Abstract
A goal of our studies is to develop a potential therapeutic for Parkinson’s disease (PD) by a human GDNF (hGDNF) expression plasmid administered to the rat striatum as a compacted DNA nanoparticle (DNP) and which will generate long-term hGDNF expression at biologically active levels. In the present study we used a DNA plasmid encoding for hGDNF and a polyubiquitin C (UbC) promoter that was previously shown to have activity in both neurons and glia, but primarily in glia. A two-fold improvement was observed at the highest plasmid dose when using hGDNF DNA incorporating sequences found in RNA splice variant 1 compared to splice variant 2; of note, the splice variant 2 sequence is used in most preclinical studies. This optimized expression cassette design includes flanking scaffold matrix attachment elements (S/MARs) as well as a CpG-depleted prokaryotic domain and, where possible, eukaryotic elements. Stable long-term GDNF activity at levels 300–400% higher than baseline was observed following a single intracerebral injection. In a previous study DNPs plasmids encoding for reporter genes had been successful in generating long-term reporter transgene activity in the striatum (>365 days) and in this study produced sustained GDNF activity at the longest assessed time point (6 months).
Keywords: plasmid optimization, Parkinson’s disease, gene therapy, splice variant
INTRODUCTION
Recent gene therapy studies have shown promising results using viral vector gene delivery systems in the central nervous system (Manfredsson and Mandel, 2010). Although striatal delivery of various neurotrophic factors have been shown to provide modest improvement in a rat model of PD, members of the GDNF family have provided the most beneficial effects and it is these factors that have moved on to clinical trials. Neuroprotective and neurorestorative effects for GDNF have been demonstrated using a variety of viral vectors: adeno-associated (AAV) in rodents (Mandel et al., 1997, Kirik et al., 2000) and non-human primates (Eslamboli et al., 2005, Elsworth et al., 2008, Eberling et al., 2009), lentivirus (LV) in rodents (Bensadoun et al., 2000, Georgievska et al., 2002, Azzouz et al., 2004) and non-human primates (Kordower et al., 2000, Palfi et al., 2002), adenovirus (Ad) in rodents (Choi-Lundberg et al., 1998, Connor et al., 2001) and herpes simplex virus (HSV) in rodents (Natsume et al., 2001, Sun et al., 2005).
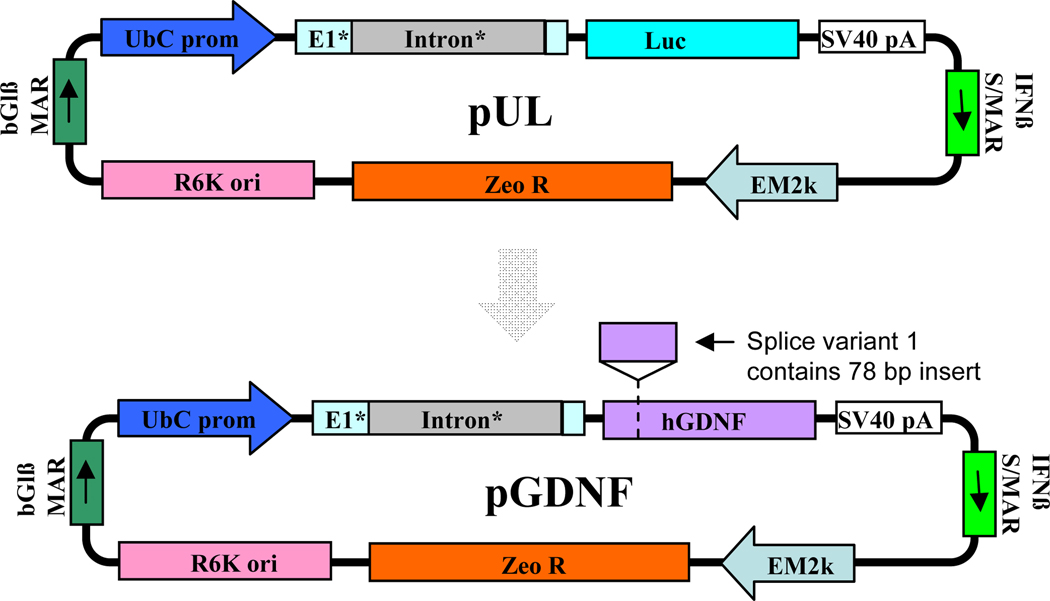
Previously, we used a plasmid (pUL; see Figure 1) compacted into DNA nanoparticles (DNPs) to achieve long-term and stable transgene activity of a reporter gene, luciferase, in the brain; transgene activity was sustained for >365 days in the mouse (Yurek et al., 2011). To develop a non-viral therapy for PD preclinical models, we developed DNPs containing a pUL derivative encoding rat GDNF cDNA. GDNF was chosen for its potent neurotrophic effect on dopaminergic neurons; firstly, to ameliorate the neurodegenerative effects of Parkinson’s disease and secondly, for morphological differentiation of dopaminergic neurons (Lin et al., 1993). We observed sustained GDNF over-expression after single injections of rat GDNF DNPs into the striatum (Yurek et al., 2009b); these same GDNF DNPs imparted a neurotrophic effect on grafted fetal ventral mesencephalic cells when injected into the brain 1 week prior to the grafting procedure (Yurek et al., 2009a). Given these encouraging results, we now develop optimized human GDNF (hGDNF) DNPs to produce high level and prolonged duration of hGDNF in the rat striatum. A panel of DNPs was formulated to evaluate expression plasmids encoding hGDNF splice variants 1 and 2: 1) splice variants of human GDNF cDNA; 2) splice variants of a custom-designed codon-optimized and CpG-depleted hGDNF; 3) splice variants of a custom-designed hGDNF additionally modified to minimize heterochromatin formation. The prokaryotic backbone and, where possible, eukaryotic elements were CpG depleted. An optimized Kozak sequence was included in every construct to ensure efficient translation initiation. Splice variants 1 and 2 of GDNF produce the same mature, secreted hGDNF protein, although the shorter splice variant 2 has a 78 bp deletion in the pro-region of the GDNF coding sequence (Figure 1) (Schaar et al., 1993, Trupp et al., 1995, Grimm et al., 1998). The motivation for testing these splice variants is a report suggesting that the shorter splice variant results in less secreted hGDNF (Wang et al., 2008). Additionally, since heterochromatin formation can reduce levels and duration of transgene expression, strategies to reduce heterochromatin formation have been proposed (Luykx et al., 2006, Radwan et al., 2008). Based on prolonged production of biologically active levels of hGDNF, an optimized hGDNF DNP has been selected for the next phase of preclinical testing.
Figure 1.
Plasmid maps for pUL and pGDNF. The pGDNF plasmid used in this study was derived from the pUL plasmid, which had been successfully tested in previous studies (Yurek et al., 2011). Six different derivatives of pGDNF were tested in this study, as described in the text.
