Abstract
Obesity and related complications are major health burdens. Almost 700 million adults are currently obese globally and the prevalence is predicted to rise towards 2030. The sudden change of lifestyle with physical inactivity and excessive calorie intake undoubtedly have a major part of the epidemic development; however, some individuals seem to be more prone to be affected by an unhealthy lifestyle than others. Hence, genetic predisposition also has an essential role in determining disease susceptibility and response to lifestyle factors. Since the introduction of genome-wide association studies (GWAS), the success of identifying obesity susceptibility variants have increased, and a total of 32 variants have been identified associating genome-wide significantly with body mass index (BMI) and 18 with measures of fat distribution during four overall obesity GWAS waves. However, the immediate success of the GWAS approach has eased off, but the proportion of explained variance for BMI by the identified obesity variants remains low. This review suggests and discusses new initiatives to take GWAS of obesity to the next level, including gene–environment interactions as modulating/masking factors, low-frequent or rare variants and ways to address such analyses, and finally reflections about the applicability of epigenetic modifications when elucidating the genetic background of obesity.
Keywords: obesity, genome-wide association studies, future perspectives, gene–environment interaction, low-frequent and rare variants, epigenetic modifications
Introduction
Obesity and the complications associated with excessive body fat accumulation has become a major global health burden. Projection estimates predict that the number of obese adults will rise from 500 million in 2008 to over 700 million in 2015, and this trend will continue towards 2030.1, 2 The rapid increase in incidence and prevalence of obesity seems to be explained predominantly by the radical change in lifestyle during the last century where high intake of energy-dense food and physical inactivity have become more common. Yet, some individuals seem to be more susceptible to this obesogenic environment, underlining an important genetic component, that also has been established in several twin, family and adoption studies, with heritability estimates ranging from 40 to 70%.3, 4, 5
Obesity is a result of positive energy balance, and biological pathways such as appetite regulation, metabolism and adipogenesis are important factors in the aetiology; however, the complete molecular background of obesity is far from understood. It is anticipated that a deeper understanding of the genetic predisposition to the disease will contribute to the identification of new biological pathways, and hence new drug targets, as well as better prediction and prevention strategies. However, common obesity is a complex, heterogeneous and multi-factorial disease and consequently the unravelling of its genetic architecture has turned out to be a challenging task.
Before 2007 where genome-wide association studies (GWAS) were introduced, obesity gene identification was facilitated using the biological candidate gene method or linkage studies. These methods have resulted in the suggestion of numerous genes; however, none which could be firmly validated.6 Retrospectively, the lack of success was linked to substantial shortcomings of both these methods. Commonly, they suffered from inadequate statistical power to detect the outlined associations, whereas a major limitation of the biological candidate method was inadequate biological and genomic knowledge. Linkage studies identified extremely broad genomic regions and the subsequent fine-mapping to pinpoint the causative gene and/or variant was virtually impossible at the time, and the only withstanding gene is PCSK1 identified using a combination of the two methods.7
The overall lack of success identifying disease-associated genes combined with the aspiration to increase the general biological knowledge and pathological understanding of complex diseases have facilitated new and innovative approaches including GWAS, where the entire genome is scanned for common disease-associating variants in a hypothesis-free manner. This review depicts the progress made within the genetic field of obesity following the introduction of GWAS, with an overview of the identified variants and the method refinements made continually through the GWAS waves. Endingly possible ways ahead and new strategies within the GWAS framework will be discussed.
Genome-wide association studies
The advent of GWAS was facilitated by technological progress and increased knowledge about the human genome, with the International HapMap Consortium (www.hapmap.org) as a major driving force. The complete outline of common single-nucleotide polymorphisms (SNPs) and the existing linkage disequilibrium enabled near-genomic coverage (∼80%) of common variation using a moderate number of SNPs (∼500 000–1 600 000). Simultaneously, progression in genotyping methods shifting to a chip-based technology made massive SNP typing with high accuracy at relatively low costs possible. The number of SNPs analysed and their hypothesis-free scattering across the genome has revolutionised the association study approach, but has also created challenges both in regards to significance threshold, replication demands and interpretation of the functionality. However, as the GWAS waves progressed most challenges have been addressed and adaptive refinements have been made continually. Stringent genome-wide significance thresholds (<10−8) have been established to overcome false-positive findings and a design that involves a discovery stage and at least one replication stage has been introduced to ensure higher validity of the findings. Moreover, imputation strategies have been applied8, 9 to allow combination of data across GWAS populations effectively enlarging the study samples through meta-analyses consequently increasing the power to detect associations. Nevertheless, as these refinements to ensure reproducibility have been an adaptive process, some non-replicable findings did emerge when the GWAS approach was first implemented in the search for obesity susceptibility variants.
GWAS suggested obesity susceptibility loci
The first GWAS of obesity phenotypes was published in 2006. Compared with the later GWAS it was small not only in respect to the number of SNPs analysed but also in respect to the sample size, as a total of 86 604 SNPs were analysed in 694 participants from family studies, and therefore it is often regarded as a pre-GWAS. One SNP, rs7566605 near INSIG2, was suggested to associate with obesity, which was validated in the independent replication stage10 (Table 1).
Table 1. Variants and loci suggested to associate with obesity and/or BMI in GWAS.
| Regional gene(s) | Chromosome | SNP ID | SNP type | RAFa | Effect size BMI (kg m−2)a | Effect size obesity (OR 95% CI)a | Discovery study |
|---|---|---|---|---|---|---|---|
| INSIG2 | 2q14 | rs7566605 | Intergenic | 0.37 | 1.00b | 1.22 (1.05–1.42)b | 10 |
| FTO | 16q12 | rs9939609 | Intronic | 0.45 | 0.36 | 1.31 (1.23–1.39) | 12 |
| PFKPc | 10p15 | rs6602024 | Intronic | 0.10 | 0.84d | — | 13 |
| CTNNBL1 | 20q11 | rs6013029 | Intronic | 0.05 | 0.12 | 1.42 (1.14–1.77)e | 22 |
| rs6020846 | Intronic | 0.07 | 0.09 | 1.32 (1.07–1.62) | |||
| FDFT1c | 8p23 | rs7001819 | Intergenic | 0.41 | — | — | 22 |
| MC4Rf | 18q21 | rs17782313g | Intergenic | 0.24 | 0.22d | 1.12 (1.08–1.16) | 26 |
| TMEM18 | 2p25 | rs6548238h | Intergenic | 0.84 | 0.26 | 1.19 (1.10–1.26) | 15 |
| rs7561317g | Intergenic | 0.84 | 0.19i | 1.20 (1.13–1.27) | 16 | ||
| SH2B1, ATP2A1 | 16p11 | rs7498665 | Coding | 0.41 | 0.15 | 1.11 (1.06–1.17) | 15 |
| 0.44 | 0.45i | 1.08 (1.03–1.13) | 16 | ||||
| KCTD15 | 19q13 | rs11084753h | Intergenic | 0.67 | 0.06 | 1.04 (0.98–1.10) | 15 |
| rs29941 | Intergenic | 0.70 | 0.45i | 1.10 (1.04–1.15) | 16 | ||
| NEGR1 | 1p31 | rs2815752h | Intergenic | 0.62 | 0.10 | 1.05 (1.01–1.11) | 15 |
| rs2568958g | Intergenic | 0.58 | 0.43i | 1.07 (1.02–1.12) | 16 | ||
| GNPDA2 | 4p13 | rs10938397h | Intergenic | 0.45 | 0.19 | 1.12 (1.07–1.17) | 15 |
| MTCH2 | 11p11 | rs10838738h | Intronic | 0.34 | 0.07 | 1.03 (0.98–1.08) | 15 |
| BDNF, LIN7C, LGR4 | 11p14 | rs925946 | Intergenic | 0.30 | 0.19j | 1.11 (1.05–1.16) | 16 |
| SEC16B, RASAL2 | 1q25 | rs10913469 | Intergenic | 0.20 | 0.50i | 1.11 (1.05–1.18) | 16 |
| FAIM2, BCDIN3D | 12q13 | rs7138803 | Intergenic | 0.37 | 0.54i | 1.14 (1.09–1.19) | 16 |
| ETV5, SFRS10, DGKG | 3q27 | rs7647305 | Intergenic | 0.77 | 0.54i | 1.11 (1.05–1.17) | 16 |
| NPC1 | 18q11 | rs1805081 | Coding | 0.44 | −0.06 | 0.71 (0.62–0.84) | 17 |
| MAF | 16q23 | rs1424233 | Intergenic | 0.43 | 0.03 | 1.39 (1.23–1.54) | 17 |
| PTER | 10p12 | rs10508503 | Intergenic | 0.09 | 0.02 | 0.68 (0.38–0.98) | 17 |
| PRL | 6p22 | rs4712652 | Intergenic | 0.41 | −0.08 | 0.83 (0.68–0.98) | 17 |
| RBJ, ADCY3, POMC | 2p23 | rs713586h | Intergenic | 0.47 | 0.14 | 1.07 (1.05–1.09) | 18 |
| GPRC5B, IQCK | 16p12 | rs12444979h | Intergenic | 0.87 | 0.17 | 1.08 (1.04–1.11) | 18 |
| MAP2K5, LBXCOR1 | 15q23 | rs2241423h | Intronic | 0.78 | 0.13 | 1.07 (1.04–1.10) | 18 |
| QPCTL, GIPR | 19q13 | rs2287019h | Intronic | 0.80 | 0.15 | 1.09 (1.05–1.12) | 18 |
| TNNI3K | 1p31 | rs1514175h | Intronic | 0.43 | 0.07 | 1.04 (1.02–1.07) | 18 |
| SLC39A8 | 4q24 | rs13107325h | Coding | 0.07 | 0.19 | 1.10 (1.05–1.15) | 18 |
| FLJ35779, HMGCR | 5q14 | rs2112347h | Intergenic | 0.63 | 0.10 | 1.05 (1.03–1.08) | 18 |
| LRRN6C | 9p21 | rs10968576h | Intronic | 0.31 | 0.11 | 1.04 (1.02–1.06) | 18 |
| TMEM160, ZC3H4 | 19q13 | rs3810291h | Intergenic | 0.67 | 0.09 | 1.06 (1.03–1.08) | 18 |
| FANCL | 2p16 | rs887912h | Intergenic | 0.29 | 0.10 | 1.06 (1.03–1.08) | 18 |
| CADM2 | 3p12 | rs13078807h | Intronic | 0.20 | 0.10 | 1.03 (1.00–1.06) | 18 |
| PRKD1 | 14q11 | rs11847697h | Intergenic | 0.04 | 0.17 | 1.10 (1.03–1.17) | 18 |
| LRP1B | 2q21 | rs2890652h | Intergenic | 0.18 | 0.09 | 1.05 (1.02–1.08) | 18 |
| PTBP2 | 1p21 | rs1555543 | Intergenic | 0.59 | 0.06 | 1.02 (0.99–1.04) | 18 |
| MTIF3, GTF3A | 13q12 | rs4771122 | Intronic | 0.24 | 0.09 | 1.05 (1.01–1.08) | 18 |
| ZNF608 | 5q23 | rs4836133 | Intergenic | 0.48 | 0.07 | 1.03 (1.01–1.05) | 18 |
| RPL27A, TUB | 11p15 | rs4929949 | Intergenic | 0.52 | 0.06 | 1.03 (1.01–1.05) | 18 |
| NUDT3 | 6p21 | rs206936 | Intronic | 0.21 | 0.06 | 1.03 (1.01–1.06) | 18 |
| NRXN3k | 14q31 | rs10150332 | Intronic | 0.21 | 0.13 | 1.09 (1.05–1.12) | 18 |
| TFAP2B | 6p12 | rs987237 | Intronic | 0.18 | 0.13 | 1.09 (1.05–1.12) | 18 |
| TNKS, MSRA | 8p23 | rs17150703 | Intergenic | 0.10 | −0.10 | 1.06 (0.86–1.30) | 19 |
| SDCCAG8g,l | 1q44 | rs12145833 | Intronic | 0.87 | 0.05 | 1.15 (0.96–1.37) | 19 |
| KCNMA1g,l | 10q22 | rs2116830 | Intronic | 0.80 | 1.00 | 1.26 (1.12–1.41) | 20 |
| OLFM4m | 13q14 | rs9568856 | Intergenic | 0.16 | — | 1.22 (1.14–1.29) | 21 |
| HOX5Bm | 17q21 | rs9299 | Coding | 0.65 | — | 1.14 (1.09–1.20) | 21 |
Abbreviations: BMI, body mass index; CI, confidence interval; GWAS, genome-wide association studies; OR, odds ratio; RAF, risk allele frequency; SNP, single-nucleotide polymorphism.
