Abstract
Small nucleolar RNAs (snoRNAs) are emerging as an important new class of genes deregulated in cancer. Orphans snoRNAs are encoded outside of ribosomal protein genes and are involved in either gene splicing or are microRNA precursors. In this issue of JCI, Chu et al. find that ACA11, an orphan snoRNA encoded in an intron of the WHSC1 gene, is aberrantly overexpressed in t(4;14)-positive patients with multiple myeloma (MM), in which it influences growth of MM cells, resistance to chemotherapy, and oxidative stress. These findings represent the first identification of a snoRNA overexpressed as a consequence of a chromosomal translocation, a potent driving force of the neoplastic process in general and hematopoietic malignancies in particular.
It is now believed that the increase in the complexity of our genome over the course of evolution has been more associated with an expansion of expressed noncoding regions of DNA than an increase in protein-coding genes. Thus, a new dimension in our understanding of gene regulation is emerging in which each RNA transcript is potentially able to influence cellular homeostasis. Indeed, it has been demonstrated that long noncoding RNAs (lncRNAs) can influence the epigenetic landscape of DNA (reviewed in ref. 1), while pseudogenes, lncRNAs, and mRNAs can all act as competing endogenous RNAs (ceRNAs) for microRNA (miRNA) binding (2). In addition, miRNAs are modulators of mRNA stability and protein synthesis, while small nucleolar RNAs (snoRNAs) are commonly involved in ribosomal biogenesis (reviewed in refs. 3, 4). Hence, a major investigational challenge in cellular and cancer biology is now to decipher the functional roles and relationships between coding and noncoding genes and how this aberrant cross talk may lead to the development of cancer. Toward this end, in this issue of JCI, Chu et al. demonstrate a novel and unexpected relationship between genetic translocations and aberrant expression of snoRNAs in the hematopoietic malignancy multiple myeloma (MM; ref. 5).
Aberrant gene fusions resulting from chromosomal translocations are particularly distinctive of hematopoietic malignancies. The chimeric gene products resulting from such fusions are often more than the sum of their two components, and Chu et al. investigate just such a case, identifying a “hidden player” involved in the t(4:14) translocation of MM: ACA11 (also known as SCARNA22), a snoRNA located in the intron of the Wolf-Hirschhorn syndrome candidate 1 (WHSC1) gene.
MM is an incurable cancer of B-lineage plasma cells that accounts for more than 13% of hematologic malignancies. It is characterized by clonal expansion of malignant cells in the bone marrow and is associated with anemia, renal failure, and cortical bone destruction (6). MM is a heterogeneous disease that results from several genetic abnormalities, including a t(4;14)(p16.3;q32.3) chromosomal translocation involving the immunoglobulin heavy chain region enhancer and the 5′ end of WHSC1. This genetic event occurs in more than 20% of patients with MM and results in the aberrant overexpression of WHSC1 (7). The oncogenic role of this translocation has been previously characterized, but Chu et al. add to the existing picture, demonstrating that ACA11, an orphan box H/ACA class snoRNA encoded within an intron of the WHSC1 gene, can act as an additional driving force in MM (and possibly in other cancers in which ACA11 is overexpressed) by cooperating with the three major isoforms of WHSC1 (5).
snoRNAs in cellular homeostasis and cancer
snoRNAs are one of the largest groups of single-stranded small noncoding RNAs (ncRNAs; 60–300 nt) and generally are involved in posttranscriptional modification of ribosomal RNAs (rRNAs) and small nuclear RNAs (snRNAs) (reviewed in refs. 3, 4). Located in the nucleolus, snoRNAs are one of the most conserved species of ncRNAs: a number of homologs of eukaryotic snoRNAs have been identified in prokaryotes as well as in archaea. Even though global alteration of snoRNA-guided posttranscriptional rRNA modifications (e.g., pseudouridylation and methylation) negatively affects cell growth and viability, inactivation of individual snoRNAs and their respective rRNA modifications was found to be dispensable for growth, indicating that the modifications are not essential, at least in yeast (3). Thus, for several years the scientific community has considered snoRNAs as housekeeping genes, with a relatively low impact on cellular homeostasis.
Several lines of evidence suggest that a dramatic change has taken place in snoRNA functions over the course of their evolution. The majority of snoRNAs are encoded in introns of host genes and are in this way transcriptionally and functionally related to their hosts. Yet, although ancient snoRNAs are predominantly located within host genes involved in ribosomal biogenesis and protein translation, functional characterization of those snoRNAs that have landed in their current genomic location more recently (8) is currently lacking. We speculate that snoRNAs are currently evolving as a new class of small regulator RNAs. This hypothesis is supported by the recent observation of different snoRNAs with no apparent sequence complementarity to either rRNA or other snoRNAs. These orphan snoRNAs are often expressed in a tissue-specific manner and can be localized either in introns of long ncRNAs or in coding genes unrelated to ribosomal biogenesis, such as WHSC1.