EXPERIMENTAL PROCEDURES
Plasmid Construction
DNA vectors were constructed using standard molecular biology techniques, followed by restriction analysis and sequencing of subcloned regions (Sambrook et al., 1989). Detailed methodology for the pUL plasmid construction is reported in a previous publication (Yurek et al., 2009b). Figure 1 shows the pUL plasmid map from which the GDNF plasmids were derived.
Preparation of Condensing Peptide
L-Cysteinyl-poly-L-lysine 30-mers (UCB Bioproducts, Inc.) were conjugated with 10-kDa polyethylene glycol (PEG) (Nektar Therapeutics) as described in (Liu et al., 2003) except that trifluoroacetate counterion was replaced with acetate by size-exclusion chromatography on Sephadex G-25 before lyophilization of the PEGylated peptide.
Formulation of DNA Nanoparticles
Compacted DNA was manufactured by adding 20.0 mL of DNA solution (0.1 mg/mL in water) to 2.0 mL of PEGylated condensing peptide (3.2 mg/mL in water) at a rate of 4.0 mL/min by a syringe pump and through sterile tubing ended with a blunt cannula. During this addition, the tube with peptide was vortexed at a controlled rate so that the two materials mixed instantaneously. Peptide and DNA were formulated at a final amine-to-phosphate ratio of 2:1. The DNP was then filtered through a vacuum-driven sterile filter with 0.2-µm polyethersulfone membrane. The filtered DNP was then processed with tangential flow filtration to remove excess peptide and exchange solvent for saline, and then was concentrated 20–30 fold using VIVASPIN centrifugal concentrators (MWCO 100k). The final concentrations of DNA were 4.1 µg/µl, 4.4 µg/µl, 4.1 µg/µl, and 4.3 µg/µl for pGDNF_1a, pGDNF_1b, pGDNF_2b, and pGDNF_3b. After formulation, the DNPs underwent several quality control tests, including sedimentation, turbidity, gel electrophoresis, and transmission electron microscopy, as described in (Liu et al., 2003, Ziady et al., 2003). Also, endotoxin levels were checked using an ENDOSAFE® PTS (Portable Test System) manufactured by Charles River Laboratories. Estimated number of transfecting nanoparticles: pGDNF_1a = 9.39 × 1011 particles/µl; pGDNF_1b = 9.88 × 1011 particles/µl; pGDNF_2b = 9.21 × 1011 particles/µl; pGDNF_3b = 9.66 × 1011 particles/µl.
Transfection of Ventral Midbrain Culture
The ventral mesencephalon from E14 fetuses were dissected on ice and placed into oxygenated calcium/magnesium-free buffer (CMF; 0.15 M NaCl, 8.0 mM Na2HPO4, 2.7 mM KCl, 1.5 mM KHPO4, 26.0 mM NaHCO3, 0.1% glucose, 100 mg/ml streptomycin, 2.5 mg/ml fungizone, pH 7.2). Embryonic tissue was pooled and rinsed with CMF; a cell suspension was prepared using a 0.125% trypsin solution (Invitrogen; Carlsbad, CA) followed by inactivation with 1.0 ml newborn calf serum (Invitrogen; Carlsbad, CA) and 500 µl of DNase [0.5 mg/ml] in 3.5 ml Hank’s Balanced Salt Solution (Invitrogen; Carlsbad, CA). Cells were triturated with a 22g needle 15 times, placed drop-wise on top of sterile fetal bovine serum (Invitrogen; Carlsbad, CA), and spun at 1000 RPM for 10 minutes. Cells were plated on 24 well poly-D-lysine coated plates at a density of 3.75 × 105 cells per well in DMEM/F12/N2 medium. At 72 hours post-plating, cultured ventral midbrain cells were treated with 1 µg of naked GDNF plasmid in the presence of Lipofectamine™ and incubated for 4 hours. After 4 hours of incubation, medium was removed and DMEM/F12/N2 was added. Cells were fed every other day until day 7. The medium was removed and stored at −80°C for analysis of secreted GDNF protein. 200 µl of 1X cell culture lysis reagent (Promega; Madison, WI) was added to each well. Plates were placed on shaker in cold room for 30 minutes and samples were transferred to microcentrifuge vials. Samples were spun at 10,000 × g for 10 minutes at 4°C. Supernatant was stored at −80°C for intracellular GDNF protein analysis.
Animals
Male Sprague-Dawley rats at ages 4–5 months were obtained from Harlan Farms and used exclusively in this study. All rat procedures were conducted in strict compliance with approved institutional protocols, and in accordance with the provisions for animal care and use described in the "Guide for the Care and Use of Laboratory Animals” (NIH publication No. 86-23, NIH, 1985).
Stereotactic Microinjection of DNPs into Brain
DNA nanoparticles or plasmids alone were suspended in sterile saline and loaded into a sterile 30 gauge Hamilton syringe needle. Injections were made stereotactically into the brain of isoflurane-anesthetized rats at a rate of 0.5 µL/min for 4 minutes per site. DNPs were delivered to the brain using two needle tracts for injections made into the striatum. For each tract the injector was lowered to the deepest DV coordinate (DV1), a 2.0 µL deposit of DNPs was made at this site, the injector was then retracted to the second site (DV2) and a second 2.0 µL deposit was made at this site. Stereotactic coordinates: Tract 1 AP +0.5, ML +3.6, DV1 −6.5, DV2 −5.0; Tract 2 AP +0.5, ML +2.4, DV1 −6.5, DV2 −5.0. All stereotaxic adjustments were made with the skull in the flat position: AP and ML adjustments were made from the intersection of the sagittal and coronal sutures (bregma), and DV adjustments were made from the top of the skull; all stereotactic coordinates used in these studies were determined using a stereotaxic atlas (Paxinos and Watson, 2007).
6-hydroxydopamine Lesion
Some rats were given unilateral 6-hydroxydopamine (6-OHDA) lesions of the left nigrostriatal pathway; 6-OHDA (Sigma) was dissolved in 0.9% saline (containing 0.2% ascorbic acid) at a concentration of 3.0 µg/µl and stereotaxically injected into the nigrostriatal pathway of anesthetized rats at a rate of 1.0 µl/min for 2 min. Each rat received two injections of 6-OHDA: one injection in the medial forebrain bundle (AP −4.4, ML 1.2, DV −7.5) area and the other in the rostral pars compacta of the substantia nigra (AP −5.3, ML 2.0, DV −7.5); all coordinates reported in this study represent millimeter adjustments from bregma (AP, ML) and below the dural surface (DV) with the top of the skull in a flat position. This technique routinely produces complete lesions of dopamine neurons in the A9 and A10 midbrain regions, and near complete denervation of dopaminergic fibers innervating the ipsilateral striatum (Thomas et al., 1994).
