SUMMARY
Evolutionary theory assumes that mutations occur randomly in the genome; however, studies performed in a variety of organisms indicate the existence of context-dependent mutation biases. Sources of mutagenesis variation across large genomic contexts (e.g. hundreds of bases) have not been identified. Here, we use high-coverage whole genome sequencing of a conditional mismatch repair mutant line of diploid yeast to identify mutations that accumulated after 160 generations of growth. The vast majority of the mutations accumulated as insertion/deletions (in-dels) in homopolymeric (poly(dA:dT)) and repetitive DNA tracts. Surprisingly, the likelihood of an in-del mutation in a given poly(dA:dT) tract is increased by the presence of nearby poly(dA:dT) tracts in up to a 1000 bp region centered on the given tract. Our work suggests that specific mutation hotspots can contribute disproportionately to the genetic variation that is introduced into populations, and provides the first long-range genomic sequence context that contributes to mutagenesis.
Keywords: DNA Mismatch Repair, homopolymeric tracts, mutation hotspot
INTRODUCTION
Mutations arising from cellular metabolism and environmental insults confer fitness defects that are either removed by natural selection, drift neutrally in the population, or provide the raw fuel of adaptive evolution (Nishant et al., 2009). A corner stone of classical evolutionary theory is that mutations occur randomly throughout the genome and that biases in mutation contribute little to the ultimate outcome of the evolutionary process. However, experiments performed over many years suggest that not all sites in the genome have an equal probability of acquiring a mutation (reviewed in Wright, 2000). Experimental and indirect methods have been used to infer mutation rates (Nishant et al., 2009; Baer et al., 2007). Through such work mutation rates have been shown to vary with respect to base composition, local recombination rate, gene density, transcription, nucleosome location, and replication timing (Hawk et al., 2005; Wolfe et al., 1989; Matassi et al., 1999; Arndt et al., 2005; Hardison et al., 2003; Datta et al., 1995; Teytelman et al., 2008; Washietl et al., 2008; Stamatoyannopoulos et al., 2009). In addition, studies have suggested that larger genomic contexts exist that can affect mutation patterns but specific sequences within such contexts have not been identified. For example, Bailey et al. (2004) obtained evidence for hotspots in mammalian chromosomal evolution by observing conserved chromosome breakpoints and argue against a random-breakage model for chromosome evolution (Eichler and Sankoff, 2003). Understanding the molecular mechanisms that lead to mutation variation is a major challenge that is likely to provide insights into human disease progression (e.g., mutation accumulation in cancer tumors) and molecular evolution.
Our goal is to determine whether broad DNA sequence contexts underlie variability in mutagenesis across the genome. To test for such a context, we focused on identifying mutations that arise during DNA replication. The rate of such errors is low, ranging from 3 × 10−10 to 2 × 10−8 mutations per base pair per generation (Nishant et al., 2009). To accelerate the accumulation of mutations in a population, we employed conditional mismatch repair (MMR) mutants. MMR is a highly conserved pathway that excises DNA replication errors arising primarily from polymerase misincorporation and slippage events (Tran et al., 1997; Denver et al., 2005; Denver et al., 2004; Gragg et al., 2002; Streisinger et al., 1966; Sia et al., 1997). In eukaryotes, two heterodimeric MutS homolog complexes, MSH2-MSH3 and MSH2-MSH6, act in mismatch recognition. Both MSH complexes interact primarily with MLH1-PMS1 to form a mismatch-MSH-MLH complex that activates downstream repair steps including strand discrimination, excision, and resynthesis (Kunkel and Erie, 2005).
We used paired-end sequencing technologies and a Bayesian genotype caller to identify mutations that accumulated in MMR-deficient lines of baker`s yeast. We identified broad sequence contexts that contribute to mutation hotspots: the likelihood of a mutation in a given poly(dA:dT) tract is increased by the presence of poly(dA:dT) tracts in a 1000 bp region centered on the given tract. The presence of mutation hotspots is expected to contribute disproportionately to the genetic variation available to natural selection and to causative mutations in genetic diseases.
RESULTS
We examined three independent conditional MMR-defective (mlh1-7ts) diploid lines (referred to as Mut) of yeast derived from a common ancestor and grown for 160 generations at the non-permissive temperature, with bottlenecks reducing the population to one cell every 20 generations. At the non-permissive temperature, the mlh1-7ts mutants show a null-like phenotype in the canavanine resistance mutation assay and a nearly null phenotype in the lys2A14 reversion assay (Heck et al., 2006b).
To identify mutations present in Mut lines, we performed whole genome sequencing of two lines, Mut2 and Mut3 to moderate coverage (Zanders et al., 2010) and paired-end whole genome sequencing of two lines, wild-type and Mut4, to very high coverage (>200X). As shown in Tables 1, S1 and S2, we detected 19 base substitutions and 73 single- and di-nucleotide insertion/deletion (in-del) mutations in Mut4, all of which were heterozygous. The mutation rate for 8- to 14-bp homopolymeric (HP) tracts was 3.2 × 10−5, within a 2 to 10-fold range of levels in reporter assays in MMR null mutants (Tran et al., 1997; Gragg et al., 2002). The mutation rate in the 6- to 17-bp di-nucleotide tracts was 6.8 × 10−5/di-nt tract/generation, also within the range seen in reporter assays in MMR null mutants (Sia et al., 1997). To estimate the efficiency of detection, we took advantage of the fact that the Mut4 line at generation 160 showed 3% spore viability (Heck et al., 2006b). In Mut4, 34 of the heterozygous mutations map to open reading frames, five of which are frameshifts in HP tracts in genes (KRR1, KRS1, RAD3, MDN1, RRP15) in which null mutations confer lethality. Genotyping analysis showed that Mut4 viable spores contained only wild-type alleles of these genes; both wild-type and mutant alleles were detected in spores for other heterozygous Mut4 mutations (Table 1 and Fig. S1). The low spore viability (3%) seen in Mut4 is consistent with five recessive lethal mutations, though a meiotic chromosome aneuploidy phenotype observed in this line provides a minor contribution to the spore viability phenotype (Fig. S1). We are confident that these mutations encompass most, if not all, mutations present in coding regions in this line.
