Abstract
We previously reported that children in the UKALL XI ALL trial with HLA-DP 1 and -DP 3 supertypes had significantly worse event-free survival (EFS) than children with other DP supertypes. As DP 1 and DP 3 share two of four key antigen-binding amino-acid polymorphisms (aspartic acid84–lysine69), we asked whether Asp84-Lys69 or Asp84 alone were independent prognostic indicators in childhood acute lymphoblastic leukemia (ALL). We analysed EFS in 798 UKALL XI patients, stratified by Asp84-Lys69 vs non-Asp84-Lys69, for a median follow-up of 12.5 years. Asp84-Lys69 was associated with a significantly worse EFS than non-Asp84-Lys69 (5-year EFS: Asp84-Lys69: 58.8% (95% CI (confidence of interval): 52.7–64.9%); non-Asp84-Lys69: 67.3% (63.4–71.2%); 2P=0.007). Post-relapse EFS was 10% less in Asp84-Lys69 than non-Asp84-Lys69 patients. EFS was significantly worse (P=0.03) and post-relapse EFS marginally worse (P=0.06) in patients with Asp84 compared with Gly84. These results suggest that Asp84-Lys69 predicted adverse EFS in the context of UKALL XI because of Asp84, and may have influenced post-relapse EFS. We speculate that this may be due to the recruitment of Asp84-Lys69-restricted regulatory T cells in the context of this regimen, leading to the re-emergence of residual disease. However, functional and molecular studies of the prognostic value of this and other HLA molecular signatures in other childhood ALL trials are needed.
Keywords: childhood ALL, HLA-DP supertype, DP molecular signature, event-free survival, relapse
Introduction
Multi-agent chemotherapy has delivered major improvements in long-term survival in childhood acute lymphoblastic leukemia (ALL) over the last four decades, from <10% to over 80%.1, 2, 3 Nevertheless, up to 20–25% of patients relapse, even on modern intensive treatment protocols, and the survival of relapsed patients remains poor.4, 5 Prediction of treatment outcome has thus become a central issue in the development of risk-adapted therapies in childhood leukaemia.6, 7
Prediction of treatment outcome using cellular (minimum residual disease), cytogenetic and gene-expression markers requires a source of leukaemic blasts.8, 9, 10, 11 Conversely, risk stratification predicted by constitutional (heritable) genotypes is independent of leukaemia burden and offers scope for outcome prediction very early in treatment. However, identification of inter-individual genetic variations associated with responses to therapy has yet to find routine application in childhood ALL.12, 13
Yang et al.14 recently identified 102 constitutional single-nucleotide polymorphisms associated with minimal residual disease in a genome-wide association study of childhood ALL. A number of single-nucleotide polymorphisms were located on chromosome 6p, close to the HLA genes in the major histocompatibility complex. An emerging body of evidence suggests that HLA alleles are associated with drug hypersensitivity,15 but the role of the major histocompatibility complex in response to chemotherapy in childhood ALL remains uncertain. Previously, we reported that two HLA-DP supertypes (DP 1, DP 3) were associated with a significantly worse event-free survival (EFS) than four other DP supertypes (DP 2, 4, 6, 8).16 DP 1 and DP 3 share two (aspartic acid 84 and lysine 69; pockets 1 and 4) but differ for two (pockets six and nine) of the four key polymorphic antigen-binding pockets that influence the specificity of peptides bound by DP molecules.17, 18 It was not clear from our previous analysis whether Asp84-Lys69 or Asp 84 alone contributed to the worse EFS of DP 1 and DP 3. Here, we re-analyse EFS in patients in relation to these two amino-acid ‘signatures' (Asp84-Lys69, and Asp84 alone) and compared their outcome with patients having neither signature (non-Asp84-Lys69 or non-Asp84, respectively). In this study, unlike our previous analysis, we included patients with two additional supertypes, DP 11 and DP 15.
Materials and methods
Patients
Details of the UKALL XI childhood ALL trial have been published elsewhere.19 Briefly, 2090 children aged 1–14 years were entered into UKALL XI between October 1990 and March 1997. Patients were treated with a four-drug induction regimen (l-asparaginase, vincristine, prednisolone and intrathecal methotrexate), two or three blocks of intensification therapy and randomisation for CNS-directed therapy using a long course of intrathecal (I/T) methotrexate with or without high-dose intravenous methotrexate for low white cell count patients (<50 × 109/l), or cranial irradiation, with short course I/T methotrexate vs high-dose methotrexate and long course I/T for high white cell count patients (>50 × 109/l).
HLA-DPB1 molecular typing and supertype classification
Patients in UKALL XI were simultaneously recruited by the UK Childhood Cancer Study,20 an independent case–control epidemiological study designed to assess the impact of environmental carcinogen exposure on the aetiology of childhood leukaemia. High resolution DP molecular typing was carried out as previously described.21 DP data from the UKCCS patients recruited by UKALL XI are used in this paper.
We previously defined structural DP supertypes as clusters of DP alleles with the same amino-acid polymorphisms in three peptide pockets (1, 3 and 6) at positions 84, 69 and 11 of the DPβ1 domain of the DP antigen-binding site.18 More than 90% of DP alleles in the UK population can be clustered into 8 supertypes (DP 1–4, DP 6, DP 8, DP 11 and DP 15). Here, we examine the polymorphic amino-acid signatures defined by residues 84 and 69 (peptide pockets 1 and 4) of the DPβ subunit (Table 1).
Table 1. DPβ1 pocket 1–4 amino-acid signature of DP supertypes.
| Supertypea | DP 1 | DP 2 | DP 3 | DP 4 | DP 6 | DP 8 | DP 11 | DP 15 |
|---|---|---|---|---|---|---|---|---|
|
Pocket 1–4 signatureb |
Asp84-Lys69 |
Gly84-Glu69 |
Asp84-Lys69 |
Gly84-Lys69 |
Asp84- Glu69 |
Asp84-Glu69 |
Asp84-Arg69 |
Val84-Arg69 |
| 0101 | 0201 | 0301 | 0401 | 0601 | 0801 | 1101 | 1501 | |
| 0501 | 0202 | 1401 | 0402 | 0901 | 1601 | |||
| DPB1 | 5001 | 3301 | 2001 | 2301 | 1001 | 1901 | ||
| alleles | 4801 | 2501 | 2401 | 1301 | ||||
| 2601 | 4901 | 1701 | ||||||
| 3501 | 5101 | 2101 | ||||||
| 3001 |
Abbreviations: Asp, aspartic acid; Arg, arginine; Gly, glycine; Glu, glutamic acid, Leu, leucine; Lys, lysine; Val, valine.