Tissue Dissection for ELISA Assays
For striatal dissections, brains were removed from euthanized animals and then placed upside down (ventral surface facing upwards) into a stainless steel brain matrix (RMB-4000C, ASI Instruments). For the following descriptions, the approximate stereotaxic coordinates are listed in parentheses. The initial coronal cut was targeted at a level just caudal to the optic chiasm (~AP −0.5) and a second cut was made 3.0 mm anterior to the initial cut. The 3.0 mm thick coronal slab formed by these two cuts was removed from the brain matrix and then divided into two pieces along the midline. Both the left and right striatum were dissected from the two pieces. A rectangular piece of tissue was dissected by making the following cuts: 1) a cut parallel to the midline was made just lateral to the lateral ventricle, 2) a cut parallel to the midline was made just medial to the lateral aspect of striatum at the striatum/corpus callosum border, 3) a cut perpendicular to the midline was made just ventral to the dorsal aspect of the striatum at the striatum/corpus callosum border, and 4) a cut perpendicular to the midline was made just dorsal to the anterior commissure. Dissected tissues were weighed, immediately frozen on dry ice and stored at −80°C until analyzed for protein content.
Quantification of GDNF by Enzyme-Linked immunosorbent Assay (ELISA)
Protein levels of GDNF were measured in culture media, VM culture cell lysates or tissue samples. Subsequently, each tissue sample was homogenized in 300 µl volumes of homogenate buffer [400 mM NaCl, 0.1% Triton-X, 2.0 mM EDTA, 0.1 mM benzethonium chloride, 2.0 mM benzamidine, 0.1 mM PMSF, Aprotinin (9.7 TIU/ml), 0.5% BSA, 0.1 M phosphate buffer, pH=7.4]. The homogenate was centrifuged for 10 minutes at 10,000 × g at 4°C. Tissue homogenates, VM culture cell lysates and culture media samples were assayed for GDNF content using a GDNF Emax™ ImmunoAssay System (Promega; Madison, WI).
Statistical Analyses
The α level of significance was set at p < 0.05. Analysis of variance (ANOVA) was used for statistical analyses; choice of test was dependent on the experimental design. Descriptive statistics: means are reported with their corresponding standard error of the mean (s.e.m.). For the in vivo plasmid dose-response study, we used a 3-way ANOVA; the 3 factors were plasmid, dose, and side (left or right hemisphere). For the longitudinal analysis of GDNF protein levels, groups of treated animals were sacrificed at each time point and the amount of GDNF in the injected striatum was normalized to the amount in the non-injected (contralateral) striatum; a log10 transformation of the normalized data was performed in order to carry out a 1-way ANOVA to analyze for the factor of time after DNP injection.
RESULTS
Plasmid Optimization for human pGDNF
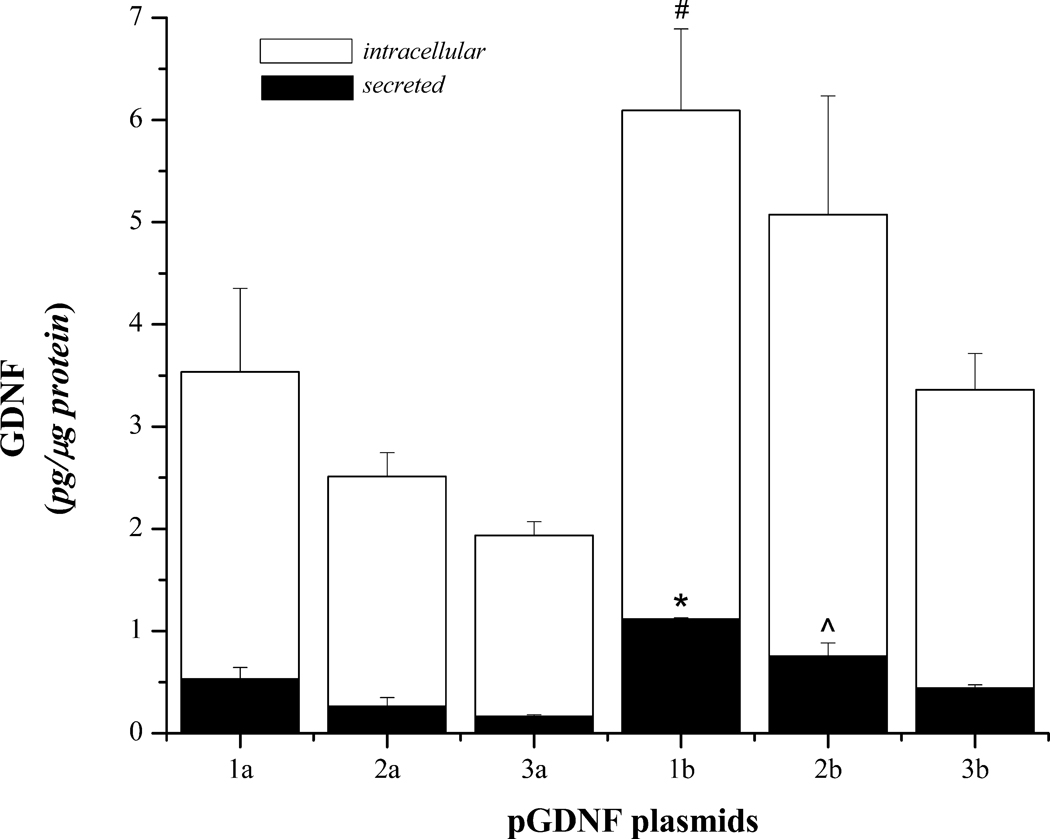
Primary ventral midbrain cultures were established and then transfected with the six different plasmids described in Table 1; all plasmids were derived from the pUL plasmid (Figure 1) and encoded for hGDNF splice variants 1 or 2. Four days after transfection, media was removed and analyzed for secreted and intracellular GDNF using ELISA (Figure 2). Statistical analyses of GDNF protein values revealed a significant treatment effect for secreted GDNF [F(5,6)=19.03, p<0.001] as well as intracellular GDNF protein [F(5,6)=5.16, p=0.035]. From the protein analyses generated from the VM culture studies, we chose the 4 best plasmids (pGDNF_1b, pGDNF_2b, pGDNF_1a, and pGDNF_3b) and then we compacted each plasmid individually into DNPs for an in vivo dose-response study.
Table 1.
| Vector | Splice Variant | Vector Description |
|---|---|---|
| pGDNF_1a | 2 (short) | pUL derivative with natural hGDNF cDNA |
| pGDNF_1b | 1 (long) | |
| pGDNF_2a | 2 | pUL derivative with codon optimized hGDNF |
| pGDNF_2b | 1 | |
| pGDNF_3a | 2 | pUL derivative with nucleosome non-friendly hGDNF |
| pGDNF_3b | 1 |
Figure 2.
Secreted and intracellular GDNF in primary ventral midbrain cultures transfected with pGDNF plasmids. Ventral midbrain cultures were established as described in the Methods section and transfected with one of the following plasmids: pGDNF_1a (1a), pGDNF_1b (1b), pGDNF_2a (2a), pGDNF_2b (2b), pGDNF_3a (3a), or pGDNF_3b (3b). Each bar represents the average (± s.e.m.; n=5) amount of GDNF protein detected in cells or secreted in the culture media. *p<0.05, 1b vs. 1a, 2a, 3a, 2b, 3b; ^p<0.05 2b vs. 3a, 2a; #p<0.05 1b vs 2a, 3a, 3b.
