Abstract
The DNA damage response (DDR) is a complex regulatory network that is critical for maintaining genome integrity. Posttranslational modifications are widely used to ensure strict spatiotemporal control of signal flow, but how the DDR responds to environmental cues, such as changes in ambient oxygen tension, remains poorly understood. We found that an essential component of the ATR/CHK1 signaling pathway, the human homolog of the Caenorhabditis elegans biological clock protein CLK-2 (HCLK2), associated with and was hydroxylated by prolyl hydroxylase domain protein 3 (PHD3). HCLK2 hydroxylation was necessary for its interaction with ATR and the subsequent activation of ATR/CHK1/p53. Inhibiting PHD3, either with the pan-hydroxylase inhibitor dimethyloxaloylglycine (DMOG) or through hypoxia, prevented activation of the ATR/CHK1/p53 pathway and decreased apoptosis induced by DNA damage. Consistent with these observations, we found that mice lacking PHD3 were resistant to the effects of ionizing radiation and had decreased thymic apoptosis, a biomarker of genomic integrity. Our identification of HCLK2 as a substrate of PHD3 reveals the mechanism through which hypoxia inhibits the DDR, suggesting hydroxylation of HCLK2 is a potential therapeutic target for regulating the ATR/CHK1/p53 pathway.
Introduction
Fluctuations in oxygen tension are associated with many disease conditions, including myocardial infarction, stroke, and cancer. During hypoxia, a set of adaptive changes, such as increased erythropoiesis and angiogenesis, occur to maintain oxygen homeostasis (1). A conserved pathway regulated by oxygen-dependent prolyl hydroxylation of the α subunit of the master transcriptional regulator HIF-1 plays an essential role in these processes (2). Hydroxylation of HIF-1α facilitates its binding with von Hippel-Lindau tumor suppressor (pVHL) and subsequent proteasomal degradation (3, 4). In mammals, there are 3 isoforms of the HIF prolyl hydroxylase domain proteins, termed PHD1–PHD3 (also called EGLN1–EGLN3) (2). Although it is generally accepted that non-HIF substrates of PHDs exist in the eukaryotic proteome, the identities and roles of these non-HIF substrates of PHDs remain largely unknown. Recently, however, it has been demonstrated that β2-AR and pyruvate kinase M2 (PKM2) are also substrates of PHD3 (5, 6). Similar to prolyl hydroxylation of HIF-1α, prolyl hydroxylation of β2-AR promotes its ubiquitylation and degradation via the pVHL pathway (5). In contrast, prolyl hydroxylation of PKM2 enhances its interaction with HIF-1α, which in turn promotes HIF-1α transactivation (6). PHD3 is recognized as a proapoptotic protein, with PHD3 being required for neuronal apoptosis induced by nerve growth factor deprivation (7). However, despite the recent identification of β2-AR and PKM2 as substrates of prolyl hydroxylation, the identities of the substrates involved in PHD3-mediated apoptosis remain unknown. Likewise, the exact nature of the involvement of PHD3-mediated prolyl hydroxylation in apoptosis and how these mechanisms might be linked to hypoxic control of apoptosis have yet to be delineated.
Results
PHD3 interacts with HCLK2.
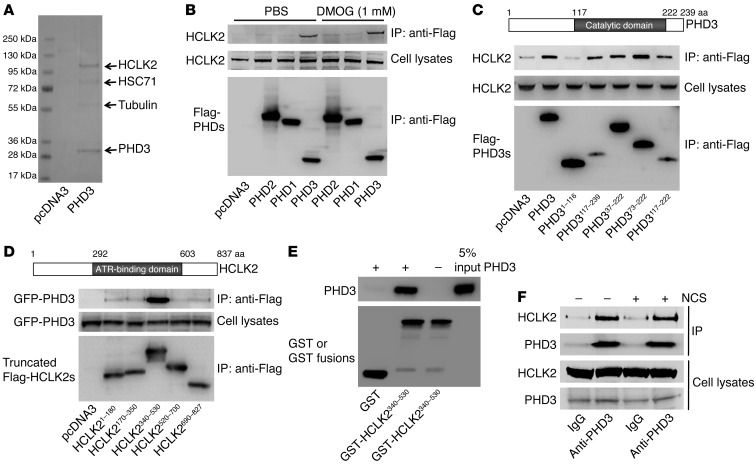
We used MALDI/TOF mass spectrometry (MS) as an inductive and unbiased method of identifying PHD3-associated proteins in HeLa cells stably transfected with Flag-PHD3. Using this approach, we identified the human homolog of the Caenorhabditis elegans biological clock protein CLK-2 (HCLK2), a critical component of the ATR/CHK1 signaling pathway of the DNA damage response (DDR) (8, 9), as one of the proteins associated with PHD3 (Figure 1A). Coimmunoprecipitation experiments revealed that PHD3 (but not PHD1 or PHD2) formed a stable complex with HCLK2 and that this association was unaffected by the hydroxylase inhibitor dimethyloxaloylglycine (DMOG) (Figure 1B and Supplemental Figure 1; supplemental material available online with this article; doi: 10.1172/JCI62374DS1). To explore the interaction between PHD3 and HCLK2 further, we engineered a series of constructs expressing fragments of PHD3 and HCLK2 to determine the domains of PHD3 and HCLK2 involved in this interaction. Coimmunoprecipitation experiments indicated that amino acids 117–222 of PHD3 (PHD3117–222), corresponding to its catalytic domain, interacted with endogenous HCLK2 and that amino acids 340–530 of HCLK2 (HCLK2340–530), comprising a region important to ATR binding (5, 10), was capable of forming a complex with PHD3 (Figure 1, C and D). Using recombinant glutathione-S-transferase–HCLK2340–530 (GST-HCLK2340–530) and PHD3 to perform an in vitro GST pull-down assay, we were able to confirm that HCLK2340–530 directly interacts with PHD3 (Figure 1E). Furthermore, we demonstrated that endogenous HCLK2 could be pulled down by endogenous PHD3 by coimmunoprecipitation (Figure 1F). Given the pivotal role that HCLK2 plays in the DDR, we explored whether the association between PHD3 and HCLK2 was affected by the induction of DNA damage. Treating cells with neocarzinostatin (NCS) to induce DNA double-strand breaks and activation of the ATR/CHK1 pathway did not alter the association between PHD3 and HCLK2 (Figure 1F), indicating that the interaction between PHD3 and HCLK2 is not affected by DNA damage. Taken together, these results demonstrate that PHD3 and HCLK2 directly associate with each other both in vitro and in vivo and that this association is independent of hydroxylation or DNA damage.
Figure 1. PHD3 interacts with HCLK2.