RAF and effect size from first discovery study.
Under a recessive model.
Replication failed in discovery study.
Absolute BMI scores assuming a s.d. of 4.3 kg m−2.
Comparing homozygotes for the risk allele with non-carriers.
Discovered by combination of GWAS data with other study samples in meta-analyses.
Lead SNP from discovery study.
Discovered in meta-analyses of GWAS.
Absolute BMI scores calculated for homozygotes for the risk allele vs non-carriers in the discovery study by Loos et al.44
BMI effect size from Loos et al.44
Previously identified as fat distribution loci.
Identified in extreme obese children and adolescent.
Identified in GWAS of common childhood obesity.
The true GWAS era was introduced a year later, and so far it constitutes of four waves. Most GWAS of obesity has used body mass index (BMI) as a continuous trait, whereas others have examined extreme obesity in children or adults, under the assumption that morbidly obese individuals might be enriched in obesity susceptibility variants. The first obesity GWAS wave resulted in the suggestion of four susceptibility loci. FTO was originally highlighted in a GWAS of type 2 diabetes;11 however, adjustment for BMI revealed that the association was mediated through obesity.12 Variants in or near FTO have since become the most replicated obesity susceptibility locus, emerging in all subsequent GWAS performed on obesity13, 14, 15, 16, 17, 18, 19, 20, 21 except one.22 In the discovery study, the lead SNP (rs9939609) showed a BMI increase of 0.36 kg m−2 and an odds ratio of 1.31 (1.23–1.39) per risk allele carried (Table 1). In the wake of the FTO discovery, a few GWAS suggested variants in or near PFKP,13 CTNNBL1 and FDFT1,22 but replication has been problematic23, 24, 25 even in the replication stage of the discovery studies.13, 22 In the second GWAS wave, the GIANT (Genetic Investigation of ANthropometric Traits) consortium performed meta-analyses of ∼17 000 Caucasian individuals and identified variants in or near MC4R associating with measures of obesity26 (Table 1) and the same variants were also shown to associate with fat distribution represented by waist circumference27 (Table 2).
Table 2. Variants and loci suggested to associate with waist circumference in GWAS.
| Regional gene(s) | Chromosome | SNP ID | SNP type | RAFa | Effect size waist (cm) | Discovery study |
|---|---|---|---|---|---|---|
| MC4R | 18q22 | rs12970134b | Intergenic | 0.30 | 1.48 | 27 |
| TFAP2B | 6p12 | rs987237 | Intronic | 0.16 | 0.46c,d | 28 |
| MSRA | 8p23 | rs545854e | Intergenic | 0.18 | 0.52c,d | 28 |
| NRXN3 | 14q31 | rs10146997 | Intronic | 0.21 | 0.65d | 29 |
Abbreviations: RAF, risk allele frequency; SNP, single-nucleotide polymorphism.
RAF and effect size from first discovery study.
Lead SNP from discovery study.
Effect size reported for the combined stage 1 and 2.
Absolute waist circumference scores assuming an s.d. of 13.1 cm.
The SNP has changed name from rs7826222.
The third obesity GWAS wave included three studies. A meta-analysis of GWAS and an independent GWAS identified variants in or near TMEM18, SH2B1, KCTD15, NEGR1,15, 16 GNPDA2, MTCH2,15 BDNF, SEC16B, FAIM2 and ETV5(ref. 16) genome-wide significantly associated with BMI. Both studies included ∼32 000 individuals and showed effect sizes ranging from 0.06 to 0.54 kg m−2 when comparing homozygous risk allele carriers with non-carriers (Table 1). The third study was performed in study samples of early-onset extreme obesity and reported four putative loci, NPC1, MAF, PTER and PRL; however, only MAF showed stringent genome-wide significance.17 In addition to GWAS performed using measures of general obesity, a parallel GWAS strategy focused on measures of fat distribution using waist circumference and waist-to-hip ratio (WHR) adjusted for BMI. Four novel loci were identified associating with fat distribution measures; LYPLAL1 with waist circumference in women (Table 2), TFAP2B, MSRA28 and NRXN3(ref.29) with WHR (Table 3).
Table 3. Variants and loci suggested to associate with WHR in GWAS.
| Regional gene(s) | Chromosome | SNP ID | SNP type | RAFa | Effect size WHR | Discovery study |
|---|---|---|---|---|---|---|
| LYPLAL1 | 1q41 | rs2605100b | Intergenic | 0.69 | 0.040 | 28 |
| rs4846567 | 0.28 | 0.034 | 30 | |||
| RSPO3 | 6q22 | rs9491696c | Intronic | 0.52 | 0.042 | 30 |
| VEGFA | 6p12 | rs6905288c | Intergenic | 0.56 | 0.036 | 30 |
| TBX15, WARS2 | 1p11 | rs984222c | Intronic | 0.37 | 0.034 | 30 |
| NFE2L3 | 7p15 | rs1055144c | Intergenic | 0.21 | 0.040 | 30 |
| GRB14 | 2q24 | rs10195252c | Intergenic | 0.60 | 0.033 | 30 |
| DNM3, PIGC | 1q24 | rs1011731c | Intronic | 0.57 | 0.028 | 30 |
| ITPR2, SSPN | 12p11 | rs718314c | Intergenic | 0.74 | 0.030 | 30 |
| LY86 | 6p25 | rs1294421c | Intergenic | 0.39 | 0.028 | 30 |
| HOXC13 | 12q13 | rs1443512c | Intergenic | 0.24 | 0.031 | 30 |
| ADAMTS9 | 3p14 | rs6795735c | Intergenic | 0.41 | 0.025 | 30 |
| ZNRF3, KREMEN1 | 22q12 | rs4823006c | Intergenic | 0.57 | 0.023 | 30 |
| NISCH, STAB1 | 3p21 | rs6784615c | Intronic | 0.94 | 0.043 | 30 |
| CPEB4 | 5q21 | rs6861681c | Intronic | 0.34 | 0.022 | 30 |
Abbreviations: GWAS, genome-wide association studies; RAF, risk allele frequency; SNP, single-nucleotide polymorphism; WHR, waist-to-hip ratio.
RAF and effect size from first discovery study.
Association restricted to women.
Identified through meta-analyses of GWAS.