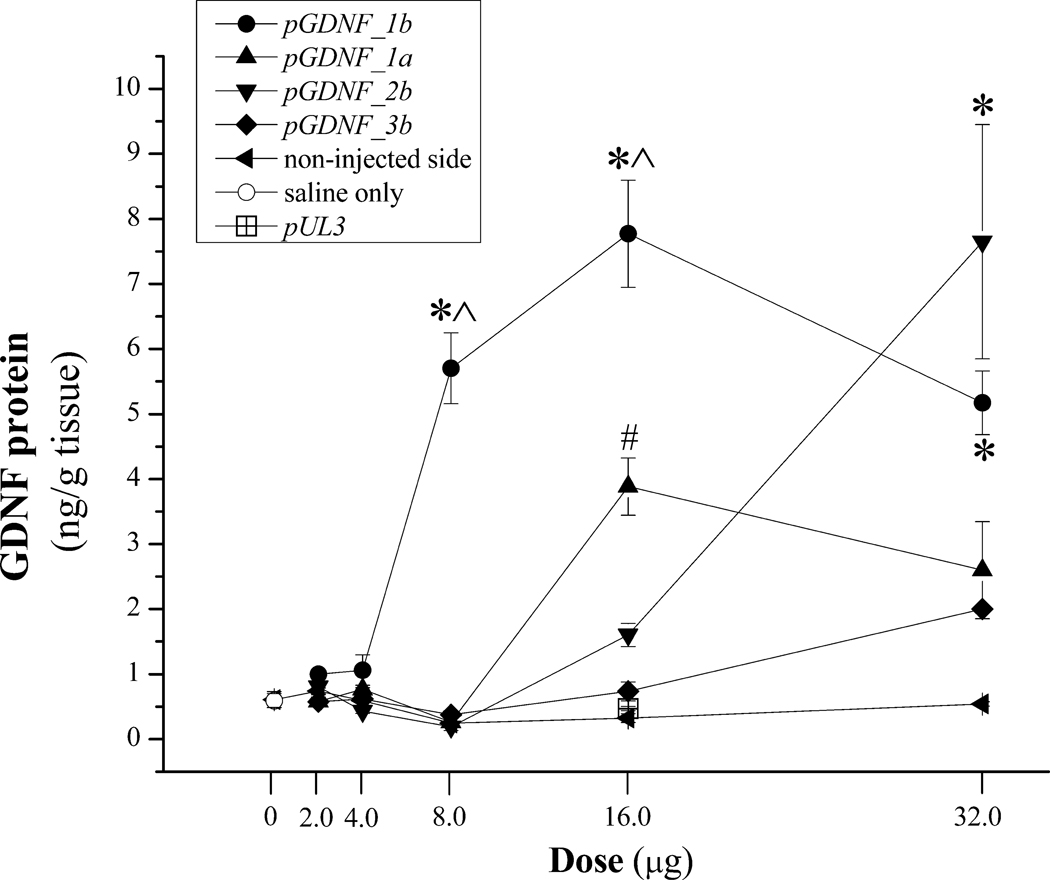
DNA nanoparticles containing pGDNF_1b, pGDNF_2b, pGDNF_1a, or pGDNF_3b were unilaterally injected into the striatum of naïve rats at the following doses: 0, 2.0, 4.0, 8.0, 16.0, or 32.0 µg; all injections were made in an equivalent volume (8.0 µl) of sterile 0.9% saline as described in the Methods section. One week later, the injected and non-injected striata were dissected from each animal and GDNF content in each sample was determined using ELISA. Figure 3 summarizes the results from the in vivo dose-response study. Three-way ANOVA of dose-response data revealed a significant Plasmid × Dose × Side interaction [F(12,154)= 11.39, p<0.001]. At the highest dose (32.0 µg), pGDNF_1b and pGDNF_2b DNPs produced the highest levels of striatal GDNF protein that were not statistically different from one another but both were significantly greater than GDNF levels produced by the two other DNPs. However, GDNF levels for pGDNF_2b DNPs dropped precipitously for the next two lower doses while GDNF levels remained fairly stable for the next two lower doses of pGDNF_1b DNPs. At the 8.0 µg dose, pGDNF_1b DNPs produced GDNF levels that were significantly greater and >10 fold higher than GDNF levels for all other DNPs; at this dose (8.0 µg), GDNF levels were not significantly greater than baseline levels for all compacted plasmids except for pGDNF_1b. The two control injections, sterile saline (vehicle; 8.0 µl) or pUL3 DNPs (16.0 µg) produced no significant change in striatal GDNF levels when compared to baseline GDNF levels. Based upon the results of this dose-response study, we chose to examine the long-term transgene expression of GDNF following intrastriatal injection of pGDNF_1b DNPs at the 16 µg dose level.
Figure 3.
Dose-response study for intracerebral injections of DNPs containing pGDNF_1a, pGDNF_1b, pGDNF_2b, or pGDNF_3b. Various doses of DNPs containing 1 of the 4 plasmids were injected in equivalent volumes (8.0 µl) of sterile saline into the left striatum. One week after DNP injection GDNF protein was measured in the injected or non-injected striatum using ELISA, and each point on the graph represents the mean (± s.e.m.; n=5) of GDNF protein for each plasmid at each dose. There is only one point for two control conditions: saline (8.0 µl; n=5) and pUL3 (16.0 µg; n=5); pUL3 is a plasmid encoding for luciferase and does not encode for GDNF. *p<0.05 vs. pGDNF_1a, pGDNF_3b, non-injected side; ^p<0.05 vs. pGDNF_2b; #p<0.05 vs. pGDNF_2b, pGDNF_3b, non-injected side.
