing more than ten blind ChIP studies at a given region (chances are one would invest heavily in many antibodies for many proteins trying to guess a specifically bound protein/PTM). In this regard, ChAP-MS is a more cost effective option for characterizing specifically bound proteins and histone PTMs relative to ChIP. Future derivations of this technology will employ targeted mass spectrometric approaches for better determination of combinatorial histone PTMs as well as identification of other regulatory PTMs on non-histone proteins from these isolated sections (Taverna et al., 2007). Given the sensitivity of the mass spectrometry analysis employed and the relatively modest biological starting material, our findings also establish a framework for applying ChAP-MS to profile across entire regions of chromosomes or investigate higher eukaryotic systems. Direct adaptation of this technology in mammalian cell lines may be difficult since targeted homologous recombination presents several challenges which may necessitate affinity agents that do not require the incorporation of a DNA affinity handle. In this regard, the use of DNA sequence-specific molecules with strand invading properties like locked nucleic acids (Dejardin and Kingston, 2009) or peptide nucleic acids (Boffa et al 1995) may circumvent these problems. However, some major hurdles for using these approaches to purify a specific locus may be the efficiency of strand invasion and the challenge of targeting these moieties in conjunction with the chemical cross-linking that is required to preserve the in vivo state of chromatin (Byrum et al., 2011a; Byrum et al., 2011b). Regardless, any advances that permit ChAP-MS analysis of in vivo untagged or unaltered samples, like tissues, will undoubtedly have valuable applications for investigating altered gene transcription mechanisms in human disease states, as this technique could provide a comprehensive way to intelligently identify targets for therapeutics.
Experimental Procedures
Construction of the LEXA::GAL1 pLexA-PrA strain
The LEXA::GAL1 pLexA-PrA strain used to affinity enrich GAL1 chromatin was designed to have a LexA DNA binding site just upstream of the GAL1 start codon and contains a plasmid constitutively expressing a LexA-PrA fusion protein. In S. cerevisiae from the W303a background, the GAL1 gene was genomically replaced with URA3 using homologous recombination. Next, the GAL1 gene (+50 base-pairs up- and downstream) was PCR amplified with primers that incorporated a LexA DNA binding site (5’-CACTTGATACTGTATGAGCATACAGTATAATTGC) immediately upstream of the GAL1 start codon. This LEXA::GAL1 cassette was transformed into the gal1::URA3 strain and selected for growth with 5-fluoroorotic acid, which is lethal in URA3 expressing cells. Positive transformants were sequenced to ensure homologous recombination of the cassette to create the LEXA::GAL1 strain. A plasmid that constitutively expresses LexA-PrA fusion protein with TRP selection was created by amplification of the PrA sequence from template pOM60 via PCR and subcloning into the Sac1/Sma1 ends of the expression plasmid pLexA-C. Transforming this plasmid into the LEXA:GAL1 strain gave rise to the LEXA:GAL1 pLexA-PrA strain. Additionally, a control used in these studies was W303a S. cerevisiae transformed only with pLexA-PrA.
Cell culture
Strains LEXA:GAL1 pLexA-PrA and pLexA-PrA were grown in yeast synthetic media lacking tryptophan to mid-log phase at 30°C. LEXA:GAL1 pLexA-PrA strain growths were done with isotopically light lysine, while strain pLexA-PrA was cultured exclusively with isotopically heavy 13C615N2-lysine. For each strain, 12 liters of media containing either 2% glucose or 3% galactose were grown to yield ~5 × 1011 cells per growth condition. At mid-log phase, the cultures were cross-linked with 1.25% formaldehyde for 5 minutes at room temperature and then quenched with 125 mM glycine for 5 minutes at room temperature. Cells were harvested by centrifugation (2,500 xg) and frozen in liquid nitrogen as pellets in suspension with 20 mM hepes, pH 7.4, 1.2% polyvinylpyrrolidone (1 mL / 10 grams of cell pellet). Frozen cell pellets were mixed as follows at 1:1 cell weight ratios: (1) LEXA:GAL1 pLexA-PrA isotopically light in glucose plus pLexA-PrA isotopically heavy control in glucose (2) LEXA:GAL1 pLexA-PrA isotopically light in galactose plus pLexA-PrA isotopically heavy control in galactose. Cell mixtures were cryogenically lysed under liquid nitrogen temperature with a Retsch MM301 ball mill (Smart et al., 2009; Tackett et al., 2005a).
