Summary
Subcellular localization of mRNA enables compartmentalized regulation within large cells. Neurons are the longest known cells, however so far evidence is lacking for an essential role of endogenous mRNA localization in axons. Localized upregulation of importin β1 in lesioned axons coordinates a retrograde injury signaling complex transported to the neuronal cell body. Here we show that a long 3′ untranslated region (3′UTR) directs axonal localization of importin β1. Conditional targeting of this 3′UTR region in mice causes subcellular loss of importin β1 mRNA and protein in axons, without affecting cell body levels or nuclear functions in sensory neurons. Strikingly, axonal knockout of importin β1 attenuates cell body transcriptional responses to nerve injury and delays functional recovery in vivo. Thus, localized translation of importin β1 mRNA enables separation of cytoplasmic and nuclear transport functions of importins, and is required for efficient retrograde signaling in injured axons.
Introduction
Subcellular localization of mRNA is now recognized as a widespread phenomenon in both prokaryotic and eukaryotic cells (Donnelly et al., 2010; Keiler, 2011). Local translation of trafficked mRNAs may allow spatial or temporal compartmentalization of cellular responses to specific stimuli, or rapid responses to environmental or developmental signals (Andreassi and Riccio, 2009; Jung et al., 2012). Such localized regulation should be of particular importance in highly polarized cells such as neurons, where the requirement for a specific protein can be at a site that is very distant from mRNA transcription in the nucleus (Donnelly et al., 2010). For example, the requirement for a specific protein in a human peripheral axon can be at a site separated by a meter of intracellular distance from mRNA transcription in the nucleus. However, the apparent paucity of ribosomes in early studies on mature vertebrate axons established a notion that axons cannot synthesize proteins (reviewed in Twiss and Fainzilber, 2009), in contrast to the well-established roles for local protein synthesis in dendrites (An et al., 2008; Miller et al., 2002; Sutton and Schuman, 2006; Swanger and Bassell, 2011). Nonetheless, an increasing number of studies over the past decade have suggested that local translation is critical for axonal maintenance and repair (Gumy et al., 2010), especially in retrograde signaling from axonal lesion sites to neuronal cell bodies. We and others have proposed that such retrograde injury signaling is mediated by a latent complex activated upon injury by local synthesis of critical components at the axonal injury site (Rishal and Fainzilber, 2010).
Importin β1 is thought to be one of the core components of the retrograde injury signaling mechanism, and its local translation in axons was suggested as a key initiation step in formation of the complex (Hanz et al., 2003). Local translation may also allow separation of cytoplasmic transport functions from the nucleocytoplasmic transport roles of importin β1, which include enhancing the affinity of its importin α partners for nuclear localization sequences (NLS) within a cargo protein, and facilitating transport through the nuclear pore (Chook and Suel, 2011; Harel and Forbes, 2004). The essential role of importin β1 in these fundamental cellular processes was highlighted by blastocyst stage lethality in homozygous embryos from a gene trap importin β1 mouse line (Miura et al., 2006). Thus, although targeting of importin β1 in axons would provide a stringent test of the validity of local axonal translation and the contribution of importins to retrograde injury signaling and other distal cytoplasmic functions, such targeting requires separation of cytoplasmic functions of importins from their essential housekeeping roles in nucleocytoplasmic transport in cell bodies. We therefore set out to identify axon-localizing elements in importin β1 transcripts in order to generate a subcellular deletion of importin β1 in the axonal compartment. Here we identify an axon-localizing region in the 3′UTR of importin β1, and show that deleting this region enables selective depletion of importin β1 from axons without perturbing its essential cell body functions. Subcellular depletion of importin β1 from axons attenuates the cell body response to neuronal injury, and delays functional recovery in vivo. Thus, localized translation of importin β1 mRNA enables separation of cytoplasmic and nuclear transport functions of importins, and is required for efficient retrograde signaling in injured axons.
Results
A Long Isoform of Importin β 3′UTR Enables Axonal Localization
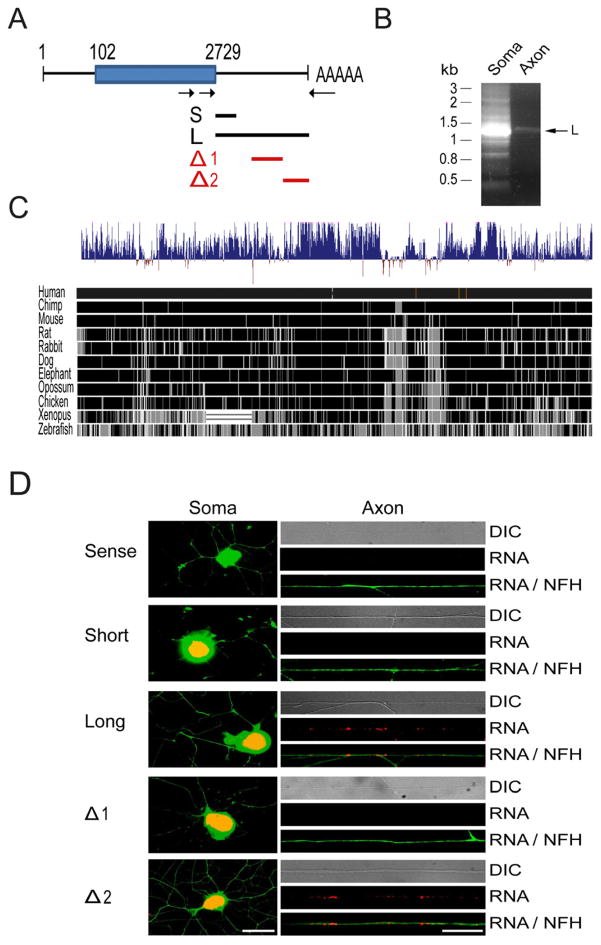
Subcellular localization and translation of mRNAs is usually determined by localization motifs in 3′UTR segments (Andreassi and Riccio, 2009; Donnelly et al., 2010), however no such motifs have been described to date for importin β1. We therefore performed nested PCRs for the 3′ end of importin β1 on cDNA prepared from adult rat DRG neuronal cell bodies versus isolated axons from insert filter cultures (Zheng et al., 2001), using forward primers at the 3′ end of the importin β1 open reading frame (ORF) and a mixture of three reverse primers anchored on polyA sequences (Figure 1A). Two major 3′UTR variants of importin β1 were obtained and sequenced, comprising a short (134 bases) isoform more prominent in cell bodies, and a long (1148 bases) isoform overlapping with the short form and more prominent in axons (Figures 1A, 1B, and S1A). These two importin β1 UTRs arise from differential usage of polyadenylation sites (Figure S1A), a widespread mechanism for defining different 3′UTRs (Proudfoot, 2011). The importin β1 UTR sequence is highly conserved in the vertebrate lineage, with 95% sequence identity between rat and mouse, and 86% identity with humans and other primates (Figure 1C).
Figure 1. 3′UTR isoforms of importin β.
(A) Schematic of the rat importin β1 transcript, with arrows indicating primers used for 3′RACE PCR. The blue box denotes the open reading frame. Lines under the schematic delineate regions subcloned for constructs containing long (L) or short (S) 3′UTR variants, or sequence regions used for deletion analysis (Δ1 and Δ2) of the long UTR.
(B) Nested 3′RACE PCR on axonal and cell body cDNA reveals different 3′UTR variants for importin β1, with a 1.15 kb L variant preferentially sorted to axons. For sequences of the two main S and L isoforms please see Genbank accession number JX096837 and Supplementary Figure S1A.
(C) Evaluation of importin β1 long 3′UTR conservation between species using the UCSC genome browser. The upper wiggle histogram summarizes conservation scores across 46 vertebrate genomes. Conserved sites with positive scores are shown in blue, while fast-evolving sites have negative scores. Pairwise alignments of 11 species to the human genome are displayed below the conservation histogram as a grey scale density plot, in which identity is shown in black.
(D) Representative images of in situ hybridization for GFP riboprobe (red) and immunohistochemistry for neurofilament (green) on cell bodies (left) and distal axon shafts (right) of rat DRG cultures transfected with the indicated importin β1 3′UTR reporter constructs. The long 3′UTR and the Δ2 region both drive axonal localization of GFP mRNA while the short 3′UTR and the Δ1 region do not. Hybridization with sense GFP riboprobes shows no signal in cell body or axons for the long UTR. Scale bars: left = 30 μm, right = 20 μm.