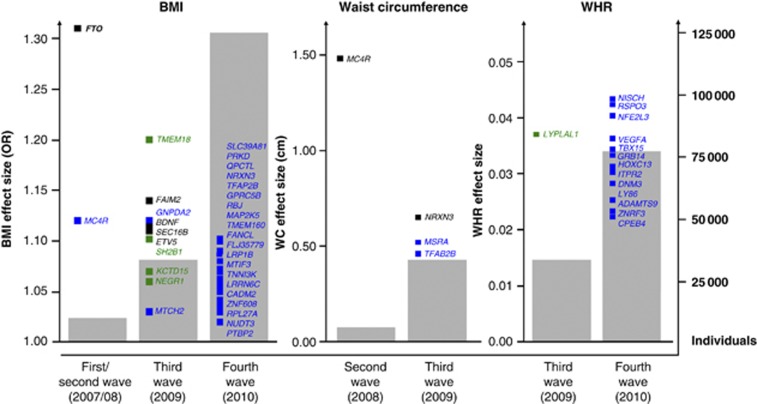
The fourth obesity GWAS wave was dominated by two meta-analyses performed by the GIANT consortium, one comprising ∼124 000 individuals in the discovery stage and ∼250 000 in total using BMI,18 and another comprising ∼77 000 individuals using WHR adjusted for BMI as obesity measure.30 These identified 18 and 13 novel loci, respectively, listed in Tables 1 and 3, respectively. Generally, the WHR variants show stronger association in women than in men, in accordance with the gender-specific difference in fat distribution. Three loci have been suggested in GWAS of extreme obesity; SDCCAG8 and TNKS observed in study samples of children and adolescent,19 KCNMA1 found in an adult population,20 and finally, two loci, OLFM4 and HOXB5, have been identified in studies of common childhood obesity21 (Table 1). Thus, a total of 43 loci have at present been suggested to predispose to overall adiposity and 18 loci to visceral fat accumulation. Of these, 32 BMI and 14 waist/WHR variants are genome-wide significant (Figure 1), as well as one variant (MAF) associating with morbid obesity. The vast majority identified in the fourth wave through extremely large meta-analyses with decreasing effect sizes as consequence (Figure 1).
Figure 1.
Development during the obesity GWAS waves. The progression of the four obesity GWAS waves (2007–2010); genome-wide significant associated loci associating with BMI, waist circumference and WHR, respectively, identified in individual GWAS (black), in both individual GWAS and meta-analysis (green) and in meta-analyses alone (blue). The number of identified genome-wide significant loci increases concurrently with an increase in individuals included in the studies (grey bars), having a decreasing effect size as a consequence (squares). Effect sizes are taken from Tables 1, 2, 3.
Replication of GWAS findings in independent studies
Replication in independent study samples was especially important in the first obesity GWAS wave, before the genome-wide significance threshold was introduced and replication demands were systematically met. Nevertheless, even after refining the GWAS approach, such studies still have their justification, as they estimate independent effect sizes not inflated by ‘winner's curse' and also often extend with analyses of additional related phenotypes, thereby contributing to the elucidation of the overall metabolic impact of identified variants/loci. FTO remains the best replicated obesity gene, as well as the strongest, and a tremendous number of studies have validated the association.31 Likewise, the relatively strong association with obesity observed for variants near MC4R has been well replicated in independent studies.32, 33 For the loci identified in the third GWAS wave, replication attempts have primarily been performed in Caucasian population with divergent results. Among the most successfully validated are TMEM18,19, 34, 35, 36 NEGR1,34, 36, 37 SH2B1,34, 36, 37 MTCH2,34, 36, 37 GNPDA2,35, 37, 38 FAIM235, 38 and BDNF,35, 38 a pattern also recognised in Asian populations.39, 40, 41 Some attempts have been made to validate the fat distribution loci identified in the third wave, however, with limited success.42, 43 These missing associations in independent studies probably reflect a lack of power due to the relatively low effect sizes.
Gained biological knowledge from the obesity GWAS waves
The potential knowledge gained through obesity GWAS findings are generally accumulating as the speed of translation into new biological insight in retrospect has been overestimated. Major impeding factors of the overall biological elucidation have been the fact that the vast majority of the identified obesity susceptibility variants are located in non-coding areas of the genome, including intronic or intergenic regions (Tables 1, 2, 3), and the obvious functionality of the SNPs is therefore difficult to establish within the frames of current genomic knowledge. Hence, the identified variants could either be linkage disequilibrium markers of the causal variant, but some could theoretically be the true causal variant, lying in unknown regulatory motifs or small coding areas of non-described regulatory molecules. Hence, a more thorough understanding of the human genome is required to label variants as functional or non-functional linkage disequilibrium markers with any certainty. In addition, the genomic location of the variants makes a precise link between SNP and affected gene difficult to establish, and consequently, no specific novel biological pathway or mechanism has yet been pinpointed. Nevertheless, it has been suggested that non-coding variants influence transcript regulation rather than gene function44 and some interesting observations are emerging when expression patterns are studied. A majority of the suggested genes harbouring variants associated with overall obesity, represented by BMI, are highly expressed in the central nervous system, whereas many of the suggested fat distribution genes are highly expressed in peripheral tissues.45
Some of the suggested genes have known functions related to obesity; MC4R that is important in appetite regulation,46 BDNF that has been linked with the reward system and eating disorders,47 SH2B1 that is implicated in leptin and insulin signalling,48 and NRXN3 also implicated in reward behaviour.29 TMEM18 is possibly responsible for neural development and NEGR1 controls neuronal outgrowth;15 however, a direct link with obesity has not been established. Several of the identified genes are specifically expressed in hypothalamic regions that could indicate important roles in controlling appetite. These include FTO,49, 50, 51 MTCH2, FAIM2, GNPDA2, KCTD15, ETV5 and NPC1;15 however, their exact biological function and link with obesity remain to be elucidated. Although overall adiposity, for a major part, seems to be mediated through the central nervous system, specific fat deposits or fat accumulation seems to be controlled peripherally, for example, by the adipose tissue itself. This is illustrated by TFAPB2 and LYLPLAL1, which both show high expression in adipose tissue28 and are responsible for lipid accumulation and lipase activity, respectively. The implication of different tissues in overall adiposity and visceral fat accumulation is thus one major biological gain from the obesity GWAS waves.
However, even though GWAS have succeeded in identifying obesity susceptibility variants, especially compared with the previous methods, the proportion of explained variance is still rather low. The GIANT consortium estimated that the confirmed obesity variants explained 1.45% of the inter-individual variation in BMI,18 and obviously a large task still exists in identifying the remaining heritability. Theoretically, a fifth GWAS wave could include even larger meta-analyses, but this would inevitable result in the identification of variants with smaller effect sizes, and it must be considered doubtful whether such knowledge can be translated into increased explained genetic variance. Hence, new strategies must be adopted to take gene identification to the next level, incorporating innovative thinking and new statistical approaches.
Beyond genetic main effects—gene–environment (G × E) interactions
One way to unravel some of the missing or hidden heritability of obesity could be by taking lifestyle factors into account. The environment has changed rapidly during a relatively short period of time, resulting in prevailing sedentary lifestyle and unhealthy dietary habits. During this time, the genetic pool has been stable, and as the obesogenic environment affects individuals at different levels, an important interplay between genes and environmental factors as causation of the obesity epidemic is indicated. This conviction is supported by studies observing an increase in the genetic contribution to BMI variance during the time the environmental changes occurred.52 Thus, a further elucidation of the genetic architecture of complex diseases could involve a comprehensive understanding of more aspects involved in its multi-factorial background, and an evaluation of plausible G × E interactions. However, several challenges supervene when implementing such interactions in genetic epidemiological studies of obesity. First, the identification and prioritisation that environmental exposures are not always straightforward. For complex diseases, such as obesity, the heterogeneous multi-factorial aetiology makes it is a rather demanding task, as numerous potential factors could be intertwining and interplaying with disease risk. Commonly accepted environmental risk factors of obesity are physical inactivity and unhealthy diet, and consequently, these are the most studied factors. Second, environmental factors can be difficult to quantify, and behavioural aspects are especially complicated to estimate. A large gap exists between the gold standard and timely and economically feasible approximations, and large-scale epidemiological studies often rely on subjective self-reports for quantification of both physical activity and dietary patterns. Problems with obesity-specific over- or underreporting have been recognised when accessing both physical activity and food intake,53, 54 but this must still be counterbalanced with the feasibility of the measuring method. Third, methodologically we are far from the ideal scenario where the statistical models used to estimate or elucidate obesity risk can include all modulating factors. This would require models of extreme complexity and the number of parameters needed to be estimated may potentially be infinite. Therefore, current statistical models are unable to fully mimic biological and environmental systems, and with concern of simplicity and practicability models are restricted to include combinations of few genetic and environmental factors. Fourth, adequate statistical power is extremely hard to achieve in G × E interaction analyses. Substantial genetic main effect is needed to obtain the statistical power to detect possible modulating effects of the environment, and even the introduction of GWAS has only resulted in the identification of few variants with sufficient impact to enter such analyses. In addition, even with adequate genetic main effects well-powered G × E interaction studies would still require extremely large study populations,55, 56, 57 only achievable through collaborations and meta-analyses.
In the post GWAS era, the most studied locus with respect to environmental influences has been FTO and especially the impact of physical activity has been evaluated. After the discovery of FTO, it was reported that the increased obesity risk associated with the rs9939609 T-allele was attenuated by physical activity.58 Comprehensive replication attempts have been made in study populations of different ethnicities and with different assessments of physical activity, and validation were achieved in some59, 60, 61, 62, 63, 64, 65, 66, 67, 68 but far from all69, 70, 71, 72, 73, 74 studies, and these inconsistencies left it unresolved whether physical activity reduced the effect of FTO on obesity. To clarify this incongruence, a large meta-analysis comprising 218 166 individuals from 48 different studies has been performed. Overall, a nominal significant interaction was observed with a per allele decreasing effect on BMI of 0.14 kg m−2 (pint=0.005) when comparing physically active and inactive individuals.75 This conclusion could be proof-of-concept in more than one sense. It indicates that well-augmented and biologically plausible G × E interactions do in fact exist, and that several studies can be combined successfully using approximations to standardised quantifications of environmental risk factors.
