The inflammatory response is a tightly regulated and exceedingly complex network of intracellular and intercellular communication events. During the initial phase of the response, soluble proinflammatory mediators are released into the affected area to cause the recruitment and activation of immune cells for clearing the offending pathogens. This is followed by an anti-inflammatory phase in which cellular recruitment ceases, soluble mediators that dampen immune cell activation are released into the surrounding tissues, and the inflammation subsides. The proper coordination of this second phase is obviously critical for the return to a healthy state, and anti-inflammatory mechanisms have been the subject of intensive research efforts for many years. In this issue of PNAS, Park et al. (1) have extended our understanding of this process by demonstrating that the inflammatory mediator nitric oxide (NO) can inhibit the activity of the JNK1 mitogen-activated protein (MAP) kinase in cultured macrophages.
NO participates in physiological processes as diverse as vasodilation, neurotransmission, and inflammation (2–4). Endogenous NO is the product of the dimeric, FAD- and FMN-containing nitric oxide synthases, which catalyze a five-electron oxidation of l-arginine (with the aid of NADPH and tetrahydrobiopterin) to yield l-citrulline and NO. In response to inflammatory stimuli, such as IFN-γ and lipopolysaccharide, expression of the inducible, Ca2+-independent form of NO synthase (iNOS) is up-regulated in macrophages (5). This causes a concomitant increase in NO production, which is a fundamental component of the cytotoxic and cytostatic activities of these cells. This radical gas can combine with molecular oxygen to produce toxic and highly reactive nitrogen oxides such as dinitrogen trioxide and peroxynitrite, and these molecules, in turn, can induce debilitating nitrosative and oxidative chemical stresses. For example, reactive nitrogen oxides can inhibit a variety of enzymes, initiate lipid peroxidation, and directly damage DNA. Moreover, certain thiol groups on the surface of endothelial cells and/or polymorphonuclear neutrophils are thought to be required for normal leukocyte-endothelial cell adhesion, and the reaction of dinitrogen trioxide with these thiol groups can form S-nitrosothiol adducts that may inhibit the adhesion process and consequently decrease leukocyte infiltration during the resolution phase of the inflammatory response (6, 7). Neutrophils from mice lacking iNOS exhibit an increased capacity for inflammation-induced lung infiltration, adhesion to endothelial tissues, and attachment to purified E-selectin ex vivo (8). Thus, NO can be considered an anti-inflammatory molecule insofar as it is instrumental in clearing foreign invaders and blocking leukocyte adhesion.
Mammalian MAP kinases are divided into the extracellular signal-regulated kinase, p38, and c-Jun N-terminal kinase (JNK) subgroups, and all have been implicated in one or more inflammatory processes. The JNKs are activated in macrophages after stimulation with lipopolysaccharide (9–11), and JNK expression is induced in peripheral CD4+ T cells during their differentiation into Th1 and Th2 effector cells (12, 13). JNK also has been shown to play a role in expression of the proinflammatory cytokine tumor necrosis factor α (TNF-α) in inflammatory cells (14–16). Furthermore, it has been demonstrated that JNK is required for the TNF-α-induced expression of E-selectin on endothelial cells, an event that is critical for leukocyte adhesion and infiltration (17, 18).