Despite the high sequence conservation we did not detect any known localization motifs in the importin β1 UTR sequences, and therefore set out to test their capacity to induce axonal localization of reporter genes. We first generated fusion constructs of importin β1 3′UTR segments and deletions thereof (Figure 1A) with a myristylated GFP ORF (Aakalu et al., 2001), and examined localization of GFP transcripts by in situ hybridization (ISH) on transfected DRG neurons in culture. Note that all the processes extended by adult DRG neurons in culture were previously shown to be axonal in nature (Vuppalanchi et al., 2010, Zheng et al., 2001). Axonal localization of GFP transcript was observed for the long UTR isoform, but not for the short UTR (Figure 1D). Two deletion constructs comprising the central and 3′ terminal segments of the long UTR (Δ1 and Δ2, Supplementary Figure S1B) were then tested for axon-localizing capacity of the GFP reporter. In situ hybridization on neurons transfected with these two constructs support the existence of an axonal localization motif only within the Δ2 region, hence towards the 3′ terminal segment of the long importin β1 UTR (Figure 1D).
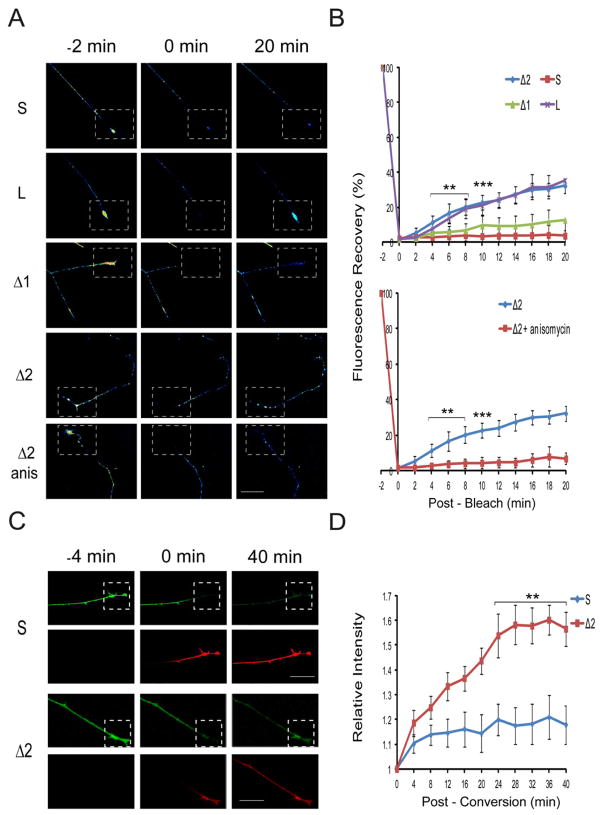
In order to further test axonal localization by a different approach, we carried out Fluorescence Recovery After Photobleaching (FRAP) experiments on axon terminals of cultured neurons transfected with importin β1 3′UTR-myrGFP fusion constructs. The myristylation domain limits diffusion for this reporter in both dendrites (Aakalu et al., 2001) and axons (Yudin et al., 2008), enabling precise visualization of local translation events. Fluorescence recovery was observed only for constructs containing the long form of importin β1 3′UTR or the 3′ end Δ2 segment, but not for the short UTR variant or the Δ1 construct (Figure 2A,B and Supplementary Figure S2). The translation inhibitor anisomycin blocked fluorescence recovery, further confirming that the reporter signal arose from locally translated axonal transcript. Additional complementary experiments with a different reporter took advantage of importin β1 3′UTR constructs fused to photoconvertible myrDendra2 (Chudakov et al., 2007). In these experiments we applied a single round of photoconversion in the region of interest and then monitored de novo appearance of Dendra2 while continuously photoconverting on proximal axonal regions to ensure that any new Dendra2 appearing in the region of interest must arise from local synthesis (Figure 2C,D and Supplementary Figure S3). The Dendra2 photoconversion experiments confirmed that the 3′ end of the long importin β1 UTR has axon-localizing capacity, as shown by FRAP and FISH with GFP reporters earlier.
Figure 2. The long importin β1 3′UTR isoform confers axon localization.
(A) Representative images from time-lapse sequences of fluorescence recovery after photobleaching (FRAP) experiments before (−2 min) and after photobleaching (0 and 20 min) of adult DRG neurons transfected with 3′ UTR constructs fused to myristylated GFP. The boxed regions indicate the area subjected to FRAP with recovery monitored over 20 min (for full time series please see Supplementary Figure S2A). Fluorescence recovery was observed only for the constructs containing the 3′ terminal segment of the long UTR isoform (L and Δ2) and this recovery was blocked upon incubation with the translation inhibitor anisomycin. Scale bar 25 μm.
(B) Quantification of fluorescence recovery over multiple time-lapse sequences with the indicated constructs. Average recoveries are shown as % of pre-bleach levels ± SD. n ≥ 6 for each series, •• denotes p < 0.01, ••• denotes p < 0.001 (Two-way ANOVA). For quantification data for additional constructs please see Supplementary Figure S2B.
(C) Representative images from time-lapse sequences of photo-conversion experiments before (−4 min) and after photo-conversion (0 and 40 min) of adult DRG neurons transfected with 3′ UTR constructs fused to myristylated Dendra2 (for full time series please see Supplementary Figure S3). Green represents unconverted or newly synthesized Dendra2; red shows photo-converted Dendra2. The boxed regions indicate the area subjected to a single round of laser-induced photo–conversion at a wavelength of 408 nm. The more proximal region was repetitively photo-converted to ensure that any green signal in the boxed region must arise from localized new synthesis of Dendra2. De novo synthesis of Dendra2 was observed for the Δ2 construct containing the 3′ end segment of the long UTR, but not for the short (S) UTR. Scale bar 25 μm.
(D) Quantification of de novo Dendra2 synthesis from fluorescence intensity in the green channel over multiple time-lapse photoconversion sequences. Average ± SEM, n = 15. ** denotes p < 0.01 (Two-way ANOVA).
Axonal Localization of Importin β1 Long 3′UTR in Transgenic Mice
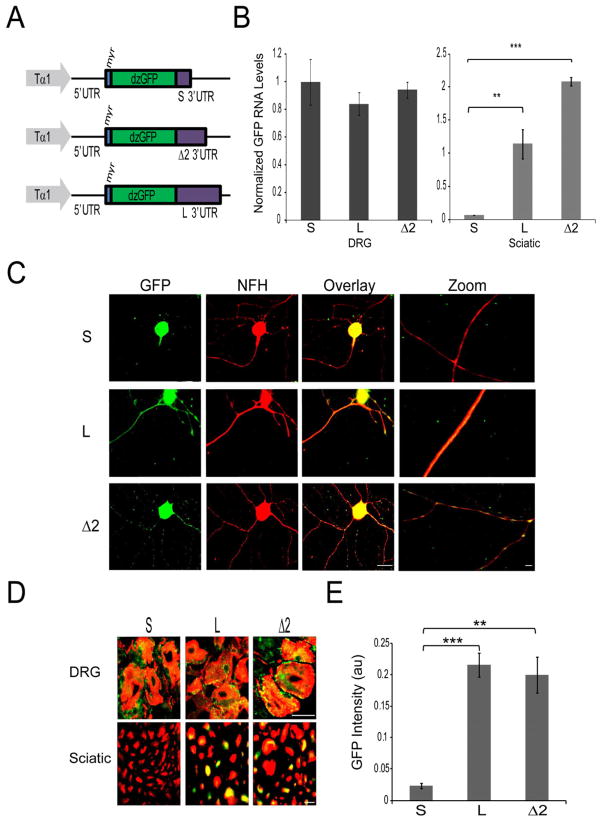
In order to test axon-localizing capacity of importin β1 3′UTRs at physiological levels of expression in vivo, we generated transgenic mice expressing either short or long UTR variants or the Δ2 region fused to myristylated GFP together with an importin β1 5′UTR segment under the control of the neuronal specific Tα1 tubulin promoter (Figure 3A), which is activated during growth and regeneration of sensory neurons (Gloster et al., 1994; Willis et al., 2011). Sensory neurons from these transgenic mice revealed differential distribution of GFP, with both cell body and axonal localization in neurons expressing reporters with the long (L) or the 3′-end fragment (Δ2) UTRs, while GFP expression was restricted to the cell body in neurons expressing the short (S) UTR reporter (Figure 3B,C). Moreover, after crush lesion of sciatic nerve in vivo, immunostaining revealed axonal GFP only in animals expressing the long or the 3′-end fragment UTRs, while no axonal expression of GFP could be detected in animals expressing the short UTR construct (Figure 3D,E and Supplementary Figure S4), despite the robust expression levels for the short UTR construct in neuronal cell bodies (Figure 3B, C and Supplementary Figure S4). Axonal expression in mouse sciatic nerve in vivo is at cm range distances from neuronal cell bodies. This fact, together with the clear differences between short and long form UTRs, strongly support active mRNA transport from the neuronal cell body and localized protein synthesis within the axon as the mechanisms involved in axonal GFP expression in these transgenic lines. Thus, the long importin β1 UTR or its 3′-end segment suffice for axonal mRNA localization in mouse sensory neurons in vivo.
