Abstract
Squamous cell carcinomas (SCCs) are heterogeneous and aggressive skin tumors for which innovative, targeted therapies are needed. Here, we identify a p53/TACE pathway that is negatively regulated by FOS and show that the FOS/p53/TACE axis suppresses SCC by inducing differentiation. We found that epidermal Fos deletion in mouse tumor models or pharmacological FOS/AP-1 inhibition in human SCC cell lines induced p53 expression. Epidermal cell differentiation and skin tumor suppression were caused by a p53-dependent transcriptional activation of the metalloprotease TACE/ADAM17 (TNF-α–converting enzyme), a previously unknown p53 target gene that was required for NOTCH1 activation. Although half of cutaneous human SCCs display p53-inactivating mutations, restoring p53/TACE activity in mouse and human skin SCCs induced tumor cell differentiation independently of the p53 status. We propose FOS/AP-1 inhibition or p53/TACE reactivating strategies as differentiation-inducing therapies for SCCs.
Introduction
Nonmelanoma skin cancer is the most common form of human cancer (1), and squamous cell carcinomas (SCCs) are the most aggressive and heterogeneous skin cancers, with alterations in several signaling pathways, such as RAS, p53, p14, and p16 (1). The stress-inducible transcription factor p53 is functionally impaired in the majority of SCCs, and inactivating mutations account for about 50% of all SCCs (2, 3), suggesting that p53 function is important for skin tumor development. How p53 is functionally suppressed in skin cancer, why its functional impairment facilitates skin tumor development, and how p53 possibly exerts skin tumor suppression are not well understood.
Members of the AP-1 transcription factor complex, such as FOS and c-Jun, were shown to facilitate tumor development by restricting the tumor-suppressive function of p53 through direct transcriptional repression (4–7). In particular, the FOS gene exhibits both oncogenic and tumor-suppressive functions, depending on the cellular context (8, 9). In skin physiology and cancer, the role of FOS as an oncogene is widely documented, such as in the RAS-dependent DMBA/TPA (where DMBA indicates 7,12-dimethylbenz[a]anthracene and TPA indicates 12-O-Tetradecanoylphorbol-13-acetate) mouse carcinogenesis model (10, 11), in the EGFR-dependent K5-SOS+ tumor-prone transgenic mouse model (12–14), and in human SCCs (11). However, the molecular mechanisms by which FOS contributes to skin tumor development are unknown. FOS expression appears dispensable for mouse skin development and homeostasis (15) while required for RAS-induced benign and malignant squamous cell lesions and in skin tumor progression (15, 16). Importantly, p53 is inactivated at early stages of skin tumor development, and its loss facilitates malignant progression of murine skin tumors (17, 18), suggesting that FOS and p53 might antagonize each other during malignant transformation.
Among many targets in keratinocytes, p53 induces NOTCH1 expression (19, 20), which acts as a tumor suppressor in the epidermis (21, 22) and importantly, induces keratinocyte differentiation through several “canonical” targets (23). NOTCH1 activation and its nuclear translocation occur subsequent to ligand-mediated NOTCH receptor unfolding, which permits shedding and intramembrane proteolysis by one of the S2 proteases, TACE/ADAM17 (TNF-α–converting enzyme) or ADAM10, and the γ-secretases, presenilin 1 and 2, respectively (24–26). TACE is a disintegrin metalloproteinase that sheds membrane-anchored proteins, such as ligands of EGFR, cytokines, cytokine receptors, and adhesion molecules involved in several cellular processes (27). TACE-knockout mice exhibit several developmental defects (28), whereas TACE activation, mostly inhibited by the tissue inhibitor of metalloproteinases TIMP-3 — a transcriptional target of Jun/AP-1 (29) — is mainly associated with proinflammatory responses, tumor promotion, and invasiveness (30). Recently, it was shown that TACE antagonizes inflammation in certain cell types (31, 32). Moreover, the rhomboid family member iRhom2 was shown to promote trafficking and activation of TACE by binding to TACE in the endoplasmic reticulum (33, 34). However, little is known about the transcriptional control of TACE, the physiological consequences of TACE-mediated NOTCH1 activation, and whether TACE plays a role in skin tumor development.
Here, we describe what we believe is a novel FOS-dependent p53 pathway exerting tumor-suppressive functions by induction of tumor cell differentiation through p53-dependent TACE induction.
Results
Skin tumor suppression by loss of FOS in epidermal cells.
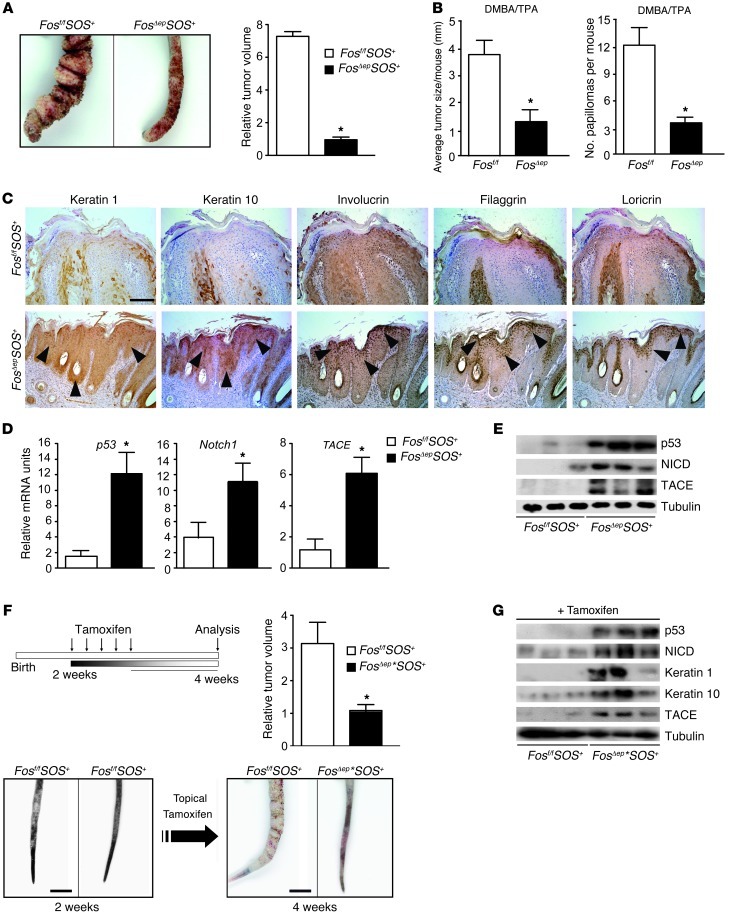
To investigate the role of FOS in skin tumor development, the K5-SOS+/RAS-dependent papilloma-prone mouse model (13) and the well-established DMBA/TPA skin carcinogenesis protocol were employed. Mice lacking epidermal Fos showed no obvious spontaneous skin phenotype and displayed normal proliferation and differentiation (Supplemental Figure 1A; supplemental material available online with this article; doi: 10.1172/JCI63103DS1). Importantly, tumor development on a WT p53 background (4) was suppressed in epidermal K5-SOS+ Fos-deficient mice, as shown by severely reduced tumor volume (Figure 1A). DMBA/TPA-induced RAS-mutated papillomas, which retain WT p53 (35), were also diminished and tumor size reduced in mice lacking epidermal Fos (Figure 1B and Supplemental Figure 2A). Notably, the frequency of Ras mutation at codon 61 was not altered upon epidermal Fos deletion (refs. 10, 36, and Supplemental Figure 2B). In both skin tumor models, no significant alterations in basal cell layer proliferation (Ki67-positive cells) and apoptosis (TUNEL-positive cells) were detected (Supplemental Figure 2C). FOS expression was reduced, and no alterations in Keratin 5 expression were observed, although p21 expression was induced upon Fos deletion (Supplemental Figure 2D and data not shown). Importantly, reduced papilloma formation correlated with increased differentiation, as measured by early and late differentiation marker expression, such as Keratin 1, 10, involucrin, filaggrin, and loricrin (Figure 1C). This also led to reduced malignant progression and diminished expression of invasion markers such as Podoplanin and MMPs (12). We next analyzed the differentiation-inducing NOTCH1 pathway and found increased levels of p53, NOTCH1, and active NOTCH1 (NICD) as well as TACE both at the mRNA and protein levels (Figure 1, D and E). In contrast, no significant changes were observed in p16 and p19 mRNA levels as well as in presenilin 1 and presenilin 2 and ADAM10 (Supplemental Figure 2, E and F). Furthermore, Wnt3 and Wnt4 downregulation in Fos-deficient papillomas confirmed NOTCH1-induced, p21-dependent suppression of Wnt signaling (ref. 37 and data not shown). These results demonstrate that Fos-deficient papillomas undergo precocious differentiation and tumor suppression through activation of the NOTCH1 pathway.
Figure 1. Fos epidermal deletion in mice suppresses skin cancer development due to increased differentiation.