In a previous study Park and colleagues (19) demonstrated that JNK pathway signaling is inhibited by a thiol redox mechanism, and that this mechanism acts at cysteine-116 in JNK1. Those authors now demonstrate that the S-nitrosylation of cysteine-116 in macrophage JNK1 follows the IFN-γ-induced production of NO, and that this modification causes a significant inhibition of JNK1 activity (1). This finding provides valuable insight into how the inflammatory response operates. Previous reports have shown that in certain inflammatory cell types (e.g., Jurkat T cells, and RAW264.7 macrophages), and under certain experimental conditions, JNK, p38, and extracellular signal-regulated kinase (ERK) MAP kinase pathways are activated in response to reactive nitrogen and reactive oxygen species (20, 21). It also has been demonstrated that NO causes the S-nitrosylation of cysteine-118 on p21(Ras), and that this modification activates Ras (22). S-nitrosylated, activated Ras can recruit and activate phosphatidylinositol 3′-kinase and Raf-1, and this causes the activation of MAP kinase pathways, particularly those mediated by ERK (23). However, there has been no reason to discount the notion that under slightly different conditions nitrosative and/or oxidative stresses might actually repress MAP kinase pathways. The inflammatory response is a temporally and spatially dynamic set of events, and at any given moment the inflammatory microenvironment is a truly Byzantine arrangement of cellular physiologies. Thus, more detailed analyses are needed that examine the effects of time of NO exposure and different NO concentrations on MAP kinase activities in a variety of inflammatory cell types. These investigations should begin to shed light on what may be an endogenous anti-inflammatory mechanism of vital importance. It is interesting to speculate that through the S-nitrosylation of both cysteine-116 in macrophage JNK1 and critical thiol groups on the surface of endothelial cells, NO may down-regulate the inflammatory response via a multifaceted approach. These actions of NO may synergistically promote anti-inflammatory sequelae.
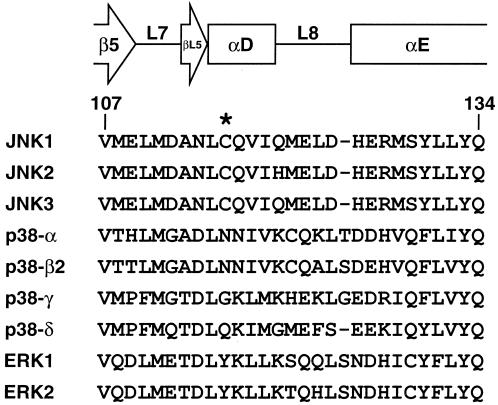
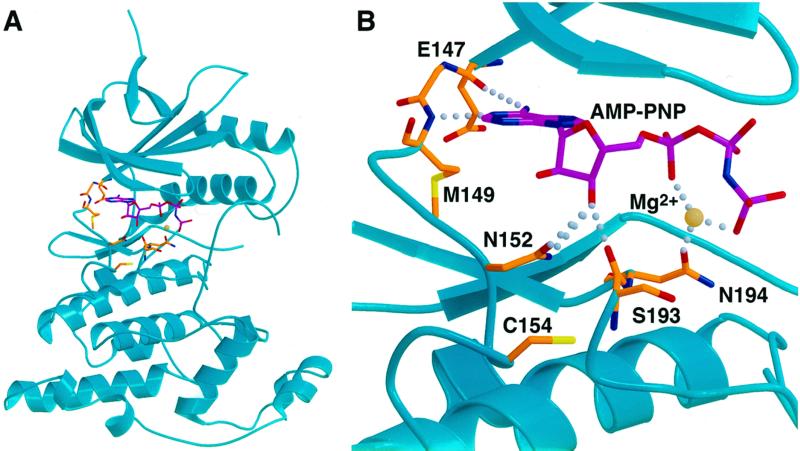
Cysteine-116 is conserved only among the JNK subgroup of MAP kinases (Fig. 1). This raises the intriguing possibility that among the various MAP kinase pathways that are activated during the inflammatory response macrophage NO may specifically repress JNK activity. By itself, this would suggest that JNK pathway inactivation is critical for the ultimate resolution of the inflammatory response, and that JNK inhibitors are indeed excellent candidate therapeutics for disorders in which dysregulated or immoderate inflammation is thought to play a role. It is currently not known precisely how the S-nitrosylation of cysteine-116 inactivates JNK1, but certain assumptions can be drawn from an inspection of the crystal structure of inactive JNK3 in a complex with the ATP analog adenylyl imidodiphosphate (AMP-PNP) (24) (Fig. 2). Cysteine-154 in JNK3, which corresponds to cysteine-116 in JNK1, lies at the amino terminus of α helix D in kinase subdomain V. Immediately amino-terminal to this helix is a linker region that contains three residues that form hydrogen bonds with the AMP-PNP molecule. Specifically, the backbone carbonyl of glutamate-147 forms a hydrogen bond with the amino group of the adenine base (at the N6 position), the backbone amide of methionine-149 forms a hydrogen bond with the nitrogen at the N1 position in the adenine base, and the ribose O3′ hydroxyl forms a hydrogen bond with the side chain of asparagine-152. In addition, cysteine-154 lies quite close to another linker region in the active site, which also contacts the AMP-PNP moiety. Within this linker region the carbonyl group of serine-193 forms a hydrogen bond with the ribose O3′ hydroxyl, and the side-chain carbonyl of asparagine-194 contacts a magnesium ion that bridges the oxygens of the α- and γ-phosphoryl groups of AMP-PNP. By analogy, then, it is possible that the S-nitrosylation of