Figure 3. Validation of the importin β1 axon localization region in transgenic mice.
(A) Schematic of DNA constructs used to generate transgenic mice. The neuronal specific Tα1 tubulin promoter was used to drive expression of a destabilized myristylated GFP fused to the 5′UTR of Importin β1, and different variants of Importin β1 3′UTR as indicated.
(B) Quantitative RT-PCR for GFP mRNA from adult transgenic mouse tissue extracts. β actin served as an internal control. Left – GFP mRNA levels in DRG from the different transgenic lines, normalized to short 3′UTR line; Right – GFP mRNA levels in sciatic nerve from the different transgenic lines, normalized to DRG levels for each line.
(C) Cultured adult sensory neurons from transgenic 3′UTR reporter mice (S, L, and Δ2 lines) after 48 hr in vitro. Robust GFP levels (green) are seen in neuronal cell bodies and in axons for the long and Δ2 UTR lines, but only in the cell body for the short 3′UTR line. Scale bar 20 μm, scale bar zoom panels 10 μm.
(D) Merged images of GFP (green) and immunostaining for NF-H (red) on DRG and sciatic nerve sections from short, long and Δ2 transgenic lines seven days following sciatic nerve crush. For individual channel images please see Supplementary Figure S4. Axonal GFP signals are clearly observed in sciatic nerve sections from the long and Δ2 transgenic lines, but not from the short UTR line, despite equivalent expression in neuronal cell bodies in ganglia sections. Scale bar 10 μm.
(E) Quantification of GFP intensity levels (au, arbitrary units) in sciatic nerve sections from short, long and 3′ terminal segment (Δ2) using CellProfiler image analysis software (average ± SEM) •• denotes p < 0.01, ••• denotes p < 0.001.
Subcellular Knockout shows that the Importin β1 Long 3′UTR is Required for Axonal Localization
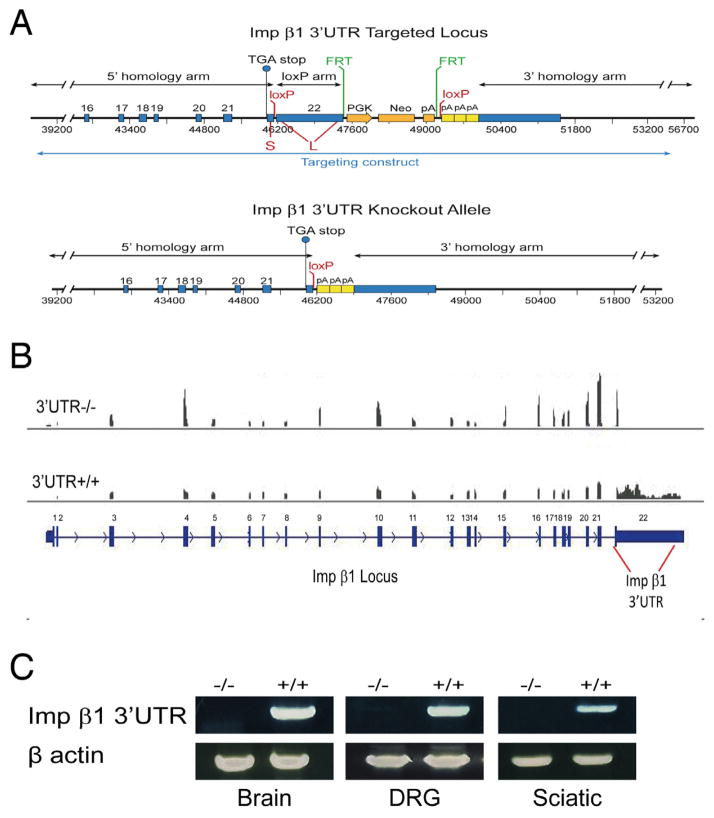
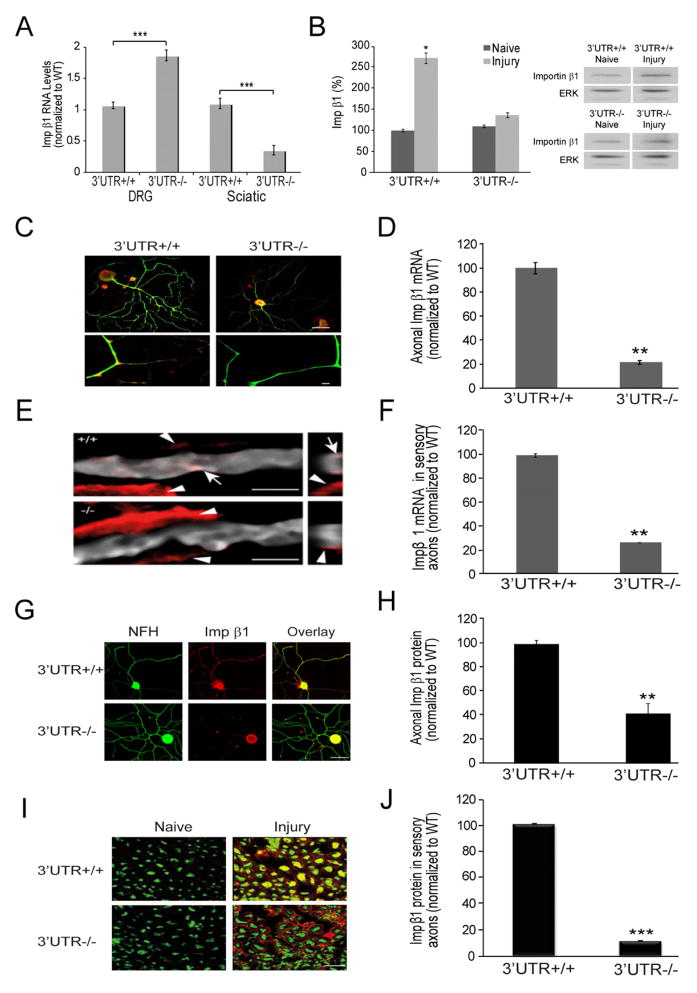
We then set out to generate a conditional knockout to determine specific functions for transcripts containing the long form of importin β1 3′UTR. A targeting construct was generated by flanking the differential sequence between short and long UTRs with loxP sites to allow for Cre-mediated deletion, with three SV40 polyA signals inserted immediately downstream of the second loxP site to ensure stability of the short UTR transcript that should be transcribed from the recombined allele (Figure 4A). Floxed allele mice were obtained, and male floxed mice were cross-bred to female PGK-Cre animals (Lallemand et al., 1998). This Cre line exhibits early and uniform expression of Cre recombinase under dominant maternal control, establishing complete recombination in all organs of the offspring. Targeted homozygous animals were viable and fertile, and complete elimination of transcripts containing the targeted 3′UTR region was confirmed in neural tissues (Figure 4B, C). Quantification of importin β1 mRNA levels revealed a significant decrease in total importin β1 mRNA in sciatic nerves of the knockout animals, with a corresponding increase in the DRG (Figures 4B and 5A). Similar results were obtained with animals in which recombination was driven by Advillin-Cre (Hasegawa et al., 2007), which is specific for sensory neurons (Supplementary Figure S5A). Importantly, these data confirm that the short UTR message transcribed from the knockout allele is robustly expressed with an apparently stable mRNA. Moreover, these results are consistent with transcript accumulation in neuronal cell bodies within the ganglia due to impaired axonal transport in the absence of the long 3′UTR.
Figure 4. Conditional targeting of the axon-localizing region in importin β1 3′UTR.
(A) Targeting strategy for importin β1 3′UTR. The loxP insertion sites are marked in red and the location of the 3′ and 5′ homology arms are in black. The PGK-neo selection cassette is inserted downstream of the region to be deleted (orange arrows) and flanked by FRT sites (green) that can be deleted using FLP recombinase. Three SV40 polyA signals are inserted immediately downstream of the floxed region (yellow boxes). For full sequences of the targeting construct and recombined locus please see Supplementary Procedures.
(B) Deep sequencing analyses on RNA from knockout and wild type DRG confirms deletion of the targeted 3′UTR region in PGK-Cre/Importin β1 3′UTR loxP mice.