(A) Papilloma macroscopic view in 6-week-old mice deficient for Fos in the epidermis (K5-SOS+) compared with controls. Relative tumor volume is shown in 6-week-old mice (n = 10). (B) Papilloma size average per mouse and numbers of papillomas per mouse upon DMBA/TPA throughout 15 weeks in mice deficient for Fos in the epidermis (K5-SOS+) (black bars) compared with littermate controls (white bars). n = 7 per genotype. (C) Immunohistochemistry of 4-week-old mice deficient for Fos in the epidermis (K5-SOS+) (lower panels) compared with K5-SOS+ controls (upper panels) demonstrates increased expression of Keratin 1, Keratin 10, and Involucrin, Filaggrin, and Loricrin keratinocyte differentiation markers. Arrowheads indicate positive staining. Scale bar: 100 μm. Original magnification, ×10. (D) qRT-PCR analyses of tumor samples from 4-week-old mice deficient for Fos in the epidermis (K5-SOS+) compared with controls. Increased expression of p53, Notch1, and TACE in mice lacking epidermal Fos. n = 4 per genotype. (E) Western blot from 4-week-old mice deficient for Fos in the epidermis (K5-SOS+) compared with controls. Increased expression of p53, NICD, and TACE in K5-SOS+ mice lacking epidermal Fos. (F) Tamoxifen application in 2-week-old mice and analysis 2 weeks later. Relative tumor volume is shown in 4-week-old mice (black bars) compared with controls (white bars), demonstrating reduced papilloma size upon inducible, epidermal Fos deletion (upper panels). Macroscopic appearance of papillomas before and after inducible epidermal Fos deletion (lower panels). n = 5 per genotype. Scale bar: 0.5 cm. *P < 0.05. (G) Western blot analyses of tumor samples from inducible Fos-deficient K5-SOS+ mice. Results show increased expression of p53, NICD, Keratin 1, Keratin 10, and TACE.
Interestingly, topical application of TPA to Fos-deficient epidermis induced precocious differentiation of keratinocytes, which was suppressed by N-((3,5-difluorophenyl)acetyl)-L-alanyl-2-phenylglycine-1,1-dimethylethyl ester (DAPT), an inhibitor of NOTCH1 activation (Supplemental Figure 2G). This indicates that precocious differentiation of interfollicular epidermis likely occurs prior to tumor development.
To investigate whether differentiation-induced skin tumor suppression can be exploited therapeutically, we deleted Fos in tumor-prone K5-SOS+ mice when tail papillomas were already macroscopically visible (Figure 1F). Tumor development was arrested and molecular analyses showed that papillomas displayed a pronounced differentiated phenotype, expressing high levels of p53, NICD, Keratin 1, Keratin 10, and TACE (Figure 1G). Therefore, constitutive and inducible deletion of Fos leads to epidermal differentiation and tumor suppression in 2 RAS-driven genetic mouse models that harbor WT p53 (4, 35).
Fos represses p53 to prevent stress-induced keratinocyte differentiation.
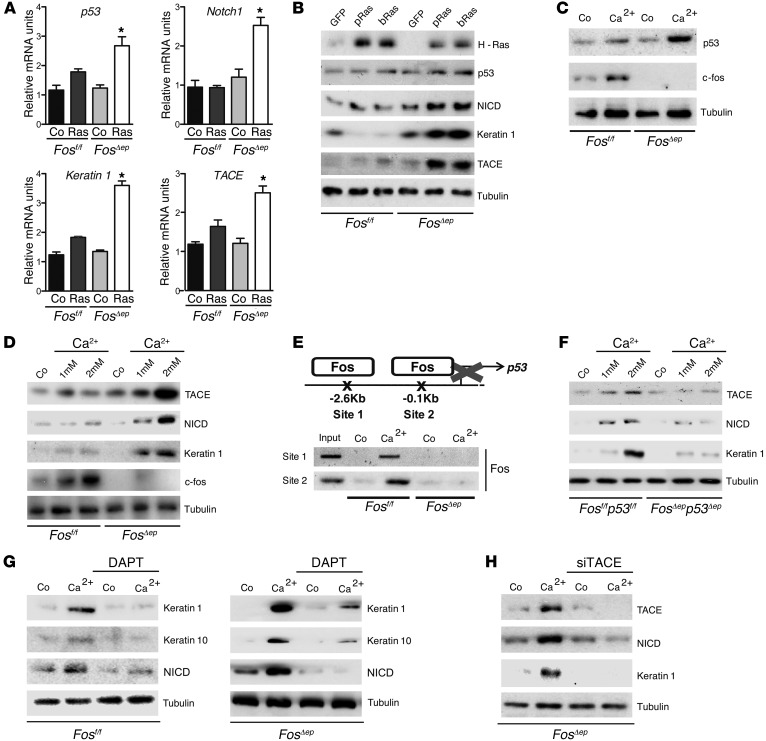
To find out whether the effect of FOS on keratinocyte differentiation is cell autonomous, we expressed oncogenic RAS in Fos-deficient keratinocytes in vitro. Expression of oncogenic RAS was shown to induce p53 in human and mouse keratinocytes, which displayed both senescence-like and differentiation-like features (38, 39). In the absence of FOS, enhanced mRNA levels of p53, Notch1, TACE, Keratin 1, Keratin 10, and p21 were observed (Figure 2A and Supplemental Figure 3A). Moreover, increased protein levels were detected for p53, NICD, Keratin 1, and TACE (Figure 2B and Supplemental Figure 3A). In addition, TACE activity, which is causal for increased NICD expression, was upregulated with no changes in TIMP-3 levels (Supplemental Figure 3B and data not shown). Interestingly, expression of senescence-associated markers such as p16 and p19 was prevented in the absence of Fos (Supplemental Figure 3A), while no differences in keratinocyte proliferation were observed, as measured by p-Ser10 Histone 3 and PCNA (Supplemental Figure 2C).
Figure 2. FOS represses p53 expression, which induces precocious TACE- and NOTCH1-dependent differentiation of Fos-deficient keratinocytes.
(A) qRT-PCR analyses of Fos-deficient primary keratinocytes 6 days after H-RasV12 (Ras) or empty vector (Co) retroviral infection. Fos deficiency induces increased expression of p53, Notch1, Keratin 1, and TACE. *P < 0.05. (B) Western blot analyses of Fos-deficient primary keratinocytes infected with pLPC H-RasV12 (pRas), pBabe H-RasV12 retroviruses (bRas), or GFP for 6 days. Fos deficiency induces increased expression of p53, NICD, Keratin 1, and TACE. (C) Western blot of Fos-deficient and control keratinocytes stimulated for 48 hours with 2 mM Ca2+. p53 expression is enhanced in Fos-deficient keratinocytes. (D) Western blot analyses of Fos-deficient and control keratinocytes treated with 1 mM and 2 mM Ca2+ for 48 hours. Fos deficiency induces increased expression of TACE, NICD, and Keratin 1. (E) Schematic of p53 promoter containing 2 AP-1–binding sites (upper panel). ChIP on 2 different AP-1 sites shows that FOS binds to p53 promoter only in control cells after 24 hours treatment with 2 mM Ca2+ (lower panel). (F) Western blot of keratinocytes concomitantly lacking Fos and p53 treated with 2 mM Ca2+. Fos and p53 deficiency impairs expression of NOTCH1, p21, Keratin 1, and TACE compared with controls. (G) Western blot of WT and Fos-deficient keratinocytes upon NOTCH1 inhibition with DAPT (γ-secretase inhibitor) in 2 mM Ca2+ conditions for 24 hours shows NOTCH1-dependent differentiation. (H) Western blot analyses of Fos-deficient primary keratinocytes upon TACE siRNA knockdown in 2 mM Ca2+ conditions for 24 hours shows TACE-dependent precocious differentiation.
Culturing keratinocytes in the presence of increased calcium (Ca2+) is a well-established way to induce keratinocyte differentiation in vitro, which also activates FOS (Figure 2C and Supplemental Figure 4A). Under these conditions, increased levels of p53 were detected in the absence of Fos, suggesting that in differentiation-inducing conditions, FOS repressed p53 (Figure 2C). Importantly, Fos-deficient keratinocytes underwent precocious differentiation, as shown by increased TACE and NICD protein expression, which correlated with increased Keratin 1 (Figure 2D). Increased mRNA levels of TACE, Notch1, Hes1 (a NOTCH1 target gene; ref. 40), Keratin 1, Keratin 10, p21, and TIMP-1 (a potent inhibitor of ADAM10) were measured (Supplemental Figure 4, B and C). In addition, reduced levels of TIMP-3 were detected, supporting an increase in TACE activity (Supplemental Figure 4, C and D), while no changes in Keratin 5 and 14 and ADAM10 were observed (Supplemental Figure 4C). Finally, other stress-inducing stimuli, such as TPA treatment, induced precocious differentiation of FOS-deficient keratinocytes (Supplemental Figure 5). These results demonstrate that precocious differentiation in response to diverse stimuli is likely due to increased p53 expression.
To analyze a possible direct transcriptional effect of FOS on p53 expression, ChIP assays were performed. Upon Ca2+ treatment, FOS bound to the p53 promoter at 2 different AP-1 sites (Figure 2E). Furthermore, c-Jun also bound the p53 promoter, but only when FOS was present (data not shown). These results demonstrate that under stress conditions, FOS suppresses p53 expression through direct binding to its promoter, which in the absence of Fos, triggers precocious differentiation.
Precocious differentiation of Fos-deficient keratinocytes is p53, TACE, and Notch1 dependent.
To investigate whether FOS prevents keratinocyte differentiation through p53 repression, keratinocytes lacking both Fos and p53 were generated. Double-deficient keratinocytes, when stimulated with Ca2+, exhibited impaired differentiation with reduced expression of TACE, NICD, and Keratin 1 protein and of mRNAs for Notch1, TACE, Hes1, p21, and the early differentiation markers Keratin 1 and Keratin 10 (Figure 2F and Supplemental Figure 6A). Furthermore, proliferation was measured using Cyclin D1 in keratinocytes overexpressing H-RasV12, but lacking Fos, p53, and Fos/p53, and upon concomitant TACE shRNA knockdown. Differentiation-induced proliferation arrest of Fos-deficient keratinocytes was rescued following TACE knockdown in a fashion similar to that of when p53 is absent, which is known to favor proliferation and transformation (Supplemental Figure 6B).