Another approach recently adopted in G × E interaction analyses is the conversion of several obesity variants into a genetic predisposition score to circumvent lack of power to detect the interactions individually. The applicability was illustrated by a study comprising 20 430 individuals, where 12 SNPs from the first two obesity GWAS waves were combined in a genetic predisposition score summarising the number of BMI increasing alleles. Each BMI increasing allele was associated with a 0.154-kg m−2 increase in BMI, more pronounced in physically inactive individuals (0.205 kg m−2 per allele) than in physically active individuals (0.131 kg m−2 per allele; pint=0.005).76 Collectively, these results indicate that a vast amount of genetic information is hidden or modulated by different lifestyle patterns, and that G × E interaction analyses likely will help improve our understanding of the pathophysiology of obesity and related phenotypes in the future.
Ideally, G × E interaction analyses should be included already in the discovery phase of future GWAS. This could lead to the identification of associations masked by environmental exposures, and hence variants with limited overall genetic main effect but pronounced effect in subgroups of the population. However, the implementations of G × E interactions in GWAS discovery phases pose a huge challenge to international collaborations, consortia and meta-analyses as it is recommended that the study samples are four-doubled when interaction terms are included in the statistical models.77
Methodologically, there is a long way before complete capability to model the complex biology of combined genetic predisposition and modulating environmental exposures is accomplished. Several methods have begun emerging with different focus areas. Some aim at implementing the G × E interaction analysis in the GWAS discovery phase using the likelihood-ratio tests,78, 79, 80 thereby increasing the power to detect associations masked by environmental exposures.81 Others refine associations of genetic variants with known main effect on disease risk, for example, using Bayesian approaches and random forest, to cope with the uncertainties in the general assumption about independence between genetics and the environmental exposure.82, 83, 84 These methods are also employed when searching for the combination of genotypes and environmental factors or interaction chains, with highest impact on disease risk.82, 84, 85 Finally, pathway-driven approaches collecting multiple genetic variants according to their biological function and pathway involvement is gaining ground in G × E interaction analyses.86 However, these innovative methods are not widely used yet, but they could withhold promises for the future for better selection of well-argued combinations of genetic variants and environmental factors in multi-factorial analyses.82, 85, 87
Missing heritability
The current GWAS design has focussed on common SNPs as the predominant type of variation. Nevertheless, a substantial part of the missing or hidden heritability could be found in other types of variants, either structural or of lower frequency.
Copy number variations (CNVs)
The implication of structural variations in common diseases as represented by SNPs in linkage disequilibrium with CNVs on GWAS arrays have been low,88, 89 which could be a result of underrepresentation of such CNV tagSNPs on genotyping chips. Nevertheless, for obesity a few examples have in fact been suggested. The obesity-associated signal rs2815752 tag a 45-kb deletion upstream of NEGR1. Hence, the deletion is a causal candidate for the association signal, but further work is needed in terms of fine-mapping and functional studies before this can be firmly determined.15 Further evidence that CNVs contribute to the genetic architecture of obesity comes from the finding that large deletions on chromosome 16q11 are associated with severe obesity,90, 91 and the deletion spans a large number of genes including SH2B1 also identified in GWAS of obesity.15, 16 A genome-wide analysis has suggested that CNVs at chromosome 11q11 are involved in early-onset extreme obesity; however, this did not reach genome-wide significance.92 Moreover, a spectacular pattern of CNVs has been observed at chromosome 16p11.2. Where deletions in this chromosomal region causes morbid obesity,93 duplications result in underweight among both children and adults94 as an impressive example of how gene dosage can be linked with extreme mirror body composition phenotypes.
Hence, implications of the involvement of structural variation in obesity are present, but due to technical challenges in identifying, quantifying and hence genotyping the CNVs, the complete impact of these types of variation is difficult to estimate with any accuracy before new and better approaches have been developed.
Low-frequent and rare variants
The risk allele frequencies of the obesity variants identified through the GWAS waves are all quite high (Tables 1, 2, 3). Much speculation about missing heritability and improvement of explained variance has focussed on detecting variants with lower frequencies but substantially higher impact on disease risk. Low frequency (∼1–5%) and rare (<1%) variants could have large effect sizes, increasing the risk two- to threefold, without demonstrating Mendelian inheritance,95 and it has been suggested that low-frequent and rare variants in fact are disease disposing,96 and that they can be used for efficient prediction in complex diseases.97 However, detection of potentially disease predisposing, low-frequency and rare variants requires sequencing of a large number of cases and controls,98 which is a demanding task both with respect to costs and the amount of data created. Nevertheless, initiatives to sequence the entire human genome,99 as well as extensive sequencing of all coding regions (the exome) in the ∼20 000 known human genes,100 are already ongoing, and the number of identified low-frequent and rare variants is excessive. It is expected that each of these variants will have a relatively low impact on the disease endpoint and in combination with the heterogeneous nature of common complex diseases, the power to detect associations when testing one variant at the time will be rather low. New analytical strategies cumulating several variants are therefore optimal to obtain adequate statistical power. A tremendous number of methods for these genetic burden tests have recently been developed.101 Some methods use simplistic collapsing of the rare variants (usually <1%) analysing them as one unit, and some weigh the variants using allele frequencies or predicted functionality. As simple pooling of variants can be hampered by associations in different directions, some methods use data-based algorithms, which allow variants to be either protective or deleterious to overcome the diminishing association signal introduced by opposing associations.101
Nevertheless, the gain of identifying a catalogue of low-frequency or rare variants with larger impact on disease risk would be tremendous. A contribution of low-frequency and rare variants in common complex diseases seems to be established,102 and it has been estimated that ∼30 variants with a frequency of 1% and an odds ratio of ∼3 putatively could explain all inherited variance of complex disease.103
Epigenetic modifications
Factors not directly changing the DNA sequence could also contribute to the missing heritability of complex diseases, for example, epigenetic alterations. Epigenetics refer to modifications that regulate gene activity and/or expression rather than its DNA sequence.104 This could be methylation of the DNA sequence, in imprinting, packing of DNA on histones or as blockage of specific gene transcription through methylation of CpG islands in promoter regions. Epigenetic modifications can be programmed already in the intrauterine environment,105, 106 and interestingly, rodent models show inheritance through generations.107 If this is validated to apply to humans, it will interfere with the accepted notion that genetic variation is the only source of heritable diseases, and could give rise to new fundamental theories about heritability of metabolic diseases.108 To what extend epigenetic modifications contribute to the total heritability of obesity is presently unknown. A complicating factor when elucidating the role of epigenetic modifications in complex diseases is the fact that they are highly dynamic and display great tissue specificity.109 As obesity in part is a central nervous system-mediated disorder, tissue samples are inaccessible, further complicating the complete understanding of the role of these modifications. However, several loci related to obesity have interestingly been shown to be subject to genetic imprinting,107, 110 indicating the importance of epigenetic modifications. Moreover, it has been suggested that epigenetics could constitute the link between genetic susceptibility and environmental factors,111 as the plasticity of methylation patterns and histone packing fits perfectly with the dynamic structure of environmental exposures.112 Future steps could therefore include linking the causally unexplained GWAS association signals with epigenetics,113 yet, major efforts lie ahead, even though new technological advances move towards the point where epigenetic can be taken to a large-scale genome-wide level.114, 115 Among other epigenetic modifications and regulators of gene expression are microRNAs, which are small non-coding RNA molecules shown to have a role in many biological and pathological processes through regulation of gene expression.116 Several microRNAs have been shown to interfere with genes in adipogenesis and lipid metabolism; however, the precise mechanisms and extent has not been clarified.117, 118
Concluding remarks and looking ahead
For human genomics research, 2007 was a banner year, where the use of genotyping platforms made GWAS feasible and lifted genetic epidemiological studies to a higher level. The breakthroughs of the HapMap project were integrated in an agnostic approach revealing SNPs located in unanticipated locations of the genome and near loci with no prior link to the disease of interest.
In obesity research, the success of GWAS has resulted in four major waves and a total of 32 validated genome-wide significant loci associated with measures of overall adiposity and 18 loci associated with visceral fat accumulation. However, the instant and immediate success seems to have eased off, and the identification of new SNPs and novel loci only proceeded through the establishment of consortia and collection of large sample sizes in meta-analyses. However, despite a reasonable number of obesity susceptibility variants identified, the proportion of explained genetic variance of BMI remains low.18 The discrimination ability between normal weight and obese individuals is likewise inadequate and far from clinically useful.18, 37, 119 Still, overall important lessons have been learned during the four obesity GWAS waves; fewer variants than expected has been identified, which could be a result of overestimated or anticipated statistical power given the effect sizes appearing. Nevertheless, it is possible that some missing heritability lies in the variants associating near genome-wide significantly in the fourth GWAS wave, but given the heterogeneous and complex nature of the disease, where a high number of common variants most likely contributes in divergent combinations in different individuals, it requires an extremely large study sample to obtain statistical power to clarify this and, the contribution of such variants to explained genetic variance and discrimination ability is, for the same reasons probably, low.18 Therefore, different and innovative strategies increasing the likelihood of identifying new obesity variants with high impact should be incorporated in future GWAS obesity waves.
Emerging strategies include a shift from focussing at common adult obesity to focus at common childhood obesity, and such an initiative has already yielded success. A GWAS meta-analysis of totally 5530 cases and 8318 controls, using age- and gender-matched measures of BMI, identified two loci, OLFM4 and HOXB5, associating genome wide significantly with common childhood obesity,21 and this and similar strategies will undoubtedly contribute to the genetical knowledge of overall obesity in the future.
However, studying the extremes of the BMI distribution still seems as a possible and reasonable way to move forward towards a further unravelling of obesity genetics. Quite some examples already exist where genes causing monogenic forms of obesity through rare, severe and often private mutations also appear in GWAS of common obesity represented by less severe and often non-coding SNPs in proximity to the gene, such examples includes variants near MC4R, POMC and BDNF.