The role of small ncRNAs in tumorigenesis has been investigated with respect to miRNAs (reviewed in ref. 9), but a recent wave of papers suggests that snoRNAs also play a pivotal role in cancer biology (reviewed in ref. 10). For example, snoRNA U50 is mutated and downregulated in prostate and breast cancer, and a homozygous 2-bp (TT) deletion identified in breast and prostate cell lines as well as patients with breast and prostate cancer abrogates the ability of U50 to inhibit anchorage-independent growth (11). Conversely, a recent deep sequencing analysis for snRNAs in prostate cancer has shown an increase in both global snoRNAs and transfer RNA expression in metastatic lymph node prostate cancer when compared with that in primary prostate cancer, suggesting a possible oncogenic role for snoRNAs, particularly in advanced tumors (12). Although these and other papers clearly point to the biological significance of snoRNAs in cancer, the mechanisms by which their loss or gain of function affects cellular homeostasis, and thus influences transformation, are yet to be defined.
Insights into unexpected mechanisms of the function of orphan snoRNAs can arise from their host genes. For instance, growth arrest-specific transcript 5 (GAS5) contains in its introns several snoRNAs constitutively expressed and involved in rRNA biogenesis. Conversely, GAS5 is practically undetected in proliferating cells, as a consequence of nonsense-mediated decay, but it rapidly accumulates in growth-arrested cells upon serum deprivation and inhibition of protein translation as well as in confluent cells (13). GAS5 acts as a decoy RNA for the glucocorticoid receptor (GR) by preventing the binding of GR to target genes. Thus, starved cells are not able to activate GR-responsive genes and are more sensitive to apoptotic stimuli (14). Hence, snoRNAs and their host genes’ RNAs could cooperatively act as a protein decoy or as ceRNAs, as we will discuss below.
ACA11 — a novel oncogene in MM
ACA11 is encoded within intron 18–19 of the WHSC1 gene overexpressed in t(4:14)-positive MM. Even though the oncogenic potential of WHSC1 isoforms has been previously reported (15), in their study, Chu et al. demonstrate that none of the Eε-WHSC1 mice have a hematopoietic phenotype and that retroviral overexpression of WHSC1 isoforms in bone marrow precursor cells does not result in transformation, even in a tumor-prone Cdkn2a–/– background (5). The authors investigated the locus using a tiling array chip spanning the translocation breakpoint and demonstrated ACA11 overexpression in t(4;14)-positive MM cells and patients. More surprisingly, ACA11 did not bind proteins of the pseudouridylation machinery but instead proteins involved in splicing, such as heterogeneous nuclear ribonucleoproteins (HNRNPs). These data suggest an alternative mechanism of action of this snoRNA in cancer. Furthermore, small nuclear ribonuclear protein ACA11 complexes were found to interact with other snoRNA intermediates located within introns of ribosomal protein (RP) host genes. Intriguingly, the authors observed a powerful association between t(4;14)-positive MM and downregulation of RP genes. Indeed, ACA11 overexpression reduced the expression of some RP proteins but did not affect normal ribosomal biogenesis. Thus, ACA11 seems not to be involved in classical rRNA modifications. Additionally, Chu and collaborators followed up on a recently described link between snoRNAs and oxidative stress. Lipotoxic stress induces snoRNAs U32A, U33, and U35A, which in turn delocalize in the cytoplasm and sensitize cells to oxidative stress (16), while ACA11 overexpression in MM cells downregulates U33 and the RPL13A host gene. Thus, when ACA11 is overexpressed in MEFs or in t(4;14)-negative MM cells, it suppresses ROS upon H2O2 treatment, and its downregulation increases ROS production, decreases cell proliferation, and sensitizes t(4;14)-positive MM cells to chemotherapy. Strikingly, ACA11 silencing impaired tumor growth of an MM cell line (5).
snoRNAs in cancer — future perspectives
The study by Chu et al. does not explicitly describe the mechanisms by which ACA11 or U32A, U33, and U35A influence cell proliferation and stress response, but the findings suggest that these snoRNAs are not working in a canonical way. Chu et al. demonstrate that ACA11 binds HNRNPs rather than the proteins involved in ribosomal biogenesis; in addition, metabolic stress increases cytoplasmic levels of U32A, U33, and U35A without affecting their nuclear level. Other recently discovered snoRNAs exhibit a similar profile; several with no specific complementarity to rRNAs but with tissue-specific expression have been identified. More surprisingly, snoRNA-derived miRNAs have been identified and validated. These snoRNAs can be processed in a Dicer-dependent manner into functional miRNAs or small RNAs with a miRNA-like function of posttranscriptional downmodulation by base pair pairing (17). It remains to be established whether this phenomenon is widespread and applies to other snoRNAs, such as ACA11.