ChAP-MS procedure
Each of the following two cell lysates were processed for purification of GAL1 chromatin: (1) LEXA:GAL1 pLexA-PrA isotopically light in glucose plus pLexA-PrA isotopically heavy control in glucose that will be referred to as the glucose ChAP-MS and (2) LEXA:GAL1 pLexA-PrA isotopically light in galactose plus pLexA-PrA isotopically heavy control in galactose that will be referred to as the galactose ChAP-MS. Twenty grams of frozen cell lysate (~5 × 1011 cells) was used for each of the glucose and galactose ChAP-MS analyses. ChAP-MS steps were performed at 4°C unless otherwise noted. Lysates were re-suspended in 20 mM hepes, pH 7.4, 1 M NaCl, 2 mM MgCl2, 1 M urea, 0.1% tween-20 and 1% Sigma fungal protease inhibitor cocktail with 5 mL buffer per gram of frozen lysate. Lysates were subjected to sonication with a Diagenode Bioruptor UCD-200 (low setting, 30 seconds on/off cycle, 12 minutes total time) in 20 mL aliquots to yield ~1 kb chromatin fragments. Supernatants from sonicated lysates were collected by centrifugation at 2,000 xg for 10 minutes. Dynabeads (80 mg) coated with rabbit IgG were added to the lysates and incubated for 4 hours with constant agitation (Byrum et al., 2012). Dynabeads were collected with a magnet and washed 5 times with the purification buffer listed above and 3 times with 20 mM hepes, pH 7.4, 2 mM MgCl2, 10 mM NaCl, 0.1% tween-20. Washed Dynabeads were treated with 0.5 N ammonium hydroxide / 0.5 mM EDTA for 5 minutes at room temperature to elute proteins. Eluants were lyophilized with a Savant SpeedVac Concentrator. Lyophilized proteins were re-suspended in Laemmli SDS-PAGE loading buffer, heated to 95°C for 20 minutes, resolved with 4–20% tris-glycine Invitrogen pre-cast gels, and visualized by colloidal Coomassie-staining.
High resolution mass spectrometry and data analysis
Gel lanes were sliced into 2 mm sections and subjected to in-gel trypsin digestion (Byrum, 2011a; Byrum, 2011b; Byrum et al., 2012; Tackett et al., 2005b). Peptides were analyzed with a Thermo Velos Orbitrap mass spectrometer coupled to a Waters nanoACQUITY liquid chromatography system (Byrum et al., 2011b). Using a data dependent mode, the most abundant 15 peaks were selected for MS2 from a high resolution MS scan. Proteins were identified and the ratio of isotopically light/heavy lysine-containing tryptic peptide intensity was determined with Mascot and Mascot Distiller. The search parameters included: precursor ion tolerance 10 ppm, fragment ion tolerance 0.65 Da, fixed modification of carbamidomethyl on cysteine, variable modification of oxidation on methionine, and 2 missed cleavages possible with trypsin. A threshold of 95% confidence for protein identification, 50% confidence for peptide identification and at least 2 identified peptides per protein was used, which gave a 2% peptide false discovery rate. All specifically associating protein identifications and ratios were manually validated.
A baseline was established for non-specifically associated proteins with non-specifically enriched ribosomal proteins (Smart et al., 2009). The average percent isotopically light peptides from 20 ribosomal proteins from the glucose and galactose growth condition were used to establish this non-specifically associated baseline. This resulted in a non-specifically associated baseline of 49.93 ± 2.12 % light for the glucose ChAP-MS and 66.8 ± 7.1 % light for the galactose ChAP-MS. Proteins were categorized as specifically associating if the percent light was greater than two standard deviations above the ribosomal level (Supplemental Tables S6 & S7) (Smart et al., 2009). Duplicate ChAP-MS procedures showed Pearson and Spearman correlation coefficient p values of < 0.001.