DP supertypes (1–4, 6, 8, 11, 15) are assigned from amino-acid residues lining peptide pockets 1, 4 and 6 of the DPβ1 domain.
Pocket 1–4 amino-acid signature shows residues at positions 84 and 69 of the DPβ1 domain.
Statistical analysis
Comparisons between categorical variables were made using the χ2-test or Fisher's exact test where appropriate. EFS and post-relapse EFS (pr-EFS) were calculated in patients grouped by the amino-acid ‘signatures' in pockets 1 and 4 (Asp84-Lys69, respectively) and by non-Asp84-Lys69 (that is, other residues in pockets 1 and 4, see Table 1), or by pocket 1 alone (either Asp84 or non-Asp84). Kaplan–Meier life tables were constructed for survival data and compared using the log-rank test. Stratified analyses were used to allow for initial patient/disease characteristics and confirmed by multivariate Cox regression. EFS was defined as time-to-relapse or death from any cause (relapse, refractory disease and infection), and pr-EFS was defined as time from first relapse to second relapse or death from any cause, excluding patients who never relapsed. Surviving patients were censored on the follow-up date, 30 April 2007, or, for the small number of patients lost to follow-up, at the date at which they were last known to be alive. Median follow-up, from commencement of treatment, for DP-typed patients is 12.5 years, range (3.5–16.3 years). All P-values quoted are two-sided.
Results
The 798 ALL patients included in this study represent 38% of the 2090 cases in the UKALL XI trial. Although potential sources of bias in the clinical characteristics and outcome of the DP-typed compared with the untyped UKALL XI ALL patients were previously excluded,16 re-analysis revealed no significant differences in age at diagnosis, sex ratio, immunophenotype, presenting white blood cell (WBC) count or leukaemia karyotype (t(12;21), high hyperdiploidy) between DP supertypes (data not shown), with one exception. The frequency of the Philadelphia translocation (t(9;22)) in DP 2 patients was five-fold higher than non-DP 2 patients (3.0% vs 0.6%, P=0.03), but numbers are too small to draw any firm conclusions about the cause of this excess.
Previously we reported that two HLA-DP supertypes (DP 1, DP 3) were separately associated with significantly worse EFS than four other DP supertypes (DP 2, 4, 6, 8).16 To determine whether the Asp84-Lys69 signature was responsible for the worse outcome, we pooled patients with these two supertypes into one group (N=248) and compared EFS with patients not having this motif (non-Asp84-Lys69; N=979). Unlike our previous study16 we included patients with DP 11 and DP 15 in the non-Asp84-Lys69 group (these patients having only one or neither of these two residues). Also, DP 11 patients were included in an Asp84 alone group and DP 15 patients in a non-Asp84 alone group (Table 1). The frequencies of patients with all possible pocket 1–4 signatures in the total series, in relapsed and in non-relapsed patients are compared in Table 2, which shows a marginal though nonsignificant excess (P=0.09) of Asp84-Lys69 in relapsed compared with non-relapsed patients (35% vs 29%) but no difference in other pocket 1–4 signatures.
Table 2. Frequency of DPβ1 pocket 1-4 amino-acid signatures in ALL patients who relapse.
| DPβ1 |
Frequency (%) of patients with signature |
||||
|---|---|---|---|---|---|
| amino-acid signature | DP supertype | Total n=798 | Relapse n=294 | Non-relapse n=504 | P (relapse vs non-relapse) |
| Asp84-Lys69 | DP 1+DP 3 | 248 (31) | 102 (35) | 146 (29) | 0.09 |
| Gly84-Glu69 | DP 2 | 161 (20) | 56 (19) | 105 (21) | 0.5 |
| Gly84-Lys69 | DP 4 | 655 (83) | 242 (83) | 413 (83) | 0.9 |
| Asp84-Glu69 | DP 6+DP 8 | 117 (15) | 42 (14) | 75 (15) | 0.8 |
| Asp84-Arg69 | DP 11 | 12 (2) | 5 (2) | 7 (1) | 0.8 |
| Val84-Arg69 | DP 15 | 34 (4) | 10 (3) | 24 (5) | 0.4 |
Abbreviations: ALL, acute lymphoblastic leukemia; Asp, aspartic acid; Arg, arginine; Gly, glycine; Glu, glutamic acid, Leu, leucine; Lys, lysine; Val, valine.
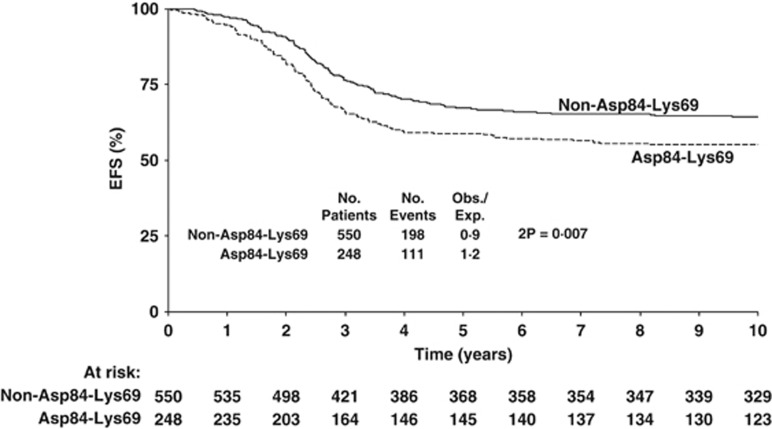
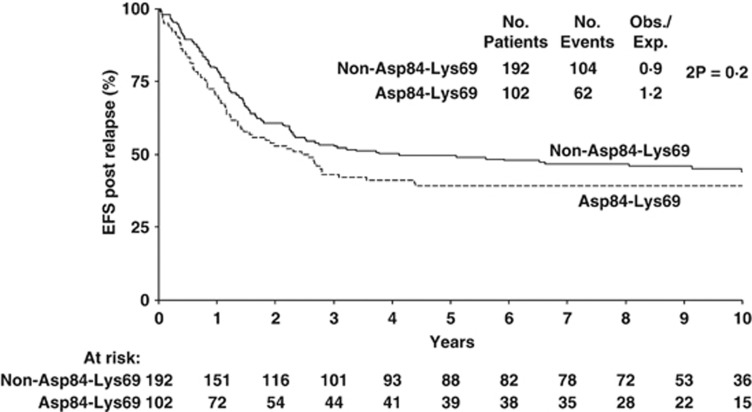
Analysis of EFS over time reveals this to be significantly worse (2P=0.007) in Asp84-Lys69 patients being almost 10% less than in non-Asp84-Lys69 patients after 5 and 10 years (Figure 1). Thus, 5-year Asp84-Lys69 EFS was 58.8% (52.7–64.9%) compared with non-Asp84-Lys69 EFS of 67.3% (63.4–71.2%). Although 10 year Asp84-Lys69 EFS is similar to 5-year EFS (55.2% (48.9–61.5%)), 10 year non-Asp84-Lys69 EFS suggests some deterioration (64.5% (60.6–68.4%)). Examination of post-relapse EFS in the two groups of amino-acid signatures (Figure 2) shows a 5-year Asp84-Lys69 pr-EFS about 10% less (39.2% (29.8–48.6%)) than non-Asp84-Lys69 (49.6% (42.5–56.7%)) but this difference is not significant (P=0.2).