To further analyze whether precocious differentiation of Fos-deficient keratinocytes is indeed NOTCH1 and TACE dependent, we inhibited either NOTCH1 or TACE. Inhibition of NOTCH1 activation by the γ-secretase inhibitor DAPT in WT and Fos-deficient keratinocytes led to impaired TACE and NOTCH1 activation and reduced Keratin 1 expression (Figure 2G). Likewise, TACE inhibition with TAPI-1, a broad MMP/ADAM inhibitor (Supplemental Figure 6C), or siRNA-mediated TACE knockdown impaired precocious differentiation (Figure 2H and Supplemental Figure 6D). Since ADAM10 also activates NOTCH1, we analyzed whether it can also affect keratinocyte differentiation. Differentiation marker expression and NOTCH1 activation were not altered upon ADAM10 knockdown, indicating that ADAM10 does not play a significant role in Ca2+-induced keratinocyte differentiation (Supplemental Figure 6E). These results demonstrate that Fos deficiency induces precocious keratinocyte differentiation due to enhanced p53 and TACE expression leading to NOTCH1 activation.
We also noted that besides NOTCH1 activation, Notch1 mRNA levels were induced in the absence of Fos following RAS expression (Figure 2A), suggesting a positive feedback loop on NOTCH1 signaling. Indeed, NICD overexpression in WT keratinocytes led to increased Notch1 mRNA and full-length NOTCH1 protein (Supplemental Figure 7, A and B). Increased expression was inhibited when MAM51 peptide, a suppressor of NOTCH1-dependent transcription, was coexpressed (Supplemental Figure 7A). On the contrary, TACE expression was not affected by NOTCH1 (Supplemental Figure 7, C and D). Moreover, when NOTCH1 activation upon Ca2+ treatment was inhibited by DAPT, Notch1 RNA expression was reduced (Supplemental Figure 7E), demonstrating a positive feedback loop between NICD and Notch1 expression, consistent with similar findings in T cells (41).
p53 induces keratinocyte differentiation through TACE induction and Notch1 activation.
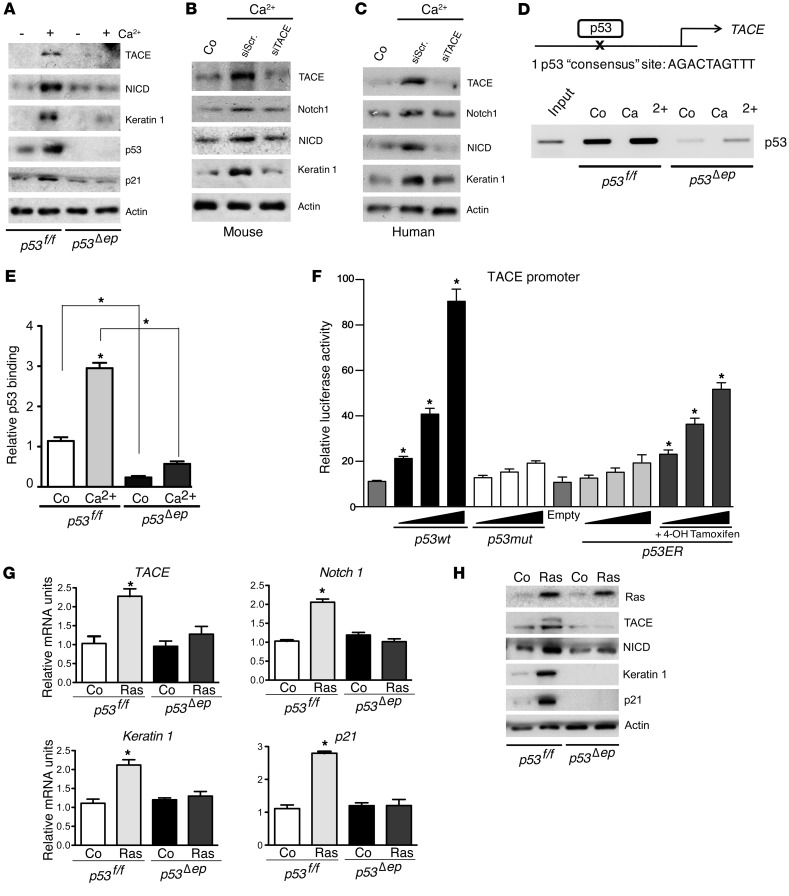
A role of p53 in keratinocyte differentiation and senescence upon diverse stimuli has been described (4, 19, 20, 38). Therefore, we next challenged p53-deficient keratinocytes with Ca2+. p53 deficiency led to impaired expression of TACE, Notch1, Hes1, Keratin 1, Keratin 10, p21, and TIMP-3, while ADAM10 mRNA levels were not altered (Supplemental Figure 8A). Similarly, protein analyses showed impaired expression of TACE, NICD, and Keratin 1 in p53-deficient cells (Figure 3A). In addition, stimulation of p53-deficient keratinocytes with TPA showed impaired mRNA expression of TACE, Notch1, and keratinocyte differentiation markers (Supplemental Figure 8B).
Figure 3. p53 induces TACE expression to promote keratinocyte differentiation.
(A) Western blot analyses of p53-deficient and control keratinocytes treated with 2 mM Ca2+ for 48 hours. Lack of p53 impairs expression of TACE, NICD, Keratin 1, and p21. (B) Western blot analyses of WT mouse keratinocytes upon siRNA-mediated TACE (siTACE) or scrambled (siScr) knockdown followed by 48 hours of 1 mM Ca2+ treatment. Expression of TACE, NICD, and Keratin 1 is TACE dependent. (C) Western blot analyses of human primary keratinocytes upon siRNA-mediated TACE or scrambled knockdown after 48 hours of 1 mM Ca2+ treatment. Expression of TACE, NOTCH1 (NICD), and Keratin 1 is TACE-dependent. (D) TACE promoter contains a p53-binding site. RT-PCR analyses from p53 ChIP in 2 mM Ca2+-induced differentiation conditions (lower panel). (E) Quantitative RT-PCR upon p53 ChIP on the TACE promoter as in Figure 1D. (F) Luciferase reporter activity assay with TACE promoter construct. Increasing concentrations of WT p53 or p53-ER plus 1 μM 4-OH Tamoxifen induce TACE reporter activity, whereas mutant p53 (p53 mut) does not.(G) qRT-PCR analyses of p53-deficient keratinocytes infected with retroviruses overexpressing H-RasV12 or empty vector after 6 days. p53 absence impairs expression of TACE, Notch1, Keratin 1, and p21. *P < 0.05. (H) Western blot analyses of p53-deficient keratinocytes infected with retroviruses overexpressing H-RasV12 or empty vector after 6 days. p53 absence impairs TACE, NICD, Keratin 1, and p21 expression.
We next investigated whether TACE is necessary and sufficient for keratinocyte differentiation. In primary mouse and human keratinocytes stimulated with Ca2+, NOTCH1 protein expression and activation and early differentiation marker expression were impaired following TACE knockdown (Figure 3, B and C). This indicates that TACE is required for NOTCH1-dependent keratinocyte differentiation. On the other hand, TACE or WT p53 overexpression, but not mutant p53, induced mRNA expression of early differentiation markers, but not late differentiation markers, similarly to ectopic expression of NICD (Supplemental Figure 9A). Protein levels of TACE, its nonprocessed form Pro-TACE, NOTCH1, and NICD were elevated upon p53 WT overexpression (Supplemental Figure 9B) and in keratinocytes treated with Nutlin-3, a murine double-minute 2 (MDM2) inhibitor that increases p53 levels (Supplemental Figure 9C).
Next, we tested to determine whether TACE is a direct transcriptional target of p53. A p53 consensus-binding site was identified within the TACE promoter (42). ChIP assays showed that p53 binds this site under basal conditions. Upon Ca2+ stimulation, p53 binding was increased, but was absent in p53-deficient keratinocytes (Figure 3, D and E). A fragment of the mouse TACE promoter harboring the p53-binding sequence was cloned into a luciferase reporter. Cotransfection with a WT p53–expressing construct induced luciferase activity, which did not occur when a DNA-binding domain mutant p53 construct was used (Figure 3F). In addition, employing a p53ER fusion protein, luciferase activity was increased specifically in the presence of 4-hydroxytamoxifen (4-OHT). These results clearly demonstrate that p53 directly activates TACE transcription and thus NOTCH1-dependent keratinocyte differentiation.
To further genetically define how p53 induces epidermal tumor suppression, the response of WT and p53-deficient keratinocytes to oncogenic RAS signaling was analyzed. Upon H-RasV12 expression, p53-deficient keratinocytes displayed impaired expression of TACE, NOTCH1, NICD, Keratin 1, Keratin 10, Involucrin, and p21 as analyzed by quantitative RT-PCR (qRT-PCR) and Western blotting (Figure 3, G and H, and Supplemental Figure 9D). Interestingly, no changes in the expression of ADAM10 and presenilin 1 and presenilin 2 were observed (Supplemental Figure 9D). Wild-type primary keratinocytes expressing H-RASV12 showed the expected expression of senescence markers, such as p16 and p19, which as expected, were not induced in p53-deficient keratinocytes (Supplemental Figure 9, D and E). These results indicate that upon oncogenic stress signaling, p53 induces keratinocyte differentiation through direct induction of TACE expression with subsequent NOTCH1 activation.
p53 restoration leads to skin tumor cell differentiation in vivo.