Table 1.
Segregation of heterozygous mutations in the Mut4 line
| locus | location | knockout recessive lethal? | WT:mutant allele |
|---|---|---|---|
| KIN82 | Chr3 275085 | no | 3:3 |
| EPLI | Chr6 88632 | no | 5:11 |
| PHO4 | Chr6 225029 | no | 4:2 |
| YBR219C | Chr2 662320 | no | 3:3 |
| AIM19 | Chr9 199795 | no | 2:4 |
| TOM70 | Chr14 399797 | no | 2:4 |
| AVT4 | Chr14 435396 | no | 2:4 |
| BIO3 | Chr14 734316 | no | 2:4 |
| FMP27 | Chr12 1047541 | no | 2:4 |
| ERV41 | Chr13 139505 | no | 4:7 |
| BUL1 | Chr13 816264 | no | 4:2 |
| YJRO12C | Chr10 460308 | no | 3:5 |
| KRR1 | Chr3 22745 | yes | 12:0 |
| KRS1 | Chr4 525612 | yes | 12:0 |
| CDC7* | Chr4 424584 | yes | 10:2 |
| RAD3 | Chr5 528691 | yes | 12:0 |
| MDN1 | Chr12 363531 | yes | 16:0 |
| FMP40 | Chr16 131383 | no | 4:2 |
| RAD1 | Chr16 509432 | no | 2:9 |
| REC8 | Chr16 569931 | no | 1:10 |
| Chr16 639362 | no | 1:11 | |
| RRP15 | Chr16 818766 | yes | 12:0 |
Spore clones obtained from tetrads dissected from Mut4 at generation 160 were genotyped by Sanger sequencing as described in the Supplemental Experimental Procedures.
non-synonymous mutation.
The 73 in-dels, representing nearly 80% of all of the mutations detected in Mut4, consisted of 65 deletions and 8 insertions, and occurred in 4- to 13-nt long HP tracts or in 6- to 13-repeat dinucleotide (di-nt) tracts (Table S1). The mutations in the HP tracts were all in An or Tn sequences, consistent with these repeats representing ~ 95% of the 5 to 20 nt HP tracts in the genome and greater than 99% of HP tracts 8 nt or larger. The predominance of nucleotide deletions over insertions and base substitutions in MMR defective strains was similar to that seen previously in a genome wide analysis (Zanders et al., 2010) and in reporter constructs (Tran et al., 1997; Gragg et al., 2002).
Identification of mutation hotspots in the genome
We examined whether broader sequence contexts were associated with mutagenesis. First, we examined the Mut4 sequencing data and that of two other lines, Mut2 and 3 (Zanders et al., 2010), to look for specific sites mutated in two of three Mut generation 160 lines. Nine such mutations were found that were single nucleotide in-dels in poly(dA:dT) tracts of 9–14 nt (Table 2, Fig. S2). The probability of identifying nine independent mutations at multiple sites by chance was low (P = 7.15×10−3). We were unable to identify any associations for the nine mutations with respect to origins of DNA replication (ORC and Mcm2 binding sites; Xu et al., 2006), centromere position, and Ty-element density (http://www.yeastgenome.org). It is possible that the small size of our data set precludes the identification of a specific pattern, or that complex non-overlapping parameters create mutation hotspots at these sites.
Table 2.
Independent mutations observed in two of three Mut generation 160 lines.