(A) Flag-PHD3 immunoprecipitated from HeLa cells was separated by SDS-PAGE and stained with Coomassie blue. Protein identities were confirmed with MALDI/TOF/TOF. (B) HeLa cells transfected with Flag–PHD1–PHD3 or empty vector were harvested after treatment with DMOG or PBS for 4 hours. Immunoprecipitation with anti-Flag beads was followed by Western blot analysis with the indicated antibodies. (C) Truncated Flag-PHD3s or (D) truncated Flag-HCLK2s and GFP-PHD3 were transfected into HeLa cells. Cell lysates were immunoprecipitated with anti-Flag beads, and Western blot analysis was performed with the indicated antibodies. (E) GST or GST-HCLK2340–530 fusion proteins (1 μg) were incubated with an equal amount of recombinant PHD3 prior to GST pull down. Pull-down product and 5% of PHD3 input were separated with SDS-PAGE and analyzed by Western blot with anti-PHD3 or anti-GST antibodies. (F) Endogenous PHD3 was immunoprecipitated with an anti-PHD3 antibody. Immunoprecipitates were analyzed by Western blot with anti-PHD3 or HCLK2 antibodies. Representative blots from at least 3 experiments are shown in B–D.
PHD3 plays a crucial role in the ATR/CHK1/p53 pathway following DNA damage.
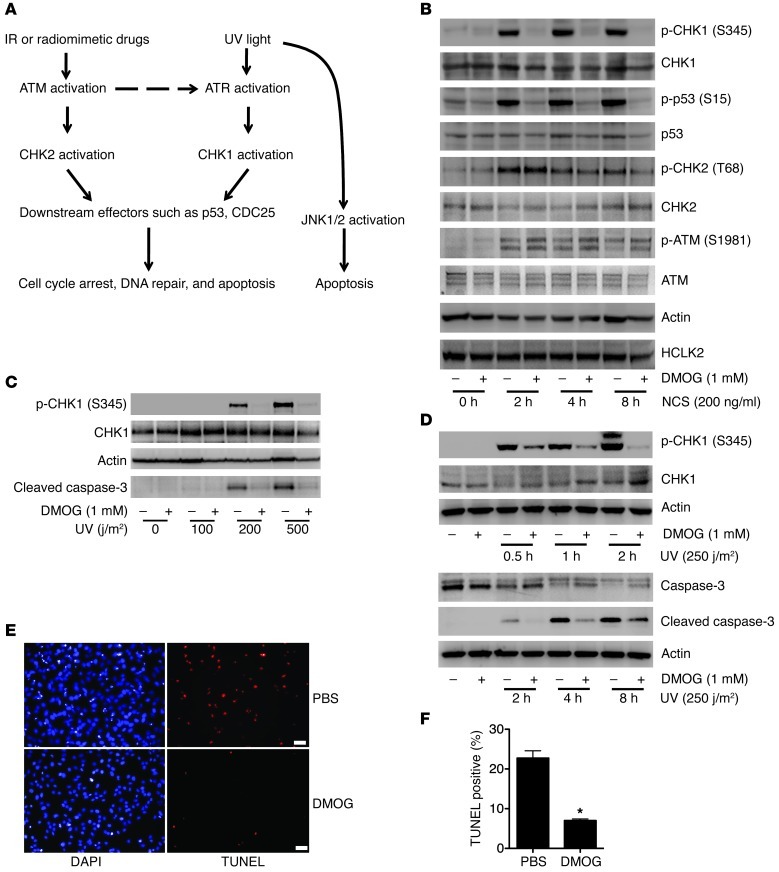
Depletion of HCLK2 compromises the DDR, including ATR/CHK1 and ATM/CHK2 pathways in response to ionizing radiation (IR), UV light, and hydroxyurea, which correlates with significantly reduced protein levels of ATM, ATR, and CHK1 (8–10). Given our data demonstrating a direct interaction between PHD3 and HCLK2, we asked whether PHD3 and its hydroxylase activity are required for the activation of the ATM/CHK2 and ATR/CHK1 pathway in response to DNA damage. To answer this question, we treated HeLa cells with DMOG (to inhibit PHD3 activity) for 4 hours, followed by NCS treatment to induce DNA double-strand breaks, thereby activating both the ATR/CHK1 and ATM/CHK2 pathways (Figure 2A and ref. 11). Treatment of cells with DMOG did not affect the steady-state protein level of HCLK2, suggesting that, unlike the PHD prolyl hydroxylation of HIF-1α and β2-AR, interaction of PHD3 with HCLK2 does not lead to HCLK2 degradation (Figure 2B). In addition, treatment of cells with DMOG for 4 hours had no effect on the protein levels of ATM, ATR, CHK1, and CHK2 (Figure 2B and as shown below). Interestingly, treatment of cells with DMOG dramatically inhibited the phosphorylation of CHK1 and p53 induced by NCS, without affecting the phosphorylation of ATM and CHK2 (Figure 2B and Supplemental Figure 2A). Similarly, DMOG also substantially inhibited the activation of CHK1 induced by hydroxyurea (Supplemental Figure 2B). Since oxygen is an essential cosubstrate of PHDs, and hypoxia significantly inhibits their enzymatic activity (12), we next examined the effect of oxygen deprivation on the ability of NCS to activate CHK1. Not surprisingly, 1% O2 markedly inhibited phosphorylation of CHK1 induced by NCS, further implying a role for PHD3 in the activation of the ATR/CHK1 pathway (Supplemental Figure 2C).
Figure 2. Prolyl hydroxylase inhibitor DMOG specifically inhibits the ATR/CHK1/p53 pathway following DNA damage.
(A) A simplified schematic of the signaling pathways induced by DNA damage. (B) HeLa cells were pretreated with DMOG for 4 hours, followed by NCS treatment as indicated. Western blots were performed with indicated antibodies. (C) HeLa cells were pretreated with DMOG for 4 hours followed by UV treatment at different doses. Cells were harvested after 2 hours, and Western blots were performed with the indicated antibodies. (D–F) HeLa cells were pretreated with DMOG for 4 hours, followed by UV treatment (250 j/m2), and harvested at different time points as indicated. (D) Western blots were performed with the indicated antibodies. TUNEL staining was performed at 8 hours after UV treatment. Representative images of cells pretreated with PBS or DMOG are shown in E. Scale bars: 20 μm. Original magnification, ×100. (F) TUNEL-positive cells (mean ± SEM) were quantified. n = 3; *P < 0.01. In B–D, representative blots from 3 similar experiments are shown.
To maintain genome integrity, DNA damage induced by either environmental agents such as UV light or agents derived from normal cellular metabolism such as reactive oxygen species must be repaired. Cells harboring nonrepairable DNA damage will be programmed to senescence or death. DDR plays a central role in these processes. Interestingly, PHD3 has long been considered as a proapoptotic protein and plays an essential role in neuronal apoptosis (13, 14). To determine whether prolyl hydroxylation mediated by PHD3 plays a crucial role in apoptosis induced by DNA damage, we treated HeLa cells with UV light to activate DDR and induce apoptosis. We found that UV treatment of HeLa cells induced activation of CHK1 and caspase-3 (Figure 2, C and D, and Supplemental Figure 2D) and led to substantial apoptotic cell death after 8 hours of treatment (Figure 2, E and F). However, when cells were treated with DMOG, activation of CHK1 and caspase-3, as well as the subsequent apoptosis induced by UV treatment, were dramatically inhibited (Figure 2, C–F, and Supplemental Figure 2D). Both the DDR and JNK pathways can be activated by UV light and play crucial roles in UV-induced apoptosis (15, 16). In contrast, treatment of cells with DMOG before UV irradiation had no effect on the activation of JNK1/2 (Supplemental Figure 2D), further confirming the specificity of the inhibitory effect of DMOG on the ATR/CHK1 pathway.