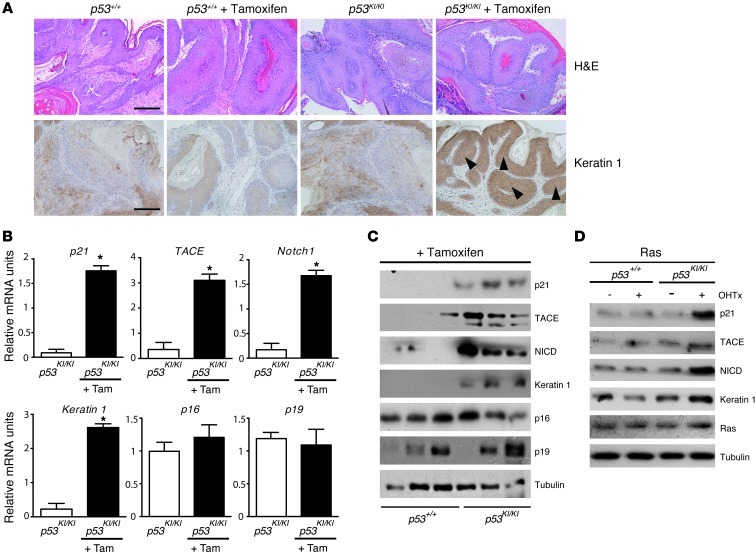
The function of p53 is frequently impaired in SCCs, although Fos deletion in mouse skin tumor models induces WT p53, leading to tumor cell differentiation. We next asked whether restoring p53 function in skin papillomas that are fully developed in a p53-deficient background could lead to tumor suppression. Thus, p53-deficient knockin mice (43) (p53KI/KI) were used, in which the endogenous p53 gene is replaced by an inducible p53ER-TAM fusion gene, where p53 activity can be restored by addition of the ligand 4-OHT. Upon 4-OHT treatment in vivo or in vitro, p53-deficient cells are rapidly converted to cells with fully functional WT p53 (ref. 43 and data not shown).
We employed again the established DMBA/TPA skin carcinogenesis tumor model, in which p53 is WT, but functionally inactivated (35), on WT and p53KI/KI mice. Comparable papilloma numbers and size developed in mice of all genotypes (Supplemental Figure 10A). Once papillomas had developed at week 15, p53 activity was restored by daily tamoxifen injection for 2 weeks. Histological analyses demonstrated that tumors differentiated following p53 restoration (Figure 4A). No effects on proliferation, apoptosis, or senescence-associated marker expression such as p16, p19, and β-galactosidase activity and expression were observed (Supplemental Figure 10, B and C, and data not shown). As expected, the mRNA and protein levels of components of the NOTCH1 differentiation pathway — including p21, TACE, Notch1, and Keratin 1 — were increased upon p53 restoration (Figure 4, B and C). Importantly, expression of keratinocyte differentiation markers, such as suprabasal Keratin 1 and Keratin 10, was found induced only in papillomas, where p53 function was restored (Figure 4, A–C, and Supplemental Figure 10C). These results convincingly demonstrate that p53 restoration induced skin tumor cell differentiation and suppression with no apparent effect on apoptosis, proliferation, or senescence.
Figure 4. p53 restoration induces tumor suppression through differentiation.
(A) Immunohistochemical analyses of papillomas of WT and p53KI/KI mice treated with DMBA/TPA for 15 weeks and tamoxifen for 15 days (3 mg/mouse/d). H&E staining (upper panels). Increased Keratin 1 expression (brown stain) is detected upon p53 restoration. Scale bar:100 μm. Original magnification, ×10. n = 7 mice per genotype. Arrowheads indicate positive staining. (B) qRT-PCR analyses of papillomas of WT and p53KI/KI mice treated with DMBA/TPA for 15 weeks and tamoxifen (Tam) for 15 days (3 mg/mouse/d). p21 expression is increased, as well as TACE, Notch1 and early differentiation markers such as Keratin 1. No induction of senescence-associated markers such as p16 and p19 is detected. *P < 0.05. n = 4 mice per genotype. (C) Western blot of the papillomas of WT and p53KI/KI mice treated with DMBA/TPA for 15 weeks and tamoxifen for 15 days (3 mg/mouse/d). p21 protein levels are increased as well as TACE, NICD, and early differentiation markers such as Keratin 1. However, no significant difference in induction of senescence-associated markers such as p16 and p19 is detected. (D) Western blot analyses of WT and p53KI/KI primary keratinocytes overexpressing H-RASV12 for 6 days after retroviral infection (Ras) and 48 hours treatment with 1 μM 4-OH tamoxifen to restore p53 expression. p21 expression is increased in p53KI/KI cells as well as TACE, NICD, and early differentiation markers such as Keratin 1.
To analyze whether the effect of p53 restoration is cell autonomous, p53+/+ and p53KI/KI primary keratinocytes expressing oncogenic RAS were treated with 4-OHT. TACE, Notch1, Hes1, Keratin 1, Keratin 10, and p21 mRNA and p21, TACE, NICD and Keratin 1 protein levels were increased upon p53 restoration (Figure 4D and Supplemental Figure 10, D and E). No changes in mRNA expression were observed for ADAM10 and the γ-secretases presenilin 1 and presenilin 2 (data not shown). Importantly, p53KI/KI keratinocytes without addition of 4-OHT behaved like p53-deficient keratinocytes (data not shown). These results strongly indicate that p53 restoration in p53-deficient H-RASV12–transformed keratinocytes in vitro as well as in DMBA/TPA-treated p53-deficient mice in vivo lead to differentiation and tumor suppression.
Human SCCs: FOS inhibition and p53/TACE activation lead to SCC cell differentiation in a p53-dependent manner.
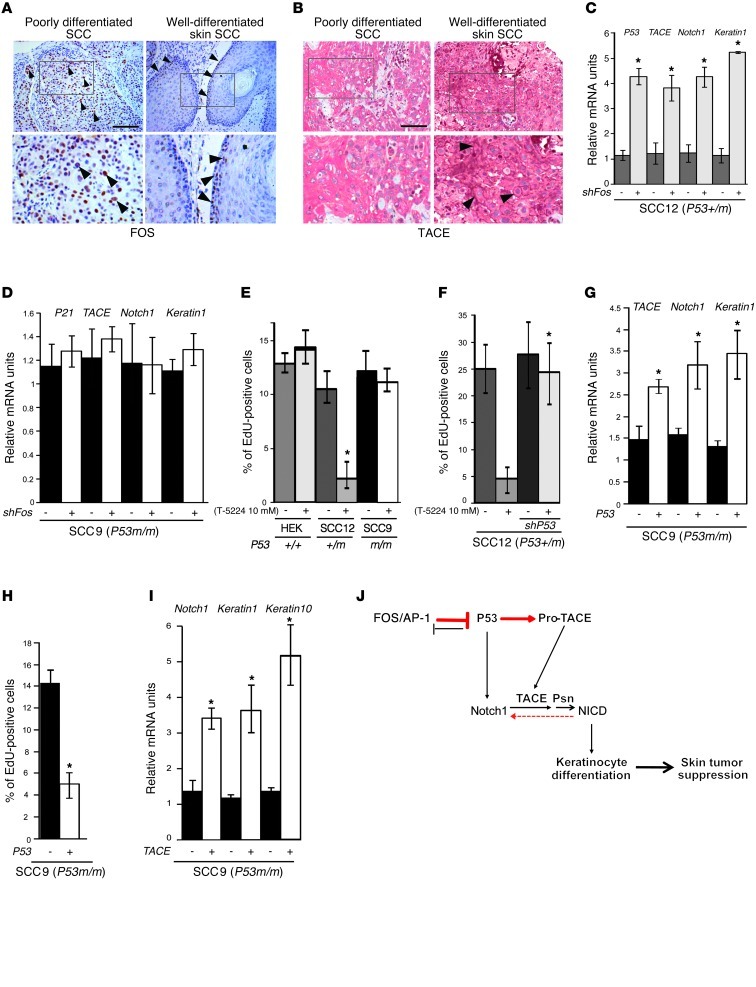
In order to validate our findings in human tumors, we analyzed FOS expression in a panel of 54 pathologically diagnosed, poorly differentiated human skin SCCs compared with 28 well-differentiated human skin SCCs. FOS nuclear staining was detected in almost all poorly differentiated, proliferating SCC tumor cells. These cells express cytoplasmic TACE and total NOTCH1 (not active), but do not express Keratin 1 and Loricrin. On the contrary, FOS expression was not detectable in well-differentiated, nonproliferative SCC tumor cells. These cells express membranous TACE (active), Keratin 1, and Loricrin, but are negative for total NOTCH1 (Figure 5A, Supplemental Figure 11, and Supplemental Table 1). This suggests an inverse correlation between FOS expression and keratinocyte differentiation in SCCs. When the TACE protein expression pattern was analyzed in a similar set of samples, membranous TACE expression was only detected in differentiated cells. While normal skin has a uniform membranous staining in basal and stratum spinosum keratinocytes, membranous TACE was found in 93% of cases of sun-exposed skin, in 86% of cases of aktinic keratosis (examples given in Supplemental Figure 12, A and B, and Supplemental Table 2), and in 20% of SCCs. With regard to SCCs, membranous TACE was restricted to keratinizing areas (Figure 5B, Supplemental Table 1, Supplemental Figure 11), whereas cytoplasmic TACE expression was detected in nonkeratinizing, poorly differentiated cells.
Figure 5. Inverse correlation in FOS and membranous TACE expression in human SCCs.