| Chromosome | SGD Position | poly(dA:dT) tract | lines:type of mutation |
|---|---|---|---|
| 2 | 92,273–92,281 | 9 | 2:del, 3:del |
| 4 | 216,494–216,503 | 10 | 3:del, 4:del |
| 4 | 314,305–314,316 | 12 | 3:del, 4:del |
| 7 | 533,997–534,006 | 10 | 3:del, 4:ins |
| 7 | 394,901–394,911 | 11 | 2:del, 3:del |
| 8 | 519,049–519,060 | 12 | 2:del, 3:del |
| 9 | 169,789–169,797 | 9 | 2:del, 3:del |
| 9 | 406,049–406,058 | 10 | 2:del, 3:del |
| 13 | 468,259–468,272 | 14 | 3:del, 4:del |
A broad sequence context for mutagenesis
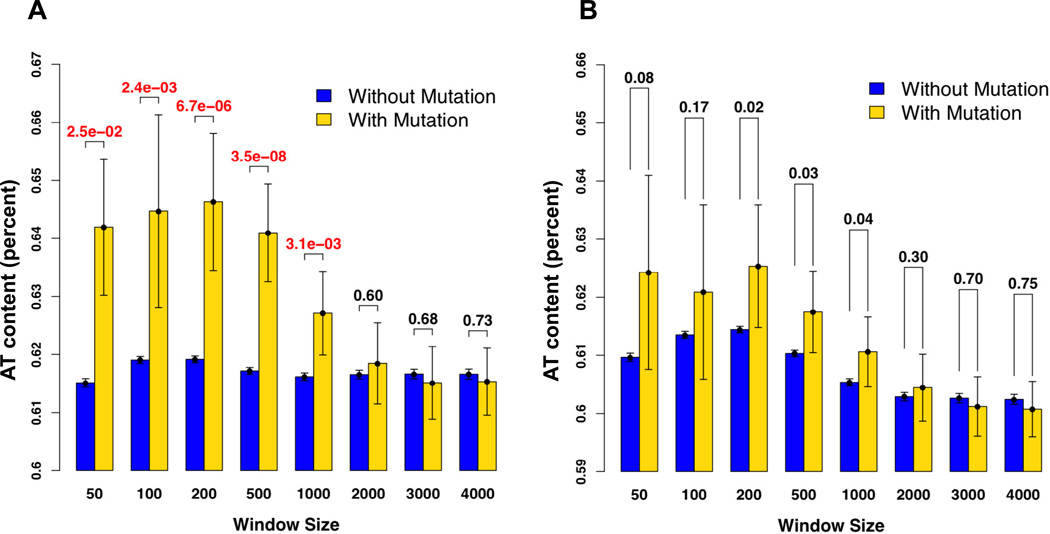
Mutation hotspots occur in repetitive DNA such as HP tracts and di-nucleotide repeats (e.g. Tran et al., 1997; Sia et al., 1997). While such mutation biases have been identified at a local sequence level (within ~80 bp), larger genomic contexts were not thought to contribute or may be difficult to find (e.g. Harfe and Jinks-Robertson, 2000; Rogozin et al., 2005; Canella and Seidman, 2000). To test for the presence of specific sequences/broader sequence contexts associated with mutagenesis in Mut4, we used a non-overlapping window analysis that involved an analysis of 50 to 4000 bp windows centered on size-matched 5–14 nt poly(dA:dT) tracts (Experimental Procedures). This was done because mutations in poly(dA:dT) tracts represented the majority (~70%) of mutations detected in the Mut4 line and would thus provide the best opportunity to find broad sequence contexts. Our statistical method accounts for the need to compare small (detected mutations) and large (potential sites in the genome) data sets. As shown in Fig. 1A, the AT content of the genomic regions surrounding the poly(dA:dT) tract mutation was significantly higher than for unmutated poly(dA:dT) tracts for 50 to 1000 bp window sizes, but not for the 2000 to 4000 bp windows.
Fig. 1. Association testing of nearby AT content in a fixed window size in the Mut4 line.
A. AT content was determined for each window (50–4000 bp) under conditions where a centered poly(dA:dT) tract, with or without an in-del mutation, was excluded. The X-axis shows the fixed window size and the Y-axis displays the mean AT content observed among all windows for the fixed window size. For a given fixed window size, we included the 5% error for each of the two distributions and also grouped the two distributions. The P-value for a Z-test used to compare the means of the two distributions is shown for each window. Red represents significance (P < 0.01) and black represents a lack of significance (P > 0.01; Experimental Procedures). B. AT content was determined as in panel A., except poly(dA:dT) tracts surrounding the mutated or unmutated centered poly(dA:dT) tract were removed.
We noted a pattern in which there was an enrichment of poly(dA:dT) tracts near mutations in poly(dA:dT) tracts (Zanders et al. 2010). To determine if this pattern is significant, we conducted two analyses, a case set in which we counted in increasing non-overlapping windows the number of poly(dA:dT) tracts surrounding a mutation in a given poly(dA:dT) tract, and a control set in which we counted the number of poly(dA:dT) tracts surrounding a given unmutated poly(dA:dT) tract. We then used statistical methods to determine if the pattern is genuine (Experimental Procedures). The analysis was performed using non-overlapping 50 to 4000 bp windows (mutated site excluded) centered on size-matched 5–14 nt poly(dA:dT) tracts. 5 to 14 nt run lengths were examined based on a visual inspection of poly(dA:dT) tracts located near a mutated site (Zanders et al. 2010) and the following criteria: 1. The upper limit was selected because it is difficult to identify in a single short read sequence (36 nt) tracts larger than 14 nt. This upper limit did not have a major effect our analysis because poly(dA:dT) tracts tracts greater than 14 nt are extremely rare in the yeast genome (< 0.4% of poly(dA:dT) tracts greater than 4 nt in size). 2. The lower limit was selected because Tran et al. (1997) observed that A5 runs appear to be at a threshold for large increases in the rate of frameshift mutations in MMR mutants. Consistent with this observation, they saw synergistic increases in frameshift mutations in A5 runs in mutants defective in both DNA MMR and polymerase proof reading.