(C) RT-PCR on RNA from knockout brain, DRG and sciatic nerve confirms complete deletion of the targeted 3′UTR region in PGK-Cre/Importin β1 3′UTR loxP mice.
Figure 5. The long 3′UTR is required for importin β1 localization to axons.
(A) Quantification of relative importin β1 transcript levels in adult DRG and sciatic nerve extracts of wild type and PGK-Cre targeted mice. Note the significant decrease in message levels in the knockout nerve (residual message is likely from glial component of the tissue) coupled with increase in knockout DRG, consistent with accumulation of importin β1 transcript in ganglia due to the lack of an axon-localizing element. β actin served as an internal control, average ± SEM, n=4, ••• denotes p < 0.001 (unpaired two sample t-test). For additional analyses using Adv-Cre targeted mice, please see Supplementary Figure S5.
(B) Representative images and quantification of Western blots for axoplasmic importin β1 in sciatic nerves of wild type and knockout animals before and 6 hours after injury, average ± SEM, n=3, * p < 0.05. Erk1/2 (ERK) was used as a loading control.
(C) In situ hybridization for importin β1 (red) on NF-H positive (green) sensory neurons in culture. Scale bars - 20μm upper panels, 10 μm lower axon zoom panels.
(D) Quantification of importin β1 mRNA in axons from neuronal cultures using CellProfiler image analysis software, average % intensity ± SEM, n=20, ** denotes p < 0.01.
(E) In situ hybridization for importin β1 (red) on longitudinal sections from wild type or knockout sciatic nerve. Sensory axons are identified with NF-H (grey). Arrows mark axonal transcript, while arrowheads mark signal in adjacent Schwann cells. Scale bar 5 μm.
(F) Quantification of importin β1 mRNA in axons from longitudinal sections, average % intensity ± SEM, n=3, ** p < 0.01.
(G) Immunostaining for Importin β1 (red) and NF-H (green) on adult DRG neurons in culture. Scale bar 20 μm.
(H) Quantification of Importin β1 immunoreactivity in axons from neuronal cultures using CellProfiler image analysis software, average % intensity ± SEM, n=20, •• denotes p < 0.01.
(I) Cross-sections of sciatic nerve taken 6 hr after crush lesion and immunostained for importin β1 (red) and the neuronal marker NF-H (green). Scale bar 20 μm.
(J) Quantification of axonal importin β1 immunoreactivity in the sciatic nerve cross sections using CellProfiler, average % intensity ± SEM, n=20, ••• denotes p < 0.001.
Western blotting of axoplasm extracts from naive and injured nerve showed the expected injury-induced increase in importin β1 in wild type nerve, but there was no such increase in nerves from knockout animals (Figure 5B). In situ hybridization for endogenous importin β1 mRNA on both cultured neurons and longitudinal sections from sciatic nerve (Figure 5C–F) confirmed the specific reduction of importin β1 transcript in axonal tracts but not in neuronal cell bodies or non-neuronal cells. Immunostaining of cultured neurons (Figure 5G, H) and of sciatic nerve sections after crush lesion (Figure 5I, J) revealed similar results at protein level, showing unequivocally that the upregulation of importin β1 protein in sensory axons after nerve injury is due to local translation of axonal mRNA. Importantly, the latter finding was also verified in sensory neuron specific knockouts generated with the Advillin Cre driver (Supplementary Figure S5B,C).
Axonal Knockout of Importin β1 Attenuates the Cell Body Transcriptional Response to Nerve Injury
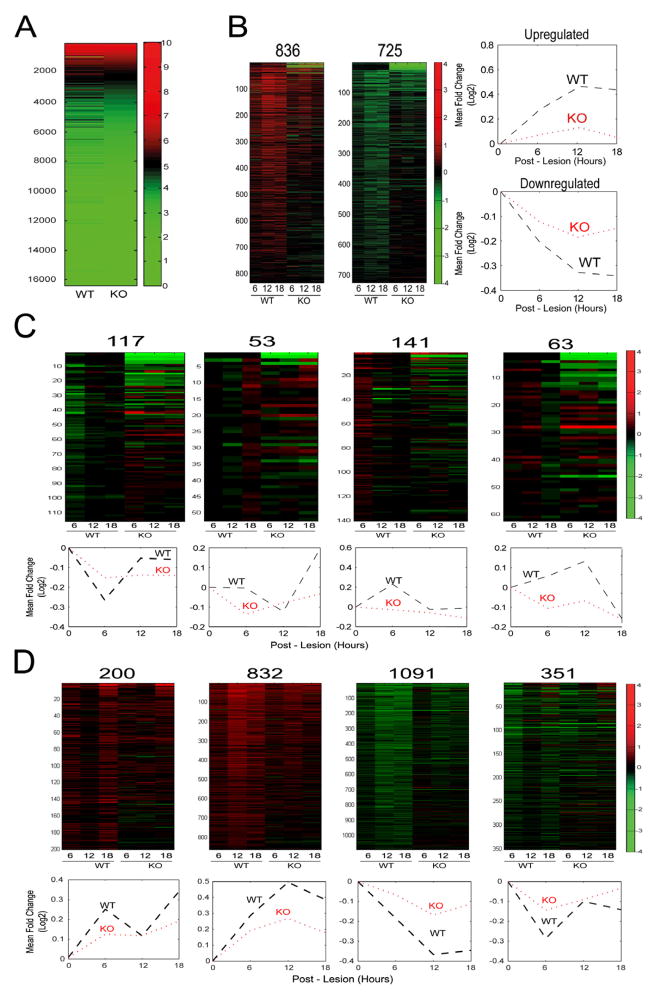
It is well established that peripheral nerve injury elicits a strong transcriptional response in the cell bodies of peripheral sensory neurons, mediated via retrograde signaling from axonal injury sites (Costigan et al., 2002; Michaelevski et al., 2010; Smith et al., 2011). Before testing the effects of axonal importin β1 knockout on the cell body response, we wanted to determine if the knockout had any effect on basal transcription profiles in the DRG. We therefore carried out RNA-seq on duplicate samples of wild type versus knockout DRG from adult PGK-Cre/Impβ1-3′UTR mice without prior nerve injury. Strikingly, the basal transcriptional profile of these sensory ganglia was essentially identical, with less than 1% of the ganglia transcriptome differing between the two genotypes (Figure 6A). These data strongly indicate that subcellular knockout of importin β1 in axons has little or no effect on nuclear functions and transcriptional output in uninjured sensory neuron cell bodies. We then carried out a comparison of gene expression in L4/L5 DRG at three time points (6, 12 and 18 hr) after sciatic nerve lesion in adult mice, using a microarray platform (Illumina, Mouse Ref v2.0) comprising approximately 25,600 well-annotated RefSeq transcripts. Similarly to the deep sequencing data from all DRG, microarrays of L4/L5 DRG showed few differences between wild type and knockout in the naive state. In contrast, there were widespread and marked differences between the two genotypes after injury, with approximately 63% of the injury-regulated transcriptome showing significantly attenuated regulation in DRG from axonal importin β1 knockout mice following sciatic nerve injury (Figure 6B-D, Supplementary Table S1). The remaining injury-regulated transcripts mostly showed similar changes in wild-type versus knockout mice (Supplementary Figure S6A), with only a small subset showing more marked regulation in knockout than in wild-type (Supplementary Figure S6B). Thus, subcellular elimination of importin β1 from axons has specific and profound effects on the cell body transcriptional response to nerve injury.
Figure 6. Lack of Axonal Importin β1 Attenuates the Cell Body Transcriptional Response to Nerve Injury.
(A) Deep sequencing data-sets from RNA isolated from knockout or wild-type DRG were analyzed with DESeq. The heat maps shown were generated using FPKM (fragments per kilobase per million sequenced reads) values for 16,383 RefSeq genes expressed in the tissue. Two biological replicates were sequenced for each sample with essentially identical results, only 154 genes out of 27,563 differed between the genotypes.
(B) Comparative L4/L5 DRG gene expression analyses between wild type and 3′UTR knockout mice after sciatic nerve lesion. RNA was extracted at three time points after injury (6, 12, and 18 hours) and analyzed using Illumina expression microarrays (n=3 for each sample). The heat maps show clusters of differentially expressed genes with log2 fold changes as indicated by the color key to the right. The traces on the right show the averaged trend of up- or down-regulation for the entire cluster for both genotypes. Numbers of genes in each cluster are indicated above the corresponding heat map. The two clusters shown in this panel are the largest (836 upregulated and 725 downregulated genes, respectively) that show highly significant (p ≤ 0.005, ANOVA) attenuation in their transcriptional regulation in knockout DRG as compared to wild type.