FOS knockdown or pharmacological inhibition in human SCC cell lines induces p53-dependent differentiation and growth inhibition. (A) Immunohistochemistry of FOS in human skin SCCs. Original magnification, ×10 (upper panels); ×20 (lower panels). Arrowheads indicate positive FOS staining. Scale bar: 100 μm. See Supplemental Table 1 for complete analyses. (B) Immunohistochemistry of TACE in human skin SCCs. Membranous TACE expression is restricted to differentiated cells only. Original magnification, ×10 (upper panels); ×20 (lower panels). Arrowheads indicate positive TACE staining. Scale bar: 100 μm. See Supplemental Table 1 for complete analyses. (C) qRT-PCR analyses upon Fos shRNA–mediated knockdown in SCCO12 (SCC12) show increased expression of p53 and TACE, Notch1, and Keratin 1. (D) qRT-PCR analyses upon Fos shRNA–mediated knockdown in SCC9. No induction of p21 and TACE, Notch1, and Keratin 1 upon Fos knockdown. (E) EdU proliferation assay performed after FOS inhibition (48 hours treatment) in HEK, SCCO12, and SCC9. Experiment was performed 2 times. (F) EdU proliferation assay performed after FOS inhibition (48 hour treatment) in SCCO12 where p53 was knocked down. Experiment was performed 2 times. (G) qRT-PCR analyses upon WT p53 overexpression in SCC9 cells show increased expression of TACE, Notch1, and Keratin 1. Experiment was performed 2 times. (H) EdU proliferation assay after p53 overexpression in SCC9 cells. Experiment was performed 2 times. (I) qRT-PCR upon TACE overexpression through transient transfection in SCC9 cells. *P < 0.05. Experiment was performed 2 times. (J) Model illustrating how the FOS/p53/TACE pathway controls keratinocyte differentiation and skin tumor suppression. p53 directly induces TACE and NOTCH1 expression, thereby promoting keratinocyte differentiation, leading to skin tumor suppression. Red lines indicate the pathways described in this manuscript. Solid lines indicate direct interaction. Dotted line indicates indirect interaction.
To mechanistically address a possible therapeutic potential of interfering with FOS, p53, and TACE in human tumor cells, keratinocyte-derived human SCC cell lines were employed. FOS mRNA levels (10 out of 11) and FOS activity (11 out of 11) were found increased in SCC cell lines when compared with human primary keratinocytes (Supplemental Figure 13, A and B, and Supplemental Table 3). Thus, the potential therapeutic value of FOS inhibition was assessed by Fos knockdown or inhibitory studies. shRNA-mediated Fos knockdown significantly reduced Fos mRNA levels in all SCC cell lines (Supplemental Figure 13C). Interestingly, all cell lines that expressed a WT p53 allele underwent differentiation (Figure 5C and Supplemental Table 1). As previously observed in mouse cells, p53 mRNA was induced and the p53-dependent TACE/Notch1 pathway was activated, leading to Keratin 1 expression (Figure 5C and Supplemental Table 1). In contrast, the p53-dependent pathway was not activated in SCC cell lines with mutant p53, and consequently cells remained undifferentiated (Figure 5D and Supplemental Table 1). In addition, EdU incorporation assays showed decreased proliferation upon FOS knockdown in SCC cells with a WT p53 allele and no significant alterations in SCC cells with mutated p53 alleles (Supplemental Figure 13D). Pharmacologic inhibition using a FOS/AP-1 inhibitor (T-5224) (44), which shows no obvious toxicity to normal epithelia (44), also led to the induction of p53, TACE, NOTCH1, and Keratin 1 (data not shown), resulting in differentiation and halted proliferation only in SCC cell lines carrying a WT p53 allele except for human primary keratinocytes (Figure 5E and Supplemental Figure 14A). Since we reasoned that this phenotype is p53 dependent, knockdown of p53 was performed in SCC cell lines carrying a WT p53 allele. The decrease in proliferation upon FOS inhibition was abolished when p53 was knocked down (Figure 5F and Supplemental Figure 14B). Thus, FOS inhibition in SCC cell lines carrying a WT p53 allele induced p53-dependent tumor cell differentiation.
Finally, the potential therapeutic value of p53 reexpression and activation was assessed in SCC cell lines carrying mutant p53 alleles, which are resistant to FOS inhibition. Forced WT p53 expression induced TACE and NOTCH1-dependent tumor cell differentiation and reduced proliferation (Figure 5, G and H, and Supplemental Figure 15A). Most importantly, similar results were obtained when TACE was overexpressed (Figure 5H and Supplemental Figure 15, B and C). These data demonstrate the potential therapeutic benefit for differentiation therapies in epidermal-derived SCCs independent of the status of p53.
Discussion
The tumor suppressor p53 is frequently lost or functionally inactivated in human skin SCCs, and thus its tumor suppressor function is compromised (2, 3). Here, we describe what we believe is a novel pathway in murine in vitro and in vivo skin tumor models, in human SCC cell lines, and in primary human skin tumors. We identify the function of WT p53 as a transcriptional activator of TACE, leading to epidermal tumor suppression through the induction of keratinocyte differentiation by NOTCH1. We further demonstrate that TACE, and not ADAM10, is required for the activation of p53-induced NOTCH1. In addition, we reveal in 2 genetic mouse models of skin tumors, in keratinocytes in vitro under oncogenic or stress conditions, and in human SCC cell lines that FOS represses p53 expression, thereby enabling tumor development (Figure 5I). Importantly, p53 restoration in murine skin tumors induced TACE-dependent tumor cell differentiation without evidence of apoptosis, altered proliferation, or senescence induction. When FOS was inhibited in human SCC cell lines, p53-dependent differentiation accompanied by impaired proliferation was observed, thus indicating potential beneficial effects of FOS inhibition in SCCs. Moreover, ectopic expression of WT p53 or TACE led to SCC cell differentiation even when p53 was mutated or functionally inactivated. This implies that the activation of this p53/TACE pathway, provided it is still intact in a given tumor cell, may offer a therapeutic potential for differentiation-inducing therapies in SCCs.
Skin tumor progression was previously shown to be FOS dependent using diseased osteopetrotic Fos-knockout mice (15). Our results in both K5-SOS+ and DMBA/TPA skin tumor models indicate that Fos is required specifically in epidermal keratinocytes prior to and for the initial steps of skin tumor development, which is likely achieved through repressing p53 expression. Skin tumor formation in the K5-SOS+ model is EGFR dependent (13). Although TACE is a sheddase for many EGFR ligands (45–47), we have not observed any alterations on homeostatic EGFR signaling. This is in contrast to what was previously reported for papilloma formation in epidermal c-Jun–deficient mice (48). In these mice, increased levels of p53, NOTCH1 (4), and TACE (data not shown) were detected, indicating that WT p53 is repressed in the K5-SOS+ model, although it can be functionally reactivated.
p53 induces tumor suppression in different tissues through induction of apoptosis, cell cycle arrest, or senescence (49). Approximately half of all human tumors express high levels of functionally inactive mutant p53. Therefore, functional restoration of WT p53 would be an extremely appealing strategy for selective removal of tumor cells. Recent studies have demonstrated a beneficial outcome of p53 restoration in certain mouse tumor models (50–52), whereas in other tumors, such as in non–small cell lung cancer, p53 restoration showed only a stage-specific beneficial effect (53, 54). Particularly in the skin, the absence of p53 does not affect the incidence of skin papillomas using the DMBA/TPA protocol, but favors their progression (18). Furthermore, one additional copy of WT p53 can confer resistance to DMBA/TPA-induced skin carcinogenesis (55), indicating that WT p53 can induce skin tumor suppression. Our results indicate that p53 limits keratinocyte transformation by inducing terminal differentiation with no effect on apoptosis and senescence.
Importantly, our work demonstrates that stress-induced p53 leads to TACE expression, which is reminiscent of p73α overexpression leading to TACE induction in lung carcinoma cells (56). We further demonstrate that TACE is a direct p53 transcriptional target in vitro and in vivo. p53-dependent TACE expression is required for activation of p53-induced NOTCH1, an epidermal tumor suppressor, but a tumor promoter in other tissues (40, 57). TACE is considered a principal target for inhibitory strategies to treat TNF-dependent pathologies (58), whereas we show that activation of epidermal TACE leads to skin tumor suppression through induction of keratinocyte differentiation. These dual roles of the TACE/NOTCH1 pathway are possibly explained by cell-context–dependent target gene expression.
Mice lacking TACE die around birth and have disorganized hair follicles, with altered skin morphology (28), whereas mice lacking epidermal TACE develop skin inflammation (32). Interestingly, epidermal-deficient ADAM10 pups also die perinatally due to impaired NOTCH1 activation (59). These results indicate that both TACE and ADAM10, known activators of NOTCH1 (60), are regulators of NOTCH1 activation in the skin, which seem to have redundant roles during development, but specific functions at later stages in adult organisms. Moreover, NOTCH1 activation by ADAM10 is ligand dependent, whereas TACE also induces ligand-independent NOTCH1 activation (61). This is consistent with our findings demonstrating a key role of TACE in inducing ligand-independent, stress-induced NOTCH1-dependent keratinocyte differentiation upon nonphysiological or oncogenic stress stimuli, while ADAM10 expression remained unchanged. We further propose that a positive feedback loop exists to induce more Notch1 following NOTCH1 activation, consistent with similar findings in T cells (41), possibly as a fail-safe mechanism to ensure irreversible keratinocyte differentiation.
In summary, we propose that FOS/AP-1 inhibition, and p53 restoration and activation together with TACE/NOTCH1-activating therapies should be considered for the differentiation of skin SCCs. Recent studies employing small molecules that specifically inhibit breast cancer stem cell proliferation through the induction of cell differentiation (62) indicate that such strategies may hold great promise for tumor cell differentiation-induced therapies to become beneficial for cancer patients.
Methods
Mice.