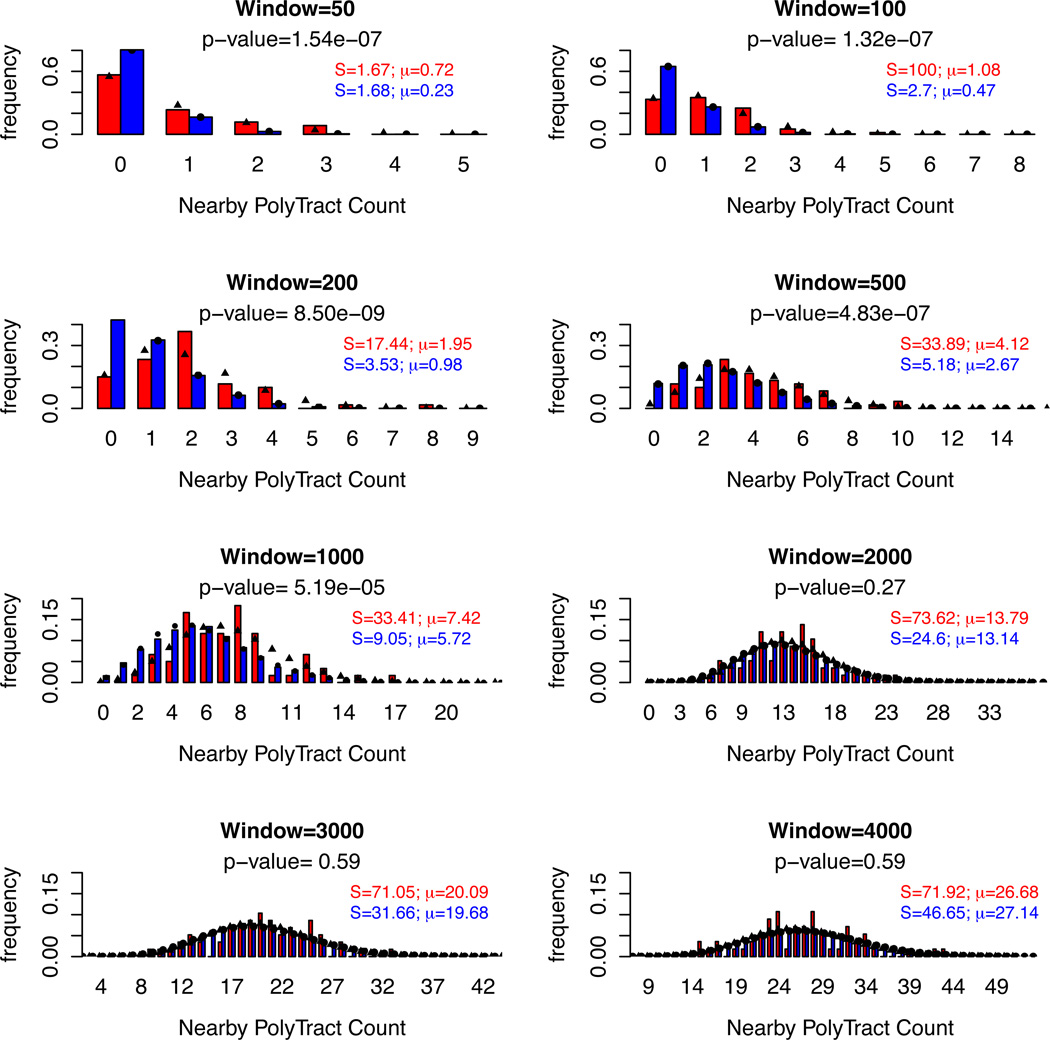
Windows either contained (64 sites) or lacked (39290, 32891, 24959, 14743, 8773, 4780, 2654, 1972 sites for 50, 100, 200, 500, 1000, 2000, 3000, 4000 bp windows, respectively) an in-del mutation in a poly(dA:dT) tract. The occurrence of a mutation in a poly(dA:dT) tract was highly associated with the number of nearby 5 to 14 nt poly(dA:dT) tracts for windows of 50 to 1000 bp (Fig. 2). An even stronger correlation was seen for the same window sizes when data for the Mut2 and Mut3 poly(dA:dT) tracts were included (data not shown). A statistical association was not seen for 2000 to 4000 bp windows. If the 50 bp surrounding the mutated poly(dA:dT) tract is excluded, the genomic context of the mutated poly(dA:dT) tracts still contains significantly higher poly(dA:dT) tracts than unmutated HP tracts for window sizes of 100 (P =1.7 × 10−3), 200 (P = 2.0 × 10−5), 500 (P = 4.8 × 10−5) and 1000 (P = 9.2 × 10−4) bp. Finally, we perform the AT content analysis presented in Fig. 1A but with surrounding poly(dA:dT) tracts removed from the analysis. As shown in Fig. 1B, AT content was no longer significantly different for mutated vs. unmutated poly(dA:dT) tracts for all window sizes (P > 0.01). These analyses show that a larger genomic context, clusters of poly(dA:dT) tracts, plays a role in the formation of mutations at a given poly(dA:dT) tract.
Fig. 2. Sliding window analysis for in-del mutations in poly(dA:dT) tracts in the Mut4 line.
The number of 5–14 nt poly(dA:dT) tracts was counted under different window sizes (50–4000 bp). This was determined for windows centered on poly(dA:dT) tracts with (red) and without (blue) an in-del mutation. The center sites were excluded from the counting analysis. The X-axis displays the number of poly(dA:dT) tracts contained within each window. The Y-axis shows the frequency for which each poly(dA:dT) tract was observed. The fitted size (S) and mean (µ) for each of the two distributions in a fixed window size is listed. The P-value of the likelihood-based method used to compare the means of two Negative Binomial distributions (Aban, 2008) is shown for each window (Experimental Procedures).