ChIP and gene transcription assays
ChIP and gene transcription assays were performed as previously reported (Gradolatto, 2008; Gradolatto et al., 2009; Tackett et al., 2005b; Taverna, 2006). Assays were performed in triplicate and analyzed by real time PCR.
Supplementary Material
Highlights.
ChAP-MS provides for the isolation of a ~1 kb section of native chromatin
Proteins and histone posttranslational modifications are identified by mass spectrometry
ChAP-MS is the first approach for isolation of a single locus for proteomic analysis
ChAP-MS will provide unprecedented insight into epigenetic regulatory mechanisms
Acknowledgments
We would like to acknowledge the UAMS Proteomics Facility for mass spectrometric support and support from NIH grants R01DA025755, F32GM093614, P30GM103450, P20RR015569, and P20RR016460. We would like to thank Aaron Storey for experimental assistance; and Nathan Avaritt, Marie Burdine and Signe Larson for critical review.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Akiyoshi B, Nelson CR, Ranish JA, biggins S. Quantitative proteomic analysis of purified yeast kinetochores identified a PP1 regulatory subunit. Genes Dev. 2009;23:2887–2899. doi: 10.1101/gad.1865909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–37. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- Bock I, Dhayalan A, Kudithipudi S, Brandt O, Rathert P, Jeltsch A. Detailed specificity analysis of antibodies binding to modified histone tails with peptide arrays. Epigenetics. 2011;6:256–263. doi: 10.4161/epi.6.2.13837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boffa LC, Carpaneto EM, Allfrey VG. Isolation of active genes containing CAG repeats by DNA strand invasion by a peptide nucleic acid. Proc Natl Acad Sci U S A. 1995;92:1901–5. doi: 10.1073/pnas.92.6.1901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrum SD, Taverna SD, Tackett AJ. Quantitative analysis of histone exchange for transcriptionally active chromatin. J Clin Bioinforma. 2011a;1:17. doi: 10.1186/2043-9113-1-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrum S, Mackintosh SG, Edmondson RD, Cheung WL, Taverna SD, Tackett AJ. Analysis of Histone Exchange during Chromatin Purification. JIOMICS. 2011b;1:61–65. doi: 10.5584/jiomics.v1i1.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrum S, Smart SK, Larson S, Tackett AJ. Analysis of stable and transient protein-protein interactions. Methods Mol Biol. 2012;833:143–152. doi: 10.1007/978-1-61779-477-3_10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi P, Allis CD, Wang GG. Covalent histone modifications--miswritten, misinterpreted and mis-erased in human cancers. Nat Rev Cancer. 2010;10:457–69. doi: 10.1038/nrc2876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dedon PC, Soults JA, Allis CD, Gorovsky MA. Formaldehyde cross-linking and immunoprecipitation demonstrate developmental changes in H1 association with transcriptionally active genes. Mol Cell Biol. 1991;11:1729–33. doi: 10.1128/mcb.11.3.1729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dejardin J, Kingston RE. Purification of proteins associated with specific genomic Loci. Cell. 2009;136:175–186. doi: 10.1016/j.cell.2008.11.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griesenbeck J, Boeger H, Strattan JS, Kornberg RD. Affinity Purification of Specific Chromatin Segments from Chromosomal Loci in Yeast. Mol Cell Biol. 2003;23:9275–9282. doi: 10.1128/MCB.23.24.9275-9282.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho L, Crabtree GR. Chromatin remodeling during development. Nature. 2010;463:474–84. doi: 10.1038/nature08911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houseley J, Rubbi L, Grunstein M, Tollervey D, Vogelauer M. A ncRNA modulates histone modification and mRNA induction in the yeast GAL gene cluster. Mol Cell. 2008;32:685–695. doi: 10.1016/j.molcel.2008.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson DS, Mortazavi A, Myers RM, Wold B. Genome-wide mapping of in vivo protein-DNA interactions. Science. 2007;316:1497–502. doi: 10.1126/science.1141319. [DOI] [PubMed] [Google Scholar]