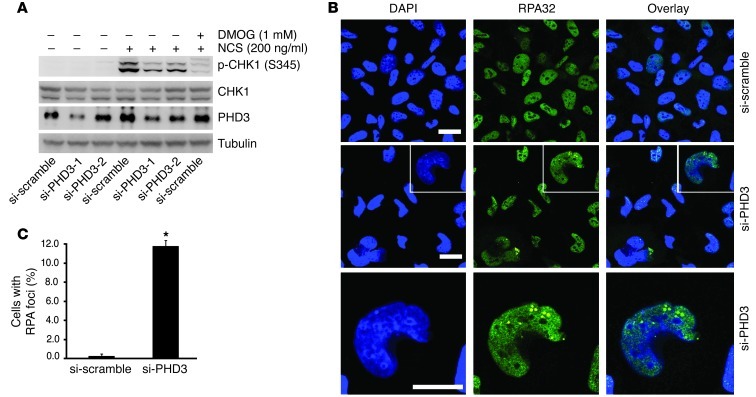
Since DMOG is a pan-inhibitor of 2-oxoglutarate–dependent dioxygenase, its inhibitory effect on CHK1 activation implied the involvement of a prolyl hydroxylase in the activation of the ATR/CHK1 pathway. However, these data did not identify which prolyl hydroxylase was involved or explain what functional role this prolyl hydroxylase was playing. Therefore, to determine whether PHD3 specifically regulates the ATR/CHK1 pathway and to assess the functional role it might play, we knocked down the expression of endogenous PHD3 in HeLa cells using PHD3-specific small, interfering RNA (si-RNA) and discovered that partial depletion of PHD3 markedly inhibited CHK1 activation induced by NCS (Figure 3A). It was demonstrated that HCLK2 prevented spontaneous DNA damage, and depletion of HCLK2 led to the accumulation of the focus formation of the single-stranded DNA-binding protein subunit RPA32 in HeLa cells under nonchallenged conditions in an ATR-dependent manner (8). Considering the important role of PHD3 in the ATR/HCLK2 pathway, we next asked whether depletion of PHD3 in HeLa cells would also result in a replication stress. Indeed, we observed a significant accumulation of RPA32 foci in HeLa cells transfected with PHD3-specific si-RNA compared with that in cells transfected with the scrambled si-RNA (si-scramble) (Figure 3, B and C). Approximately 10% of cells transfected with si-RNA specific for PHD3 (si-PHD3) exhibited RPA32 foci under nonchallenged conditions, compared with less than 0.5% of cells transfected with si-scramble (Figure 3, B and C). Collectively, these data suggest that PHD3 and its hydroxylase activity play an essential role in activating the ATR/CHK1 pathway in HeLa cells.
Figure 3. PHD3 plays a crucial role in DDR.
(A) HeLa cells transfected with si-scramble or si-PHD3 for 3 days were treated with NCS for 2 hours. si-PHD3-1 and si-PHD3-2, si-RNA specific for PHD3 construct 1 and 2, respectively. Cell lysates were analyzed by Western blot with the indicated antibodies. Representative blots from 2 experiments are shown. (B and C) HeLa cells transfected with si-scramble or si-PHD3 for 3 days were immunostained with anti-RPA32 antibody. Representative images are shown in B. The bottom panel in B shows magnified images of the white boxes in the middle panel. Scale bars: 10 μm. Original magnification, ×200. (C) Quantification of the number of cells positive for RPA32 (mean ± SEM) from 3 independent experiments. *P < 0.01.
PHD3 hydroxylation of HCLK2 mediates the interaction between HCLK2 and ATR.
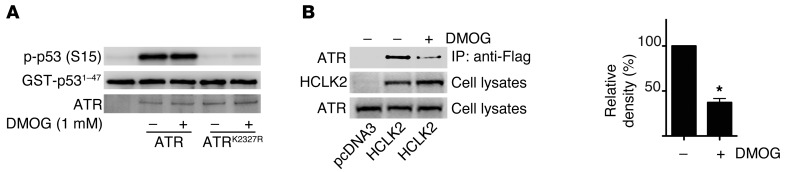
To refine our understanding of the role that PHD3 has on the activation of the ATR/CHK1 pathway, we dissected out the individual components of this pathway and analyzed the effect that hydroxylation has on each component. Treatment of cells with DMOG had no effect on the level of ATR kinase activity as determined by an in vitro kinase assay, ruling out the possibility that DMOG directly inhibits ATR kinase activity (Figure 4A). Given that the formation of an ATR/HCLK2 complex is crucial for CHK1 activation (10), we next examined the effect of DMOG on the association between ATR and HCLK2. Interestingly, pretreatment of cells with DMOG significantly inhibited the interaction of ATR with HCLK2 (Figure 4B).
Figure 4. DMOG inhibits the interaction between HCLK2 and ATR but not ATR activity.
(A) An in vitro kinase assay was performed with GST-p531–47 and wild-type ATR or mutant ATRK2327R lacking the kinase activity immunoprecipitated from HEK293 cells in the presence or absence of DMOG. Western blots were then performed with the indicated antibodies. Representative blots from 3 similar experiments are shown. (B) After 6 hours of treatment with DMOG, HeLa cells transfected with pcDNA3 or Flag-HCLK2 were harvested and immunoprecipitated using anti-Flag beads. Western blots were then performed with the indicated antibodies. Densitometry analysis (mean ± SEM) was performed for the coimmunoprecipitated ATR. n = 3; *P < 0.01.