Promoter regions often contain long poly(dA:dT) tracts that serve as constitutive promoters (Struhl, 1985; Iyer and Struhl, 1995). Tran et al. (1997) showed that larger poly(dA:dT) tracts (e.g. 8–14 nt) undergo significantly higher rates of DNA slippage compared to smaller (5–7 nt) tracts. 8–14 nt poly(dA:dT) tracts are present at nearly three-fold higher levels in non-coding (4,139 in haploid S288c reference genome) compared to coding (1,563) regions; in contrast, 5–7 nt tracts appear at higher frequency in coding regions (42,005 tracts in coding, 27,128 in non-coding). Consistent with larger poly(dA:dT) tracts undergoing frameshift mutations in MMR mutants at higher frequency compared to smaller tracts, we found that the majority (84%) of mutations in 8–14 nt poly(dA:dT) tracts were in non-coding regions (Table S1). These data match reasonably well with the overall distribution of 8–14 nt poly(dA:dT) tracts in non-coding regions (73%). One hypothesis for the non-coding region bias is that poly(dA:dT) tracts in promoter regions are more tolerant to changes in size compared to tracts in coding regions where frameshift mutations would likely disrupt gene function and affect fitness. Lastly, the broad sequence context patterns observed for the entire genome (Figs. 1 and 2) were also seen for mutations in poly(dA:dT) tracts in non-coding regions (data not shown).
The larger genomic context identified above cannot be explained by a clustering of sites in a small window (~1 KB ) that each mutate at high frequency. First, we did not observe any apparent clustering of mutations at HP sites and no overlap (within 3 KB) was observed between mutated sites (Table S1). Second, we performed a statistical analysis on one given HP tract at a time-this excludes influences from other sites. Third, we observed a significant hotspot pattern for only a discrete distance from a mutated site. It is important to note that no significant association was found for the Mut4 line when a window analysis (window sizes 50, 100, 200) was performed to examine a correlation between single base change mutations and nearby poly(dA:dT) tracts (P > 0.1 for all windows).
DISCUSSION
Individual HP tracts are known to be sensitive to in-del mutations, which are caused primarily by DNA slippage during DNA replication (e.g. Tran et al., 1997). These slippage events are not thought to be influenced by local sequence context, including adjacent HP tracts (Harfe and Jinks-Robertson 2000). We show that the likelihood of a mutation in a given poly(dA:dT) tract is increased by the presence of poly(dA:dT) tracts in a 1 KB region centered on the given tract. Due to the size of the hotspot region, ~ 1 KB, it would have been very difficult to identify such a broad DNA sequence context by creating specific reporter constructs or searching for the association of mutations with unique DNA sequence motifs. Our work is distinct from bioinformatic studies of Denver et al. (2004), who observed that the C. elegans genome contains distinct clusters of HP tracts in autosomal arms. They hypothesized that such sites could be hotspots for recombination but also suggested that certain types of nearly tandem repeat clusters could serve as hotspots for slippage-mediated deletions.
Molecular, population genetic, and bioinformatic studies have shown that mutation rate varies across the eukaryotic genome (see Introduction). For example, Hawk et al. (2005) showed in baker’s yeast that the mutation rate of a microsatellite reporter placed at different chromosomal positions could vary by 16-fold; however, they were unable to identify a specific motif/chromosomal signature associated with shared mutations. Why might clusters of poly(dA:dT) tracts create mutation hotspots? One possibility is that clusters of these tracts form a secondary structure such as bent or flexible DNA that would predispose DNA polymerase to slippage (Hile and Eckert, 2008). If such structures exist, they are likely to be unstable, because we were unable to detect in acrylamide gels a change in the expected mobility of ~400 bp DNA fragments containing the DNA sequence in which in-dels were detected (data not shown). Alternatively, poly(dA:dT) tracts have been shown to be stiff, resist bending, and could affect mutagenesis by excluding nucleosomes (Washietl et al., 2008; Segal and Widom, 2009). A third possibility is that DNA polymerase stalling at HP tracts facilitates polymerase switching, perhaps to a DNA polymerase that replicates adjoining HP tracts with lower fidelity (Lovett, 2007). Work by Kim et al. (2007) support such an idea. They found that in-del mutations in HP tracts under high-transcription conditions were partially dependent on the function of polymerase zeta, an error-prone translesion DNA polymerase. A fourth possibility is that a cluster of HP tracts confers an increased mutation rate through increased transcription-it is known that poly(dA:dT) tracts serve as ubiquitous promoters (Struhl, 1985; Iyer and Struhl, 1995). It will be important to develop model systems to distinguish between these possible mechanisms.
Our work supports the idea that the primary role of MMR is to remove in-del mutations in HP tracts (Tran et al., 1997; Zanders et al., 2010). Such in-del mutations occur during DNA replication primarily as the result of slippage by DNA polymerases (Tran et al., 1997; Gragg et al., 2002; Streisinger et al., 1966; Hile and Eckert, 2008). In wild-type yeast DNA slippage events are rarely detected in HP tracts due to the detection and removal of slippage intermediates by MMR (Nishant et al., 2010). Based on the observation that 25% of yeast ORFs have HP tracts 8 nt or longer and 56% of ORFs have HP tracts 5 nt or longer (S288c reference genome), Tran et al. (1997) hypothesized that the high rate of mutation in HP tracts could explain “the high rates of recessive lethal mutations that accumulate in diploid Mmr− (pms1 and msh2) yeast.” They also suggested that “the lack of MMR in cancer tissue could lead to inactivation of genes with long homonucleotide runs that are important for cancer progression and for secondary effects of cancer.” Mutations in four MMR genes confer predisposition to hereditary, nonpolyposis colorectal cancer (HNPCC; Kunkel and Erie, 2005; Lynch et al., 2009). Our genome-wide analysis of mutations observed in MMR defective lines, coupled with the detection of recessive lethal mutations seen as frameshift mutations in HP tracts (Table S1), confirms the Tran et al. (1997) hypothesis and supports the idea that inactivation of genes with HP tracts is critical for cancer progression in MMR deficient tumors. Genes with long HP runs are mutated in MMR deficient tumors (reviewed in Shah et al., 2010) and thus are likely to contribute to the cancer specificity observed in MMR mutants.