More general initiatives can be made to increase the probability of identifying novel obesity susceptibility variants. The use of refined and more accurate phenotypes could entail more precise classification of existing obesity subtypes, thereby increasing the statistical power to detect distinctive associations. Several approaches and directions could be pursued, one being the improvement of body composition measures. BMI is an accessible measure but dependent on both fat mass and lean mass, and it has been shown to provide misleading information about overall fat content.120 If, for example, the use of skinfold measures and bioimpedance measurements, which gives more accurate estimate of body fat content, were implemented in the GWAS strategy, it would probably increase the likelihood of detecting novel and more specific obesity variants as in the case of rs2943650 near IRS1, which was identified in a GWAS of body fat percentage.121 Another approach could be the identification of serum biomarkers, such as adipokines, potentially able to differentiate between various fat deposits, such as visceral and omental fat.122 Finally, a complementary phenotyping approach could be innovative reflections about the obesity phenotype, for example, focussing on the central nervous system-controlled part of obesity and the neurobiological mechanisms that override the tightly controlled energy homeostasis. Such information on individual addictive behaviour including food preferences could be gained from questionnaires and from functional neuroimaging.
Within genomics, the possibilities of developing and improving the GWAS approach are many. One obvious way to move forward is by focussing on low-frequent and/or rare variants. Novel reference genomes and newly developed algorithms123 make more accurate imputation a plausible gateway to the analyses of low-frequent variants in GWAS settings and this may very well be the next step forward in the unravelling of the genetic background of complex diseases, including obesity. Nevertheless, rare variants are currently not covered by such imputation strategies and initiatives using deep next generation sequencing approaches have, as discussed, already been applied to identify disease predisposing variants with frequencies below 5%. Where whole genome sequencing continues the GWAS outline, with no a priori hypothesis as to genomic location, whole exome sequencing is based on the anticipation that the majority of functional variants will be located in regions presently known to be coding, which also makes the interpretation of functionality more straightforward with the current genomic understanding. Both approaches rely on sequencing cases and controls; however, when studying obesity, this setup and consequently statistical power to identify predisposing variants could be compromised by the fact that the disease theoretically consists of a large number of subtypes with phenotypic distinctions that could be at an almost personal level. No obvious solution exist to circumvent this, but genetics could turn out to be an important contributor when identifying obesity subtypes, as the general subdivision or classification could be predicted by the underlying genetic architecture.
However, substantial challenges emerge when association studies shift focus from common to low-frequent and/or rare variants. Single SNP analyses will be statistically underpowered even in extremely large study populations and hence, large efforts are being put into the development of genetic burden tests were the combined and weighted effect of multiple risk and susceptibility variants in a single gene, a restricted genomic region or in genes involved a biological pathway can be analysed. Some of the developed methods even allow inclusion of interaction terms and this way G × E or gene–gene (G × G) interactions, or in theory even longer interaction chains, could therefore be incorporated into these collapsing methods making this an interesting avenue for future studies.
Even though progress and innovation is important, the bulge of work that has accumulated during the first four obesity GWAS waves cannot be dismissed. It is argued that the non-coding association signals are markers and not the actual causal variants, and this is a highly plausible explanation in the context of the current knowledge about the human genome; however, this is far from complete. Furthermore, the function of most of the human genes, as well their regulation, is unknown; therefore, important transcription factors, and hence also transcription factor binding sites could theoretically exist. Such undiscovered regulatory motifs and coding sequences for small regulatory molecules could justify the theory of the identified association signals being positioned in functional regions. A deeper understanding regarding the genomic location of the identified variants could be an important indicator of where to search for genomic variation in future GWAS and whole genome sequencing waves. One approach that has been used to narrow down the functional variant is resequencing of flanking regions; however, even this can be a daunting task as the distance between the association signal and a causative variant is unknown and in theory can be quite substantial.
Although deep imputation strategies and genetic burden test combining multiple common, low-frequent and rare variants identified through sequencing are realistic approaches in the near future, long-term strategies could include taking large parts of the human genomic sequence into consideration as a personal ‘barcode'. This could instead of focussing on single-nucleotide exchanges also include a more precise mapping of structural variation such as insertions/deletions or CNVs, as well as non-coding RNAs and CpG islands, which could bring the determination of epigenetic modifications much further compared with what is possible today.
Conclusively, the success in genetic epidemiology studies introduced by GWAS has started a scientific avalanche that hopefully will lead to the development of new statistical tools, more detailed genomic insight, deeper biological understanding of disease pathology and translation into clinical use. Eventually, these efforts may have great impact on the treatment strategies for common metabolic disorders like obesity. Moreover, they may at an early stage enable prediction of individuals at high risk of developing obesity making more effective prevention strategies feasible, which could be one of the turning points for the current metabolic health crisis.
The authors declare no conflict of interest.
References
- Kelly T, Yang W, Chen CS, Reynolds K, He J. Global burden of obesity in 2005 and projections to 2030. Int J Obes (Lond) 2008;32:1431–1437. doi: 10.1038/ijo.2008.102. [DOI] [PubMed] [Google Scholar]
- World Health Organization Obesity and overweight, Fact sheet #311, May 2012.
- Maes HH, Neale MC, Eaves LJ. Genetic and environmental factors in relative body weight and human adiposity. Behav Genet. 1997;27:325–351. doi: 10.1023/a:1025635913927. [DOI] [PubMed] [Google Scholar]
- Stunkard AJ, Harris JR, Pedersen NL, McClearn GE. The body-mass index of twins who have been reared apart. N Engl J Med. 1990;322:1483–1487. doi: 10.1056/NEJM199005243222102. [DOI] [PubMed] [Google Scholar]
- Allison DB, Kaprio J, Korkeila M, Koskenvuo M, Neale MC, Hayakawa K. The heritability of body mass index among an international sample of monozygotic twins reared apart. Int J Obes Relat Metab Disord. 1996;20:501–506. [PubMed] [Google Scholar]
- Rankinen T, Zuberi A, Chagnon YC, Weisnagel SJ, Argyropoulos G, Walts B, et al. The human obesity gene map: the 2005 update. Obesity (Silver Spring) 2006;14:529–644. doi: 10.1038/oby.2006.71. [DOI] [PubMed] [Google Scholar]
- Benzinou M, Creemers JW, Choquet H, Lobbens S, Dina C, Durand E, et al. Common nonsynonymous variants in PCSK1 confer risk of obesity. Nat Genet. 2008;40:943–945. doi: 10.1038/ng.177. [DOI] [PubMed] [Google Scholar]
- Marchini J, Howie B. Genotype imputation for genome-wide association studies. Nat Rev Genet. 2010;11:499–511. doi: 10.1038/nrg2796. [DOI] [PubMed] [Google Scholar]
- Li Y, Willer C, Sanna S, Abecasis G. Genotype imputation. Annu Rev Genomics Hum Genet. 2009;10:387–406. doi: 10.1146/annurev.genom.9.081307.164242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herbert A, Gerry NP, McQueen MB, Heid IM, Pfeufer A, Illig T, et al. A common genetic variant is associated with adult and childhood obesity. Science. 2006;312:279–283. doi: 10.1126/science.1124779. [DOI] [PubMed] [Google Scholar]
- Scott LJ, Mohlke KL, Bonnycastle LL, Willer CJ, Li Y, Duren WL, et al. A genome-wide association study of type 2 diabetes in Finns detects multiple susceptibility variants. Science. 2007;316:1341–1345. doi: 10.1126/science.1142382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frayling TM, Timpson NJ, Weedon MN, Zeggini E, Freathy RM, Lindgren CM, et al. A common variant in the FTO gene is associated with body mass index and predisposes to childhood and adult obesity. Science. 2007;316:889–894. doi: 10.1126/science.1141634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scuteri A, Sanna S, Chen WM, Uda M, Albai G, Strait J, et al. Genome-wide association scan shows genetic variants in the FTO gene are associated with obesity-related traits. PLoS Genet. 2007;3:e115. doi: 10.1371/journal.pgen.0030115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinney A, Nguyen TT, Scherag A, Friedel S, Bronner G, Muller TD, et al. Genome wide association (GWA) study for early onset extreme obesity supports the role of fat mass and obesity associated gene (FTO) variants. PLoS One. 2007;2:e1361. doi: 10.1371/journal.pone.0001361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willer CJ, Speliotes EK, Loos RJ, Li S, Lindgren CM, Heid IM, et al. Six new loci associated with body mass index highlight a neuronal influence on body weight regulation. Nat Genet. 2009;41:25–34. doi: 10.1038/ng.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorleifsson G, Walters GB, Gudbjartsson DF, Steinthorsdottir V, Sulem P, Helgadottir A, et al. Genome-wide association yields new sequence variants at seven loci that associate with measures of obesity. Nat Genet. 2009;41:18–24. doi: 10.1038/ng.274. [DOI] [PubMed] [Google Scholar]