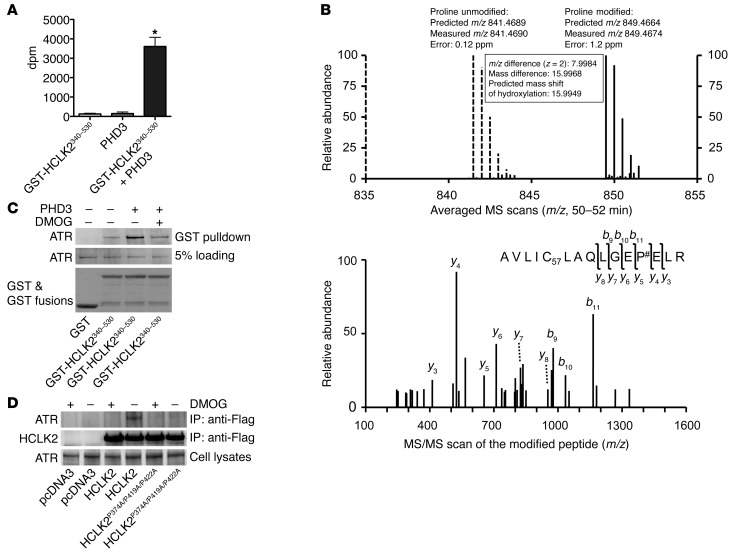
It is well documented that prolyl hydroxylation potentiates the interaction between HIF-1α and pVHL (3, 4), and we have recently demonstrated that prolyl hydroxylation of β2-AR also facilitates its interaction with pVHL (5). Having demonstrated that PHD3 directly interacts with HCLK2 and is required for the activation of the ATR/CHK1 pathway, we hypothesized that HCLK2 is also a hydroxylation substrate of PHD3 and that hydroxylation of HCLK2 potentiates its association with ATR. To test this hypothesis, we performed an in vitro quantitative hydroxylation assay using 2-oxoglutarate [1-14C] as the cosubstrate and measured the radioactivity of 14CO2 formed during the hydroxylation-coupled decarboxylation of 2-oxoglutarate [1-14C] (17). A significant level of hydroxylation activity occurred with PHD3 in the presence of GST-HCLK2340–530 (Figure 5A), indicating that PHD3 can hydroxylate HCLK2 in vitro. To identify the prolyl hydroxylation sites, Flag-HCLK2340–530 immunoprecipitated from HeLa cells was trypsin digested and analyzed by liquid chromatography–MS (LC-MS/MS). Modified peptide sequence AVLIC57LAQLGEP#ELR (C57 indicating mass shift of 57.0215 Da by iodoacetamide alkylation on the Cys residue; # indicating proline hydroxylation), which contains Pro374, was identified based on the high-resolution MS survey scan comparison between the unmodified form and the hydroxylated form as well as the MS/MS scan of the hydroxylated peptide. The unmodified form was detected as a doubly charged monoisotopic ion of 841.4690 m/z, whereas the modified form was identified as 849.4674 m/z, which is consistent with the exact mass difference caused by hydroxylation (Figure 5B). The modification was further validated by multiple product ions (b and y ions) in the MS/MS scan by collision-induced dissociation (Figure 5B). In addition, 2 more hydroxylation sites in HCLK2 at Pro419 and Pro422 were also identified with similar approach (Supplemental Figure 3). Thus, our data demonstrating that PHD3 can hydroxylate HCLK2 on multiple proline residues provides a mechanistic link for the involvement of PHD3 in the activation of the ATR/CHK1 pathway and subsequent apoptosis induced by DNA damage.
Figure 5. PHD3 hydroxylation of HCLK2 mediates the interaction between HCLK2 and ATR.
(A) In vitro hydroxylation assays using 2-oxo-glutarate [1-14C] as the cosubstrate were performed in the presence of GST-HCLK2340–530 or PHD3 as indicated. The released 14CO2 (mean ± SEM) was trapped and measured. n = 3; *P < 0.01 versus PHD3 alone. (B) Flag-HCLK2340–530 immunoprecipitated from HeLa cells was trypsin digested and analyzed by LC-MS/MS. Modified peptide sequence AVLIC57LAQLGEP#ELR (C57 indicating mass shift of 57.0215 Da by iodoacetamide alkylation on the Cys residue; # indicating proline hydroxylation) was identified based on the high-resolution MS/MS survey scan comparison between the unmodified form (dashed lines) and the hydroxylated form (solid lines) as well as the MS/MS scan of the hydroxylated peptide. The unmodified form was detected as a doubly charged monoisotopic ion of 841.4690 m/z, whereas the modified form was identified as 849.4674 m/z. The modification was further validated by multiple product ions (b and y ions) in the MS/MS scan by collision-induced dissociation. (C) GST or GST-HCLK2340–530 immobilized on GSH beads were incubated with Flag-ATR purified from HEK293 cells, either directly or after 60 minutes of incubation with PHD3 in the presence or absence of DMOG. The pull-down products were analyzed by Western blot analysis. (D) HeLa cells transfected with pcDNA3, Flag-HCLK2, or Flag-HCLK2P374A/P419A/P422A were treated with DMOG for 6 hours. Coimmunoprecipitation and Western blot analysis were then performed with the indicated antibodies. In C and D, representative blots from 2 similar experiments are shown.
Our hypothesis that prolyl hydroxylation of HCLK2 potentiates its interaction with ATR was further strengthened by an in vitro hydroxylation assay using GST-HCLK2340–530 as a substrate with recombinant PHD3. As shown in Figure 5C, the interaction between recombinant GST-HCLK2340–530 and ATR increased markedly after incubation with PHD3 but was abolished by DMOG, verifying that PHD3-mediated prolyl hydroxylation of HCLK2 regulates its interaction with ATR. In addition, the interaction between HCLK2 and ATR in HeLa cells was increased by overexpression of PHD3 and markedly inhibited by hypoxia (Supplemental Figure 4). Furthermore, mutation of Pro374, Pro419, and Pro422 on HCLK2 to Ala dramatically reduced the interaction between HCLK2P374A/P419A/P422A and endogenous ATR (Figure 5D). Interestingly, although the interaction between ATR and either of the single mutants HCLK2P374A or HCLK2P419A was also partially inhibited, DMOG could still further attenuate their interactions (Supplemental Figure 5). In contrast, mutations at Pro400 and Pro401 of HCLK2, which are not hydroxylated, had no effect on the interaction between HCLK2 and ATR (Supplemental Figure 5). Taken together, these experiments provide substantial support for a model in which PHD3 hydroxylates HCLK2 on multiple, yet specific, proline residues and indicate that this is a necessary step in order for optimal association of HCLK2 and ATR and subsequent activation of the ATR/CHK1 pathway.
Knockout of endogenous PHD3 inhibits the activation of the ATR/CHK1/p53 pathway following DNA damage and blocks G0/G1 to S phase transition in primary embryonic fibroblasts.