Figure 1.
Event-free survival in ALL patients with Asp84-Lys69 compared with non-Asp84-Lys69.
Figure 2.
Post-relapse event-free survival in ALL patients with Asp84-Lys69 compared with non-Asp84-Lys69.
Summarising, EFS for each of the six DP pocket 1–4 signatures (rather than DP supertypes) confirms a significantly worse EFS in patients with Asp84-Lys69 (Table 3), and suggests possible beneficial effects of having a Gly84-Glu69, Gly84-Lys69 or Asp84-Arg69, though these were not significant. There was no significant effect of Asp84-Lys69 copy number (1 or 2 copies) on EFS (data not shown). The difference in EFS between Asp84-Lys69 and non-Asp84-Lys69 patients remained significant even after allowance for baseline patient characteristics (age, sex, WBC count, immunophenotype and karyotype). Multivariate Cox analysis confirmed the independent significance of Asp84-Lys69, sex, WBC count and t(9;22) on EFS. Univariate hazard ratio (1.38 (95% CI: 1.10–1.75)) for Asp84-Lys69 was not materially altered after inclusion of these variables (hazard ratio=1.33 (1.03–1.72)). However, stratification for factors known to significantly affect post-relapse EFS (duration of remission, site of relapse, age, immunophenotype) reduced the size of the Asp84-Lys69 effect (O/E=1.1).
Table 3. Overall and post-relapse EFS in ALL patients by DPβ1 pocket 1–4 molecular signature.
| Amino-acid signature | DP supertype |
Overall EFS |
Post-relapse EFS |
||||||
|---|---|---|---|---|---|---|---|---|---|
| No. of events/no. patients | % (se) at 5 years | % (se) at 10 years | P-value (log-rank test) | No. of events/no. of patients | % (se) at 5 years | % (se) at 10 years | P-value (log-rank test) | ||
| Asp84-Lys69 | DP 1, 3 | 111/248 | 58.8 (3.1) | 55.2 (3.2) | 0.007 | 62/102 | 39.2 (4.8) | 39.2 (4.8) | 0.2 |
| Non-Asp84-Lys69 | DP 2, 4, 6, 8, 11, 15 | 198/550 | 67.3 (2.0) | 64.5 (2.0) | 104/192 | 49.6 (3.6) | 44.0 (3.8) | ||
| Gly84-Glu69 | DP 2 | 57/161 | 68.9 (3.6) | 65.1 (3.8) | 0.3 | 28/56 | 53.3 (6.7) | 48.4 (6.9) | 0.2 |
| Non-Gly84-Glu69 | DP 1, 3, 4, 6, 8, 11, 15 | 252/637 | 63.6 (1.9) | 60.7 (1.9) | 138/238 | 44.2 (3.2) | 40.7 (3.3) | ||
| Gly84-Lys69 | DP 4 | 252/655 | 65.0 (1.9) | 61.9 (1.9) | 0.6 | 133/242 | 48.0 (3.2) | 43.6 (3.3) | 0.2 |
| Non-Gly84-Lys69 | DP 1, 2, 3, 6, 8, 11, 15 | 57/143 | 62.9 (4.0) | 60.0 (4.1) | 33/52 | 36.3 (6.7) | 36.3 (6.7) | ||
| Asp84-Glu69 | DP 6, 8 | 45/117 | 62.4 (4.5) | 61.4 (4.5) | 1.0 | 27/42 | 40.3 (7.6) | 35.3 (7.4) | 0.3 |
| Non-Asp84-Glu69 | DP 1, 2, 3, 4, 11, 15 | 264/681 | 65.0 (1.8) | 61.6 (1.9) | 139/252 | 46.9 (3.2) | 43.5 (3.3) | ||
| Asp84-Arg69 | DP 11 | 11/34 | 67.6 (8.0) | 67.6 (8.0) | 0.6 | 5/10 | 50.0 (15.8) | 50.0 (15.8) | 0.7 |
| Non-Asp84-Arg69 | DP 1, 2, 3, 4, 6, 8, 15 | 298/764 | 64.5 (1.7) | 61.3 (1.8) | 161/284 | 45.8 (3.0) | 42.0 (3.1) | ||
| Val84-Arg69 | DP 15 | 5/12 | 58.3 (14.2) | 58.3 (14.2) | 0.9 | 3/5 | 40.0 (21.9) | 40.0 (21.9) | 0.9 |
| Non-Val84-Arg69 | DP 1, 2, 3, 4, 6, 8, 11 | 304/786 | 64.7 (1.7) | 61.6 (1.8) | 163/289 | 46.1 (2.9) | 42.3 (3.0) | ||
Abbreviations: ALL, acute lymphoblastic leukemia; Asp, aspartic acid; Arg, arginine; EFS, event-free survival; Gly, glycine; Glu, glutamic acid, Leu, leucine; Lys, lysine; Val, valine.