cysteine-116 in JNK1 causes either a steric constraint or unfavorable packing within the active site, which ultimately disrupts this network of hydrogen bonds between JNK1 and the ATP molecule. JNK then would be inhibited by a resulting inability to use ATP for the phosphotransfer reaction. An important caveat to this conclusion, of course, is that these structural determinations were made with an inactive JNK3 molecule. Thus, it is possible that after JNK3 activation via the phosphorylation of threonine-221 and tyrosine-223 (or upon binding of JNK3 to its cognate MAPK kinase), the hydrogen bonds described above between the ATP molecule and the JNK3 active site are naturally disrupted and other contacts take their place. This, in turn, might render the S-nitrosylation of cysteine-154 ineffectual in terms of inhibiting the binding of ATP within the active site and/or inhibiting the subsequent utilization of ATP in the phosphotransfer reaction. The S-nitrosylation of cysteine-116 may simply disrupt intramolecular contacts within JNK1, which are required for normal levels of kinase function. Structural determinations of both inactive and active JNK molecules are needed to clarify these issues.
Figure 1.
Alignment of amino acid sequences of human MAP kinases that correspond to the region surrounding cysteine-116 in JNK1. Secondary structure was taken from ref. 24. Numbers indicate residue positions in JNK1.
Figure 2.
The atomic structure of JNK3 (24) is illustrated. (A) Ribbon representation of the inactive JNK3 structure (cyan) in complex with AMP-PNP (magenta). Specific residues involved in nucleotide binding and Cys-154 are shown in orange. Oxygens are shown in red, nitrogens in blue, sulfurs in yellow, and magnesium in off-white. (B) Close-up view of the active site of JNK3. Colors and orientation are the same as in A. Specific nucleotide contacts and Cys-154 are labeled, and the hydrogen bonds to the nucleotide are shown. The Protein Data Bank entry (1JNK) indicates that the hydrogen bonding contacts of N152 are with the O3′ hydroxyl of the ribose (compare with ref. 24). The images were generated with molscript (31) and rendered with Raster3D (32).
Many signal transduction pathways that promote resolution of the inflammatory response are initiated by cytokine-receptor interactions. Examples include the anti-inflammatory signals promulgated by IL-10 (25), and the down-regulation of the immunomodulatory effects of transforming growth factor β that is initiated by IFN-γ (26). It is becoming increasingly clear, however, that other types of soluble mediators are unleashed during the resolution phase that exert potent anti-inflammatory effects through the covalent modification of signaling proteins. For example, cyclooxygenase-2 (COX2) expression is up-regulated in response to inflammatory stimuli, and at the early stages of the inflammatory response COX2 synthesizes proinflammatory prostaglandins from arachidonic acid. However, at the later stages of inflammatory episodes COX2 directs the production of anti-inflammatory cyclopentenone prostaglandins (cyPGs) such as PGA1 and 15-deoxy-Δ12–14-PGJ2 (27). As reported earlier this year by Rossi et al. (28), these A- and J-type cyPGs covalently modify and inhibit the IκB kinase-β (IKKβ) subunit of the IKK complex both in vitro and in vivo. This subunit is required for the proper regulation of NF-κB-dependent signaling (29, 30). During the inflammatory response, the IKK complex phosphorylates the NF-κB inhibitor IκBα and thereby promotes its ubiquitin-mediated degradation. This causes a concomitant increase in the translocation of NF-κB to the nucleus and an up-regulation of NF-κB-dependent, proinflammatory gene expression. Subsequent to this, and in the later stages of the response, COX2-derived cyPGs inhibit IKKβ and thus inhibit NF-κB activity by decreasing the phosphorylation and degradation of IκBα. In a manner analagous to the NO-dependent inhibition of JNK activity, these actions of cyPGs contribute, ultimately, to the progression of anti-inflammatory sequelae and the denouement of the response.