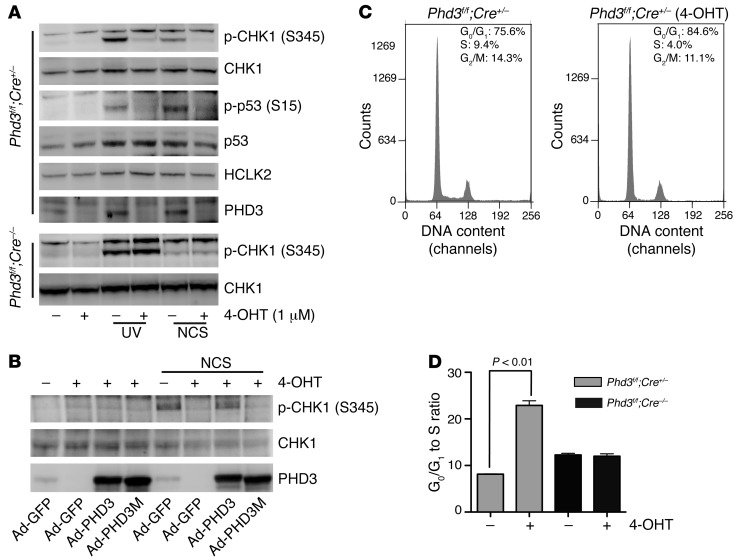
To determine whether the hydroxylation of HCLK2 mediated by PHD3 holds any physiological significance in vivo, we generated Phd3f/f;Cre/Esr1+/– (Phd3Β/Β;Cre+/–) mice by crossing Phd3f/f mice with CAG-Cre/Esr1 transgenic mice (The Jackson Laboratory), which have a tamoxifen-inducible and Cre-mediated recombination system driven by the chicken β-actin promoter (18). Primary mouse embryonic fibroblasts (MEFs) were isolated from embryos on day 13.5 and genotyped to identify MEF populations that were either Phd3Β/Β;Cre+/– or Phd3Β/Β;Cre–/– (Supplemental Figure 6A). Phd3Β/Β;Cre+/– MEFs were subsequently treated with 4-hydroxy-tamoxifen (4-OHT) to deplete the expression of PHD3 at both the mRNA and protein levels (Supplemental Figure 6B and Figure 6A). MEFs from PHD3-containing or PHD3-depleted populations were treated with either UV irradiation or NCS to induce DNA damage, and the activation of CHK1 and p53 was assessed. As shown in Figure 6A and Supplemental Figure 6C, depletion of PHD3 in MEFs resulted in a dramatic inhibition of CHK1 and p53 activation, whereas phosphorylation of JNK1/2 was not affected (Supplemental Figure 6C). Similarly, CHK1 activation induced by IR was also inhibited when PHD3 was depleted from Phd3Β/Β;Cre+/– MEFs (Supplemental Figure 6D). The inhibitory effect of 4-OHT on CHK1 and p53 activation was not due to nonspecific activity, since CHK1 and p53 were phosphorylated normally in Phd3Β/Β;Cre–/– MEFs in the presence of 4-OHT (Figure 6A and Supplemental Figure 6C). To ensure that the inhibitory effect of PHD3 depletion on CHK1 activation was not related to some unique characteristic of Phd3Β/Β;Cre+/– MEFs, we infected Phd3Β/Β;Cre–/– MEFs with adenovirus-expressing Cre recombinase (Ad-Cre) to render these cells PHD3-deficient. Western blot analysis demonstrated that the Ad-Cre–induced reduction in the protein level of endogenous PHD3 correlated with a substantial inhibition in the activation of CHK1 (but not CHK2) induced by NCS in Phd3Β/Β;Cre–/– MEFs (Supplemental Figure 6E). Finally, we infected Phd3Β/Β;Cre+/– MEFs with Ad-PHD3 and Ad-PHD3H135A/D137A/H196A (PHD3M, a hydroxylase-inactive form of PHD3), in which the crucial HxD.H motif for Fe2+ binding is mutated (19), and demonstrated that only wild-type PHD3 but not PHD3M restored the activation of CHK1 induced by NCS in Phd3Β/Β;Cre+/– MEFs after treatment with 4-OHT (Figure 6B).
Figure 6. PHD3 plays a crucial role in the activation of the ATR/CHK1/p53 pathway following DNA damage and blocks G0/G1 to S phase transition in primary MEFs.
(A) After 3 days of treatment with 4-OHT, Phd3Β/Β;Cre+/– or Phd3Β/Β;Cre–/– MEFs were exposed to UV light (250 j/m2) or NCS (200 ng/ml) as indicated. Two hours later, cells were harvested, and Western blots were performed with the indicated antibodies. Representative blots from 3 similar experiments are shown. (B) Phd3Β/Β;Cre+/– MEFs were treated with 4-OHT for 3 days. On the second day, cells were infected with Ad-PHD3 or Ad-PHD3M as indicated. Cells were then treated with NCS (200 ng/ml) for 2 hours, and cell lysates were analyzed by Western blot with the indicated antibodies. Representative experiments from 2 experiments are shown. (C and D) Phd3Β/Β;Cre+/– or Phd3Β/Β;Cre–/– MEFs were treated with 4-OHT (1 μM) or solvent only for 3 days. Cells were then stained with propidium iodide (PI) and analyzed with FACS. The representative histograms of Phd3Β/Β;Cre+/– MEFs treated with or without 4-OHT are shown in C. (D) The relative sizes of G0/G1 to S phase ratio of the indicated MEFs (mean ± SEM). n = 3; *P < 0.01.
It was reported that depletion of HCLK2 from primary MEFs induced cell cycle arrest with reduced S phase index (9). Interestingly, PHD3 was recently shown to play a role in cell cycle regulation, and knockdown of PHD3 inhibited G1 phase to S phase transition in carcinoma cells (20). Consistently, treatment of Phd3Β/Β;Cre+/– MEFs but not Phd3Β/Β;Cre–/– MEFs with 4-OHT led to a significant increase of cells in G1/G0 phase and decrease of cells in S phase, which resulted in a dramatic increase of the G1/G0 to S ratio (Figure 6, C and D). Taken together, these results corroborate our results obtained in cells in which endogenous PHD3 activity had been inhibited (either by DMOG treatment or PHD3-specific si-RNA) and validated the use of transgenic mice of this genotype in studying the physiological ramifications of PHD3 deletion in vivo.
PHD3 plays a crucial role in thymocyte apoptosis following DNA damage in vivo.
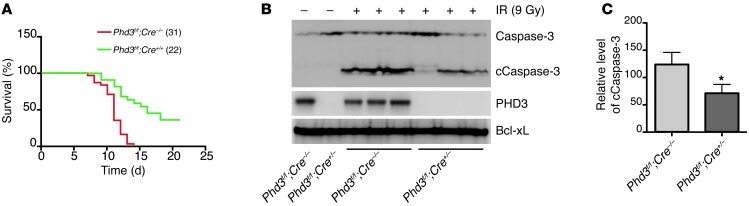
To examine whether the molecular and cellular defects in the ATR/CHK1 pathway caused by deletion of PHD3 discovered in our in vitro studies resulted in an alteration in the in vivo response to DNA damage, we treated Phd3Β/Β;Cre+/– mice and Phd3Β/Β;Cre–/– littermates with tamoxifen, thereby depleting PHD3 expression in Phd3Β/Β;Cre+/– mice (Supplemental Figure 7A). Subsequently, mice were irradiated with 9 Gy of X-rays and monitored for survival. All Phd3Β/Β;Cre–/– mice (n = 31) died within 2 weeks after irradiation, with a 50% survival time of 11 days, which is similar to previous reports (21). In contrast, approximately half of the Phd3Β/Β;Cre+/– mice (n = 22) survived beyond 2 weeks, with a 50% survival time of 16 days (Figure 7A). Since thymocyte apoptosis and depletion are important biomarkers of DNA damage and toxicity associated with IR (21), we were interested in determining whether there was a difference in the degree of apoptotic markers in the thymuses of Phd3Β/Β;Cre+/– mice compared with that in Phd3Β/Β;Cre–/– mice following irradiation. Therefore, we performed Western blot analysis to examine the activation of caspase-3 in the thymus after IR. Activation of caspase-3 and CHK1 was significantly inhibited in thymuses from Phd3Β/Β;Cre+/– mice (Figure 7, B and C, and Supplemental Figure 7C). In addition, TUNEL staining revealed that there was a substantial decrease in apoptosis in the thymuses from Phd3Β/Β;Cre+/– mice (Supplemental Figure 7B). Taken together, these results indicate that PHD3 plays a crucial role in thymocyte apoptosis induced by DNA damage in vivo and that deletion of PHD3 increases the survival of mice upon IR.
Figure 7. Depletion of PHD3 inhibits caspase-3 activation induced by IR in thymuses in vivo.