(C) Additional and smaller clusters of differentially expressed genes that show highly significant (p ≤ 0.005) attenuation in their transcriptional regulation in knockout DRG as compared to wild type. Numbers of genes in each cluster are indicated above the corresponding heat map.
(D) Clusters of differentially expressed genes that show significant (0.005 ≤ p ≤ 0.05) attenuation in their transcriptional regulation in knockout DRG as compared to wild type. Numbers of genes in each cluster are indicated above the corresponding heat map.
Additional gene clusters regulated by injury that did not differ significantly between knockout and wild type are shown in Supplementary Figure S6. Gene-lists for all the clusters are provided in Supplementary Table S1.
Axonal Importin β1 Knockout Delays Recovery from Nerve Injury
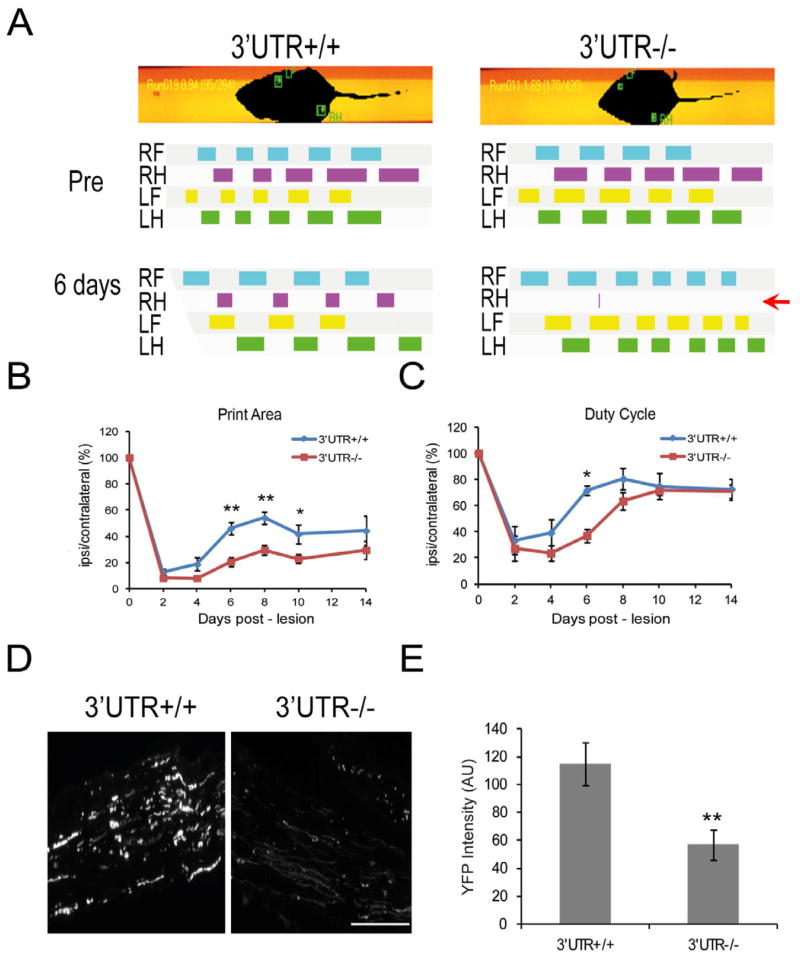
In order to determine whether the attenuated cell body response in axonal importin β1 knockouts has functional consequences for nerve regeneration, we examined the recovery profile of wild type and PGK-Cre/Impβ1–3′UTR knockout mice following crush lesion of the sciatic nerve using CatWalk gait analysis (Bozkurt et al., 2008). In this system animals are trained to cross a glass runway that enables video recording of gait and locomotion, and subsequent analyses of both dynamic and static gait parameters (Figure 7A). Behavioral consequences, recovery and outcome of injury can therefore be tested in a comprehensive manner. Mice underwent two weeks of daily training on the apparatus before injury, and were then monitored at 2–4 day intervals in the month following unilateral sciatic nerve crush in the right hind leg. There were no apparent differences in basal gait parameters between wild type and knockout mice before injury (Figure 7A). Two days after the injury there were significant reductions in both static and dynamic gait parameters for the injured limb in both genotypes (Figure 7B,C). The injured mice exhibited reductions in print area (the area of the paw that touches the surface when stepping) and in duty cycle (the participation of the limb in the walking sequence) for the injured limb. Recovery, manifested by improvement in both these parameters over the following month, was evident in both genotypes but at significantly different rates (Figure 7B,C and Supplementary Figure S7). Knockout mice exhibited a clear delay in recovery, lagging behind the wild type animals over the first 10 days after injury (Figure 7B,C) until reaching the same level of functionality in the injured limb (Supplementary Figure S7). The differences between the genotypes were most prominent at six days post-injury, when the wild type animals were already making appreciable use of the injured limb, while the knockout mice were clearly not doing so (Figure 7A, note red arrow).
Figure 7. Lack of Axonal Importin β1 Delays Recovery from Sciatic Nerve Injury.
(A) Representative images of CatWalk gait analysis (Noldus, version 9.0) with analyses of footprints and gait from wild type and PGK-Cre targeted mice before and six days after unilateral (right) sciatic nerve lesion. Six days after injury, the knockout is making less use of the right hind limb (ipsilateral to the injury) than the wild type (red arrow).
(B) Quantification of recovery time course for print area in injured right hind limb. The print area of the ipsilateral hind paw is expressed in relation to the print area of the contralateral hind paw over subsequent days (in percent). Data are expressed as average ± SEM, n=12, * denotes p < 0.05, ** denotes p < 0.01 (Two-way ANOVA). For a complete time course up until Day 26 and more extensive statistical analyses please see Supplementary Figure S7A.
(C) Quantification of recovery for the duty cycle parameter in injured right hind limb. The duty cycle of the ipsilateral hind paw is expressed in relation to the duty cycle of the contralateral hind paw over subsequent days (in percent). Data are expressed as average ± SEM, n=12, * denotes p < 0.05 (Two-way ANOVA). For a complete time course up until Day 26 and more extensive statistical analyses please see Supplementary Figure S7B.
(D) YFP-expressing sensory axons after sciatic nerve lesion. Representative images of longitudinal sections 2 mm distal to the injury site, from YFP/wild type or YFP/knockout sciatic nerve, six days after sciatic lesion. Scale bar 100 μm.
(E) Quantification of YFP levels in axonal fibers reveals significant differences between wild-type and importin β1 3′UTR null nerves six days after lesion. Average intensity ± SEM, n=6, ** denotes p < 0.01 (independent sample t-test).
In order to corroborate the behavioral recovery data we then examined axonal morphologies in sciatic nerve using PGK-Cre/Impβ1–3′UTR mice crossbred with a transgenic line that expresses YFP in sensory axons (Feng et al., 2000). We observed clear differences between wild type and knockout mice six days after crush injury, in distal segments of the nerve. Wild type nerve revealed robust axonal YFP at 2 mm distal to the crush site, while the signal in knockout nerve was much reduced (Figure 7D,E). Thus, both behavioral and histological parameters show delayed regeneration of sensory neurons that specifically lack importin β1 in the axonal compartment.
Discussion
A Central Role for Importins in Retrograde Axonal Signaling after Nerve Injury
Our results reveal a central role for locally translated importin β1 in retrograde axonal signaling after nerve injury. The cell body response to axonal injury in sensory neurons is dependent on the transport of injury signals from lesion site to soma (Rishal and Fainzilber, 2010). Three different types of signaling modalities have been suggested to act in this pathway, including growth factor and receptor complexes (Brock et al., 2010), jun kinase and associated molecules together with the adaptor Sunday Driver (Cavalli et al., 2005), and importin-dependent signals (Rishal and Fainzilber, 2010). The complexity and robustness of this system was recently emphasized by a study implicating approximately hundreds of signaling proteins and thousands of genes in the retrograde injury response in rat sciatic nerve (Michaelevski et al., 2010). The fact that axonal loss of importin β1 affects over 60% of the genes activated in the cell body response to injury is striking, and supports a major role for importin-dependent transport in the injury response mechanism, as is indeed reflected in the delayed recovery from peripheral nerve lesion seen in the knockout mice. Although injury-regulated expression of the affected genes and subsequent regeneration are not completely repressed in the importin β1 long 3′UTR knockout, the largely attenuated gene regulation and delayed functional recovery we observe most likely reflects the fact that cargo proteins can still bind importin α’s at lower affinity in the absence of importin β1 (Lott and Cingolani, 2011). Partial redundancy of multiple retrograde signaling pathways might also play a role (Abe and Cavalli, 2008; Ibanez, 2007; Michaelevski et al., 2010), and the fact that approximately one third of the injury-responsive transcripts in our arrays were regulated similarly in wild-type and knockout animals highlights the participation of both importin β1-dependent and independent pathways in retrograde injury signaling.