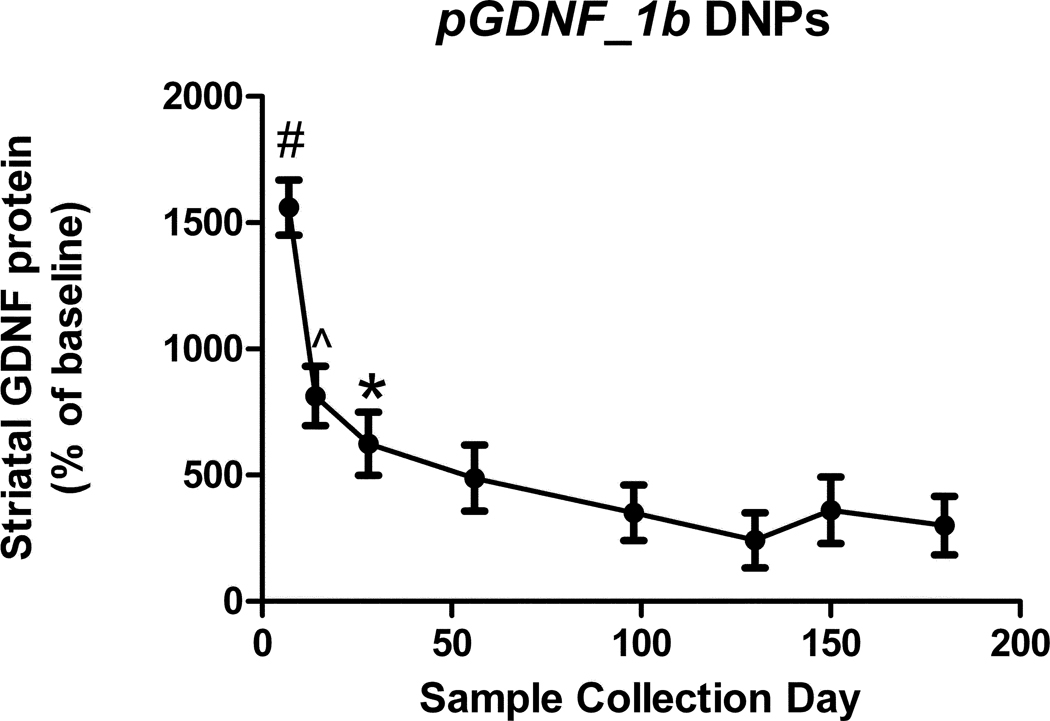
DNA nanoparticles containing pGDNF_1b were injected in the left striatum of naïve young adult rats and animals were euthanatized at 7, 14, 28, 56, 98, 130, 150, or 180 days post-injection, at which time tissue from the right (non-injected) and left (injected) striata was dissected and subsequently analyzed for GDNF protein content. At initial time points (7 and 14 days), GDNF protein values in the injected striatum were 8–16 fold higher than in the non-injected striatum (Figure 4). One month following the injection, GDNF protein levels in the injected striatum stabilized at values that were 3–6 fold higher than in the non-injected striatum for the remaining 5 months of the study.
Figure 4.
Time course of GDNF protein expression in the striatum during a six month period following a single administration of DNPs containing pGDNF_1b (16.0 µg). Animals were euthanized in groups of 5 at each time point and the injected and non-injected striatum were dissected from the brain. In each dissected sample GDNF protein content was determined by ELISA and data are graphed as a percent of baseline; that is, GDNF protein values for the injected side were divided by the values for the non-injected side (baseline). ^p<0.05 vs. Days 98, 130, or 180; #p<0.05 vs. all other days; *p<0.05 vs. Day 130.
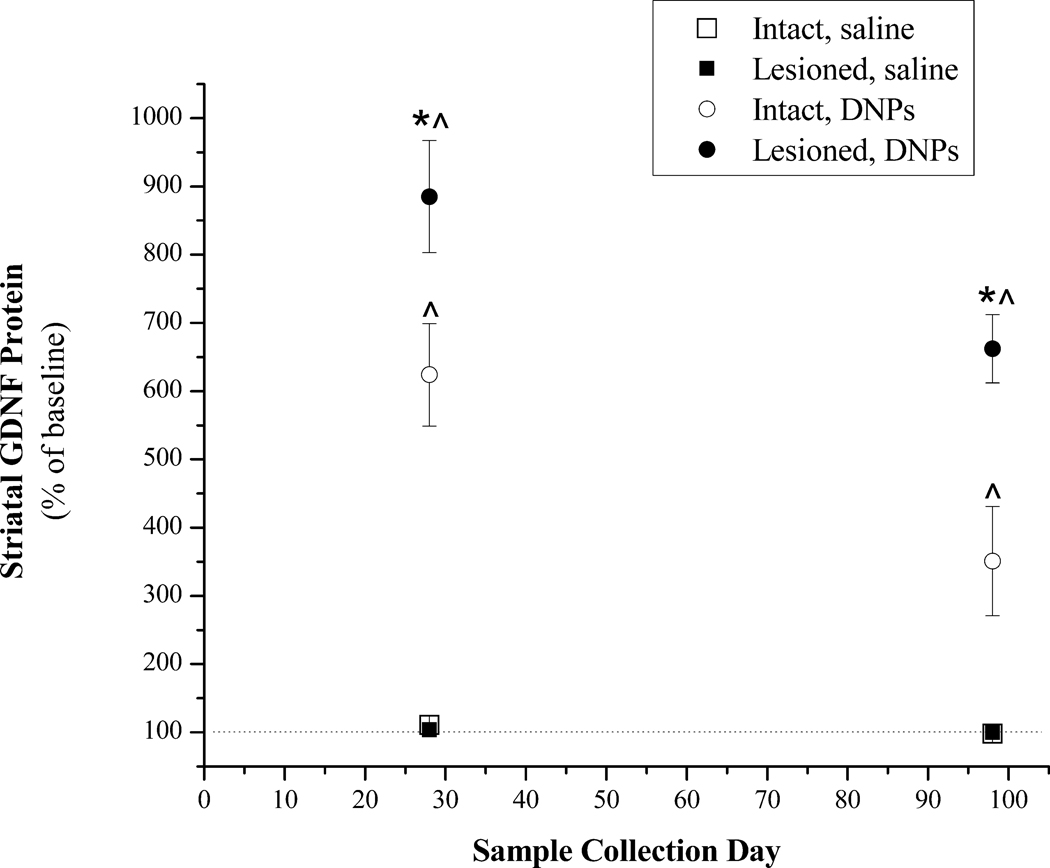
A second group of animals received 6-OHDA lesions 4 weeks before the injection of either DNPs or saline and were euthanatized at 28 and 98 days post-injection. Statistical analysis of striatal GDNF protein values revealed a significant treatment effect [F(3,26) = 136.0, p<0.001]. DNA nanoparticles injected into the intact or lesioned striatum produced significantly greater GDNF levels than in the intact or lesioned striatum of saline treated animals (Figure 5). Injections of DNPs into the lesioned striatum produced significantly higher GDNF levels than those measured in the intact striatum of DNP treated animals.
Figure 5.
Striatal GDNF protein levels at 28 or 98 days following a single injection of pGDNF_1b DNPs or saline into the intact or lesioned striatum. This study used naïve rats or rats with unilateral 6-OHDA lesions. Saline (8.0 µl) was injected into the left striatum of naïve rats (Intact, saline; n=5) or into the denervated striatum 4 weeks after 6-OHDA treatment (Lesioned, saline; n=5). pGDNF_1b DNPs (16.0 µg) was injected into the left striatum of naïve rats (Intact, DNPs; n=5) or into the denervated striatum 4 weeks after 6-OHDA treatment (Lesioned, DNPs; n=5). Animals were euthanized either 28 or 98 days following the saline or DNP injection and the injected and non-injected striatum were dissected from the brain. In each dissected sample GDNF protein content was determined by ELISA and data are graphed as a percent of baseline; that is, GDNF protein values for the injected side were divided by the values for the non-injected side (baseline). * p<0.05 Lesioned, DNPs vs. Intact, DNPs at 28 and 98 days; ^p<0.05 Lesioned, DN Ps or Intact, DNPs vs. Intact, saline or Lesioned, saline at 28 or 98 days.