To generate FosΔep mice, the Fos floxed (Fosf/f) mice in a 129/C57BL/6 genetic background (63) were crossed to transgenic mice expressing Cre recombinase under the control of the keratinocyte-specific Keratin 5 promoter (K5-cre2) in a B6C3F1 background (29). To obtain K5-SOS-F FosΔep transgenic mice and control littermates, offspring from the Fosf/f EGFRwa2/wa2, previously intercrossed with K5-SOS-F EGFRwa2/wa2 transgenic mice in a LVC background (13), were intercrossed with the K5-cre2 line. In order to obtain K5-SOS-F FosΔep* (where asterisk indicates tamoxifen-inducible deletion) transgenic mice and control littermates, similar intercrosses were performed with the K5CreERT transgenic mouse line (64). To generate p53Δep, p53 floxed mice in a 129/C57BL/6 genetic background, provided by A. Berns (Netherlands Cancer Institute, Amsterdam, Netherlands) (65), were crossed to transgenic K5-cre2 mice. p53KI/KI mice were provided by G. Evan (University of Cambridge, Cambridge, United Kingdom) (43). Littermate mice were always used as controls.
Chemical carcinogenesis.
For DMBA/TPA-induced skin carcinogenesis, cohorts of Fosf/f, FosΔep, p53+/+, and p53KI/KI mice at 4 weeks of age received a single application of 100 μl of an acetone solution containing 0.5% DMBA (Sigma-Aldrich) applied to the dorsal surface. 10-4 M TPA in acetone was applied twice a week to the dorsal surface.
TACE enzymatic assay.
Epidermis and dermis were separated, and cells were plated as specified in keratinocyte culture procedures. Enzymatic assay was performed as previously described (29).
RAS mutation analysis.
DMBA-induced CAA to CTA H-Ras gene mutation at codon 61 analyses were performed as previously described (36).
Fos activity assay.
Human primary keratinocytes and SCC cell lines were used in FOS activity assays (TransAm AP-1 FOS; Active Motif) following the manufacturer’s instructions.
Histology, immunohistochemistry, and immunofluorescence.
Tissues were fixed in PBS-buffered 3.7% formalin or frozen in OCT at –80°C. Formalin-fixed tissues were dehydrated, and 4-μm sections were used; 7-μm cryosections were cut from OCT-fixed and frozen tissues. Tissue arrays from paraffin blocks were produced as 0.5-mm punches taken out of the paraffin-embedded material by parallel viewing of H&E-stained sections. H&E staining was performed according to standard procedures (Sigma-Aldrich). Ki67 (Novocastra) immunohistochemistry was performed using an automated Discovery XT Immunohistochemistry System (Ventana Medical Systems). Immunohistochemistry was performed using Elite ABC Kit (Vectastain) and DAB (Vector Laboratories) or alkaline phosphatase (Roche) following the manufacturers’ instructions. TUNEL assay was performed following the manufacturer’s instructions (Roche). Immunofluorescence was performed as described (66). Antibodies used were Keratin 1, Keratin 10, Involucrin, Filaggrin, and Loricrin (Covance); FOS (Santa Cruz Biotechnology Inc.); NOTCH1 (Cell Signaling Technology); TACE (Millipore); and Alexa Fluor 488 dye–labeled secondary antibodies (Invitrogen). Counterstainings were performed with DAPI and hematoxylin (Sigma-Aldrich).
Plasmids.
Retroviral vectors pLPC-RASV12 (pRAS), pBabe-H-RASV12 (bRAS), and pBabe-GFP (pGFP) were provided by M. Serrano (Spanish National Cancer Research Centre-CNIO, Madrid, Spain). MAM51 peptide–overexpressing retroviral vector was provided by S.C. Blacklow (Dana Farber Cancer Institute, Boston, Massachusetts, USA). Fos shRNA lentiviral constructs were purchased from Sigma-Aldrich. p53 shRNA lentiviral construct was kindly provided by O. Fernández-Capetillo (Spanish National Cancer Research Centre-CNIO). WT, mutant (nontranscriptionally active), and fusion protein p53 plasmids were provided by J. C. Marine (Vlaams Instituut voor Biotechnologie–Katholieke Universiteit [VIB-KU], Leuven, Leuven, Belgium). TACE expression vector was provided by J. Arribas (Vall d’Hebrón Institute of Oncology [VHIO], Barcelona, Spain).
Cell culture, and retroviral, lentiviral, and adenoviral infection.
Human primary keratinocytes were purchased and cultured in CnT-57 medium following the manufacturer’s instructions (CellNTec). Isolation and culture of mouse primary tail keratinocytes was performed as described (48). Twelve hours after plating, keratinocyte medium was changed to K-SFM (Gibco; Invitrogen). Ca2+ (1 M or 2 M CaCl2) stimulation of keratinocytes was performed in equally semiconfluent cultures for proper comparison. AdNICD was provided by G. P. Dotto (University of Lausanne, Lausanne, Switzerland). Primary keratinocytes were infected with 300 particles per cell in K-SFM medium. Cells were collected 96 hours after infection. Retroviral and lentiviral constructs were produced in 293T cells after transient transfection. Epidermal-derived SCC4, SCC9, and A253 were provided by M. Sibilia (67) (Institute for Cancer Research, Vienna, Austria); SCCO11, SCCO12, SCCO13 (68), SCCO22, and SCCO28 were provided by G.P. Dotto (20, 69) (University of Lausanne); A431, Detroit 562, and FaDu were provided by A. Cano and M. Quintanilla (Instituto de Investigaciones Biomedicas, Madrid, Spain). All SCC cells were grown in DMEM, 10% FBS.
siRNA.
Dharmacon ON-TARGETplus SMARTpool was used for the knockdown of mouse or human TACE (siTACE) and mouse ADAM10 (siADAM10) in mouse/human primary keratinocytes following the manufacturer’s instructions. As a control, ON-TARGETplus Non-Targeting Pool (scramble siRNA [siScr.]) was used. The analysis was performed after 96 hours of RNAi-Max (Invitrogen) transfection.
Protein isolation.
Backskin epidermis was separated from dermis after incubation in 0.75% trypsin for 45 minutes at 37°C. Protein isolation for Western blot was performed in RIPA buffer containing a protease inhibitor cocktail (0.1 mM Na3VO4, 40 mM β-glycerophosphate, 40 mM NaPPi, 1 mM NaF; Sigma-Aldrich), and tissues were disrupted using Precellys 24 (Bertin Technologies).
qRT-PCR.
RNA was isolated using Trizol reagent (Invitrogen). cDNA synthesis was performed with 1 μg of total RNA using Ready-To-Go You-Prime-It First-Strand-Beads (Amersham Pharmacia Biotech) and random primers (Invitrogen). qPCR reactions were performed with qPCR Mix (Promega). PCR products were quantified by real-time PCR analysis using ep-realplex (Eppendorf) and the Ct method. Primer sequences obtained from the qPrimer Depot website ( http://primerdepot.nci.nih.gov/; http://mouseprimerdepot.nci.nih.gov/; NCI, NIH) can be found in Supplemental Methods.
Western blot.
Western blot analysis was performed according to standard procedures using the following antibodies: TACE (Millipore); NOTCH1, p21, and FOS (BD Biosciences — Pharmingen); active NOTCH1 and p53 (Cell Signaling Technology); actin and tubulin (Sigma-Aldrich); Keratin 1 and Keratin 10 (Covance); and p16, p19, and NOTCH1 (Santa Cruz Biotechnology Inc.). H-RASV12 antibody was provided by H. Beug (institute of Molecular Pathology, Vienna, Austria). HRP-linked secondary antibodies were from Amersham and DAKO. Western blots were developed with Crescendo (Millipore) and ECL Hyperfilm (Amersham).
ChIP.
ChIP was performed with a ChIP Assay Kit (Upstate) according to the manufacturer’s instructions. Antibodies used were FOS (Santa Cruz Biotechnology Inc.) and p53 (Cell Signaling Technology). Cells were plated as for keratinocyte culture in order to exclude any other cell type in the sample. Isotype antibodies were used as controls (BD Biosciences). Bound and unbound fragments were run on agarose gels or quantified by real-time PCR when indicated.
Luciferase assay.
Mouse TACE promoter was PCR cloned into pGL.4.23 (Promega), and luciferase assays were performed following the manufacturer’s instructions (Promega) by cotransfecting TACE promoter construct and increasing concentrations of WT and mutant p53 and p53ER fusion protein, whose activation was induced with 4-OH–tamoxifen.
Inhibitors.
Cells were treated with DAPT, an inhibitor of γ-secretase activity, for 24 hours using a 10-μM concentration. Cells were treated with TAPI-1 (an MMP and TACE inhibitor from Peptides International) in DMSO for 24 hours before calcium treatment. T-5224, a FOS inhibitor, was chemically synthesized at CNIO Experimental Therapeutics (Spanish National Cancer Research Centre-CNIO) and dissolved in DMSO. Cells were treated at a 10-μM final concentration.
Tamoxifen treatment.
Tamoxifen citrate (Sigma-Aldrich) used for topical treatment was dissolved in DMSO at 50 mg/ml. Mouse tail was painted with this solution every day for 5 consecutive days. For i.p. injection, 2 mg of tamoxifen (Sigma-Aldrich) dissolved in sunflower seed oil (Sigma-Aldrich) was injected per mouse per day. 4–hydroxy-tamoxifen (4-OH) (10 μmol/l) was used for in vitro cell cultures.
EdU proliferation.
EdU (Invitrogen) was added to cell culture medium, and 2 hours later, cells were processed following the manufacturer’s instructions. FACS was performed to quantify EdU-positive cells.