Axonal Protein Synthesis in Neuronal Responses
Local protein synthesis in axons has been proposed as a critical aspect of importin-dependent retrograde injury signaling. At least four components or regulators of the complex are thought to be locally translated in axons, including importin β1 itself (Hanz et al., 2003), vimentin, which acts as an accessory binding adaptor (Perlson et al., 2004; Perlson et al., 2005), Ran-binding protein 1 (RanBP1), which serves as a regulator of the complex (Yudin et al., 2008), and the cargo transcription factor STAT3 (Ben-Yaakov et al., 2012). More broadly, local translation was implicated in regenerative growth of injured axons in adulthood (Donnelly et al., 2011; Gumy et al., 2010) and in axon guidance decisions during development (Jung et al., 2012). These findings have been met skeptically in some quarters, since an apparent paucity of ribosomes in early microscopy studies had established a long-standing dogma that axons are not capable of synthesizing proteins (reviewed in Twiss and Fainzilber, 2009). Our current findings however unequivocally establish mRNA localization and local protein synthesis as functionally important mechanisms in mature axons. We identified a 3′UTR axonal localization element in importin β1 and validated it both in vitro and in vivo in transgenic mice. We then showed that targeting of this region depleted importin β1 at both mRNA and protein levels in sensory axons, without reducing its cell body levels or nuclear functions. Functional effects of this subcellular knockout on the retrograde injury response (this study), together with very recent work showing dominant negative effects of beta actin localization motifs in transgenic mice (Donnelly et al., 2011), confirm a central role for local protein synthesis in axonal regeneration in sensory neurons. Given the numerous roles suggested for local protein synthesis in axonal physiology (Donnelly et al., 2010; Jung et al., 2012), this definitive confirmation of axonal protein synthesis by a subcellular-targeted knockout will likely have broad implications beyond injury response mechanisms.
Cytoplasmic versus Nuclear Roles of Importins and Spatial Discrimination in Signaling Pathways
The highly specific effects of long 3′UTR targeting of importin β1 are all the more remarkable given its central roles in nuclear import (Chook and Suel, 2011; Harel and Forbes, 2004), and the fact that a complete knockout is lethal very early in embryogenesis, already at the blast ocyst stage (Miura et al., 2006). The pleiotropy and critical importance of importin β1 pose a significant challenge for dissecting its specific functions (Soderholm et al., 2011). Our findings provide a genetic model to discriminate between importin β1 functions in nuclear import versus roles in distal cytoplasm, and moreover suggest that cells can take advantage of mRNA localization mechanisms for multi-tasking of critical protein machineries. Export of importin β1 and other retrograde complex components out to axons as mRNAs protects them from diversion to nuclear import roles in the cell body. Moreover, maintenance of critical components of a signaling complex locally as mRNAs enables exquisite spatiotemporal control over their recruitment upon need, allowing rapid regulation of latent mechanisms. In addition to the injury-signaling roles described above, importins have been implicated in transport of plasticity signals in dendrites (Behnisch et al., 2011; Dieterich et al., 2008; Lai et al., 2008; Thompson et al., 2004), in synapse to nucleus signaling during development (Higashi-Kovtun et al., 2010; Mosca and Schwarz, 2010; Ting et al., 2007), and even in cytoplasmic transport of nucleic acid cargos in non-neuronal cells (Mesika et al., 2005; Salman et al., 2005). The mechanisms and models established in this study should be useful in future scrutiny of these additional specific roles of importins. Finally, application of similar subcellular targeting approaches to other transcripts in distal cytoplasm will help to determine the scope of contribution of mRNA transport and translation mechanisms to spatial discrimination in cellular functions.
Experimental Procedures
Animals, Preparations and Cultures
Adult (8–12 weeks old) male Wistar rats were from Harlan Laboratories (Rehovot, Israel). All mouse strains were bred and maintained at the Veterinary Resources Department of the Weizmann Institute. Sciatic nerve crush and DRG culture preparations were as previously described (Hanz et al., 2003).
Generation of Transgenic and Knockout Mice
GFP transgenic mice were generated as previously described (Willis et al., 2011) with modifications as detailed in supplementary procedures. A conditional Cre/lox mediated knockout in the 3′ UTR region of importin β1 was generated by flanking the targeted 1.1 kb sequence segment with loxP sites to allow for Cre-mediated deletion. Three SV40 polyadenylation signals were inserted immediately downstream of the floxed region in the constructs, to ensure stability of the truncated mRNA. For further details please see supplementary procedures.
Quantitative PCR (QPCR)
QPCR was performed as described (Nilsson et al., 2005) using Taqman primer kits for β-actin (normalization control), GFP and Importin β1.
Immunofluorescence
Immunostaining was carried out as previously described (Hanz et al., 2003; Perlson et al., 2005), for antibody details please see supplementary procedures.
Fluorescence In-Situ Hybridization
Antisense oligonucleotide probes for importin β1 were designed using Oligo 6 software and checked for homology and specificity by BLAST. To detect GFP mRNA in transfected DRGs, antisense and sense GFP riboprobes were generated by in vitro transcription using digoxigenin-labeled nucleotide mixture (Roche). Hybridization was performed at 55°C for 4 hours and then samples were processed for immunodetection with Cy3 conjugated mouse anti-digoxigenin (1:200; Jackson Immunoresearch). Hybridization to tissue sections was performed as previously published (Muddashetty et al., 2007), with minor modifications as detailed in supplementary procedures.
Pure Axonal Preps, RT-PCRs, and 3′ RACE RT-PCR
Isolation of DRG axons was carried out as previously described (Zheng et al., 2001). Two hundred nanograms of RNA from cell body and axons were used as template for reverse transcription and PCR. For details of reaction conditions and primers please see supplementary procedures.
Fluorescence Recovery after Photobleaching (FRAP)
Dissociated DRG cultures were transfected with reporter constructs fused to importin β1 3′ UTR axonal and cell body variants using Amaxa nucleofection. Axon terminals were subjected to FRAP as previously described (Yudin et al., 2008) with minor modifications. Prior to bleaching, neurons were imaged every 30 sec for 2 min at 15% laser power. For photobleaching, the region of interest (ROI) was exposed to 75% laser power every 1.6 sec for 40 frames. Recovery was monitored every 60 sec over 20 min at 15% laser power. To test for translation dependence, cultures were pretreated with 50 μM anisomycin for 30 min before the photobleaching sequence. FRAP quantification and statistical tests are detailed in supplementary procedures.
Photoconversion of Dendra2 Reporters
Dissociated DRG cultures were transfected with Dendra2 reporter constructs fused to importin β1 3′ UTR axonal and cell body variants using Amaxa nucleofection. Dendra2 was photoconverted using a 405 nm laser at 5% energy power and 40X oil objective for 30 sec. Images were captured every 4 min under the same conditions using 488 nm (0.1% energy) and 559 nm (4% energy) lasers. The proximal region was photoconverted using a 405 laser at 0.5% energy power for 2 min every 4 min. Dendra2 quantification and statistical tests are detailed in supplementary procedures.
Microarrays
L4/L5 DRGs were dissected from crush-lesioned or control animals at the indicated time points. Total RNA pooled from three animals was extracted using Trizol (TRI, Sigma-Aldrich). Total RNA (200ng) was amplified, labeled and hybridized on Illumina arrays (MouseRef-8 v2.0 Expression BeadChip Kit). Data analysis was performed in the R environment using Bioconductor packages (www.bioconductor.org). Briefly, log2-transformed data was normalized using quantile normalization and differential expression analysis was performed using the LIMMA package as previously described (Coppola, 2011).
Deep RNA Sequencing
Total RNA was extracted from total DRGs pooled from three adult animals per replicate, using the Trizol reagent (TRI, Sigma-Aldrich) according to manufacturer’s instructions. Replicates consisting of at least 10 μg of total RNA each were processed for RNA expression analysis (RNA-Seq) on an Illumina Genome Analyzer IIx at the High-Throughput Sequencing Unit in the Weizmann Institute of Science. RNA-Seq data was analyzed using DESeq (Anders and Huber, 2010).