In light of these results we surmised that the pocket 1 residue at position 84 of DPβ alone might have exerted a key influence on the outcome of UKALL XI. We therefore stratified patients into those with Asp84 and those with non-Asp84 (that is, Gly84 or Val84; see Table 1). We compared overall and pr-EFS in the two series (Table 4), and found that 5-year EFS was marginally less frequent (61.2% vs 67.6% P=0.06) in the Asp84 compared with the non-Asp84 patients. The difference in pr-EFS was slightly greater, but this lacked significance (P=0.08). We analysed EFS and pr-EFS again in the same series of patients but this time stratified into Asp84 and Gly84 by excluding patients with Val84 (DP 15), and obtained a significantly worse EFS (%, 95% CI: 60.4, 55.3–65.5 vs 67.8, 63.5–72.1; P=0.03) and marginal pr-EFS (39.6, 31.4–47.8 vs 51.5, 43.7–59.3; P=0.06), supporting an effect of Asp84 on EFS and possibly, though not conclusively on EFS after relapse. If this is the case we might expect to see an effect of Asp84 copy number on EFS. As Table 5 shows, the trend is suggestive (P=0.09) but not conclusive. Five year EFS with increasing Asp84 copy number is: 0 copies (that is, X/X where X is either Gly84 or Val84) (n=432): 67.6% (63.1–72.1%); 1 copy (Asp84/X heterozygotes) (n=286): 61.2% (55.5–66.9), 2 copies (Asp84/Asp84 homozygotes) (n=80): 61.3% (50.7–71.9%)). Again, a similar trend (P=0.09) was observed for 5-year pr-EFS: 0 copies being 51.6% (43.6–59.6%), 1 copy, 40.7% (31.7–49.7%) and 2 copies 37.6% (19.8–55.4%). No significant effect could be discerned for Lys69 copy number, suggesting that of the two residues, Asp84 is more likely than Lys69 to be influencing EFS.
Table 4. EFS in childhood ALL by Asp84 pocket 1 residue.
| DPβ1 pocket 1 residue | DP supertype |
Overall EFS |
Post-relapse EFS |
||||
|---|---|---|---|---|---|---|---|
| N | 5-year EFS (%, 95% CI) | P-value for trenda | N | 5-year EFS (%, 95% CI) | P-value for trenda | ||
| Asp84 | DP 1, 3, 6, 8, 11 | 366 | 61.2 (56.3–66.1) | 0.06 | 142 | 40.1 (32.1–48.1) | 0.08 |
| Non-Asp84 | DP 2, 4, 15 | 432 | 67.6 (63.1–72.1) | 152 | 51.6 (43.6–59.6) | ||
Abbreviations: ALL, acute lymphoblastic leukemia; Asp, aspartic acid; CI, confidence of interval; EFS, event-free survival.
P-values are two-sided.
Table 5. Overall and post-relapse EFS in ALL patients by DP peptide pocket 1 and 4 residue.
| Pocket 1 residue |
Overall EFS |
Post-relapse EFS |
||||||
|---|---|---|---|---|---|---|---|---|
| No. of events/no. of patients | % (se) at 5 years | % (se) at 10 years | P-value (log-rank test) | No. of events/no. of patients | % (se) at 5 years | % (se) at 10 years | P-value (log-rank test) | |
| Asp84/Asp84 | 33/80 | 61.3 (5.4) | 58.6 (5.5) | 18/29 | 37.6 (9.1) | 37.6 (9.1) | ||
| Asp84/Xa | 119/286 | 61.2 (2.9) | 58.3 (2.9) | 69/113 | 40.7 (4.6) | 38.8 (4.6) | ||
| X/X | 157/432 | 67.6 (2.3) | 64.3 (2.3) | 0.09 | 79/152 | 51.6 (4.1) | 46.0 (4.3) | 0.09 |
| Lys69/Lys69 | 201/500 | 63.8 (2.1) | 60.1 (2.2) | 109/191 | 44.7 (3.6) | 41.5 (3.7) | ||
| Lys69/Yb | 93/249 | 65.0 (3.0) | 63.0 (3.1) | 48/88 | 49.8 (5.3) | 44.4 (5.4) | ||
| Y/Y | 15/49 | 71.4 (6.5) | 69.3 (6.6) | 0.2 | 9/15 | 40.0 (12.6) | 40.0 (12.6) | 0.7 |
Abbreviations: ALL, acute lymphoblastic leukemia; Asp, aspartic acid; EFS, event-free survival; Lys, lysine.
X is either glycine or valine 84 (pocket 1)
Y is either glutamic acid or arginine 69 (pocket 4).
We previously reported a strong association between DPB1*0601 (DP 6) and susceptibility to childhood leukaemia.22 Patients with DP 6 have an aspartic acid 84 residue in pocket 1, but glutamic acid not lysine, in pocket 4 (Table 1). Analysis of outcome in patients with Asp84-Glu69-Leu11 (DP 6) showed no significant difference in the frequency of relapsed and non-relapsed patients (11.6% vs 12.3% P=0.8), or any effect on EFS. A suggestion that post-relapse EFS was worse in Asp84-Glu69-Leu11 than non-Asp84-Glu69-Leu11 patients (35.3% vs 47.4% at 5 years; P=0.07) could not be attributed to differences in the clinical characteristics of patients with or without DP 6 (Table 6).
Table 6. Characteristics of relapsed ALL patients with DP 6.
| Asp84-Glu69-Leu11 | Non-Asp84-Glu69-Leu11 | P-value | |
|---|---|---|---|
| Sex | |||
| Male | 19 (56%) | 166 (64%) | 0.4 |
| Female | 15 (44%) | 94 (36%) | |
| Age | |||
| <2 | 2 (6%) | 14 (5%) | 0.7 (trend) |
| 2–9 | 26 (76%) | 193 (74%) | |
| 10+ | 6 (18%) | 53 (20%) | |
| WBC | |||
| <10 | 11 (32%) | 117 (45%) | 0.3 (trend) |
| 10− | 4 (12%) | 32 (12%) | |
| 20− | 9 (26%) | 39 (15%) | |
| 50− | 4 (12%) | 29 (11%) | |
| 100+ | 6 (18%) | 43 (16%) | |
| Immunophenotype | |||
| Pro B ALL | 3 (9%) | 6 (3%) | 0.1 |
| BCP ALL | 26 (79%) | 205 (86%) | |
| T ALL | 4 (12%) | 27 (11%) | |
| t(9;22)a | |||
| No | 30 (97%) | 211 (98%) | 0.6 |
| Yes | 1 (3%) | 4 (2%) | |
| t(12;21) | |||
| No | 16 (80%) | 108 (78%) | 0.9 |
| Yes | 4 (20%) | 30 (22%) | |
| Heh | |||
| No | 21 (68%) | 154 (72%) | 0.7 |
| Yes | 10 (32%) | 61 (28%) | |
Abbreviations: ALL, acute lymphoblastic leukemia; Asp, aspartic acid; BCP, B-cell precursor; Glu, glutamic acid; Heh, high hyperdiploid ALL; Leu, leucine; WBC, white blood cell.