- Meyre D, Delplanque J, Chevre JC, Lecoeur C, Lobbens S, Gallina S, et al. Genome-wide association study for early-onset and morbid adult obesity identifies three new risk loci in European populations. Nat Genet. 2009;41:157–159. doi: 10.1038/ng.301. [DOI] [PubMed] [Google Scholar]
- Speliotes EK, Willer CJ, Berndt SI, Monda KL, Thorleifsson G, Jackson AU, et al. Association analyses of 249,796 individuals reveal 18 new loci associated with body mass index. Nat Genet. 2010;42:937–948. doi: 10.1038/ng.686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherag A, Dina C, Hinney A, Vatin V, Scherag S, Vogel CI, et al. Two new loci for body-weight regulation identified in a joint analysis of genome-wide association studies for early-onset extreme obesity in French and german study groups. PLoS Genet. 2010;6:e1000916. doi: 10.1371/journal.pgen.1000916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao H, Arner P, Hoffstedt J, Brodin D, Dubern B, Czernichow S, et al. Genome wide association study identifies KCNMA1 contributing to human obesity. BMC Med Genomics. 2011;4:51. doi: 10.1186/1755-8794-4-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradfield JP, Taal HR, Timpson NJ, Scherag A, Lecoeur C, Warrington NM, et al. A genome-wide association meta-analysis identifies new childhood obesity loci. Nat Genet. 2012;44:526–531. doi: 10.1038/ng.2247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu YJ, Liu XG, Wang L, Dina C, Yan H, Liu JF, et al. Genome-wide association scans identified CTNNBL1 as a novel gene for obesity. Hum Mol Genet. 2008;17:1803–1813. doi: 10.1093/hmg/ddn072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreasen CH, Mogensen MS, Borch-Johnsen K, Sandbaek A, Lauritzen T, Almind K, et al. Lack of association between PKLR rs3020781 and NOS1AP rs7538490 and type 2 diabetes, overweight, obesity and related metabolic phenotypes in a Danish large-scale study: case-control studies and analyses of quantitative traits. BMC Med Genet. 2008;9:118. doi: 10.1186/1471-2350-9-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreasen CH, Mogensen MS, Borch-Johnsen K, Sandbaek A, Lauritzen T, Almind K, et al. Studies of CTNNBL1 and FDFT1 variants and measures of obesity: analyses of quantitative traits and case-control studies in 18,014 Danes. BMC Med Genet. 2009;10:17. doi: 10.1186/1471-2350-10-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel CI, Greene B, Scherag A, Muller TD, Friedel S, Grallert H, et al. Non-replication of an association of CTNNBL1 polymorphisms and obesity in a population of Central European ancestry. BMC Med Genet. 2009;10:14. doi: 10.1186/1471-2350-10-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loos RJ, Lindgren CM, Li S, Wheeler E, Zhao JH, Prokopenko I, et al. Common variants near MC4R are associated with fat mass, weight and risk of obesity. Nat Genet. 2008;40:768–775. doi: 10.1038/ng.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers JC, Elliott P, Zabaneh D, Zhang W, Li Y, Froguel P, et al. Common genetic variation near MC4R is associated with waist circumference and insulin resistance. Nat Genet. 2008;40:716–718. doi: 10.1038/ng.156. [DOI] [PubMed] [Google Scholar]
- Lindgren CM, Heid IM, Randall JC, Lamina C, Steinthorsdottir V, Qi L, et al. Genome-wide association scan meta-analysis identifies three Loci influencing adiposity and fat distribution. PLoS Genet. 2009;5:e1000508. doi: 10.1371/journal.pgen.1000508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heard-Costa NL, Zillikens MC, Monda KL, Johansson A, Harris TB, Fu M, et al. NRXN3 is a novel locus for waist circumference: a genome-wide association study from the CHARGE Consortium. PLoS Genet. 2009;5:e1000539. doi: 10.1371/journal.pgen.1000539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heid IM, Jackson AU, Randall JC, Winkler TW, Qi L, Steinthorsdottir V, et al. Meta-analysis identifies 13 new loci associated with waist-hip ratio and reveals sexual dimorphism in the genetic basis of fat distribution. Nat Genet. 2010;42:949–960. doi: 10.1038/ng.685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loos RJ, Bouchard C. FTO: the first gene contributing to common forms of human obesity. Obes Rev. 2008;9:246–250. doi: 10.1111/j.1467-789X.2008.00481.x. [DOI] [PubMed] [Google Scholar]
- Zobel DP, Andreasen CH, Grarup N, Eiberg H, Sorensen TI, Sandbaek A, et al. Variants near MC4R are associated with obesity and influence obesity-related quantitative traits in a population of middle-aged people: studies of 14,940 Danes. Diabetes. 2009;58:757–764. doi: 10.2337/db08-0620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kring SI, Holst C, Toubro S, Astrup A, Hansen T, Pedersen O, et al. Common variants near MC4R in relation to body fat, body fat distribution, metabolic traits and energy expenditure. Int J Obes (Lond) 2009;34:182–189. doi: 10.1038/ijo.2009.215. [DOI] [PubMed] [Google Scholar]
- Haupt A, Thamer C, Heni M, Machicao F, Machann J, Schick F, et al. Novel obesity risk loci do not determine distribution of body fat depots: a whole-body MRI/MRS study. Obesity (Silver Spring) 2009;18:1212–1217. doi: 10.1038/oby.2009.413. [DOI] [PubMed] [Google Scholar]
- Hotta K, Nakamura M, Nakamura T, Matsuo T, Nakata Y, Kamohara S, et al. Association between obesity and polymorphisms in SEC16B, TMEM18, GNPDA2, BDNF, FAIM2 and MC4R in a Japanese population. J Hum Genet. 2009;54:727–731. doi: 10.1038/jhg.2009.106. [DOI] [PubMed] [Google Scholar]
- Holzapfel C, Grallert H, Huth C, Wahl S, Fischer B, Doring A, et al. Genes and lifestyle factors in obesity: results from 12,462 subjects from MONICA/KORA. Int J Obes (Lond) 2010;34:1538–1545. doi: 10.1038/ijo.2010.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renstrom F, Payne F, Nordstrom A, Brito EC, Rolandsson O, Hallmans G, et al. Replication and extension of genome-wide association study results for obesity in 4923 adults from northern Sweden. Hum Mol Genet. 2009;18:1489–1496. doi: 10.1093/hmg/ddp041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer F, Elbers CC, Adan RA, Loos RJ, Onland-Moret NC, Grobbee DE, et al. Obesity genes identified in genome-wide association studies are associated with adiposity measures and potentially with nutrient-specific food preference. Am J Clin Nutr. 2009;90:951–959. doi: 10.3945/ajcn.2009.27781. [DOI] [PubMed] [Google Scholar]
- Dorajoo R, Blakemore AIF, Sim X, Ong RTH, Ng DPK, Seielstad M, et al. Replication of 13 obesity loci among Singaporean Chinese, Malay and Asian-Indian populations. Int J Obes. 2012;36:159–163. doi: 10.1038/ijo.2011.86. [DOI] [PubMed] [Google Scholar]
- Hong KW, Oh B.Recapitulation of genome-wide association studies on body mass index in the Korean population Int J Obes (Lond) 2011. e-pub ahead of print 1 November 2011doi: 10.1038/ijo.2011.202 [DOI] [PubMed]
- Ng MC, Tam CH, So WY, Ho JS, Chan AW, Lee HM, et al. Implication of genetic variants near NEGR1, SEC16B, TMEM18, ETV5/DGKG, GNPDA2, LIN7C/BDNF, MTCH2, BCDIN3D/FAIM2, SH2B1, FTO, MC4R, and KCTD15 with obesity and type 2 diabetes in 7705 Chinese. J Clin Endocrinol Metab. 2010;95:2418–2425. doi: 10.1210/jc.2009-2077. [DOI] [PubMed] [Google Scholar]
- Bille DS, Banasik K, Justesen JM, Sandholt CH, Sandbaek A, Lauritzen T, et al. Implications of central obesity-related variants in LYPLAL1, NRXN3, MSRA, and TFAP2B on quantitative metabolic traits in adult Danes. PLoS One. 2011;6:e20640. doi: 10.1371/journal.pone.0020640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hotta K, Nakamura M, Nakamura T, Matsuo T, Nakata Y, Kamohara S, et al. Polymorphisms in NRXN3, TFAP2B, MSRA, LYPLAL1, FTO and MC4R and their effect on visceral fat area in the Japanese population. J Hum Genet. 2010;55:738–742. doi: 10.1038/jhg.2010.99. [DOI] [PubMed] [Google Scholar]
- Loos RJ. Recent progress in the genetics of common obesity. Br J Clin Pharmacol. 2009;68:811–829. doi: 10.1111/j.1365-2125.2009.03523.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shmueli O, Horn-Saban S, Chalifa-Caspi V, Shmoish M, Ophir R, Benjamin-Rodrig H, et al. GeneNote: whole genome expression profiles in normal human tissues. Comptes Rendus Biologies. 2003;326:1067–1072. doi: 10.1016/j.crvi.2003.09.012. [DOI] [PubMed] [Google Scholar]
- Farooqi IS. Monogenic human obesity. Front Horm Res. 2008;36:1–11. doi: 10.1159/000115333. [DOI] [PubMed] [Google Scholar]
- Gratacos M, Gonzalez JR, Mercader JM, de Cid R, Urretavizcaya M, Estivill X. Brain-derived neurotrophic factor Val66Met and psychiatric disorders: meta-analysis of case-control studies confirm association to substance-related disorders, eating disorders, and schizophrenia. Biol Psychiatry. 2007;61:911–922. doi: 10.1016/j.biopsych.2006.08.025. [DOI] [PubMed] [Google Scholar]
- Maures TJ, Kurzer JH, Carter-Su C. SH2B1 (SH2-B) and JAK2: a multifunctional adaptor protein and kinase made for each other. Trends Endocrinol Metab. 2007;18:38–45. doi: 10.1016/j.tem.2006.11.007. [DOI] [PubMed] [Google Scholar]