DISCUSSION
Our initial studies examined in vitro GDNF expression in cells that were treated with plasmids encoding for splice variants 1 or 2 of human GDNF cDNA and further modified to encode for either a codon-optimized and CpG-depleted synthetic ‘gene’ and a variant with nucleosome non-friendly elements. In theory, insertion of nucleosome non-friendly elements into the plasmid should reduce the number of DNA-histone complexes and thus provide a more favorable state for DNA transcription. Likewise, a reduction in the number of CpG dinucleotides in coding and non-coding regions of the plasmid can improve the rate of gene transcription because methylation of CpGs located close to the promoter can increase the likelihood of gene silencing (Kass et al., 1997). All vector types were screened in our in vitro studies and at least one natural cDNA, codon-optimized, or nucleosome non-friendly hGDNF plasmid was compacted into DNPs and tested in vivo. In terms of GDNF transgene expression in both the in vitro and in vivo studies, plasmids encoding for natural cDNA or custom designed synthetic hGDNF ‘genes’ as longer splice variant 1 performed better than the analogous plasmids encoding for the shorter splice variant 2. These findings are consistent with an earlier report suggesting the longer splice variant generates improved levels of secreted GDNF protein (Wang et al., 2008). At the 8 and 16 µg in vivo doses, DNPs containing plasmids encoding for natural hGDNF cDNA for splice variant 1 produced statistically significant higher levels of GDNF than DNPs containing plasmids encoding for natural hGDNF cDNA for splice variant 2. Even though the highest in vivo dose (32.0 µg) of pGDNF_2b DNPs produced comparable levels of GDNF to pGDNF_1b DNPs, GDNF levels dropped precipitously for the next two lower doses of pGDNF_2b DNPs while GDNF levels remained stable for pGDNF_1b DNPs at these same doses. It is noteworthy that the hGDNF cDNA produced higher GDNF levels than the two synthetic GDNF DNA 'genes' tested. The lack of effectiveness of a codon-optimized GDNF compared to the natural cDNA is perhaps not too surprising, since the ability of codon optimization to improve gene expression is variable (Welch et al., 2009). The pGDNF_1b DNPs appeared to have the best in vivo GDNF expression profile and for this reason we moved forward with pGDNF_1b DNPs in our longitudinal studies.
While we observed significantly higher GDNF transgene expression in the striatum transfected with pGDNF_1b DNPs when compared to the striatum transfected with pGDNF_1b DNA alone or basal levels in the non-transfected striatum, the tissue level of transgene activity is lower than that achieved with viral vectors [18]. We observed sustained GDNF transgene activity at levels 300–400% above basal levels for a 6 month period after a single dose of DNPs while AAV-GDNF can increase GDNF tissue levels to 100–1,000 fold higher than baseline over the same period. However, it is not known exactly what tissue levels of GDNF need to be achieved for therapeutic purposes, and this level may be close to that achieved by DNPs. Furthermore, continuous high level expression of GDNF is undesired and may lead to unwanted side effects, such as down-regulation of tyrosine hydroxylase (Georgievska et al., 2004). Levels of GDNF transgene expression at several fold above baseline may be sufficient for phenotypic correction. For example, AAV-GDNF treatments that produced a 300% increase of GDNF protein levels in striatal tissue over basal levels were shown to be effective in protecting the nigrostriatal pathway against 6-OHDA neurotoxicity (Eslamboli et al., 2005). This is the same level of GDNF over-expression that we observed at the end point of our experiment using DNPs; of note, we observed fairly stable luciferase transgene activity in the striatum of mice for a period greater than 1 year following a single dose of DNPs (Yurek et al., 2011). In a previous study we used rat GDNF DNPs to transfect the striatum prior to engrafting fetal dopaminergic neurons and observed enhanced survival of grafted TH+ cells when compared to grafted cell survival in the non-transfected striatum (Yurek et al., 2009a); whether or not the increased neurotrophic activity was limited to the injection site of the DNPs remains to be determined. Secondly, we also observe that intrastriatal injections of DNPs do not result in transport of the vector and/or transgene protein to the SN (Yurek et al., 2009b). On the other hand, intrastriatal injections of AAV can increase transgene protein levels in the SN as well as in the striatum. This may be simply due to the fact that AAV produces high-level transgene expression, and excess protein is transported via striatonigral neurons to the SN while low-level production of GDNF using DNPs is not transported to the SN to any significant extent. Alternatively; it may also be the case that AAV itself is transported to the ventral midbrain and transduce cells in this region. For some clinical indications, such as PD, only focal therapeutic gene expression may be desired, whereas multi-region spread of expression may lead to toxicities.
We observed that the same dose of pGDNF_1b DNPs injected into the lesioned striatum of young adult rats produces significantly higher levels of GDNF transgene expression than when injected into the intact striatum of young adult rats. While we have shown in previous studies that the lesion itself may induce a compensatory increase in endogenous GDNF levels, the lesion-induced increase is transient and typically returns to baseline levels several weeks following the lesion (Yurek and Fletcher-Turner, 2001); therefore, it is unlikely that endogenous GDNF contributed to the increased GDNF levels observed in the lesioned/transfected striatum 4–14 weeks post-lesion. It is not exactly clear why the lesioned striatum would show a higher rate of transfection than the intact striatum; however, one distinct possibility may be related to how the target cells of DNPs might be altered by the degeneration process itself. In our previous studies using DNPs to deliver reporter genes to the brain, we observed long-term transgene activity occurred primarily in astrocytes (Yurek et al., 2009b). Several studies have shown degeneration of the nigrostriatal pathway can lead to profound increases in GFAP [astrocyte] expression in the striatum and ventral midbrain. For instance, Strömberg et al. (Stromberg et al., 1986) was first to demonstrate a significant up-regulation of astrocytes in the striatum and substantia nigra following lesions of the nigrostriatal pathway using 6-OHDA or MPTP in rats or mice, respectively, that persisted for at least one month post-lesion. Subsequently, Sheng et al. (Sheng et al., 1993) reported 6-OHDA lesions produced a persistent up-regulation of GFAP+ cells in the lesioned striatum that was 170% greater than in the control striatum at 28 days post-lesion while Dervan et al. (Dervan et al., 2004) reported a >200% increase in the number of astrocytes in the striatum of mice 6–8 weeks following MPTP administration. Similarly, Rodriques et al. (Rodrigues et al., 2004) reported 166% increase in GFAP IHC in the ipsilateral ventral midbrain of 6-OHDA treated rats at a 22 days post-lesion time point. In a rat model of PD, Gordon et al. (Gordon et al., 1997) demonstrated that there is an exaggerated astrocyte reactivity in striatum of aged animals treated with 6-OHDA when compared to younger lesioned animals. Clearly, these results provide intriguing evidence that the cells targeted by DNPs, astrocytes, increase significantly as a result of nigrostriatal pathway degeneration, and it may be the case that the observed increase in GDNF levels in the lesioned striatum treated with DNPs is related to this increase in GFAP activity. This is significant because the pathological state of neurodegeneration may actually lay a foundation that is actually beneficial for this particular type of gene therapy while other gene therapy techniques, e.g, AAV, that target neurons are actually transducing cells that are decreasing as the disease progresses. From this standpoint, viral and non-viral gene therapies may actually complement one another in that they can approach the same disease state by affecting different cell types.