Ni et al. (1999) sequenced mutations in the CAN1 gene that conferred resistance in baker’s yeast to canavanine. They found that 20–35% of the mutations were frameshifts in mono-nucleotide runs, with the remainder either being base substitutions (~55–70%) or complex mutations (~10%). Thus it will be interesting to see if the long-range sequence context for mutation hotspots identified in this study is also seen in MMR proficient strains. Given that the frameshift mutation rate in wild-type is several orders of magnitude lower than in MMR mutants, we decided to use MMR deficient lines. If the MMR system is unbiased in the way it operates, this should be equivalent to looking at natural mutations accumulating over a much longer time period. On the other hand, the MMR may be biased, perhaps acting more efficiently on some substitutions or some classes of mutation compared with others, in which case the relative rates we report may not reflect the rates that occur naturally. More extensive studies will be required to determine whether such biases exist and, if so, how they affect the mutations that arise.
In summary, we found a new pattern for mutational hotspots in which the likelihood of an in-del mutation in a given poly(dA:dT) tract is increased by the presence of nearby poly(dA:dT) tracts. This work supports the idea that natural selection occurs in a landscape where certain sequences and regions of the genome are mutated at higher frequency, and reinforces the idea that mutation rates vary across different regions of the genome and that a large sequence context can affect the mutability of a given nucleotide. It also provides experimental evidence to support population genetic studies that aim to identify sequence context correlations with mutation rate (e.g. Tian et al., 2008). Such information provides important clues on targets for evolvability in cell types that are mutators due to defects in specific repair processes (Heck et al., 2006a; Demogines et al., 2008; Taddei et al., 1997; Loeb, 2011).
EXPERIMENTAL PROCEDURES
Detailed methods are in the Supplemental Information.
Statistical test to examine association of a mutation in a given poly(dA:dT) tract with nearby poly(dA:dT) tracts
To test if DNA sequences surrounding in-del mutations in the Mut4 generation 160 line were enriched for poly(dA:dT) tracts, non-overlapping 50 to 4000 bp windows, centered around size-matched 5–14 nt poly(dA:dT) tracts, were analyzed. A Negative Binomial model was fitted, where the number of poly(dA:dT) tracts in a fixed window size was counted, excluding the center site, to account for the reasonable small mean and over-dispersion that cannot be predicted by a simple Poisson model. A goodness of fit test was performed for the two distributions where nearby bins were combined so that they have an expected value of at least five for a fixed window size. This was done to make sure the negative binomial distribution is appropriate. We then tested if there was a difference between nearby poly(dA:dT) tract occurrences for windows with and without a poly(dA:dT) tract mutation (the difference2 between the mean parameters equals to 0), using likelihood-based methods. This performed well for testing equality of mean counts modeled by a negative binomial distribution, even when the over-dispersion parameter of one group was twice that of the other group (Aban, 2008; Fig. 2).
We carried out an association test of nearby AT content in fixed window sizes with and without an in-del mutations (Fig. 1A). For each fixed window size, we computed the percentage of AT content excluding the center poly(dA:dT) tract. For each of the two distributions in a fixed window size, we first fitted a normal distribution, and then used the Bootstrap Kolmogorov-Smirnov test, which executes a bootstrap version of the univariate Kolmogorov-Smirnov test to correct coverage when distributions compared are not entirely continuous (Sekhon, 2011). This assessed the fitness for each of the distributions (all P-values were > 0.1 and were not significant). Furthermore, we used an F-test to make sure that the two distributions in the fixed window size have equal variance (all P-values were > 0.1, not significant). Lastly, we used a Z-test to compare the mean between the two distributions. For Fig. 1B, the distributions of the data were not normally distributed as in Fig. 1A; they were log-normal distributed and were analyzed by a method of mean comparison for log-normal distribution (Zhou et al., 1997).
Highlights.
The presence of an insertion/deletion mutation in a given homopolymeric tract is increased by nearby homopolymeric tracts.
We provide the first long-range genomic sequence context that contributes to mutagenesis.
Mutation hotspots can contribute disproportionately to the genetic variation that is introduced into populations.