Karyotype: chromosome translocation (t)9;22; translocation (t)12;21.
Discussion
In our previous study, we reported that ALL patients with the HLA-DP 1 and DP 3 supertypes had significantly worse outcomes than those with four other DP supertypes.16 Of the four key DP polymorphic antigen-binding residues lining pockets 1, 4, 6 and 9,17 DP 1 and DP 3 are the only two supertypes that share aspartic acid in pocket 1 at position 84 linked to lysine in pocket 4 at position 69 of DPβ, (though they differ for residues in pockets 6 and 9). We therefore wondered whether the antigen-binding molecular signature influenced by aspartic acid in pocket 1, and lysine in pocket 4 (Asp84-Lys69) of DPβ1, rather than the motif determined by pocket 1, 4, 6 and 9 might provide a more specific functional surrogate of outcome in UKALL XI.
Although the DP-typed cases represent only a proportion (38%) of total UKALL XI patients, lack of significant differences in clinical characteristics between typed and non-typed patients suggest that no bias was introduced by analysing the association of DP molecular signatures with outcome in two-fifths of UKALL XI cases. Our analysis confirmed that 5 and 10 year EFS in patients with Asp84-Lys69 was significantly (∼10%) worse than non-Asp84-Lys69. Of the DP-typed patients, 44% of Asp84-Lys69 cases had events, while only 36% of non-Asp84-Lys69 cases had events. The difference in EFS between the Asp84-Lys69 and non-Asp84-Lys69 patients remained significant even after adjustment for other prognostic factors, strongly suggesting that Asp84-Lys69 is an independent predictor of adverse outcome in UKALL XI. A total of 294 of the 798 DP-typed patients presented in this paper survived relapse. Of the patients that typed for Asp84-Lys69, 60% had post-relapse events, compared with 54% of the patients with non-Asp84-Lys69, this difference being nonsignificant.
We previously clustered DP alleles into six structural DP supertypes (DP 1–4, 6, 8) defined by polymorphic amino-acid residues lining pockets 1, 4 and 6, at positions 84, 69 and 11 of the DPβ subunit.18 In the present study we included patients with two additional supertypes (DP 11, 15), and classified patients only by the presence of polymorphic amino-acid residues at positions 84 and 69. Position 84 of the DPβ subunit lines pocket 1 of the DP antigen-binding site, and is equivalent to position 86 of DRβ.17 In DPβ, residues 84–87 form a linked unit, which influences the size and charge of the amino-acid side chains of antigens preferred by pocket 1. Thus, the aspartic acid 84–glutamic acid 85–alanine 86–valine 87 (DEAV) peptide binding motif was present in 20 (64%) of the 31 DP alleles in the current patient series, while the remainder had either glycine 84–glycine 85–proline 86–methionine 87 (GGPM) in 10 (32%) alleles, or valine 84–glycine 85–proline 86–methionine 87 (VGPM) in one (3%) DP allele (DPB1*1501). Structure-function studies of DP molecules have shown that DEAV differs from GGPM by modifying the contact area between the DPα1 and DPβ1 subunits of the DPβ heterodimer, as well as influencing the binding of amino acids at the P1 and P2 positions of antigenic peptides by increasing the negative charge of the P1/P2 pockets.17 In HLA-DR alleles, DRβ86 is occupied by glycine or valine and peptides may bind to Gly86, but not to Val86 owing to steric effects.23, 24 Castelli et al.25 identified pockets 1 and 6 of DP as accommodating the main anchor residues of DP-binding peptides, but our results suggest that the outcome of UKALL XI may have been influenced by the peptide binding motif (DEAV or GGPM) of DP pocket 1 alone.
Previous studies of HLA and outcome in childhood ALL have generally involved small patient numbers, limited HLA allele resolution, inadequate diagnostic detail or different treatment regimens to enable clear overall conclusions to be drawn.26, 27, 28, 29, 30, 31 A typical example of the confusion that this has engendered is the reported association of HLA-DR5 with long remission,30 no impact on remission31 and an increased incidence of relapse.32 It is possible that the reason for this lack of agreement is that HLA associations with treatment outcome can only be evaluated in the context of specific trial regimens, due to differences in the effects of therapy on HLA-mediated functions. Despite indications of major histocompatibility complex loss33 and HLA class I downregulation by ALL cells34, 35 no recent attempt has been made to establish the baseline contribution of heritable major histocompatibility complex variation to the clinical outcome of childhood ALL. Although prospective HLA typing of children presenting with ALL using serological methods previously presented a significant technical challenge, the availability of DNA-based molecular techniques now affords a rapid and reliable means of determining HLA genotypes. Although a fully prospective study has yet to be carried out, the availability of remission samples from patients recruited by a large UK population-based case–control epidemiological study of childhood ALL aetiology (the UKCCS20) most of whom were randomised to the UKALL XI trial, has provided a timely opportunity to redress this deficiency and to establish the contribution of HLA-DP supertypes to outcome, preparatory to an analysis of HLA association with outcome in more recent childhood ALL trials using different treatment regimens.