Highlights.
-
➢
We tested a non-viral gene transfer technique in brain.
-
➢
Plasmids were optimized to achieve long-term expression in brain tissue.
-
➢
Optimized plasmids were compacted into DNA nanoparticles and injected into brain.
-
➢
One plasmid/DNP complex achieved long-term transgene activity in brain.
-
➢
DNA nanoparticles may be a potential therapeutic for Parkinson’s disease.
ACKNOWLEDGEMENTS
This research was supported in part by the National Institutes of Health NS50311 (DMY) and NS75871 (DMY), the Michael J. Fox Foundation for Parkinson’s Research (DMY, MJC), the Jelm Foundation (DMY), and the State of Ohio Biomedical Research Commercialization Program (MJC).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Azzouz M, Ralph S, Wong LF, Day D, Askham Z, Barber RD, Mitrophanous KA, Kingsman SM, Mazarakis ND. Neuroprotection in a rat Parkinson model by GDNF gene therapy using EIAV vector. Neuroreport. 2004;15:985–990. doi: 10.1097/00001756-200404290-00011. [DOI] [PubMed] [Google Scholar]
- Bensadoun JC, Deglon N, Tseng JL, Ridet JL, Zurn AD, Aebischer P. Lentiviral vectors as a gene delivery system in the mouse midbrain: cellular and behavioral improvements in a 6-OHDA model of Parkinson's disease using GDNF. Exp Neurol. 2000;164:15–24. doi: 10.1006/exnr.2000.7409. [DOI] [PubMed] [Google Scholar]
- Choi-Lundberg DL, Lin Q, Schallert T, Crippens D, Davidson BL, Chang Y-N, Chiang YL, Qian J, Bardwaj L, Bohn MC. Behavioral and cellular protection of rat dopaminergic neurons by an adenoviral vector encoding glial cell line-derived neurotrophic factor. Exp Neurol. 1998;154:261–275. doi: 10.1006/exnr.1998.6887. [DOI] [PubMed] [Google Scholar]
- Connor B, Kozlowski DA, Unnerstall JR, Elsworth JD, Tillerson JL, Schallert T, Bohn MC. Glial cell line-derived neurotrophic factor (GDNF) gene delivery protects dopaminergic terminals from degeneration. Exp Neurol. 2001;169:83–95. doi: 10.1006/exnr.2001.7638. [DOI] [PubMed] [Google Scholar]
- Dervan AG, Meshul CK, Beales M, McBean GJ, Moore C, Totterdell S, Snyder AK, Meredith GE. Astroglial plasticity and glutamate function in a chronic mouse model of Parkinson's disease. Exp Neurol. 2004;190:145–156. doi: 10.1016/j.expneurol.2004.07.004. [DOI] [PubMed] [Google Scholar]
- Eberling JL, Kells AP, Pivirotto P, Beyer J, Bringas J, Federoff HJ, Forsayeth J, Bankiewicz KS. Functional Effects of AAV2-GDNF on the Dopaminergic Nigrostriatal Pathway in Parkinsonian Rhesus Monkeys. Hum Gene Ther. 2009;20:511–518. doi: 10.1089/hum.2008.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elsworth JD, Redmond DE, Jr, Leranth C, Bjugstad KB, Sladek JR, Jr, Collier TJ, Foti SB, Samulski RJ, Vives KP, Roth RH. AAV2-mediated gene transfer of GDNF to the striatum of MPTP monkeys enhances the survival and outgrowth of co-implanted fetal dopamine neurons. Exp Neurol. 2008;211:252–258. doi: 10.1016/j.expneurol.2008.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eslamboli A, Georgievska B, Ridley RM, Baker HF, Muzyczka N, Burger C, Mandel RJ, Annett L, Kirik D. Continuous low-level glial cell line-derived neurotrophic factor delivery using recombinant adeno-associated viral vectors provides neuroprotection and induces behavioral recovery in a primate model of Parkinson's disease. J Neurosci. 2005;25:769–777. doi: 10.1523/JNEUROSCI.4421-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgievska B, Kirik D, Bjorklund A. Overexpression of glial cell line-derived neurotrophic factor using a lentiviral vector induces time- and dose-dependent downregulation of tyrosine hydroxylase in the intact nigrostriatal dopamine system. J Neurosci. 2004;24:6437–6445. doi: 10.1523/JNEUROSCI.1122-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgievska B, Kirik D, Rosenblad C, Lundberg C, Bjorklund A. Neuroprotection in the rat Parkinson model by intrastriatal GDNF gene transfer using a lentiviral vector. Neuroreport. 2002;13:75–82. doi: 10.1097/00001756-200201210-00019. [DOI] [PubMed] [Google Scholar]
- Gordon MN, Schreier WA, Ou X, Holcomb LA, Morgan DG. Exaggerated astrocyte reactivity after nigrostriatal deafferentation in the aged rat. J Comp Neurol. 1997;388:106–119. [PubMed] [Google Scholar]
- Grimm L, Holinski-Feder E, Teodoridis J, Scheffer B, Schindelhauer D, Meitinger T, Ueffing M. Analysis of the human GDNF gene reveals an inducible promoter, three exons, a triplet repeat within the 3'-UTR and alternative splice products. Hum Mol Genet. 1998;7:1873–1886. doi: 10.1093/hmg/7.12.1873. [DOI] [PubMed] [Google Scholar]
- Kass SU, Pruss D, Wolffe AP. How does DNA methylation repress transcription? Trends Genet. 1997;13:444–449. doi: 10.1016/s0168-9525(97)01268-7. [DOI] [PubMed] [Google Scholar]
- Kirik D, Rosenblad C, Bjorklund A, Mandel RJ. Long-term rAAV-mediated gene transfer of GDNF in the rat Parkinson's model: intrastriatal but not intranigral transduction promotes functional regeneration in the lesioned nigrostriatal system. J Neurosci. 2000;20:4686–4700. doi: 10.1523/JNEUROSCI.20-12-04686.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kordower JH, Emborg ME, Bloch J, Ma SY, Chu Y, Leventhal L, McBride J, Chen EY, Palfi S, Roitberg BZ, Brown WD, Holden JE, Pyzalski R, Taylor MD, Carvey P, Ling Z, Trono D, Hantraye P, Deglon N, Aebischer P. Neurodegeneration prevented by lentiviral vector delivery of GDNF in primate models of Parkinson's disease. Science. 2000;290:767–773. doi: 10.1126/science.290.5492.767. [DOI] [PubMed] [Google Scholar]
- Lin L-F, Doherty D, Lile JD, Bektesh S, Collins F. GDNF: a glial cell line-derived neurotrophic factor for midbrain dopaminergic neurons. Science. 1993;260:1130–1132. doi: 10.1126/science.8493557. [DOI] [PubMed] [Google Scholar]
- Liu G, Li D, Pasumarthy MK, Kowalczyk TH, Gedeon CR, Hyatt SL, Payne JM, Miller TJ, Brunovskis P, Fink TL, Muhammad O, Moen RC, Hanson RW, Cooper MJ. Nanoparticles of compacted DNA transfect postmitotic cells. J Biol Chem. 2003;278:32578–32586. doi: 10.1074/jbc.M305776200. [DOI] [PubMed] [Google Scholar]