(A) Age- and sex-matched 6- to 8-week-old Phd3Β/Β;Cre+/– and Phd3Β/Β;Cre–/– mice were injected intraperitoneally with tamoxifen (1 mg/20 g body weight/d) for 5 consecutive days. A week later, mice were treated with 9 Gy X-ray. The survival of mice was monitored. Parenthetical numbers indicate the number of mice analyzed. P < 0.001. (B and C) Thymuses were harvested from mice 8 hours after exposure to IR (9 Gy), and (B) Western blots were performed with the indicated antibodies. (C) Densitometry analysis of the protein level of cleaved caspase-3 (cCaspase-3) (mean ± SEM). Bcl-xL was used as the loading control. n = 3; *P < 0.05 versus Phd3Β/Β;Cre–/–.
Discussion
The results presented here demonstrate that hydroxylation of HCLK2 by PHD3 regulates the activation of the ATR/CHK1 pathway and subsequent apoptosis in response to DNA damage. Our experiments also provide evidence that HCLK2 is a bone fide substrate of PHD3. In addition to HIF-1α, a short list of PHD3 substrates, including β2-AR and PKM2, have been identified, suggesting that prolyl hydroxylation is a posttranslational modification that regulates a unique set of cellular events (5, 6, 12). The first identified role for prolyl hydroxylation, catalyzed by collagen prolyl-4 hydroxylase (C-P4H), was to assist in the proper folding and stability of the collagen triple helix structure (22). C-P4H and the PHDs belong to the same superfamily of Fe+2 and 2-oxoglutarate–dependent dioxygenases (19). However, unlike C-P4H, which catalyzes prolyl hydroxylation in conserved X-Pro-Gly or X-Pro-Ala motifs, PHDs do not require a strict motif for substrate hydroxylation. Although the hydroxylation of HIF-1α occurs in a conserved LXXLAP motif, substitution of the flanking Leu or Ala residues has little effect on prolyl hydroxylation (12, 23). In addition, this motif is not required for the prolyl hydroxylation of β2-AR and PKM2 (5, 6). The crystal structure of the catalytic domain of PHD2, with a peptide corresponding to HIF-1α c-terminal oxygen-dependent degradation (CODD) domain, reveals multiple hydrophobic interactions between PHD2 and the Leu residues flanking Pro564 in CODD (24). Although there are multiple Leu residues near Pro374 of human HCLK2, sequence alignment indicates that this proline residue is not conserved in mammals (Supplemental Figure 8A). Conversely, Pro419 and Pro422, which lack the flanking Leu residues, are highly conserved in vertebrates (Supplemental Figure 8B), suggesting that hydroxylation of these 2 Pro residues may be conserved in vertebrates and, therefore, that the flanking Leu residues are not crucial for prolyl hydroxylation mediated by PHDs.
Solid tumors often contain hypoxic regions, and the presence of these hypoxic regions within tumor masses is associated clinically with a poor prognosis (25). Protein levels of HIF-1α, the most well-characterized substrate of the PHDs, are significantly increased in many solid tumors, especially in those regions farthest from blood vessels that are more hypoxic (25). It is believed that this increased expression of HIF-1α provides a survival advantage for tumor cells under hypoxic conditions, with HIF-1α mediating adaptive responses to hypoxia by regulating target genes, such as angiogenic factors, glucose transporters, and glycolytic enzymes (25). However, the causal role of HIF-1α in tumor development has not been firmly established, apart from a few cases, such as pVHL-defective tumors (26). In addition to the HIF-1α–mediated adaptive changes seen in hypoxic tumors, accumulated evidence supports the notion that hypoxia can attenuate DNA repair, increase mutation rates, and drive genetic instability, suggesting that hypoxia may directly impact the DDR in a HIF-1α–independent fashion (27). Likewise, under hypoxic conditions, tumor cells can acquire radioresistance or chemoresistance both in vitro and in vivo (27, 28). Clinical trials over the last four decades have demonstrated that the origin of radioresistance can be eliminated by normoxic or hyperoxic modification of tumor environment (29), implying that radioresistance of tumor cells may be a direct result of lack of oxygen, rather than just genetic mutations. More pertinent to this study is the recent report that activation of PHDs by esterified 2-oxoglutarate is associated with increased apoptosis in the hypoxic region of tumors and inhibition of tumor growth in a HIF-1α–independent and PHD3-dependent way (30). Furthermore, silencing of PHD3 by abnormal hypermethylation of its promoter region is evident in a wide range of cancer types (31), suggesting that the inhibition or suppression of PHD3 may provide a selective advantage for tumors. The studies presented here provide a mechanism through which such effects might occur.
In summary, the data we present here advance our understanding of the regulation of the DDR, linking PHD3-mediated prolyl hydroxylation of HCLK2 to activation of the ATR/CHK1 pathway and revealing a potential mechanism for how hypoxia inhibits the DDR. The discovery that HCLK2 is a substrate for PHD3-dependent prolyl hydroxylation defines the hydroxylation of HCLK2 as a potential druggable mechanism for regulating the ATR/CHK1/p53 pathway to modulate events that endanger genomic integrity and determine the successful treatment of tumorigenesis.
Methods
Animals.
Phd3f/f mice were described previously (32). The CAG-Cre/Esr1 transgenic mice, which have a tamoxifen-inducible and Cre-mediated recombination system driven by the chicken β-actin promoter, were obtained from The Jackson Laboratory and mated with Phd3f/f mice to generate Phd3f/f;Cre+/– mice. Phd3f/f;Cre+/– mice were then mated with Phd3f/f;Cre–/– mice to generate Phd3f/f;Cre+/– and Phd3f/f;Cre–/– mice for experiments. The genotypes of these mice were determined by PCR as described previously with a protocol provided by The Jackson Laboratory (32).
Tamoxifen injection and IR.
Tamoxifen (Sigma-Aldrich) in corn oil (20 mg/ml) was aliquoted and frozen at –80°C for later use. Phd3f/f;Cre+/– mice, at approximately 6 to 8 weeks old, were injected intraperitoneally with tamoxifen (1 mg/20 g body weight/d) for 5 consecutive days to generate Phd3–/– mice (18). The deletion of the Phd3 gene was confirmed by real-time PCR 1 week after the first injection of tamoxifen, using primers described previously (32). Littermates that were injected with tamoxifen but which did not harbor Cre-Esr1 (Phd3f/f;Cre–/–) were used as controls. One week after the first injection of tamoxifen, mice of the indicated genotypes received a single dose of 9 Gy of X-rays (X-RAD 320, Precision X-Ray).
Cell culture.
HeLa cells were maintained in MEM with 10% FBS. PO2 was controlled by incubating cells at 37°C in humidified, O2/CO2-regulated incubators adjusted to 5% CO2 and the indicated PO2 (balanced by N2). Primary MEFs were isolated from embryos in the same uterus on day 13.5 following the standard protocol and maintained in MEM supplemented with 10% FBS. Genotypes of these MEFs were determined by PCR as described above. The primary MEFs used in the experiments were passaged no more than 4 times.
Knockdown of PHD3 by si-RNA.
si-RNA duplexes designed to suppress the expression of human PHD3 (sense, 5′-GCAAAUACUACGUCAAGGAUU-3′; antisense, 5′-UCCUUGACGUAGUAUUUGCUU-3′ and sense, 5′-UUCAGGAAUUUAACUAGGAUU-3′; antisense, 5′-UCCUAGUUAAAUUCCUGAAUU-3′; Genbank accession no., PHD3 NM_022073) and scrambled si-RNA were purchased from Dharmacon. HeLa cells at 40% to 50% confluence were transfected with si-RNAs (50 nM) using Lipofectamine 2000 (Invitrogen) for 72 hours. The efficiency of suppression was validated with Western blot analysis.