- Gerken T, Girard CA, Tung YC, Webby CJ, Saudek V, Hewitson KS, et al. The obesity-associated FTO gene encodes a 2-oxoglutarate-dependent nucleic acid demethylase. Science. 2007;318:1469–1472. doi: 10.1126/science.1151710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fredriksson R, Hagglund M, Olszewski PK, Stephansson O, Jacobsson JA, Olszewska AM, et al. The obesity gene, FTO, is of ancient origin, up-regulated during food deprivation and expressed in neurons of feeding-related nuclei of the brain. Endocrinology. 2008;149:2062–2071. doi: 10.1210/en.2007-1457. [DOI] [PubMed] [Google Scholar]
- Sanchez-Pulido L, Andrade-Navarro MA. The FTO (fat mass and obesity associated) gene codes for a novel member of the non-heme dioxygenase superfamily. BMC Biochem. 2007;8:23. doi: 10.1186/1471-2091-8-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rokholm B, Silventoinen K, Angquist L, Skytthe A, Kyvik KO, Sorensen TI. Increased genetic variance of BMI with a higher prevalence of obesity. PLoS One. 2011;6:e20816. doi: 10.1371/journal.pone.0020816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lissner L. Measuring food intake in studies of obesity. Public Health Nutr. 2002;5:889–892. doi: 10.1079/phn2002388. [DOI] [PubMed] [Google Scholar]
- Pietilainen KH, Korkeila M, Bogl LH, Westerterp KR, Yki-Jarvinen H, Kaprio J, et al. Inaccuracies in food and physical activity diaries of obese subjects: complementary evidence from doubly labeled water and co-twin assessments. Int J Obes (Lond) 2009;34:437–445. doi: 10.1038/ijo.2009.251. [DOI] [PubMed] [Google Scholar]
- Dempfle A, Scherag A, Hein R, Beckmann L, Chang-Claude J, Schafer H. Gene-environment interactions for complex traits: definitions, methodological requirements and challenges. Eur J Hum Genet. 2008;16:1164–1172. doi: 10.1038/ejhg.2008.106. [DOI] [PubMed] [Google Scholar]
- Luan JA, Wong MY, Day NE, Wareham NJ. Sample size determination for studies of gene-environment interaction. Int J Epidemiol. 2001;30:1035–1040. doi: 10.1093/ije/30.5.1035. [DOI] [PubMed] [Google Scholar]
- Wareham NJ, Young EH, Loos RJ. Epidemiological study designs to investigate gene-behavior interactions in the context of human obesity. Obesity (Silver Spring) 2008;16 (Suppl 3:S66–S71. doi: 10.1038/oby.2008.521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreasen CH, Stender-Petersen KL, Mogensen MS, Torekov SS, Wegner L, Andersen G, et al. Low physical activity accentuates the effect of the FTO rs9939609 polymorphism on body fat accumulation. Diabetes. 2008;57:95–101. doi: 10.2337/db07-0910. [DOI] [PubMed] [Google Scholar]
- Cauchi S, Stutzmann F, Cavalcanti-Proenca C, Durand E, Pouta A, Hartikainen AL, et al. Combined effects of MC4R and FTO common genetic variants on obesity in European general populations. J Mol Med (Berl) 2009;87:537–546. doi: 10.1007/s00109-009-0451-6. [DOI] [PubMed] [Google Scholar]
- Jacobsson JA, Riserus U, Axelsson T, Lannfelt L, Schioth HB, Fredriksson R. The common FTO variant rs9939609 is not associated with BMI in a longitudinal study on a cohort of Swedish men born 1920-1924. BMC Med Genet. 2009;10:131. doi: 10.1186/1471-2350-10-131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HJ, Kim IK, Kang JH, Ahn Y, Han BG, Lee JY, et al. Effects of common FTO gene variants associated with BMI on dietary intake and physical activity in Koreans. Clinica Chimica Acta. 2010;411:1716–1722. doi: 10.1016/j.cca.2010.07.010. [DOI] [PubMed] [Google Scholar]
- Rampersaud E, Mitchell BD, Pollin TI, Fu M, Shen H, O'Connell JR, et al. Physical activity and the association of common FTO gene variants with body mass index and obesity. Arch Intern Med. 2008;168:1791–1797. doi: 10.1001/archinte.168.16.1791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz JR, Labayen I, Ortega FB, Legry V, Moreno LA, Dallongeville J, et al. Attenuation of the effect of the FTO rs9939609 polymorphism on total and central body fat by physical activity in adolescents: the HELENA study. Arch Pediatrics Adolescent Med. 2010;164:328–333. doi: 10.1001/archpediatrics.2010.29. [DOI] [PubMed] [Google Scholar]
- Scott RA, Bailey ME, Moran CN, Wilson RH, Fuku N, Tanaka M, et al. FTO genotype and adiposity in children: physical activity levels influence the effect of the risk genotype in adolescent males. Eur J Hum Genet. 2010;18:1339–1343. doi: 10.1038/ejhg.2010.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonestedt E, Roos C, Gullberg B, Ericson U, Wirfalt E, Orho-Melander M. Fat and carbohydrate intake modify the association between genetic variation in the FTO genotype and obesity. Am J Clin Nutr. 2009;90:1418–1425. doi: 10.3945/ajcn.2009.27958. [DOI] [PubMed] [Google Scholar]
- Vimaleswaran KS, Li S, Zhao JH, Luan J, Bingham SA, Khaw KT, et al. Physical activity attenuates the body mass index-increasing influence of genetic variation in the FTO gene. Am J Clin Nutr. 2009;90:425–428. doi: 10.3945/ajcn.2009.27652. [DOI] [PubMed] [Google Scholar]
- Sonestedt E, Gullberg B, Ericson U, Wirfalt E, Hedblad B, Orho-Melander M. Association between fat intake, physical activity and mortality depending on genetic variation in FTO. Int J Obes (Lond) 2010;35:1041–1049. doi: 10.1038/ijo.2010.263. [DOI] [PubMed] [Google Scholar]
- Ahmad T, Lee IM, Pare G, Chasman DI, Rose L, Ridker PM, et al. Lifestyle interaction with fat mass and obesity-associated (FTO) genotype and risk of obesity in apparently healthy U.S. women. Diabetes Care. 2011;34:675–680. doi: 10.2337/dc10-0948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liem ET, Vonk JM, Sauer PJ, van der Steege G, Oosterom E, Stolk RP, et al. Influence of common variants near INSIG2, in FTO, and near MC4R genes on overweight and the metabolic profile in adolescence: the TRAILS (TRacking Adolescents' Individual Lives Survey) Study. Am J Clin Nutr. 2009;91:321–328. doi: 10.3945/ajcn.2009.28186. [DOI] [PubMed] [Google Scholar]
- Jonsson A, Renstrom F, Lyssenko V, Brito EC, Isomaa B, Berglund G, et al. Assessing the effect of interaction between an FTO variant (rs9939609) and physical activity on obesity in 15,925 Swedish and 2,511 Finnish adults. Diabetologia. 2009;52:1334–1338. doi: 10.1007/s00125-009-1355-2. [DOI] [PubMed] [Google Scholar]
- Kaakinen M, Laara E, Pouta A, Hartikainen AL, Laitinen J, Tammelin TH, et al. Life-course analysis of a fat mass and obesity-associated (FTO) gene variant and body mass index in the Northern Finland Birth Cohort 1966 using structural equation modeling. Am J Epidemiol. 2010;172:653–665. doi: 10.1093/aje/kwq178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lappalainen TJ, Tolppanen AM, Kolehmainen M, Schwab U, Lindstrom J, Tuomilehto J, et al. The common variant in the FTO gene did not modify the effect of lifestyle changes on body weight: the Finnish Diabetes Prevention Study. Obesity (Silver Spring) 2009;17:832–836. doi: 10.1038/oby.2008.618. [DOI] [PubMed] [Google Scholar]
- Liu G, Zhu H, Lagou V, Gutin B, Stallmann-Jorgensen IS, Treiber FA, et al. FTO variant rs9939609 is associated with body mass index and waist circumference, but not with energy intake or physical activity in European- and African-American youth. BMC Med Genet. 2010;11:57. doi: 10.1186/1471-2350-11-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan JT, Dorajoo R, Seielstad M, Sim XL, Ong RT, Chia KS, et al. FTO variants are associated with obesity in the Chinese and Malay populations in Singapore. Diabetes. 2008;57:2851–2857. doi: 10.2337/db08-0214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilpelainen TO, Qi L, Brage S, Sharp SJ, Sonestedt E, Demerath E, et al. Physical activity attenuates the influence of FTO variants on obesity risk: a meta-analysis of 218,166 adults and 19,268 children. PLoS Med. 2011;8:e1001116. doi: 10.1371/journal.pmed.1001116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Zhao JH, Luan J, Ekelund U, Luben RN, Khaw KT, et al. Physical activity attenuates the genetic predisposition to obesity in 20,000 men and women from EPIC-Norfolk prospective population study. PLoS Med. 2010;7:8. doi: 10.1371/journal.pmed.1000332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas D. Gene-environment-wide association studies: emerging approaches. Nat Rev Genet. 2010;11:259–272. doi: 10.1038/nrg2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murcray CE, Lewinger JP, Gauderman WJ. Gene-environment interaction in genome-wide association studies. Am J Epidemiol. 2009;169:219–226. doi: 10.1093/aje/kwn353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraft P, Yen YC, Stram DO, Morrison J, Gauderman WJ. Exploiting gene-environment interaction to detect genetic associations. Hum Hered. 2007;63:111–119. doi: 10.1159/000099183. [DOI] [PubMed] [Google Scholar]
- Lindstrom S, Yen YC, Spiegelman D, Kraft P. The impact of gene-environment dependence and misclassification in genetic association studies incorporating gene-environment interactions. Hum Hered. 2009;68:171–181. doi: 10.1159/000224637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murcray CE, Lewinger JP, Conti DV, Thomas DC, Gauderman WJ. Sample size requirements to detect gene-environment interactions in genome-wide association studies. Genet Epidemiol. 2011;35:201–210. doi: 10.1002/gepi.20569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albrechtsen A, Castella S, Andersen G, Hansen T, Pedersen O, Nielsen RA. Bayesian multilocus association method: allowing for higher-order interaction in association studies. Genetics. 2007;176:1197–1208. doi: 10.1534/genetics.107.071696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee B, Ahn J, Gruber SB, Ghosh M, Chatterjee N. Case-control studies of gene-environment interaction: Bayesian design and analysis. Biometrics. 2009;66:934–948. doi: 10.1111/j.1541-0420.2009.01357.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun YV, Jay CD, Jason HM.Multigenic Modeling of Complex Disease by Random Forests Advances in Geneticsvol. 72, Academic Press, Elsevier: Amsterdam, Netherlands; 201073–99. [DOI] [PubMed] [Google Scholar]