Park and colleagues (1) have provided a potentially important insight into the biochemistry of JNK signaling during inflammation. However, all too often experimental in vitro settings cannot replicate the physiological environment, and future studies regarding the effect of NO on JNK signaling must confirm that JNK is in fact inhibited by S-nitrosylation during inflammatory events in vivo. Investigations of endogenous servomechanisms such as these undoubtedly will provide therapeutically valuable information, while they broaden and enrich our understanding of the inflammatory response.
Acknowledgments
We thank Dr. David Lambright for assistance with the JNK3 structural analysis. R.J.D. is an Investigator of the Howard Hughes Medical Institute.
Footnotes
See companion article on page 14382.
References
- 1.Park H-S, Huh S-H, Kim M-S, Lee S H, Choi E-J. Proc Natl Acad Sci USA. 2000;97:14382–14387. doi: 10.1073/pnas.97.26.14382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nathan C. FASEB J. 1992;6:3051–3064. [PubMed] [Google Scholar]
- 3.Nathan C, Xie Q W. J Biol Chem. 1994;269:13725–13728. [PubMed] [Google Scholar]
- 4.Grisham M B, Jourd'Heuil D, Wink D A. Am J Physiol. 1999;276:G315–G321. doi: 10.1152/ajpgi.1999.276.2.G315. [DOI] [PubMed] [Google Scholar]
- 5.Nathan C. J Clin Invest. 1997;100:2417–2423. doi: 10.1172/JCI119782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Grisham M B, Granger D N, Lefer D J. Free Radical Biol Med. 1998;25:404–433. doi: 10.1016/s0891-5849(98)00094-x. [DOI] [PubMed] [Google Scholar]
- 7.Granger D N, Kubes P. Methods Enzymol. 1996;269:434–442. doi: 10.1016/s0076-6879(96)69044-2. [DOI] [PubMed] [Google Scholar]
- 8.Hickey M J, Sharkey K A, Sihota E G, Reinhardt P H, Macmicking J D, Nathan C, Kubes P. FASEB J. 1997;11:955–964. doi: 10.1096/fasebj.11.12.9337148. [DOI] [PubMed] [Google Scholar]
- 9.Chan E D, Winston B W, Jarpe M B, Wynes M W, Riches D W. Proc Natl Acad Sci USA. 1997;94:13169–13174. doi: 10.1073/pnas.94.24.13169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hambleton J, Weinstein S L, Lem L, DeFranco A L. Proc Natl Acad Sci USA. 1996;93:2774–2778. doi: 10.1073/pnas.93.7.2774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sanghera J S, Weinstein S L, Aluwalia M, Girn J, Pelech S L. J Immunol. 1996;156:4457–4465. [PubMed] [Google Scholar]
- 12.Weiss L, Whitmarsh A J, Yang D D, Rincon M, Davis R J, Flavell R A. J Exp Med. 2000;191:139–146. doi: 10.1084/jem.191.1.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rincon M, Derijard B, Chow C W, Davis R J, Flavell R A. Genes Funct. 1997;1:51–68. doi: 10.1046/j.1365-4624.1997.00007.x. [DOI] [PubMed] [Google Scholar]