- Luykx P, Bajic IV, Khuri S. NXSensor web tool for evaluating DNA for nucleosome exclusion sequences and accessibility to binding factors. Nucleic Acids Res. 2006;34:W560–W565. doi: 10.1093/nar/gkl158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandel RJ, Spratt SK, Snyder RO, Leff SE. Midbrain injection of recombinant adeno-associated virus encoding rat glial cell line-derived neurotrophic factor protects nigral neurons in a progressive 6-hydroxydopamine-induced degeneration model of Parkinson's disease in rats. PNAS. 1997;94:14083–14088. doi: 10.1073/pnas.94.25.14083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manfredsson FP, Mandel RJ. Development of gene therapy for neurological disorders. Discov Med. 2010;9:204–211. [PubMed] [Google Scholar]
- Natsume A, Mata M, Goss J, Huang S, Wolfe D, Oligino T, Glorioso J, Fink DJ. Bcl-2 and GDNF delivered by HSV-mediated gene transfer act additively to protect dopaminergic neurons from 6-OHDA-induced degeneration. Exp Neurol. 2001;169:231–238. doi: 10.1006/exnr.2001.7671. [DOI] [PubMed] [Google Scholar]
- Palfi S, Leventhal L, Chu Y, Ma SY, Emborg M, Bakay R, Deglon N, Hantraye P, Aebischer P, Kordower JH. Lentivirally delivered glial cell line-derived neurotrophic factor increases the number of striatal dopaminergic neurons in primate models of nigrostriatal degeneration. J Neurosci. 2002;22:4942–4954. doi: 10.1523/JNEUROSCI.22-12-04942.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paxinos G, Watson C. The Rat Brain in Stereotaxic Coordinates. New York: Academic Press; 2007. [Google Scholar]
- Radwan A, Younis A, Luykx P, Khuri S. Prediction and analysis of nucleosome exclusion regions in the human genome. BMC Genomics. 2008;9:186. doi: 10.1186/1471-2164-9-186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigues RW, Gomide VC, Chadi G. Astroglial and microglial activation in the wistar rat ventral tegmental area after a single striatal injection of 6-hydroxydopamine. Int J Neurosci. 2004;114:197–216. doi: 10.1080/00207450490249338. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. New York: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Schaar DG, Sieber B-A, Dreyfus CF, Black IB. Regional and cell-specific expression of GDNF in rat brain. Exp Neurol. 1993;124:368–371. doi: 10.1006/exnr.1993.1207. [DOI] [PubMed] [Google Scholar]
- Sheng JG, Shirabe S, Nishiyama N, Schwartz JP. Alterations in striatal glial fibrillary acidic protein expression in response to 6-hydroxydopamine-induced denervation. Exp Brain Res. 1993;95:450–456. doi: 10.1007/BF00227138. [DOI] [PubMed] [Google Scholar]
- Stromberg I, Bjorklund H, Dahl D, Jonsson G, Sundstrom E, Olson L. Astrocyte responses to dopaminergic denervations by 6-hydroxydopamine and 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine as evidenced by glial fibrillary acidic protein immunohistochemistry. Brain Res Bull. 1986;17:225–236. doi: 10.1016/0361-9230(86)90119-x. [DOI] [PubMed] [Google Scholar]
- Sun M, Kong L, Wang X, Lu XG, Gao Q, Geller AI. Comparison of the capability of GDNF, BDNF, or both, to protect nigrostriatal neurons in a rat model of Parkinson's disease. Brain Res. 2005;1052:119–129. doi: 10.1016/j.brainres.2005.05.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas J, Wang J, Takubo H, Sheng J, de Jesus S, Bankiewicz KS. A 6-hydroxydopamine-induced selective parkinsonian rat model: further biochemical and behavioral characterization. Exp Neurol. 1994;126:159–167. doi: 10.1006/exnr.1994.1054. [DOI] [PubMed] [Google Scholar]
- Trupp M, Ryden M, Jornvall H, Funakoshi H, Timmusk T, Arenas E, Ibanez CF. Peripheral expression and biological activities of GDNF, a new neurotrophic factor for avian and mammalian peripheral neurons. J Cell Biol. 1995;130:137–148. doi: 10.1083/jcb.130.1.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Geng Z, Zhao L, Huang SH, Sheng AL, Chen ZY. GDNF isoform affects intracellular trafficking and secretion of GDNF in neuronal cells. Brain Res. 2008;1226:1–7. doi: 10.1016/j.brainres.2008.05.087. [DOI] [PubMed] [Google Scholar]
- Welch M, Villalobos A, Gustafsson C, Minshull J. You're in a googol: optimizing genes for protein expression. J R Soc Interface. 2009;6:S467–S476. doi: 10.1098/rsif.2008.0520.focus. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yurek DM, Flectcher AM, Kowalczyk TH, Padegimas L, Cooper MJ. Compacted DNA nanoparticle gene transfer of GDNF to the rat striatum enhances the survival of grafted fetal dopamine neurons. Cell Transplant. 2009a;18:1183–1196. doi: 10.3727/096368909X12483162196881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yurek DM, Fletcher-Turner A. Differential expression of GDNF, BDNF, and NT-3 in the aging nigrostriatal system following a neurotoxic lesion. Brain Res. 2001;891:228–235. doi: 10.1016/s0006-8993(00)03217-0. [DOI] [PubMed] [Google Scholar]
- Yurek DM, Fletcher AM, McShane M, Kowalczyk TH, Padegimas L, Weatherspoon MR, Kaytor MD, Cooper MJ, Ziady AG. DNA Nanoparticles: Detection of long-term transgene activity in brain using bioluminescence imaging. Mol Imaging. 2011 doi: 10.2310/7290.2010.00053. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yurek DM, Fletcher AM, Smith GM, Seroogy KB, Ziady AG, Molter J, Kowalczyk TH, Padegimas L, Cooper MJ. Long-term transgene expression in the central nervous system using DNA nanoparticles. Mol Ther. 2009b;17:641–650. doi: 10.1038/mt.2009.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziady AG, Gedeon CR, Miller T, Quan W, Payne JM, Hyatt SL, Fink TL, Muhammad O, Oette S, Kowalczyk T, Pasumarthy MK, Moen RC, Cooper MJ, Davis PB. Transfection of airway epithelium by stable PEGylated poly-L-lysine DNA nanoparticles in vivo. Mol Ther. 2003;8:936–947. doi: 10.1016/j.ymthe.2003.07.007. [DOI] [PubMed] [Google Scholar]