- Li L, Yu M, Jason RD, Shen C, Azzouz F, McLeod HL, et al. A mixture model approach in gene-gene and gene-environmental interactions for binary phenotypes. J Biopharm Stat. 2008;18:1150–1177. doi: 10.1080/10543400802369038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzeng JY, Zhang D, Pongpanich M, Smith C, McCarthy MI, Sale MM, et al. Studying gene and gene-environment effects of uncommon and common variants on continuous traits: a marker-set approach using gene-trait similarity regression. Am J Hum Genet. 2011;89:277–288. doi: 10.1016/j.ajhg.2011.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heidema AG, Feskens EJ, Doevendans PA, Ruven HJ, van Houwelingen HC, Mariman EC, et al. Analysis of multiple SNPs in genetic association studies: comparison of three multi-locus methods to prioritize and select SNPs. Genet Epidemiol. 2007;31:910–921. doi: 10.1002/gepi.20251. [DOI] [PubMed] [Google Scholar]
- Conrad DF, Pinto D, Redon R, Feuk L, Gokcumen O, Zhang Y, et al. Origins and functional impact of copy number variation in the human genome. Nature. 2010;464:704–712. doi: 10.1038/nature08516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarroll SA, Kuruvilla FG, Korn JM, Cawley S, Nemesh J, Wysoker A, et al. Integrated detection and population-genetic analysis of SNPs and copy number variation. Nat Genet. 2008;40:1166–1174. doi: 10.1038/ng.238. [DOI] [PubMed] [Google Scholar]
- Walters RG, Jacquemont S, Valsesia A, de Smith AJ, Martinet D, Andersson J, et al. A new highly penetrant form of obesity due to deletions on chromosome 16p11.2. Nature. 2010;463:671–675. doi: 10.1038/nature08727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bochukova EG, Huang N, Keogh J, Henning E, Purmann C, Blaszczyk K, et al. Large, rare chromosomal deletions associated with severe early-onset obesity. Nature. 2010;463:666–670. doi: 10.1038/nature08689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarick I, Vogel CI, Scherag S, Schafer H, Hebebrand J, Hinney A, et al. Novel common copy number variation for early onset extreme obesity on chromosome 11q11 identified by a genome-wide analysis. Hum Mol Genet. 2011;20:840–852. doi: 10.1093/hmg/ddq518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walters RG, Jacquemont S, Valsesia A, de Smith AJ, Martinet D, Andersson J, et al. A new highly penetrant form of obesity due to deletions on chromosome 16p11.2. Nature. 2010;463:671–675. doi: 10.1038/nature08727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacquemont S, Reymond A, Zufferey F, Harewood L, Walters RG, Kutalik Z, et al. Mirror extreme BMI phenotypes associated with gene dosage at the chromosome 16p11.2 locus. Nature. 2011;478:97–102. doi: 10.1038/nature10406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy MI, Abecasis GR, Cardon LR, Goldstein DB, Little J, Ioannidis JP, et al. Genome-wide association studies for complex traits: consensus, uncertainty and challenges. Nat Rev Genet. 2008;9:356–369. doi: 10.1038/nrg2344. [DOI] [PubMed] [Google Scholar]
- Gorlov IP, Gorlova OY, Sunyaev SR, Spitz MR, Amos CI. Shifting paradigm of association studies: value of rare single-nucleotide polymorphisms. Am J Hum Genet. 2008;82:100–112. doi: 10.1016/j.ajhg.2007.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janssens AC, Aulchenko YS, Elefante S, Borsboom GJ, Steyerberg EW, van Duijn CM. Predictive testing for complex diseases using multiple genes: fact or fiction. Genet Med. 2006;8:395–400. doi: 10.1097/01.gim.0000229689.18263.f4. [DOI] [PubMed] [Google Scholar]
- Cirulli ET, Goldstein DB. Uncovering the roles of rare variants in common disease through whole-genome sequencing. Nat Rev Genet. 2010;11:415–425. doi: 10.1038/nrg2779. [DOI] [PubMed] [Google Scholar]
- 1000 Genomes Project Consortium A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–1073. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Vinckenbosch N, Tian G, Huerta-Sanchez E, Jiang T, Jiang H, et al. Resequencing of 200 human exomes identifies an excess of low-frequency non-synonymous coding variants. Nat Genet. 2010;42:969–972. doi: 10.1038/ng.680. [DOI] [PubMed] [Google Scholar]
- Basu S, Pan W. Comparison of statistical tests for disease association with rare variants. Genet Epidemiol. 2011;35:606–619. doi: 10.1002/gepi.20609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kryukov GV, Pennacchio LA, Sunyaev SR. Most rare missense alleles are deleterious in humans: implications for complex disease and association studies. Am J Hum Genet. 2007;80:727–739. doi: 10.1086/513473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy MI, Hirschhorn JN. Genome-wide association studies: potential next steps on a genetic journey. Hum Mol Genet. 2008;17:R156–R165. doi: 10.1093/hmg/ddn289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bird A. Perceptions of epigenetics. Nature. 2007;447:396–398. doi: 10.1038/nature05913. [DOI] [PubMed] [Google Scholar]
- Waterland RA, Michels KB. Epigenetic epidemiology of the developmental origins hypothesis. Annu Rev Nutr. 2007;27:363–388. doi: 10.1146/annurev.nutr.27.061406.093705. [DOI] [PubMed] [Google Scholar]
- Waterland RA, Jirtle RL. Early nutrition, epigenetic changes at transposons and imprinted genes, and enhanced susceptibility to adult chronic diseases. Nutrition. 2004;20:63–68. doi: 10.1016/j.nut.2003.09.011. [DOI] [PubMed] [Google Scholar]
- Morgan HD, Sutherland HG, Martin DI, Whitelaw E. Epigenetic inheritance at the agouti locus in the mouse. Nat Genet. 1999;23:314–318. doi: 10.1038/15490. [DOI] [PubMed] [Google Scholar]
- Stoger R. The thrifty epigenotype: an acquired and heritable predisposition for obesity and diabetes. Bioessays. 2008;30:156–166. doi: 10.1002/bies.20700. [DOI] [PubMed] [Google Scholar]
- Barros SP, Offenbacher S. Epigenetics: connecting environment and genotype to phenotype and disease. J Dent Res. 2009;88:400–408. doi: 10.1177/0022034509335868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campion J, Milagro FI, Martinez JA. Individuality and epigenetics in obesity. Obes Rev. 2009;10:383–392. doi: 10.1111/j.1467-789X.2009.00595.x. [DOI] [PubMed] [Google Scholar]
- Ober C, Vercelli D. Gene-environment interactions in human disease: nuisance or opportunity. Trends Genet. 2011;27:107–115. doi: 10.1016/j.tig.2010.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vercelli D.Genetics, epigenetics, and the environment: switching, buffering, releasing J Allergy Clin Immunol 2004113381–386.quiz 387. [DOI] [PubMed] [Google Scholar]
- Relton CL, Davey Smith G. Epigenetic epidemiology of common complex disease: prospects for prediction, prevention, and treatment. PLoS Med. 2010;7:e1000356. doi: 10.1371/journal.pmed.1000356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khulan B, Thompson RF, Ye K, Fazzari MJ, Suzuki M, Stasiek E, et al. Comparative isoschizomer profiling of cytosine methylation: the HELP assay. Genome Res. 2006;16:1046–1055. doi: 10.1101/gr.5273806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oda M, Glass JL, Thompson RF, Mo Y, Olivier EN, Figueroa ME, et al. High-resolution genome-wide cytosine methylation profiling with simultaneous copy number analysis and optimization for limited cell numbers. Nucleic Acids Res. 2009;37:3829–3839. doi: 10.1093/nar/gkp260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo H, Ingolia NT, Weissman JS, Bartel DP. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature. 2010;466:835–840. doi: 10.1038/nature09267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heneghan HM, Miller N, Kerin MJ. Role of microRNAs in obesity and the metabolic syndrome. Obes Rev. 2009;11:354–361. doi: 10.1111/j.1467-789X.2009.00659.x. [DOI] [PubMed] [Google Scholar]
- Alexander R, Lodish H, Sun L. MicroRNAs in adipogenesis and as therapeutic targets for obesity. Expert Opin Ther Targets. 2011;15:623–636. doi: 10.1517/14728222.2011.561317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandholt CH, Sparso T, Grarup N, Albrechtsen A, Almind K, Hansen L, et al. Combined analyses of 20 common obesity susceptibility variants. Diabetes. 2010;59:1667–1673. doi: 10.2337/db09-1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller MJ, Bosy-Westphal A, Krawczak M. Genetic studies of common types of obesity: a critique of the current use of phenotypes. Obes Rev. 2010;11:612–618. doi: 10.1111/j.1467-789X.2010.00734.x. [DOI] [PubMed] [Google Scholar]
- Kilpelainen TO, Zillikens MC, Stancakova A, Finucane FM, Ried JS, Langenberg C, et al. Genetic variation near IRS1 associates with reduced adiposity and an impaired metabolic profile. Nat Genet. 2011;43:753–760. doi: 10.1038/ng.866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng Y, Scherer PE. Adipokines as novel biomarkers and regulators of the metabolic syndrome. Ann N Y Acad Sci. 2011;1212:E1–E19. doi: 10.1111/j.1749-6632.2010.05875.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jostins L, Morley KI, Barrett JC. Imputation of low-frequency variants using the HapMap3 benefits from large, diverse reference sets. Eur J Hum Genet. 2011;19:662–666. doi: 10.1038/ejhg.2011.10. [DOI] [PMC free article] [PubMed] [Google Scholar]