In vitro hydroxylation and GST pull-down assay for HCLK2 and ATR interaction.
Five μg GST-HCLK2340–530 captured on GSH-sepharose beads (Sigma-Aldrich) was incubated with 1 μg recombinant PHD3 in a reaction buffer containing 10 μM FeSO4, 100 μM 2-oxo-glutarate, 1 mM ascorbate, 60 μg catalase, 100 μM dithiothreitol, 2 mg bovine serum albumin, and 50 μM Tris-HCl buffer, adjusted to pH 7.8. The enzyme reaction was carried out at 37°C for 30 minutes. To assay ATR capture, GST-HCLK2340–530 was mixed with recombinant Flag-ATR purified from HEK293 cells at 4°C for 2 hours, either directly or following incubation with PHD3 as above. Bound ATR was analyzed by SDS-PAGE and Western blot. For the quantitative hydroxylation assay, 1 μM GST-HCLK2340–530 and 3 μg recombinant PHD3 were incubated in the same reaction buffer as described above, except using 40 μM 2-oxo-glutarate [1-14C] as the cosubstrate. The released 14CO2 was trapped and measured in triplicate.
In vitro kinase assay for ATR.
Wild-type Flag-ATR or kinase-dead Flag-ATRK2327R overexpressed in HEK293 cells was immunoprecipitated with anti-Flag beads overnight at 4°C. Immunoprecipitates were washed 3 times in wash buffer (500 mM NaCl, 50 mM Tris, pH 7.5, 1% NP-40, plus protease and phosphatase inhibitors), followed by 3 washes in in vitro kinase buffer (50 mM Tris, pH 7.5, 10 mM MgCl2, 1 mM dithiothreitol, plus protease and phosphatase inhibitors). Beads were then incubated with 50 μl of in vitro kinase buffer containing 1 μM ATP and 1 μg GST-p531–47 for 1 hour at 30°C. Resulting products were separated by SDS-PAGE and detected by Western blot analysis with anti–phospho-p53 (Ser15), anti-GST, or anti-ATR antibodies.
Coimmunoprecipitation.
Cells were lysed in cell lysis buffer (50 mM Tris, pH 7.4, 150 mM NaCl, 5 mM EDTA, 1% NP-40, 10% glycerol, and protease inhibitor cocktail), and lysates were clarified by centrifugation at 14,000 g. Following addition of 1 to 2 μg of antibody, lysates were incubated with rotation overnight at 4°C and for an additional hour after the addition of 25 μl of 50% protein A or G agarose. Beads were washed 5 times with lysis buffer, and proteins were analyzed by Western blot.
Immunofluorescence, TUNEL staining, and microscopy.
After 3 days of transfection with si-RNA, cells were washed with PBS, fixed, and permeabilized in phosphate-buffered 2% paraformaldehyde/0.2% Triton X-100 for 30 minutes at 4°C. Immunofluorescence labeling was carried out with a mouse anti-RPA32 antibody (ab2175, Abcam) followed by a FITC-conjugated goat anti-mouse secondary antibody (Jackson ImmunoResearch Laboratories Inc.) and DAPI (Sigma-Aldrich). Images were acquired by confocal laser-scanning microscopy (Zeiss 710). Thymuses from Phd3f/f;Cre+/– and Phd3f/f;Cre–/– mice were fixed with a 10% formalin solution, embedded in paraffin prior to sectioning, and stained with hematoxylin and eosin. TUNEL staining was performed following the protocol provided by manufacturer (S7100, Millipore).
LC-MS/MS analysis.
HCLK2340–530 immunoprecipitated from HeLa cells was excised from a Coomassie blue–stained SDS gel followed by dithiothreitol reduction, iodoacetamide alkylation, and in-gel digestion (12.5 ng/μl trypsin overnight). Resulting tryptic peptides were concentrated and analyzed by a C18 capillary reverse-phase liquid chromatography coupled with tandem MS (LC-MS/MS) using a LTQ Orbitrap Velos (ThermoFisher Scientific) under optimized conditions (33). The acquired MS/MS data were searched against a database to identify hydroxylated proline sites in HCLK2 through a dynamic mass shift for the modified proline (+15.9949 Da). The identified modified peptides were accepted after manually examining the MS/MS spectra. Moreover, an additional LC-MS/MS run was performed to confirm the presence of the hydroxylated peptides, in which high-resolution MS/MS data were collected in the Orbitrap for validation.
Statistics.
Data are shown as mean ± SEM for 3 to 4 separate experiments. Differences were analyzed with 2-tailed Student’s t test. Values of P ≤ 0.05 were considered statistically significant.
Study approval.
All animal procedures were approved by the Institutional Animal Care and Use Committee of University of North Carolina at Chapel Hill.
Supplementary Material
Acknowledgments
We thank Aziz Sancar (University of North Carolina at Chapel Hill, Chapel Hill, North Carolina, USA) for providing plasmids expressing wild-type ATR and its kinase-dead mutant (K2327R). We are grateful to Andrea L. Portbury for discussion and editing. This work was supported by NIH grants R01 HL61656, R01 GM61728, and R37 65619 and by the Fondation Leduq (to C. Patterson). L. Xie is supported by an AHA NCRP Scientist Development Grant (10SDG3860014).
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Citation for this article: J Clin Invest. 2012;122(8):2827–2836. doi:10.1172/JCI62374.