Catwalk Gait analysis
CatWalk training was carried out as previously described (Deumens et al., 2007). Motivation was achieved by a combination of food restriction during the initial training and placing of palatable rewards at runway ends. Data were collected and analyzed with CatWalk software version 9.0 at days 0, 2, 4, 6, 8, 10, 14, 18, 22 and 26 post-injury. The analyzed indices are shown as a ratio between the ipsilateral (right) hind paw and contralateral (left) hind paw and are expressed as mean ± standard-error-of-the-mean (SEM). Quantification and statistical tests are detailed in supplementary procedures.
Supplementary Material
Highlights.
Knockout of a localizing UTR region selectively depletes importin β1 from axons
Axon-specific knockout of importin β1 perturbs cell body responses to axotomy
Localized axonal translation is required for efficient retrograde injury signaling
mRNA localization allows compartmentalization of distinct importin functions
Acknowledgments
We thank Erin Schuman for the myristylated GFP reporter, Freda Miller for the Tα1 tubulin promoter, and Fan Wang for the Advilin-Cre mice. We are indebted to Zehava Levy, Vladimir Kiss, Tamara Berkutzki, Golda Damari, and Dena Leshkowitz for excellent professional assistance, and to Ozgene Pty (Australia) for assistance in generating the conditional mutant mouse. This work was supported by the International Foundation for Research in Paraplegia, the Dr. Miriam and Sheldon G. Adelson Medical Research Foundation, the Minerva Foundation, the Israel Science Foundation, the Christopher and Dana Reeve Foundation, and the NIH (R01-NS041596). M.F. is the incumbent of the Chaya Professorial Chair in Molecular Neuroscience at the Weizmann Institute of Science.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aakalu G, Smith WB, Nguyen N, Jiang C, Schuman EM. Dynamic visualization of local protein synthesis in hippocampal neurons. Neuron. 2001;30:489–502. doi: 10.1016/s0896-6273(01)00295-1. [DOI] [PubMed] [Google Scholar]
- Abe N, Cavalli V. Nerve injury signaling. Curr Opin Neurobiol. 2008;18:276–283. doi: 10.1016/j.conb.2008.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- An JJ, Gharami K, Liao GY, Woo NH, Lau AG, Vanevski F, Torre ER, Jones KR, Feng Y, Lu B, et al. Distinct role of long 3′ UTR BDNF mRNA in spine morphology and synaptic plasticity in hippocampal neurons. Cell. 2008;134:175–187. doi: 10.1016/j.cell.2008.05.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11:R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreassi C, Riccio A. To localize or not to localize: mRNA fate is in 3′UTR ends. Trends Cell Biol. 2009;19:465–474. doi: 10.1016/j.tcb.2009.06.001. [DOI] [PubMed] [Google Scholar]
- Behnisch T, Yuanxiang P, Bethge P, Parvez S, Chen Y, Yu J, Karpova A, Frey JU, Mikhaylova M, Kreutz MR. Nuclear translocation of jacob in hippocampal neurons after stimuli inducing long-term potentiation but not long-term depression. PLoS One. 2011;6:e17276. doi: 10.1371/journal.pone.0017276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Yaakov K, Dagan S, Segal-Ruder Y, Shalem O, Vuppalanchi D, Willis DE, Yudin D, Rishal I, Rother F, Bader M, et al. Axonal transcription factors signal retrogradely In lesioned peripheral nerve. Embo J. 2012;31:1350–1363. doi: 10.1038/emboj.2011.494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bozkurt A, Deumens R, Scheffel J, O’Dey DM, Weis J, Joosten EA, Fuhrmann T, Brook GA, Pallua N. CatWalk gait analysis in assessment of functional recovery after sciatic nerve injury. J Neurosci Methods. 2008;173:91–98. doi: 10.1016/j.jneumeth.2008.05.020. [DOI] [PubMed] [Google Scholar]
- Brock JH, Rosenzweig ES, Blesch A, Moseanko R, Havton LA, Edgerton VR, Tuszynski MH. Local and remote growth factor effects after primate spinal cord injury. J Neurosci. 2010;30:9728–9737. doi: 10.1523/JNEUROSCI.1924-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavalli V, Kujala P, Klumperman J, Goldstein LS. Sunday Driver links axonal transport to damage signaling. J Cell Biol. 2005;168:775–787. doi: 10.1083/jcb.200410136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chook YM, Suel KE. Nuclear import by karyopherin-betas: recognition and inhibition. Biochim Biophys Acta. 2011;1813:1593–1606. doi: 10.1016/j.bbamcr.2010.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chudakov DM, Lukyanov S, Lukyanov KA. Tracking intracellular protein movements using photoswitchable fluorescent proteins PS-CFP2 and Dendra2. Nat Protoc. 2007;2:2024–2032. doi: 10.1038/nprot.2007.291. [DOI] [PubMed] [Google Scholar]
- Coppola G. Designing, performing, and interpreting a microarray-based gene expression study. Methods Mol Biol. 2011;793:417–439. doi: 10.1007/978-1-61779-328-8_28. [DOI] [PubMed] [Google Scholar]
- Costigan M, Befort K, Karchewski L, Griffin RS, D’Urso D, Allchorne A, Sitarski J, Mannion JW, Pratt RE, Woolf CJ. Replicate high-density rat genome oligonucleotide microarrays reveal hundreds of regulated genes in the dorsal root ganglion after peripheral nerve injury. BMC Neurosci. 2002;3:16. doi: 10.1186/1471-2202-3-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deumens R, Jaken RJ, Marcus MA, Joosten EA. The CatWalk gait analysis in assessment of both dynamic and static gait changes after adult rat sciatic nerve resection. J Neurosci Methods. 2007;164:120–130. doi: 10.1016/j.jneumeth.2007.04.009. [DOI] [PubMed] [Google Scholar]
- Dieterich DC, Karpova A, Mikhaylova M, Zdobnova I, Konig I, Landwehr M, Kreutz M, Smalla KH, Richter K, Landgraf P, et al. Caldendrin-Jacob: a protein liaison that couples NMDA receptor signalling to the nucleus. PLoS Biol. 2008;6:e34. doi: 10.1371/journal.pbio.0060034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donnelly CJ, Fainzilber M, Twiss JL. Subcellular communication through RNA transport and localized protein synthesis. Traffic. 2010;11:1498–1505. doi: 10.1111/j.1600-0854.2010.01118.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donnelly CJ, Willis DE, Xu M, Tep C, Jiang C, Yoo S, Schanen NC, Kirn-Safran CB, van Minnen J, English A, et al. Limited availability of ZBP1 restricts axonal mRNA localization and nerve regeneration capacity. EMBO J. 2011;30:4665–4677. doi: 10.1038/emboj.2011.347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng G, Mellor RH, Bernstein M, Keller-Peck C, Nguyen QT, Wallace M, Nerbonne JM, Lichtman JW, Sanes JR. Imaging neuronal subsets in transgenic mice expressing multiple spectral variants of GFP. Neuron. 2000;28:41–51. doi: 10.1016/s0896-6273(00)00084-2. [DOI] [PubMed] [Google Scholar]
- Gloster A, Wu W, Speelman A, Weiss S, Causing C, Pozniak C, Reynolds B, Chang E, Toma JG, Miller FD. The T alpha 1 alpha-tubulin promoter specifies gene expression as a function of neuronal growth and regeneration in transgenic mice. J Neurosci. 1994;14:7319–7330. doi: 10.1523/JNEUROSCI.14-12-07319.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gumy LF, Tan CL, Fawcett JW. The role of local protein synthesis and degradation in axon regeneration. Exp Neurol. 2010;223:28–37. doi: 10.1016/j.expneurol.2009.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanz S, Perlson E, Willis D, Zheng JQ, Massarwa R, Huerta JJ, Koltzenburg M, Kohler M, van-Minnen J, Twiss JL, et al. Axoplasmic importins enable retrograde injury signaling in lesioned nerve. Neuron. 2003;40:1095–1104. doi: 10.1016/s0896-6273(03)00770-0. [DOI] [PubMed] [Google Scholar]
- Harel A, Forbes DJ. Importin Beta; conducting a much larger cellular symphony. Mol Cell. 2004;16:319–330. doi: 10.1016/j.molcel.2004.10.026. [DOI] [PubMed] [Google Scholar]