Human skin SCC samples.
Human skin SCC samples obtained after informed consent and approval were evaluated and provided by Peter Petzelbauer (Medical University of Vienna), S. Ramón y Cajal and J. Hernández (Vall d’ Hebrón Hospital, Barcelona, Spain), and S. Wagner (Medical University of Vienna).
Statistics.
Unless stated differently, all experiments were performed 3 times. Data in bar graphs represent mean ± SD of triplicate experiments. Statistical analysis was performed using nondirectional 2-tailed Student’s t test or 1-way ANOVA tests, depending on the number of groups compared. P < 0.05 was considered significant.
Study approval.
Mice were kept in the animal facility in accordance with institutional policies and federal guidelines for animals used in biomedical research following the recommendations of the Ethics Committee and Animal Welfare of the Instituto de Salud Carlos III, following the Royal Decree 10th October 1201/2005. Animal experiments were approved by the Animal Experimental Ethics Committee of the Instituto de Salud Carlos III (Madrid, Spain). Human tumor samples were obtained after approval from the Ethics Committee of the Medical University of Vienna and the Vall d’ Hebrón Hospital. Tissue arrays from paraffin blocks were produced according to ethics committee permission 405/2006 and extension 1930/2010 of the Ethics Committeee of the Medical University of Vienna. Handling, supervision, and experimentation with mice was done in accordance with the Guidelines for Humane Endpoints for Animals Used in Biomedical Research (Instituto de Salud Carlos III, Madrid, Spain. 2012).
Supplementary Material
Acknowledgments
We are very grateful to M. Oren, M. Pérez-Moreno, F. Radtke, F. Real, M. Serrano, M. Sibilia, and members of the Wagner lab for critical reading of the manuscript and helpful discussions. The project was initiated at the IMP, which is funded by Boehringer Ingelheim (BI). E.F. Wagner is funded by the Banco Bilbao Vizcaya Argentaria (BBVA) Foundation and a European Research Council Advanced Grant (ERC FCK/2008/37). J. Guinea-Viniegra is a Ramón y Cajal award recipient.
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Citation for this article: J Clin Invest. 2012;122(8):2898–2910. doi:10.1172/JCI63103.
References
- 1.Madan V, Lear JT, Szeimies RM. Non-melanoma skin cancer. Lancet. 2010;375(9715):673–685. doi: 10.1016/S0140-6736(09)61196-X. [DOI] [PubMed] [Google Scholar]
- 2.Ratushny V, Gober MD, Hick R, Ridky TW, Seykora JT. From keratinocyte to cancer: the pathogenesis and modeling of cutaneous squamous cell carcinoma. J Clin Invest. 2012;122(2):464–472. doi: 10.1172/JCI57415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sarasin A, Giglia-Mari G. p53 gene mutations in human skin cancers. Exp Dermatol. 2002;11(suppl 1):44–47. doi: 10.1034/j.1600-0625.11.s.1.11.x. [DOI] [PubMed] [Google Scholar]
- 4.Kolev V, et al. EGFR signalling as a negative regulator of Notch1 gene transcription and function in proliferating keratinocytes and cancer. Nat Cell Biol. 2008;10(8):902–911. doi: 10.1038/ncb1750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Eferl R, et al. Liver tumor development. c-Jun antagonizes the proapoptotic activity of p53. Cell. 2003;112(2):181–192. doi: 10.1016/S0092-8674(03)00042-4. [DOI] [PubMed] [Google Scholar]
- 6.Schreiber M, et al. Control of cell cycle progression by c-Jun is p53 dependent. Genes Dev. 1999;13(5):607–619. doi: 10.1101/gad.13.5.607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stepniak E, et al. c-Jun/AP-1 controls liver regeneration by repressing p53/p21 and p38 MAPK activity. Genes Dev. 2006;20(16):2306–2314. doi: 10.1101/gad.390506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Durchdewald M, Angel P, Hess J. The transcription factor Fos: a Janus-type regulator in health and disease. Histol Histopathol. 2009;24(11):1451–1461. doi: 10.14670/HH-24.1451. [DOI] [PubMed] [Google Scholar]
- 9.Eferl R, Wagner EF. AP-1: a double-edged sword in tumorigenesis. Nat Rev Cancer. 2003;3(11):859–868. doi: 10.1038/nrc1209. [DOI] [PubMed] [Google Scholar]
- 10.Quintanilla M, Brown K, Ramsden M, Balmain A. Carcinogen-specific mutation and amplification of Ha-ras during mouse skin carcinogenesis. Nature. 1986;322(6074):78–80. doi: 10.1038/322078a0. [DOI] [PubMed] [Google Scholar]
- 11.Urabe A, Nakayama J, Taniguchi S, Terao H, Hori Y. Expression of the c-fos oncogene in chemically-induced mouse tumours and in human skin tumours. J Pathol. 1992;168(3):281–286. doi: 10.1002/path.1711680307. [DOI] [PubMed] [Google Scholar]
- 12.Durchdewald M, et al. Podoplanin is a novel fos target gene in skin carcinogenesis. Cancer Res. 2008;68(17):6877–6883. doi: 10.1158/0008-5472.CAN-08-0299. [DOI] [PubMed] [Google Scholar]
- 13.Sibilia M, et al. The EGF receptor provides an essential survival signal for SOS-dependent skin tumor development. Cell. 2000;102(2):211–220. doi: 10.1016/S0092-8674(00)00026-X. [DOI] [PubMed] [Google Scholar]
- 14.Bakiri L, et al. Functions of Fos phosphorylation in bone homeostasis, cytokine response and tumourigenesis. Oncogene. 2011;30(13):1506–1517. doi: 10.1038/onc.2010.542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Saez E, et al. c-fos is required for malignant progression of skin tumors. Cell. 1995;82(5):721–732. doi: 10.1016/0092-8674(95)90469-7. [DOI] [PubMed] [Google Scholar]
- 16.Gerdes MJ, et al. Activator protein-1 activity regulates epithelial tumor cell identity. Cancer Res. 2006;66(15):7578–7588. doi: 10.1158/0008-5472.CAN-06-1247. [DOI] [PubMed] [Google Scholar]
- 17.Brash DE. Roles of the transcription factor p53 in keratinocyte carcinomas. Br J Dermatol. 2006;154(suppl 1):8–10. doi: 10.1111/j.1365-2133.2006.07230.x. [DOI] [PubMed] [Google Scholar]
- 18.Kemp CJ, Donehower LA, Bradley A, Balmain A. Reduction of p53 gene dosage does not increase initiation or promotion but enhances malignant progression of chemically induced skin tumors. Cell. 1993;74(5):813–822. doi: 10.1016/0092-8674(93)90461-X. [DOI] [PubMed] [Google Scholar]
- 19.Yugawa T, Handa K, Narisawa-Saito M, Ohno S, Fujita M, Kiyono T. Regulation of Notch1 gene expression by p53 in epithelial cells. Mol Cell Biol. 2007;27(10):3732–3742. doi: 10.1128/MCB.02119-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lefort K, et al. Notch1 is a p53 target gene involved in human keratinocyte tumor suppression through negative regulation of ROCK1/2 and MRCKalpha kinases. Genes Dev. 2007;21(5):562–577. doi: 10.1101/gad.1484707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Demehri S, Turkoz A, Kopan R. Epidermal Notch1 loss promotes skin tumorigenesis by impacting the stromal microenvironment. Cancer Cell. 2009;16(1):55–66. doi: 10.1016/j.ccr.2009.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nicolas M, et al. Notch1 functions as a tumor suppressor in mouse skin. Nat Genet. 2003;33(3):416–421. doi: 10.1038/ng1099. [DOI] [PubMed] [Google Scholar]
- 23.Watt FM, Estrach S, Ambler CA. Epidermal Notch signalling: differentiation, cancer and adhesion. Curr Opin Cell Biol. 2008;20(2):171–179. doi: 10.1016/j.ceb.2008.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kopan R, Ilagan MX. The canonical Notch signaling pathway: unfolding the activation mechanism. Cell. 2009;137(2):216–233. doi: 10.1016/j.cell.2009.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Murphy G, Murthy A, Khokha R. Clipping, shedding and RIPping keep immunity on cue. Trends Immunol. 2008;29(2):75–82. doi: 10.1016/j.it.2007.10.009. [DOI] [PubMed] [Google Scholar]
- 26.van Tetering G, van Diest P, Verlaan I, van der Wall E, Kopan R, Vooijs M. Metalloprotease ADAM10 is required for Notch1 site 2 cleavage. J Biol Chem. 2009;284(45):31018–31027. doi: 10.1074/jbc.M109.006775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Le Gall SM, et al. ADAM17 is regulated by a rapid and reversible mechanism that controls access to its catalytic site. J Cell Sci. 2010;123(pt 22):3913–3922. doi: 10.1242/jcs.069997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Peschon JJ, et al. An essential role for ectodomain shedding in mammalian development. Science. 1998;282(5392):1281–1284. doi: 10.1126/science.282.5392.1281. [DOI] [PubMed] [Google Scholar]
- 29.Guinea-Viniegra J, et al. TNFalpha shedding and epidermal inflammation are controlled by Jun proteins. Genes Dev. 2009;23(22):2663–2674. doi: 10.1101/gad.543109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Saftig P, Reiss K. The “A Disintegrin And Metalloproteases” ADAM10 and ADAM17: novel drug targets with therapeutic potential? Eur J Cell Biol. 2011;90(6–7):527–535. doi: 10.1016/j.ejcb.2010.11.005. [DOI] [PubMed] [Google Scholar]