Although a functional explanation for the association of Asp84-Lys69 with adverse outcome in childhood ALL in UKALL XI is a matter for speculation, we favour a role for the activation of CD4+ T cells by Asp84-Lys69-restricted (that is, bound) peptides. Sidney et al.36 recently reported an extensive overlap in the peptide binding specificity of DP alleles, but peptide binding by Asp84 (DP 1) alleles could clearly be distinguished from Gly84 (DP 2) alleles by a preference for positively charged side-chain residues, notably arginine and lysine. The precise identity of any bound peptides is a matter for further investigation, but we note the presence of arginine at the P1 position of a 9-mer TEL-AML1 core junctional peptide, RIAECILGM, which elicited a DP 1-restricted CD4+ T response in vitro, and that DP 1 frequency is reduced in patients with TEL-AML1 ALL.37, 38
Our results were obtained by analysing patients in the UKALL XI trial, which is known to have had a higher rate of relapse than concurrent childhood ALL trials. Extrapolation to more recent trials, such as ALL97, with significantly improved EFS39 thus requires caution, and further study. The higher relapse rate in UKALL XI may be partly attributable to the lower anti-leukaemic toxicity of prednislone (PRED) than dexamethasone,40 but this does not explain why this should selectively involve DP alleles with an Asp84 residue. Currently, there is only limited information on changes in HLA class II expression by relapsed ALL cells41, 42 but this seems to exclude loss of HLA class II expression at relapse as an explanation for escape from T-cell control. In contrast, expression of HLA class II molecules including DP by relapsed ALL cells might lead to the binding of leukaemia-associated peptides leading to the recruitment of regulatory CD4+ T (Treg) cells.43 This might lead to a loss of effector (CD8+) T-cell control of minimal residual disease. Although direct evidence of such an effect is currently lacking, the ability of established tumours to induce Treg-associated immune tolerance associated with tumour metastasis44 and reduced survival45 has been well documented in other tumours. In the context of the UKALL XI trial, a key point may be that the use of PRED was insufficiently toxic for leukaemia cells, but exacerbated a tolerogenic Treg response, as PRED is known to promote the differentiation of Tregs and to ameliorate autoimmune disease and allergy.46, 47
We recently reported that the DP 6 supertype, in which Asp84 is linked to Glu69, was strongly associated with susceptibility to childhood leukaemia. We found that this was due to a single infrequent DPB1 allele, DPB1*0601.28 In the present analysis, DP 6 was associated with a slightly worse post-relapse EFS (P=0.07), but it was not associated with overall outcome. Of the 34 relapsed DP 6 patients included in the outcome analysis, one third typed for DPB1*0601, but there was no discernable difference in their clinical characteristics compared with non-relapsed DP 6 patients (age, sex, presenting white cell count, immunophenotype, karyotype of cells at diagnosis), indicating that DP 6 does not have independent prognostic value in UKALL XI.
In summary, we report that patients in UKALL XI with a DP Asp84-Lys69 antigen-binding molecular signature are more likely to relapse and probably have a worse post-relapse outcome than patients with a non-Asp84-Lys69 molecular signature. We suggest that this might be due to the pocket 1 residue, Asp84, a marker for the DEAV peptide binding motif of DPβ1. We propose a mechanistic explanation for this association in which recruitment of Tregs, possibly exacerbated by PRED (glucocorticoid) therapy, in response to the recognition of Asp84-bound auto- or leukaemia-associated peptides by CD4+ T cells, leads to loss of minimal residual disease control. As we find no association between HLA-A supertype and outcome in UKALL XI48 we suggest provisionally that Asp84 may be an independent prognostic factor at least in this trial. It will nonetheless be important to analyse the contribution of this and other HLA amino-acid signatures in the outcome of more recent childhood ALL trials, and specifically to determine how they might affect risk-adapted therapy. The technical simplicity of single HLA amino-acid genotyping suggests that this could be done at ALL diagnosis.
Acknowledgments
This study was funded by grants from the Kay Kendall Leukaemia Fund and Children with Leukaemia to GMT, from the UK MRC (G0901509) to SR and RW and CRUK salary support to SR. The UKALL XI childhood ALL trial database was funded by the Medical Research Council. We thank Professor E Roman and Jill Simpson at the Epidemiology and Genetics Unit, University of York for providing diagnostic and other information about patients in the UKCCS, Mrs R Carter for sample documentation, and MD Robinson, Dr C Watson, Dr D A Gokhale and S P Dearden for sample processing and DP typing. A complete list of UKCCS investigators and sponsors is given in: UK Childhood Cancer Study Investigators. The United Kingdom Childhood Cancer Study: objectives, Materials and methods. Br J Cancer 2000; 82, 1073-1102. We are grateful to the members of MRC/NCRI Childhood Leukaemia Group for their contribution to the UKCSS and for allowing us to analyse data from the UKALL XI trial.
Author Contributions
M Taylor designed the research and wrote the paper. RW and SR analysed the data. AH and PT performed the research. IH, BG and TE co-ordinated the UKALL XI trial and commented on the paper.
The authors declare no conflict of interest.
References
- Kersey JH. Fifty years of studies of the biology and therapy of childhood leukemia. Blood. 1997;90:4243–4251. [PubMed] [Google Scholar]
- Pui C-H, Robison LL, Look AT. Acute lymphoblastic leukaemia. Lancet. 2008;371:1030–1043. doi: 10.1016/S0140-6736(08)60457-2. [DOI] [PubMed] [Google Scholar]
- Stanulla M, Schrappe M. Treatment of childhood acute lymphoblastic leukemia. Semin Hematol. 2009;46:52–63. doi: 10.1053/j.seminhematol.2008.09.007. [DOI] [PubMed] [Google Scholar]