- Hasegawa H, Abbott S, Han BX, Qi Y, Wang F. Analyzing somatosensory axon projections with the sensory neuron-specific Advillin gene. J Neurosci. 2007;27:14404–14414. doi: 10.1523/JNEUROSCI.4908-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higashi-Kovtun ME, Mosca TJ, Dickman DK, Meinertzhagen IA, Schwarz TL. Importin-beta11 regulates synaptic phosphorylated mothers against decapentaplegic, and thereby influences synaptic development and function at the Drosophila neuromuscular junction. J Neurosci. 2010;30:5253–5268. doi: 10.1523/JNEUROSCI.3739-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibanez CF. Message in a bottle: long-range retrograde signaling in the nervous system. Trends Cell Biol. 2007;17:519–528. doi: 10.1016/j.tcb.2007.09.003. [DOI] [PubMed] [Google Scholar]
- Jung H, Yoon BC, Holt CE. Axonal mRNA localization and local protein synthesis in nervous system assembly, maintenance and repair. Nat Rev Neurosci. 2012;13:308–324. doi: 10.1038/nrn3210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keiler KC. RNA localization in bacteria. Current opinion in microbiology. 2011;14:155–159. doi: 10.1016/j.mib.2011.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai KO, Zhao Y, Ch’ng TH, Martin KC. Importin-mediated retrograde transport of CREB2 from distal processes to the nucleus in neurons. Proc Natl Acad Sci U S A. 2008;105:17175–17180. doi: 10.1073/pnas.0803906105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lallemand Y, Luria V, Haffner-Krausz R, Lonai P. Maternally expressed PGK-Cre transgene as a tool for early and uniform activation of the Cre site-specific recombinase. Transgenic Res. 1998;7:105–112. doi: 10.1023/a:1008868325009. [DOI] [PubMed] [Google Scholar]
- Lott K, Cingolani G. The importin beta binding domain as a master regulator of nucleocytoplasmic transport. Biochim Biophys Acta. 2011;1813:1578–1592. doi: 10.1016/j.bbamcr.2010.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mesika A, Kiss V, Brumfeld V, Ghosh G, Reich Z. Enhanced intracellular mobility and nuclear accumulation of DNA plasmids associated with a karyophilic protein. Hum Gene Ther. 2005;16:200–208. doi: 10.1089/hum.2005.16.200. [DOI] [PubMed] [Google Scholar]
- Michaelevski I, Segal-Ruder Y, Rozenbaum M, Medzihradszky KF, Shalem O, Coppola G, Horn-Saban S, Ben-Yaakov K, Dagan SY, Rishal I, et al. Signaling to transcription networks in the neuronal retrograde injury response. Sci Signal. 2010;3:ra53. doi: 10.1126/scisignal.2000952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller S, Yasuda M, Coats JK, Jones Y, Martone ME, Mayford M. Disruption of dendritic translation of CaMKIIalpha impairs stabilization of synaptic plasticity and memory consolidation. Neuron. 2002;36:507–519. doi: 10.1016/s0896-6273(02)00978-9. [DOI] [PubMed] [Google Scholar]
- Miura K, Yoshinobu K, Imaizumi T, Haruna K, Miyamoto Y, Yoneda Y, Nakagata N, Araki M, Miyakawa T, Yamamura K, et al. Impaired expression of importin/karyopherin beta1 leads to post-implantation lethality. Biochem Biophys Res Commun. 2006;341:132–138. doi: 10.1016/j.bbrc.2005.12.151. [DOI] [PubMed] [Google Scholar]
- Mosca TJ, Schwarz TL. The nuclear import of Frizzled2-C by Importins-beta11 and alpha2 promotes postsynaptic development. Nat Neurosci. 2010;13:935–943. doi: 10.1038/nn.2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muddashetty RS, Kelic S, Gross C, Xu M, Bassell GJ. Dysregulated metabotropic glutamate receptor-dependent translation of AMPA receptor and postsynaptic density-95 mRNAs at synapses in a mouse model of fragile X syndrome. J Neurosci. 2007;27:5338–5348. doi: 10.1523/JNEUROSCI.0937-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson A, Moller K, Dahlin L, Lundborg G, Kanje M. Early changes in gene expression in the dorsal root ganglia after transection of the sciatic nerve; effects of amphiregulin and PAI-1 on regeneration. Brain Res Mol Brain Res. 2005;136:65–74. doi: 10.1016/j.molbrainres.2005.01.008. [DOI] [PubMed] [Google Scholar]
- Perlson E, Medzihradszky KF, Darula Z, Munno DW, Syed NI, Burlingame AL, Fainzilber M. Differential proteomics reveals multiple components in retrogradely transported axoplasm after nerve injury. Mol Cell Proteomics. 2004;3:510–520. doi: 10.1074/mcp.M400004-MCP200. [DOI] [PubMed] [Google Scholar]
- Perlson E, Hanz S, Ben-Yaakov K, Segal-Ruder Y, Seger R, Fainzilber M. Vimentin-dependent spatial translocation of an activated MAP kinase in injured nerve. Neuron. 2005;45:715–726. doi: 10.1016/j.neuron.2005.01.023. [DOI] [PubMed] [Google Scholar]
- Proudfoot NJ. Ending the message: poly(A) signals then and now. Genes Dev. 2011;25:1770–1782. doi: 10.1101/gad.17268411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rishal I, Fainzilber M. Retrograde signaling in axonal regeneration. Exp Neurol. 2010;223:5–10. doi: 10.1016/j.expneurol.2009.08.010. [DOI] [PubMed] [Google Scholar]
- Salman H, Abu-Arish A, Oliel S, Loyter A, Klafter J, Granek R, Elbaum M. Nuclear localization signal peptides induce molecular delivery along microtubules. Biophys J. 2005;89:2134–2145. doi: 10.1529/biophysj.105.060160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith RP, Lerch-Haner JK, Pardinas JR, Buchser WJ, Bixby JL, Lemmon VP. Transcriptional profiling of intrinsic PNS factors in the postnatal mouse. Mol Cell Neurosci. 2011;46:32–44. doi: 10.1016/j.mcn.2010.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soderholm JF, Bird SL, Kalab P, Sampathkumar Y, Hasegawa K, Uehara-Bingen M, Weis K, Heald R. Importazole, a small molecule inhibitor of the transport receptor importin-beta. ACS Chemical Biology. 2011;6:700–708. doi: 10.1021/cb2000296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutton MA, Schuman EM. Dendritic protein synthesis, synaptic plasticity, and memory. Cell. 2006;127:49–58. doi: 10.1016/j.cell.2006.09.014. [DOI] [PubMed] [Google Scholar]
- Swanger SA, Bassell GJ. aking and breaking synapses through local mRNA regulation. Curr Opin Genet Dev. 2011;21:414–421. doi: 10.1016/j.gde.2011.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson KR, Otis KO, Chen DY, Zhao Y, O’Dell TJ, Martin KC. Synapse to nucleus signaling during long-term synaptic plasticity; a role for the classical active nuclear import pathway. Neuron. 2004;44:997–1009. doi: 10.1016/j.neuron.2004.11.025. [DOI] [PubMed] [Google Scholar]
- Ting CY, Herman T, Yonekura S, Gao S, Wang J, Serpe M, O’Connor MB, Zipursky SL, Lee CH. Tiling of r7 axons in the Drosophila visual system is mediated both by transduction of an activin signal to the nucleus and by mutual repulsion. Neuron. 2007;56:793–806. doi: 10.1016/j.neuron.2007.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Twiss JL, Fainzilber M. Ribosomes in axons--scrounging from the neighbors? Trends Cell Biol. 2009;19:236–243. doi: 10.1016/j.tcb.2009.02.007. [DOI] [PubMed] [Google Scholar]
- Vuppalanchi D, Coleman J, Yoo S, Merianda TT, Yadhati AG, Hossain J, Blesch A, Willis DE, Twiss JL. Conserved 3′-untranslated region sequences direct subcellular localization of chaperone protein mRNAs in neurons. J Biol Chem. 2010;285:18025–18038. doi: 10.1074/jbc.M109.061333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willis DE, Xu M, Donnelly CJ, Tep C, Kendall M, Erenstheyn M, English AW, Schanen NC, Kirn-Safran CB, Yoon SO, et al. Axonal Localization of Transgene mRNA in Mature PNS and CNS Neurons. J Neurosci. 2011;31:14481–14487. doi: 10.1523/JNEUROSCI.2950-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yudin D, Hanz S, Yoo S, Iavnilovitch E, Willis D, Gradus T, Vuppalanchi D, Segal-Ruder Y, Ben-Yaakov K, Hieda M, et al. Localized regulation of axonal RanGTPase controls retrograde injury signaling in peripheral nerve. Neuron. 2008;59:241–252. doi: 10.1016/j.neuron.2008.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng JQ, Kelly TK, Chang B, Ryazantsev S, Rajasekaran AK, Martin KC, Twiss JL. A functional role for intra-axonal protein synthesis during axonal regeneration from adult sensory neurons. J Neurosci. 2001;21:9291–9303. doi: 10.1523/JNEUROSCI.21-23-09291.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.