- 31.Chalaris A, et al. Critical role of the disintegrin metalloprotease ADAM17 for intestinal inflammation and regeneration in mice. J Exp Med. 2010;207(8):1617–1624. doi: 10.1084/jem.20092366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Murthy A, Shao YW, Narala SR, Molyneux SD, Zuniga-Pflucker JC, Khokha R. Notch activation by the metalloproteinase ADAM17 regulates myeloproliferation and atopic barrier immunity by suppressing epithelial cytokine synthesis. Immunity. 2012;36(1):105–119. doi: 10.1016/j.immuni.2012.01.005. [DOI] [PubMed] [Google Scholar]
- 33.Adrain C, Zettl M, Christova Y, Taylor N, Freeman M. Tumor necrosis factor signaling requires iRhom2 to promote trafficking and activation of TACE. Science. 2012;335(6065):225–228. doi: 10.1126/science.1214400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McIlwain DR, et al. iRhom2 regulation of TACE controls TNF-mediated protection against Listeria and responses to LPS. Science. 2012;335(6065):229–232. doi: 10.1126/science.1214448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ruggeri B, DiRado M, Zhang SY, Bauer B, Goodrow T, Klein-Szanto AJ. Benzo[a]pyrene-induced murine skin tumors exhibit frequent and characteristic G to T mutations in the p53 gene. Proc Natl Acad Sci U S A. 1993;90(3):1013–1017. doi: 10.1073/pnas.90.3.1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nagase H, Mao JH, Balmain A. Allele-specific Hras mutations and genetic alterations at tumor susceptibility loci in skin carcinomas from interspecific hybrid mice. Cancer Res. 2003;63(16):4849–4853. [PubMed] [Google Scholar]
- 37.Devgan V, Mammucari C, Millar SE, Brisken C, Dotto GP. p21WAF1/Cip1 is a negative transcriptional regulator of Wnt4 expression downstream of Notch1 activation. Genes Dev. 2005;19(12):1485–1495. doi: 10.1101/gad.341405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lin AW, Lowe SW. Oncogenic ras activates the ARF-p53 pathway to suppress epithelial cell transformation. Proc Natl Acad Sci U S A. 2001;98(9):5025–5030. doi: 10.1073/pnas.091100298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wu X, et al. Opposing roles for calcineurin and ATF3 in squamous skin cancer. Nature. 2010;465(7296):368–372. doi: 10.1038/nature08996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dotto GP. Crosstalk of Notch with p53 and p63 in cancer growth control. Nat Rev Cancer. 2009;9(8):587–595. doi: 10.1038/nrc2675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yashiro-Ohtani Y, et al. Pre-TCR signaling inactivates Notch1 transcription by antagonizing E2A. Genes Dev. 2009;23(14):1665–1676. doi: 10.1101/gad.1793709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mizui Y, Yamazaki K, Sagane K, Tanaka I. cDNA cloning of mouse tumor necrosis factor-alpha converting enzyme (TACE) and partial analysis of its promoter. Gene. 1999;233(1–2):67–74. doi: 10.1016/S0378-1119(99)00155-9. [DOI] [PubMed] [Google Scholar]
- 43.Christophorou MA, et al. Temporal dissection of p53 function in vitro and in vivo. Nat Genet. 2005;37(7):718–726. doi: 10.1038/ng1572. [DOI] [PubMed] [Google Scholar]
- 44.Aikawa Y, et al. Treatment of arthritis with a selective inhibitor of c-Fos/activator protein-1. Nat Biotechnol. 2008;26(7):817–823. doi: 10.1038/nbt1412. [DOI] [PubMed] [Google Scholar]
- 45.Scheller J, Chalaris A, Garbers C, Rose-John S. ADAM17: a molecular switch to control inflammation and tissue regeneration. Trends Immunol. 2011;32(8):380–387. doi: 10.1016/j.it.2011.05.005. [DOI] [PubMed] [Google Scholar]
- 46.Maretzky T, et al. Migration of growth factor-stimulated epithelial and endothelial cells depends on EGFR transactivation by ADAM17. Nat Commun. 2011;2:229. doi: 10.1038/ncomms1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Blobel CP. ADAMs: key components in EGFR signalling and development. Nat Rev Mol Cell Biol. 2005;6(1):32–43. doi: 10.1038/nrm1548. [DOI] [PubMed] [Google Scholar]
- 48.Zenz R, et al. c-Jun regulates eyelid closure and skin tumor development through EGFR signaling. Dev Cell. 2003;4(6):879–889. doi: 10.1016/S1534-5807(03)00161-8. [DOI] [PubMed] [Google Scholar]
- 49.Lowe SW, Cepero E, Evan G. Intrinsic tumour suppression. Nature. 2004;432(7015):307–315. doi: 10.1038/nature03098. [DOI] [PubMed] [Google Scholar]
- 50.Martins CP, Brown-Swigart L, Evan GI. Modeling the therapeutic efficacy of p53 restoration in tumors. Cell. 2006;127(7):1323–1334. doi: 10.1016/j.cell.2006.12.007. [DOI] [PubMed] [Google Scholar]
- 51.Ventura A, et al. Restoration of p53 function leads to tumour regression in vivo. Nature. 2007;445(7128):661–665. doi: 10.1038/nature05541. [DOI] [PubMed] [Google Scholar]
- 52.Xue W, et al. Senescence and tumour clearance is triggered by p53 restoration in murine liver carcinomas. Nature. 2007;445(7128):656–660. doi: 10.1038/nature05529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Feldser DM, et al. Stage-specific sensitivity to p53 restoration during lung cancer progression. Nature. 2010;468(7323):572–575. doi: 10.1038/nature09535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Junttila MR, et al. Selective activation of p53-mediated tumour suppression in high-grade tumours. Nature. 2010;468(7323):567–571. doi: 10.1038/nature09526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Matheu A, et al. Delayed ageing through damage protection by the Arf/p53 pathway. Nature. 2007;448(7151):375–379. doi: 10.1038/nature05949. [DOI] [PubMed] [Google Scholar]
- 56.Fontemaggi G, et al. Identification of direct p73 target genes combining DNA microarray and chromatin immunoprecipitation analyses. J Biol Chem. 2002;277(45):43359–43368. doi: 10.1074/jbc.M205573200. [DOI] [PubMed] [Google Scholar]
- 57.Xu P, Derynck R. Direct activation of TACE-mediated ectodomain shedding by p38 MAP kinase regulates EGF receptor-dependent cell proliferation. Mol Cell. 2010;37(4):551–566. doi: 10.1016/j.molcel.2010.01.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Overall CM, Blobel CP. In search of partners: linking extracellular proteases to substrates. Nat Rev Mol Cell Biol. 2007;8(3):245–257. doi: 10.1038/nrm2120. [DOI] [PubMed] [Google Scholar]
- 59.Weber S, et al. The disintegrin/metalloproteinase Adam10 is essential for epidermal integrity and Notch-mediated signaling. Development. 2011;138(3):495–505. doi: 10.1242/dev.055210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Brou C, et al. A novel proteolytic cleavage involved in Notch signaling: the role of the disintegrin-metalloprotease TACE. Mol Cell. 2000;5(2):207–216. doi: 10.1016/S1097-2765(00)80417-7. [DOI] [PubMed] [Google Scholar]
- 61.Bozkulak EC, Weinmaster G. Selective use of ADAM10 and ADAM17 in activation of Notch1 signaling. Mol Cell Biol. 2009;29(21):5679–5695. doi: 10.1128/MCB.00406-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Beug H. Breast cancer stem cells: eradication by differentiation therapy? Cell. 2009;138(4):623–625. doi: 10.1016/j.cell.2009.08.007. [DOI] [PubMed] [Google Scholar]
- 63.Fleischmann A, et al. Impaired long-term memory and NR2A-type NMDA receptor-dependent synaptic plasticity in mice lacking c-Fos in the CNS. J Neurosci. 2003;23(27):9116–9122. doi: 10.1523/JNEUROSCI.23-27-09116.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Brocard J, et al. Spatio-temporally controlled site-specific somatic mutagenesis in the mouse. Proc Natl Acad Sci U S A. 1997;94(26):14559–14563. doi: 10.1073/pnas.94.26.14559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Jonkers J, Meuwissen R, van der Gulden H, Peterse H, van der Valk M, Berns A. Synergistic tumor suppressor activity of BRCA2 and p53 in a conditional mouse model for breast cancer. Nat Genet. 2001;29(4):418–425. doi: 10.1038/ng747. [DOI] [PubMed] [Google Scholar]
- 66.Perez-Moreno M, Davis MA, Wong E, Pasolli HA, Reynolds AB, Fuchs E. p120-catenin mediates inflammatory responses in the skin. Cell. 2006;124(3):631–644. doi: 10.1016/j.cell.2005.11.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lichtenberger BM, Tan PK, Niederleithner H, Ferrara N, Petzelbauer P, Sibilia M. Autocrine VEGF signaling synergizes with EGFR in tumor cells to promote epithelial cancer development. Cell. 2010;140(2):268–279. doi: 10.1016/j.cell.2009.12.046. [DOI] [PubMed] [Google Scholar]
- 68.Rheinwald JG, Beckett MA. Tumorigenic keratinocyte lines requiring anchorage and fibroblast support cultures from human squamous cell carcinomas. Cancer Res. 1981;41(5):1657–1663. [PubMed] [Google Scholar]
- 69.Rocco JW, Leong CO, Kuperwasser N, DeYoung MP, Ellisen LW. p63 mediates survival in squamous cell carcinoma by suppression of p73-dependent apoptosis. Cancer Cell. 2006;9(1):45–56. doi: 10.1016/j.ccr.2005.12.013. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.