- Gaynon PS. Childhood acute lymphoblastic leukaemia and relapse. Br J Haematol. 2005;131:579–587. doi: 10.1111/j.1365-2141.2005.05773.x. [DOI] [PubMed] [Google Scholar]
- Nguyen K, Devidas M, Cheng S-C, La M, Raetz EA, Carroll WL, et al. Factors influencing survival after relapse from acute lymphoblastic leukemia: a Children's Oncology Group study. Leukemia. 2008;22:2142–2150. doi: 10.1038/leu.2008.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pui C-H, Evans WE. Treatment of acute lymphoblastic leukemia. N Engl J Med. 2006;354:166–178. doi: 10.1056/NEJMra052603. [DOI] [PubMed] [Google Scholar]
- Mitchell C, Payne J, Wade R, Vora A, Kinsey S, Richards S, et al. The impact of risk stratification by early bone marrow response in childhood lymphoblastic leukaemia: results from the United Kingdom Medical Research Council trial ALL97 and ALL97/99. Br J Haematol. 2009;146:424–436. doi: 10.1111/j.1365-2141.2009.07769.x. [DOI] [PubMed] [Google Scholar]
- Vrooman LM, Silverman LB. Childhood acute lymphoblastic leukemia: update on prognostic factors. Curr Opin Pediatr. 2009;21:1–8. doi: 10.1097/MOP.0b013e32831f1f24. [DOI] [PubMed] [Google Scholar]
- Moorman AV, Ensor HM, Richards SM, Chilton L, Schwab C, Kinsey SE, et al. Prognostic effect of chromosomal abnormalities in childhood B-cell precursor acute lymphoblastic leukaemia: results from the UK Medical Research Council ALL97/99 randomised trial. Lancet Oncol. 2010;11:429–438. doi: 10.1016/S1470-2045(10)70066-8. [DOI] [PubMed] [Google Scholar]
- Campana D. Status of minimal residual disease testing in childhood haematological malignancies. Br J Haematol. 2008;143:481–489. doi: 10.1111/j.1365-2141.2008.07350.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhojwani D, Kang H, Moskowitz NP, Min DJ, Lee H, Potter JW, et al. Biologic pathways associated with relapse in childhood acute lymphoblastic leukemia: a Children's Oncology Group study. Blood. 2006;108:711–717. doi: 10.1182/blood-2006-02-002824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheok MH, Pottier N, Kager L, Evans WE. Pharmacogenetics in acute lymphoblastic leukemia. Semin Hematol. 2009;46:39–51. doi: 10.1053/j.seminhematol.2008.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Relling MV, Altman RB, Goetz MP, Evans WE. Clinical implementation of pharmacogenomics: overcoming genetic exceptionalism. Lancet Oncol. 2010;11:507–509. doi: 10.1016/S1470-2045(10)70097-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang JJ, Cheng C, Yang W, Pei D, Cao X, Fan Y, et al. Genome-wide interrogation of germline genetic variation associated with treatment response in childhood acute lymphoblastic leukemia. J Am Med Assoc. 2009;301:393–403. doi: 10.1001/jama.2009.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatanaga H, Honda H, Shinichi O. Pharmacogenetic information derived from analysis of HLA alleles. Pharmacogenomics. 2008;9:207–214. doi: 10.2217/14622416.9.2.207. [DOI] [PubMed] [Google Scholar]
- Taylor GM, Richards S, Wade R, Hussain A, Simpson J, Hill F, et al. Relationship between HLA-DP supertype and survival in childhood acute lymphoblastic leukaemia: evidence for selective loss of immunological control of residual disease. Br J Haematol. 2009;145:87–95. doi: 10.1111/j.1365-2141.2008.07571.x. [DOI] [PubMed] [Google Scholar]
- Diaz G, Amicosante M, Jaraquemada D, Butler RH, Guillen MV, Sanchez M, et al. Functional analysis of HLA-DP polymorphism: a crucial role for DPβ residues 9, 11, 35, 55, 69, and 84-87 in T cell allorecognition and peptide binding. Int Immunol. 2003;15:565–576. doi: 10.1093/intimm/dxg057. [DOI] [PubMed] [Google Scholar]
- Taylor GM, Hussain A, Lightfoot TJ, Birch JM, Eden TOB, Greaves MF, et al. HLA-associated susceptibility to childhood B cell precursor ALL: definition and role of HLA-DPB1-supertypes. Br J Cancer. 2008;98:1125–1131. doi: 10.1038/sj.bjc.6604257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards S, Burrett J, Hann I, Chessells J, Hill F, Bailey C. for the Medical Research Council Working Party on Childhood Leukaemia. Improved survival with early intensification: combined results from the Medical Research Council childhood ALL randomised trials, UKALL X and UKALL XI. Leukemia. 1998;12:1031–1036. doi: 10.1038/sj.leu.2401065. [DOI] [PubMed] [Google Scholar]
- UK Childhood Cancer Study Investigators The United Kingdom Childhood Cancer Study: objectives, materials and methods. Br J Cancer. 2000;82:1073–1102. doi: 10.1054/bjoc.1999.1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor GM, Dearden S, Ravetto P, Ayres M, Watson P, Hussain A, et al. Genetic susceptibility to childhood common acute lymphoblastic leukaemia is associated with polymorphic peptide-binding pocket profiles in HLA-DPB1*0201. Hum Mol Genet. 2002;11:1585–1597. doi: 10.1093/hmg/11.14.1585. [DOI] [PubMed] [Google Scholar]
- Taylor GM, Hussain A, Verhage V, Thompson PD, Fergusson WD, Watkins G, et al. Strong association of the HLA-DP6 supertype with childhood leukaemia is due to a single allele, DPB1*0601. Leukemia. 2009;23:863–869. doi: 10.1038/leu.2008.374. [DOI] [PubMed] [Google Scholar]
- Ong B, Willcox N, Wordsworth P, Beeson D, Vincent A, Altmann D, et al. Critical role for the Val/Gly86 HLA-DR beta dimorphism in autoantigen presentation to human T cells. Proc Natl Acad Sci. 1991;88:7343–7347. doi: 10.1073/pnas.88.16.7343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busch R, Hill CM, Hayball JD, Lamb JR, Rothbard JB. Effect of natural polymorphism at residue 86 of the HLA-DR beta chain on peptide binding. J Immunol. 1991;147:1292–1298. [PubMed] [Google Scholar]
- Castelli FA, Buhot C, Sanson A, Zarour H, Pouvelle-Moratille S, Nonn C, et al. HLA-DP4, the most frequent HLA II molecule, defines a new supertype of peptide-binding specificity. J Immunol. 2002;169:6928–6934. doi: 10.4049/jimmunol.169.12.6928. [DOI] [PubMed] [Google Scholar]