Supplementary Material
ACKNOWLEDGMENTS
We thank the Cornell CLC and Muh-Ching Yee from the Stanford Genome Center for preparing samples for Illumina GA sequencing, Harmit Malik and the Alani and Bustamante laboratories for comments, and Brandon Barker and Zhenglong Gu for technical advice. X.M. and C.D.B. were supported by NSF grants 0606461 and 0701382. M.R., S.Z., K.T.N. and E.A. were supported by NIH GM53085. S.Z. was also supported by a Cornell Presidential Fellowship and an NIH training grant in Genetics and Development. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute Of General Medical Sciences or the National Institutes of Health.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Aban IB. Inferences and power analysis concerning two negative binomial distributions with an application to MRI lesion counts data. Comput. Stat. Data. Anal. 2008;53:820–833. doi: 10.1016/j.csda.2008.07.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arndt PF, Hwa T, Petrov DA. Substantial regional variation in substitution rates in the human genome: importance of GC content, gene density, and telomere-specific effects. J. Mol. Evol. 2005;60:748–763. doi: 10.1007/s00239-004-0222-5. [DOI] [PubMed] [Google Scholar]
- Baer CF, Miyamoto MM, Denver DR. Mutation rate variation in multicellular eukaryotes: causes and consequences. Nat. Rev. Genet. 2007;8:619–631. doi: 10.1038/nrg2158. [DOI] [PubMed] [Google Scholar]
- Bailey JA, Baertsch R, Kent WJ, Haussler D, Eichler EE. Hotspots of mammalian chromosomal evolution. Genome Biol. 2004;5:R23. doi: 10.1186/gb-2004-5-4-r23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canella KA, Seidman MM. Mutation spectra in supF: approaches to elucidating sequence context effects. Mut. Res. 2000;450:61–73. doi: 10.1016/s0027-5107(00)00016-6. [DOI] [PubMed] [Google Scholar]
- Datta A, Jinks-Robertson S. Association of increased spontaneous mutation rates with high levels of transcription in yeast. Science. 1995;268:1616–1619. doi: 10.1126/science.7777859. [DOI] [PubMed] [Google Scholar]
- Demogines A, Wong A, Aquadro C, Alani E. Incompatibilities involving yeast mismatch repair genes: a role for genetic modifiers and implications for disease penetrance and variation in genomic mutation rates. PLoS Genet. 2008;4:e1000103. doi: 10.1371/journal.pgen.1000103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denver DR, Feinberg S, Estes S, Thomas WK, Lynch M. Mutation rates, spectra and hotspots in mismatch repair-deficient Caenorhabditis elegans. Genetics. 2005;170:107–113. doi: 10.1534/genetics.104.038521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denver DR, Morris K, Kewalramani A, Harris KE, Chow A, Estes S, Lynch M, Thomas WK. Abundance, distribution, and mutation rates of homopolymeric nucleotide runs in the genome of Caenorhabditis elegans. J. Mol. Evol. 2004;58:584–595. doi: 10.1007/s00239-004-2580-4. [DOI] [PubMed] [Google Scholar]
- Eichler EE, Sankoff D. Structural dynamics of eukaryotic chromosome evolution. Science. 2003;301:793–797. doi: 10.1126/science.1086132. [DOI] [PubMed] [Google Scholar]
- Gragg H, Harfe BD, Jinks-Robertson S. Base composition of mononucleotide runs affects DNA polymerase slippage and removal of frameshift intermediates by mismatch repair in Saccharomyces cerevisiae. Mol. Cell. Biol. 2002;22:8756–8762. doi: 10.1128/MCB.22.24.8756-8762.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardison RC, Roskin KM, Yang S, Diekhans M, Kent WJ, et al. Covariation in frequencies of substitution, deletion, transposition, and recombination during eutherian evolution. Genome Res. 2003;13:13–26. doi: 10.1101/gr.844103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harfe BD, Jinks-Robertson S. Sequence composition and context effects on the generation and repair of frameshift intermediates in mononucleotide runs in Saccharomyces cerevisiae. Genetics. 2000;156:571–578. doi: 10.1093/genetics/156.2.571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawk JD, Stefanovic L, Boyer JC, Petes TD, Farber RA. Variation in efficiency of DNA mismatch repair at different sites in the yeast genome. Proc. Natl. Acad. Sci. USA. 2005;102:8639–8643. doi: 10.1073/pnas.0503415102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heck JA, Argueso JL, Gemici Z, Reeves RG, Bernard A, Aquadro CF, Alani E. Negative epistasis between natural variants of the Saccharomyces cerevisiae MLH1 and PMS1 genes results in a defect in mismatch repair. Proc. Natl. Acad. Sci. USA. 2006a;103:3256–3261. doi: 10.1073/pnas.0510998103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heck JA, Gresham D, Botstein D, Alani E. Accumulation of recessive lethal mutations in Saccharomyces cerevisiae mlh1 mismatch repair mutants is not associated with gross chromosomal rearrangements. Genetics. 2006b;174:519–523. doi: 10.1534/genetics.106.059311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hile SE, Eckert KA. DNA polymerase kappa produces interrupted mutations and displays polar pausing within mononucleotide microsatellite sequences. Nucl. Acids. Res. 2008;36:688–696. doi: 10.1093/nar/gkm1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer V, Struhl K. Poly(dA:dT), a ubiquitous promoter element that stimulates transcription via its intrinsic DNA structure. EMBO. J. 1995;14:2570–2579. doi: 10.1002/j.1460-2075.1995.tb07255.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim N, Abdulovic AL, Gealy R, Lippert MJ, Jinks-Robertson S. Transcription- associated mutagenesis in yeast is directly proportional to the level of gene expression and influenced by the direction of DNA replication. DNA. Repair. 2007;6:1285–1296. doi: 10.1016/j.dnarep.2007.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkel TA, Erie DA. DNA mismatch repair. Annu. Rev. Biochem. 2005;74:681–710. doi: 10.1146/annurev.biochem.74.082803.133243. [DOI] [PubMed] [Google Scholar]