- 14.Swantek J L, Cobb M H, Geppert T D. Mol Cell Biol. 1997;17:6274–6282. doi: 10.1128/mcb.17.11.6274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ishizuka T, Terada N, Gerwins P, Hamelmann E, Oshiba A, Fanger G R, Johnson G L, Gelfand E W. Proc Natl Acad Sci USA. 1997;94:6358–6363. doi: 10.1073/pnas.94.12.6358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kontoyiannis D, Pasparakis M, Pizarro T T, Cominelli F, Kollias G. Immunity. 1999;10:387–398. doi: 10.1016/s1074-7613(00)80038-2. [DOI] [PubMed] [Google Scholar]
- 17.Read M A, Whitley M Z, Gupta S, Pierce J W, Best J, Davis R J, Collins T. J Biol Chem. 1997;272:2753–2761. doi: 10.1074/jbc.272.5.2753. [DOI] [PubMed] [Google Scholar]
- 18.Min W, Pober J S. J Immunol. 1997;159:3508–3518. [PubMed] [Google Scholar]
- 19.Park H S, Park E, Kim M S, Ahn K, Kim I Y, Choi E J. J Biol Chem. 2000;275:2527–2531. doi: 10.1074/jbc.275.4.2527. [DOI] [PubMed] [Google Scholar]
- 20.Lander H M, Jacovina A T, Davis R J, Tauras J M. J Biol Chem. 1996;271:19705–19709. doi: 10.1074/jbc.271.33.19705. [DOI] [PubMed] [Google Scholar]
- 21.Jun C D, Oh C D, Kwak H J, Pae H O, Yoo J C, Choi B M, Chun J S, Park R K, Chung H T. J Immunol. 1999;162:3395–3401. [PubMed] [Google Scholar]
- 22.Lander H M, Hajjar D P, Hempstead B L, Mirza U A, Chait B T, Campbell S, Quilliam L A. J Biol Chem. 1997;272:4323–4326. doi: 10.1074/jbc.272.7.4323. [DOI] [PubMed] [Google Scholar]
- 23.Deora A A, Hajjar D P, Lander H M. Biochemistry. 2000;39:9901–9908. doi: 10.1021/bi992954b. [DOI] [PubMed] [Google Scholar]
- 24.Xie X, Gu Y, Fox T, Coll J T, Fleming M A, Markland W, Caron P R, Wilson K P, Su M S. Structure (London) 1998;6:983–991. doi: 10.1016/s0969-2126(98)00100-2. [DOI] [PubMed] [Google Scholar]
- 25.Stordeur P, Goldman M. Int Rev Immunol. 1998;16:501–522. doi: 10.3109/08830189809043006. [DOI] [PubMed] [Google Scholar]
- 26.Ulloa L, Doody J, Massague J. Nature (London) 1999;397:710–713. doi: 10.1038/17826. [DOI] [PubMed] [Google Scholar]
- 27.Gilroy D W, Colville-Nash P R, Willis D, Chivers J, Paul-Clark M J, Willoughby D A. Nat Med. 1999;5:698–701. doi: 10.1038/9550. [DOI] [PubMed] [Google Scholar]
- 28.Rossi A, Kapahi P, Natoli G, Takahashi T, Chen Y, Karin M, Santoro M G. Nature (London) 2000;403:103–108. doi: 10.1038/47520. [DOI] [PubMed] [Google Scholar]
- 29.Tanaka M, Fuentes M E, Yamaguchi K, Durnin M H, Dalrymple S A, Hardy K L, Goeddel D V. Immunity. 1999;10:421–429. doi: 10.1016/s1074-7613(00)80042-4. [DOI] [PubMed] [Google Scholar]
- 30.Li Q, Van Antwerp D, Mercurio F, Lee K F, Verma I M. Science. 1999;284:321–325. doi: 10.1126/science.284.5412.321. [DOI] [PubMed] [Google Scholar]
- 31.Kraulis P J. J Appl Crystallogr. 1991;24:946–950. [Google Scholar]
- 32.Merritt E A, Bacon D J. Methods Enzymol. 1997;277:505–524. doi: 10.1016/s0076-6879(97)77028-9. [DOI] [PubMed] [Google Scholar]