- Jones PA, Baylin SB. The epigenomics of cancer. Cell. 2007;128:683–692. doi: 10.1016/j.cell.2007.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin GR. Isolation of a pluripotent cell line from early mouse embryos cultured in medium conditioned by teratocarcinoma stem cells. Proc Natl Acad Sci U S A. 1981;78:7634–8. doi: 10.1073/pnas.78.12.7634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkelsen TS, Ku M, Jaffe DB, Issac B, Lieberman E, et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448:553–60. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platt A, Reece RJ. The yeast galactose genetic switch is mediated by the formation of a Gal4p-Gal80p-Gal3p complex. Embo J. 1998;17:4086–4091. doi: 10.1093/emboj/17.14.4086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pokholok DK, Harbison CT, Levine S, Cole M, Hannett NM, Lee TI, Bell GW, Walker K, Rolfe PA, Herbolsheimer E, et al. Genome-wide map of nucleosome acetylation and methylation in yeast. Cell. 2005;122:517–527. doi: 10.1016/j.cell.2005.06.026. [DOI] [PubMed] [Google Scholar]
- Ren B, Robert F, Wyrick JJ, Aparicio O, Jennings EG, et al. Genome-wide location and function of DNA binding proteins. Science. 2000;290:2306–9. doi: 10.1126/science.290.5500.2306. [DOI] [PubMed] [Google Scholar]
- Robertson G, Hirst M, Bainbridge M, Bilenky M, Zhao Y, et al. Genome-wide profiles of STAT1 DNA association using chromatin immunoprecipitation and massively parallel sequencing. Nat Methods. 2007;4:651–7. doi: 10.1038/nmeth1068. [DOI] [PubMed] [Google Scholar]
- Rossant J. Stem cells and early lineage development. Cell. 2008;132:527–31. doi: 10.1016/j.cell.2008.01.039. [DOI] [PubMed] [Google Scholar]
- Shukla A, Bajwa P, Bhaumik SR. SAGA-associated Sgf73p facilitates formation of the preinitiation complex assembly at the promoters either in a HAT-dependent or independent manner in vivo. Nucleic Acids Res. 2006;34:6225–32. doi: 10.1093/nar/gkl844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smart SK, Mackintosh SG, Edmondson RD, Taverna SD, Tackett AJ. Mapping the local protein interactome of the NuA3 histone acetyltransferase. Protein Sci. 2009;18:1987–1997. doi: 10.1002/pro.212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tackett AJ, DeGrasse JA, Sekedat MD, Oeffinger M, Rout MP, Chait BT. I-DIRT, a general method for distinguishing between specific and nonspecific protein interactions. J Proteome Res. 2005a;4:1752–1756. doi: 10.1021/pr050225e. [DOI] [PubMed] [Google Scholar]
- Tackett AJ, Dilworth DJ, Davey MJ, O'Donnell M, Aitchison JD, Rout MP, Chait BT. Proteomic and genomic characterization of chromatin complexes at a boundary. J Cell Biol. 2005b;169:35–47. doi: 10.1083/jcb.200502104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taverna SD, Ilin S, Rogers RS, Tanny JC, Lavender H, Li H, Baker L, Boyle J, Blair LP, Chait BT, Patel DJ, Aitchison JD, Tackett AJ, Allis CD. Yng1 PHD finger binding to histone H3 trimethylated at lysine 4 promotes NuA3 HAT activity at lysine 14 of H3 and transcription at a subset of targeted ORFs. Mol Cell. 2006;24:785–796. doi: 10.1016/j.molcel.2006.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taverna SD, Ueberheide BM, Liu Y, Tackett AJ, Diaz RL, Shabanowitz J, Chait BT, Hunt DF, Allis CD. Long-distance combinatorial linkage between methylation and acetylation on histone H3 N termini. Proc Natl Acad Sci U S A. 2007;104:2086–2091. doi: 10.1073/pnas.0610993104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unnikrishnan A, Gafken PR, Tsukiyama T. Dynamic changes in histone acetylation regulate origins of DNA replication. Nat Struct Mol Biol. 2010;17:430–437. doi: 10.1038/nsmb.1780. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.