- Loeb LA. Human cancers express mutator phenotypes:origin, consequences and targeting. Nat. Rev. Cancer. 2011;11:450–457. doi: 10.1038/nrc3063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovett ST. Polymerase switching in DNA replication. Mol. Cell. 2007;27:523–526. doi: 10.1016/j.molcel.2007.08.003. [DOI] [PubMed] [Google Scholar]
- Lynch HT, Lynch PM, Lanspa SJ, Snyder CL, Lynch JF, Boland CR. Review of Lynch syndrome; history, molecular genetics, screening differential diagnosis, and medicolegal ramifications. Clin. Genet. 2009;76:1–18. doi: 10.1111/j.1399-0004.2009.01230.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matassi G, Sharp PM, Gautier C. Chromosomal location effects on gene sequence evolution in mammals. Curr. Biol. 1999;9:786–791. doi: 10.1016/s0960-9822(99)80361-3. [DOI] [PubMed] [Google Scholar]
- Ni TT, Marsischky GT, Kolodner RD. MSH2 and MSH6 are required for removal of adenine misincorporated opposite 8-oxo-guanine in S. cerevisiae. Mol. Cell. 1999;4:439–444. doi: 10.1016/s1097-2765(00)80346-9. [DOI] [PubMed] [Google Scholar]
- Nishant KT, Singh ND, Alani E. Genomic mutation rates: What high-throughput methods can tell us. Bioessays. 2009;31:912–920. doi: 10.1002/bies.200900017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishant KT, Wei W, Mancera E, Argueso JL, Schlattl A, Delhomme N, Ma X, Bustamante CD, Korbel JO, Gu Z, Steinmetz LM, Alani E. The baker`s yeast diploid genome is remarkably stable in vegetative growth and meiosis. PLoS Genet. 2010;6:e1001109. doi: 10.1371/journal.pgen.1001109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogozin IB, Malyarchuk BA, Pavlov YI, Milanesi L. From Context- Dependence of Mutations to Molecular Mechanisms of Mutagenesis. Pacific Symposium on Biocomputing. 2005;10:409–420. [PubMed] [Google Scholar]
- Segal E, Widom J. Poly(dA:dT) tracts: major determinants of nucleosome organization. Curr. Opin. Struct. Biol. 2009;19:65–71. doi: 10.1016/j.sbi.2009.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekhon JS. Multivariate and propensity score matching software with automated balance optimization. J. Stat. Software. 2011;42:1–52. [Google Scholar]
- Shah SN, Hile SE, Eckert KA. Defective mismatch repair, microsatellite mutation bias, and variability in clinical cancer phenotypes. Cancer Res. 2010;70:431–435. doi: 10.1158/0008-5472.CAN-09-3049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sia EA, Kokoska RJ, Dominska M, Greenwell P, Petes TD. Microsatellite instability in yeast: dependence on repeat unit size and DNA mismatch repair genes. Mol. Cell Biol. 1997;17:2851–2858. doi: 10.1128/mcb.17.5.2851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatoyannopoulos JA, Adzhubei I, Thurman RE, Kryukov GV, Mirkin SM, Sunyaev SR. Human mutation rate associated with DNA replication timing. Nat. Genet. 2009;41:393–395. doi: 10.1038/ng.363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Streisinger G, Okada Y, Emrich J, Newton J, Tsugita A, Terzaghi E, Inouye M. Frameshift mutations and the genetic code. This paper is dedicated to Professor Theodosius Dobzhansky on the occasion of his 66th birthday. Cold Spring Harb Symp Quant. Biol. 1966;31:77–84. doi: 10.1101/sqb.1966.031.01.014. [DOI] [PubMed] [Google Scholar]
- Struhl K. Naturally occurring poly(dA-dT) sequences are upstream promoter elements for constitutive transcription in yeast. Proc. Natl. Acad. Sci. USA. 1985;82:8419–8423. doi: 10.1073/pnas.82.24.8419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taddei F, Radman M, Maynard-Smith J, Toupance B, Gouyon PH, Godelle B. Role of mutator alleles in adaptive evolution. Nature. 1997;387:700–702. doi: 10.1038/42696. [DOI] [PubMed] [Google Scholar]
- Teytelman L, Eisen MB, Rine J. Silent but not static: accelerated base-pair substitution in silenced chromatin of budding yeasts. PLoS Genet. 2008;4:e1000247. doi: 10.1371/journal.pgen.1000247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian D, Wang Q, Zhang P, Araki H, Yang S, Kreitman M, Nagylaki T, Hudson R, Bergelson J, Chen JQ. Single-nucleotide mutation rate increases close to insertions/deletions in eukaryotes. Nature. 2008;455:105–108. doi: 10.1038/nature07175. [DOI] [PubMed] [Google Scholar]
- Tran HT, Keen JD, Kricker M, Resnick MA, Gordenin DA. Hypermutability of homonucleotide runs in mismatch repair and DNA polymerase proofreading yeast mutants. Mol. Cell Biol. 1997;17:2859–2865. doi: 10.1128/mcb.17.5.2859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Washietl S, Machné R, Goldman N. Evolutionary footprints of nucleosome positions in yeast. Trends Genet. 2008;24:583–587. doi: 10.1016/j.tig.2008.09.003. [DOI] [PubMed] [Google Scholar]
- Wolfe KH, Sharp PM, Li WH. Mutation rates differ among regions of the mammalian genome. Nature. 1989;337:283–285. doi: 10.1038/337283a0. [DOI] [PubMed] [Google Scholar]
- Wright BE. A Biochemical Mechanism for Nonrandom Mutations and Evolution. J. Bact. 2000;182:2993–3001. doi: 10.1128/jb.182.11.2993-3001.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu W, Aparicio JG, Aparicio OM, Tavaré S. Genome-wide mapping of ORC and Mcm2p binding sites on tiling arrays and identification of essential ARS consensus sequences in S cerevisiae. BMC Genomics. 2006;7:276. doi: 10.1186/1471-2164-7-276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanders S, Ma X, Roychoudhury A, Hernandez RD, Demogines A, Barker B, Gu Z, Bustamante CD, Alani E. Detection of heterozygous mutations in the genome of mismatch repair defective diploid yeast using a Bayesian approach. Genetics. 2010;186:493–503. doi: 10.1534/genetics.110.120105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X-H, Gao S, Hui SL. Methods for comparing the means of two independent log-normal samples. Biometrics. 1997;53:1129–1135. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.