References
- 1.Semenza GL. HIF-1: mediator of physiological and pathophysiological responses to hypoxia. J Appl Physiol. 2000;88(4):1474–1480. doi: 10.1152/jappl.2000.88.4.1474. [DOI] [PubMed] [Google Scholar]
- 2.Kaelin WG, Jr, Ratcliffe PJ. Oxygen sensing by metazoans: the central role of the HIF hydroxylase pathway. Mol Cell. 2008;30(4):393–402. doi: 10.1016/j.molcel.2008.04.009. [DOI] [PubMed] [Google Scholar]
- 3.Ivan M, et al. HIFalpha targeted for VHL-mediated destruction by proline hydroxylation: implications for O2 sensing. Science. 2001;292(5516):464–468. doi: 10.1126/science.1059817. [DOI] [PubMed] [Google Scholar]
- 4.Jaakkola P, et al. Targeting of HIF-alpha to the von Hippel-Lindau ubiquitylation complex by O2-regulated prolyl hydroxylation. Science. 2001;292(5516):468–472. doi: 10.1126/science.1059796. [DOI] [PubMed] [Google Scholar]
- 5.Xie L, et al. Oxygen-regulated beta(2)-adrenergic receptor hydroxylation by EGLN3 and ubiquitylation by pVHL. Sci Signal. 2009;2(78):ra33. doi: 10.1126/scisignal.2000444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Luo W, et al. Pyruvate kinase M2 is a PHD3-stimulated coactivator for hypoxia-inducible factor 1. Cell. 2011;145(5):732–744. doi: 10.1016/j.cell.2011.03.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lipscomb EA, Sarmiere PD, Crowder RJ, Freeman RS. Expression of the SM-20 gene promotes death in nerve growth factor-dependent sympathetic neurons. J Neurochem. 1999;73(1):429–432. doi: 10.1046/j.1471-4159.1999.0730429.x. [DOI] [PubMed] [Google Scholar]
- 8.Collis SJ, Barber LJ, Clark AJ, Martin JS, Ward JD, Boulton SJ. HCLK2 is essential for the mammalian S-phase checkpoint and impacts on Chk1 stability. Nat Cell Biol. 2007;9(4):391–401. doi: 10.1038/ncb1555. [DOI] [PubMed] [Google Scholar]
- 9.Takai H, Wang RC, Takai KK, Yang H, de Lange T. Tel2 regulates the stability of PI3K-related protein kinases. Cell. 2007;131(7):1248–1259. doi: 10.1016/j.cell.2007.10.052. [DOI] [PubMed] [Google Scholar]
- 10.Rendtlew Danielsen JM, et al. HCLK2 is required for activity of the DNA damage response kinase ATR. J Biol Chem. 2009;284(7):4140–4147. doi: 10.1074/jbc.M808174200. [DOI] [PubMed] [Google Scholar]
- 11.Jazayeri A, et al. ATM- and cell cycle-dependent regulation of ATR in response to DNA double-strand breaks. Nat Cell Biol. 2006;8(1):37–45. doi: 10.1038/ncb1337. [DOI] [PubMed] [Google Scholar]
- 12.Epstein AC, et al. C. elegans EGL-9 and mammalian homologs define a family of dioxygenases that regulate HIF by prolyl hydroxylation. Cell. 2001;107(1):43–54. doi: 10.1016/S0092-8674(01)00507-4. [DOI] [PubMed] [Google Scholar]
- 13.Bishop T, et al. Abnormal sympathoadrenal development and systemic hypotension in PHD3–/– mice. . Mol Cell Biol. 2008;28(10):3386–3400. doi: 10.1128/MCB.02041-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lipscomb EA, Sarmiere PD, Freeman RS. SM-20 is a novel mitochondrial protein that causes caspase-dependent cell death in nerve growth factor-dependent neurons. J Biol Chem. 2001;276(7):5085–5092. doi: 10.1074/jbc.M008407200. [DOI] [PubMed] [Google Scholar]
- 15.Oda K, et al. p53AIP1, a potential mediator of p53-dependent apoptosis, and its regulation by Ser-46-phosphorylated p53. Cell. 2000;102(6):849–862. doi: 10.1016/S0092-8674(00)00073-8. [DOI] [PubMed] [Google Scholar]
- 16.Tournier C, et al. Requirement of JNK for stress-induced activation of the cytochrome c-mediated death pathway. Science. 2000;288(5467):870–874. doi: 10.1126/science.288.5467.870. [DOI] [PubMed] [Google Scholar]
- 17.Hirsila M, Koivunen P, Gunzler V, Kivirikko KI, Myllyharju J. Characterization of the human prolyl 4-hydroxylases that modify the hypoxia-inducible factor. J Biol Chem. 2003;278(33):30772–30780. doi: 10.1074/jbc.M304982200. [DOI] [PubMed] [Google Scholar]
- 18.Moresi V, et al. Myogenin and class II HDACs control neurogenic muscle atrophy by inducing E3 ubiquitin ligases. Cell. 2010;143(1):35–45. doi: 10.1016/j.cell.2010.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Aravind L, Koonin EV. The DNA-repair protein AlkB, EGL-9, and leprecan define new families of 2-oxoglutarate- and iron-dependent dioxygenases. Genome Biol. 2001;2(3):RESEARCH0007. doi: 10.1186/gb-2001-2-3-research0007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hogel H, Rantanen K, Jokilehto T, Grenman R, Jaakkola PM. Prolyl hydroxylase PHD3 enhances the hypoxic survival and G1 to S transition of carcinoma cells. PLoS One. 2011;6(11):e27112. doi: 10.1371/journal.pone.0027112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Takai H, et al. Chk2-deficient mice exhibit radioresistance and defective p53-mediated transcription. EMBO J. 2002;21(19):5195–5205. doi: 10.1093/emboj/cdf506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Myllyharju J, Kivirikko KI. Collagens, modifying enzymes and their mutations in humans, flies and worms. Trends Genet. 2004;20(1):33–43. doi: 10.1016/j.tig.2003.11.004. [DOI] [PubMed] [Google Scholar]
- 23.Huang J, Zhao Q, Mooney SM, Lee FS. Sequence determinants in hypoxia-inducible factor-1alpha for hydroxylation by the prolyl hydroxylases PHD1, PHD2, and PHD3. J Biol Chem. 2002;277(42):39792–39800. doi: 10.1074/jbc.M206955200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chowdhury R, et al. Structural basis for binding of hypoxia-inducible factor to the oxygen-sensing prolyl hydroxylases. Structure. 2009;17(7):981–989. doi: 10.1016/j.str.2009.06.002. [DOI] [PubMed] [Google Scholar]
- 25.Semenza GL. Targeting HIF-1 for cancer therapy. Nat Rev Cancer. 2003;3(10):721–732. doi: 10.1038/nrc1187. [DOI] [PubMed] [Google Scholar]
- 26.Kaelin WG. Von Hippel-Lindau disease. Annu Rev Pathol. 2007;2:145–173. doi: 10.1146/annurev.pathol.2.010506.092049. [DOI] [PubMed] [Google Scholar]
- 27.Bristow RG, Hill RP. Hypoxia and metabolism. Hypoxia, DNA repair and genetic instability. Nat Rev Cancer. 2008;8(3):180–192. doi: 10.1038/nrc2344. [DOI] [PubMed] [Google Scholar]
- 28.Shannon AM, Bouchier-Hayes DJ, Condron CM, Toomey D. Tumour hypoxia, chemotherapeutic resistance and hypoxia-related therapies. Cancer Treat Rev. 2003;29(4):297–307. doi: 10.1016/S0305-7372(03)00003-3. [DOI] [PubMed] [Google Scholar]
- 29.Overgaard J. Hypoxic radiosensitization: adored and ignored. J Clin Oncol. 2007;25(26):4066–4074. doi: 10.1200/JCO.2007.12.7878. [DOI] [PubMed] [Google Scholar]
- 30.Tennant DA, Gottlieb E. HIF prolyl hydroxylase-3 mediates alpha-ketoglutarate-induced apoptosis and tumor suppression. J Mol Med. 2010;88(8):839–849. doi: 10.1007/s00109-010-0627-0. [DOI] [PubMed] [Google Scholar]
- 31.Place TL, et al. Aberrant promoter CpG methylation is a mechanism for impaired PHD3 expression in a diverse set of malignant cells. PLoS One. 2011;6(1):e14617. doi: 10.1371/journal.pone.0014617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Takeda K, et al. Regulation of adult erythropoiesis by prolyl hydroxylase domain proteins. Blood. 2008;111(6):3229–3235. doi: 10.1182/blood-2007-09-114561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xu P, Duong DM, Peng J. Systematical optimization of reverse-phase chromatography for shotgun proteomics. J Proteome Res. 2009;8(8):3944–3950. doi: 10.1021/pr900251d. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.