- Tursz T, Hors J, Lipinski M, Amiel J-L. HLA phenotypes in long term survivors treated with BCG immunotherapy for childhood ALL. Br Med J. 1978;1 (6122:1250–1251. doi: 10.1136/bmj.1.6122.1250-a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Bruyere M, Cornu G, Heremans-Bracke T, Malchaire J, Sokal G. HLA haplotypes and long survival in childhood acute lymphoblastic leukaemia treated with transfer factor. Br J Haematol. 1980;44:243–251. doi: 10.1111/j.1365-2141.1980.tb01206.x. [DOI] [PubMed] [Google Scholar]
- Leikin S, Miller D, Sather H, Albo V, Esber E, Johnson A, et al. Immunologic evaluation in the prognosis of acute lymphoblastic leukemia. A report from children's cancer study group. Blood. 1981;58:501–508. [PubMed] [Google Scholar]
- Davey F, Lachant NA, Dock NL, Hubbell C, Stockman JA, Henry JB. HLA antigens and childhood acute lymphocytic leukaemia. Br J Haematol. 1981;47:211–220. doi: 10.1111/j.1365-2141.1981.tb02781.x. [DOI] [PubMed] [Google Scholar]
- Révész T, Benczúr M, Gyódi E, Petrányi GG, Schuler D. The association of HLA-DR5 antigen with longer survival in childhood leukaemia. Br J Haematol. 1981;48:508–510. doi: 10.1111/j.1365-2141.1981.tb02744.x. [DOI] [PubMed] [Google Scholar]
- De Jong B, Van der Does-van den Berg A, Schreuder GMT. Random HLA-DR distribution in children with acute lymphocytic leukaemia in long-term remission. Br J Haematol. 1982;52:161–169. doi: 10.1111/j.1365-2141.1982.tb03872.x. [DOI] [PubMed] [Google Scholar]
- Casper JT, Marrari M, Piaskowski V, Lauer SJ, Duquesnoy RJ. Association between HLA-D region antigens and disease-free survival in childhood non-T, non-B acute lymphoblastic leukaemia. Blood. 1982;60:698–702. [PubMed] [Google Scholar]
- McEvoy CRE, Morley AA, Firgaira FA. Evidence for whole chromosome 6 loss and duplication of the remaining chromosome in acute lymphoblastic leukemia. Genes, Chromosomes Cancer. 2003;37:321–325. doi: 10.1002/gcc.10214. [DOI] [PubMed] [Google Scholar]
- Masuda K, Hiraki A, Fujii N, Watanabe T, Tanaka M, Matsue K, et al. Loss or down-regulation of HLA class I expression at the allelic level in freshly isolated leukemic blasts. Cancer Sci. 2007;98:102–108. doi: 10.1111/j.1349-7006.2006.00356.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demanet C, Mulder A, Deneys V, Worsham MJ, Maes P, Claas FH, et al. Down-regulation of HLA-A and HLA-Bw6, but not HLA-Bw4, allospecificities in leukemic cells: an escape mechanism from CTL and NK attack. Blood. 2004;103:3122–3130. doi: 10.1182/blood-2003-07-2500. [DOI] [PubMed] [Google Scholar]
- Sidney J, Steen A, Moore C, Ngo S, Chung J, Peters B, et al. Five HLA-DP molecules frequently expressed in the worldwide human population share a common HLA supertypic binding specificity. J Immunol. 2010;184:2492–2503. doi: 10.4049/jimmunol.0903655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yun C, Senju S, Fujita H, Tsuji Y, Irie A, Matsushita S, et al. Augmentation of immune responses by altered peptide ligands of the antigenic peptide in a human CD4+ T-cell clone reacting to TEL/AML1 fusion protein. Tissue Antigens. 1999;54:153–161. doi: 10.1034/j.1399-0039.1999.540206.x. [DOI] [PubMed] [Google Scholar]
- Taylor M, Harrison C, Eden T, Birch J, Greaves M, Lightfoot T, et al. HLA-DPB1 supertype-associated protection from childhood leukaemia: relationship to leukaemia karyotype and implications for prevention. Cancer Immunol Immunother. 2008;57:53–61. doi: 10.1007/s00262-007-0349-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell CD, Richards SM, Kinsey SE, Lilleyman J, Vora A, Eden TOB. Benefit of dexamethasone compared with prednisolone for childhood acute lymphoblastic leukaemia: results of the UK Medical Research Council ALL 97 randomised trial. Br J Haematol. 2005;129:734–745. doi: 10.1111/j.1365-2141.2005.05509.x. [DOI] [PubMed] [Google Scholar]
- Kaspers GJ, Veerman AJ, Popp-Snijders C, Lomecky M, Van Zantwijk CH, Swinkels LM, et al. Comparison of the anti-leukemic activity in vitro of dexamethasone and prednisolone in childhood acute lymphoblastic leukemia. Med Pediatr Oncol. 1996;27:114–121. doi: 10.1002/(SICI)1096-911X(199608)27:2<114::AID-MPO8>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- Van Wering ER, Beishuizen A, Roeffen ET, Van der Linden-Schrever BE, Verhoeven MA, Hahlen K, et al. Immunophenotypic changes between diagnosis and relapse in childhood acute lymphoblastic leukemia. Leukemia. 1995;9:1523–1533. [PubMed] [Google Scholar]
- Guglielmi C, Cordone I, Boecklin F, Masi S, Valentini T, Vegna ML, et al. Immunophenotype of adult and childhood acute lymphoblastic leukemia: changes at first relapse and clinico-prognostic implications. Leukemia. 1997;11:1501–1507. doi: 10.1038/sj.leu.2400772. [DOI] [PubMed] [Google Scholar]
- Zou W. Regulatory T cells, tumour immunity, and immunotherapy. Nat Rev Immunol. 2006;6:295–307. doi: 10.1038/nri1806. [DOI] [PubMed] [Google Scholar]
- Viguier M, Lemaitre F, Verloa O, Cho M-S, Gorochov G, Dubertret L, et al. Foxp3 expressing CD4+CD25high regulatory T cells are overrepresented in human metastatic melanoma lymph nodes and inhibit the function of infiltrating T cells. J Immunol. 2004;173:1444–1453. doi: 10.4049/jimmunol.173.2.1444. [DOI] [PubMed] [Google Scholar]
- Curiel TJ, Coukos G, Zou L, Alvarez X, Cheng P, Mottram P, et al. Specific recruitment of regulatory T cells in ovarian cancer fosters immune privilege and predicts reduced survival. Nat Med. 2004;10:942–949. doi: 10.1038/nm1093. [DOI] [PubMed] [Google Scholar]
- Karagiannidis C, Akdis M, Holopainen P, Woolley NJ, Hense G, Rückert B, et al. Glucocorticoids upregulate FOXP3 expression and regulatory T cells in asthma. J Allergy Clin Immunol. 2004;114:1425–1433. doi: 10.1016/j.jaci.2004.07.014. [DOI] [PubMed] [Google Scholar]
- Ryanna K, Stratigou V, Safinia N, Hawrylowicz C. Regulatory T cells in bronchial asthma. Allergy. 2009;64:335–347. doi: 10.1111/j.1398-9995.2009.01972.x. [DOI] [PubMed] [Google Scholar]
- Thompson PD, Wade R, Richards S, Gibson BES, Hann I, Eden T, et al. No association of HLA-A supertype with outcome in childhood acute lymphoblastic leukaemia: results of the UKALL XI trial. Br J Haematol. 2011;153:131–133. doi: 10.1111/j.1365-2141.2010.08531.x. [DOI] [PubMed] [Google Scholar]