Base pair complementarity, however, seems to be an intrinsic property of all RNA molecules from ancestral to more recently evolved species, and this feature is conserved in both long and small transcripts. The mechanism of action is relatively simple and intuitive, but the biological consequences are enormous: the fluctuation or the dislocation of one RNA species that is complementary to another can affect the biological function of the partner, by titrating out the latter from the system when the two components are bound. We and other groups have shown that RNA molecules that share miRNA-responsive elements can regulate each other by competing for miRNA binding (18–22). On this basis, we have proposed that RNA transcripts are not merely targets of miRNAs, but also can communicate using miRNAs as letters of a new RNA code, expanding the functional role of all RNA transcripts (2). Recently, we have extended this ceRNA logic to both protein-coding and ncRNA transcripts. We have likewise demonstrated the existence of a broader PTEN ceRNA network (ceRNET) deregulated in cancer (18, 19). The cross-talking ceRNAs as well as ceRNETs represent a novel level of complexity in biology that is governed by relative abundance and localization of all players (long RNA transcripts and small RNA transcripts). Hence, it is plausible that overexpression and/or subcellular delocalization of snoRNAs may represent an unexpected mechanism by which oncogenic events, in this case a genetic translocation, can perturb functional interactions among snoRNA-ceRNA species and ceRNETs, thereby resulting in the acquisition of oncogenic properties (Figure 1). Future studies will disentangle the complexity of these networks in cancer and physiological conditions. Importantly, the therapeutic modulation of these aberrant cancer RNA networks through the use of small ncRNAs species (e.g., LNA molecules; ref. 23) will represent an exciting therapeutic venue in the years to come.
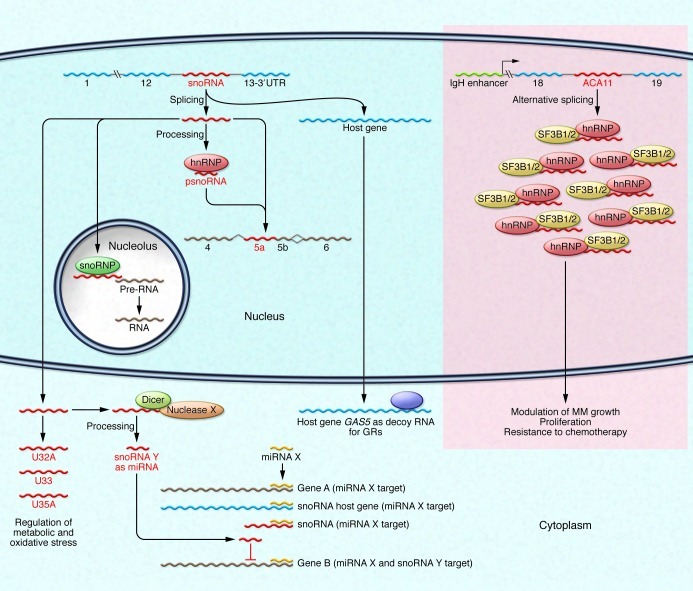
Figure 1. snoRNAs are often localized in introns of RP genes.
snoRNAs are generally involved in rRNA modifications (methylation and pseudouridylation), but orphan snoRNAs are encoded in genes not enriched in specific functional categories. ACA11 orphan snoRNA is encoded within intron 18–19 of the WHSC1 gene and is overexpressed in t(4;14)-positive patients with MM. ACA11 modulates the growth of MM cells, resistance to chemotherapy, and oxidative stress. Other orphan snoRNAs (U32A, U33, U35A) localize in the cytoplasm and regulate metabolic and oxidative stress. Interestingly, some snoRNA host genes are lncRNAs and act as decoy RNAs for GRs. Orphan snoRNAs can be involved in splicing or can be processed into smaller snoRNAs (psnoRNAs) and miRNAs. psnoRNAs modulate alternative splicing, whereas snoRNAs with miRNA-like function downmodulate specific mRNAs at the posttranscriptional level. snoRNAs with miRNA-like function can increase the number of miRNAs able to silence genes containing that specific miRNA binding site. In parallel, overexpression of snoRNAs and snoRNAs host genes with miRNA responsive elements can oppose miRNA-dependent silencing by increasing the amount of target RNAs. Thus, orphan snoRNAs and their host genes can act as novel players in the network of ceRNAs in the cell. Delocalization, overexpression, deletions, or point mutations of specific snoRNAs can profoundly deregulate cellular homeostasis and result in transformation via various mechanisms, including cellular stress and posttranscriptional gene silencing. The mechanism by which ACA11 and U32A, U33, and U35A influence metabolic and oxidative stress is still unknown. snoRNP, small nucleolar ribonucleoprotein; SF3B1/2, RNA splicing factors.
Acknowledgments
We thank Francesca Bersani, John G. Clohessy for helpful discussions and comments, and Thomas Garvey for critical editing of the manuscript. Our work is supported by NIH/NCI. R. Taulli is supported by a Marie Curie International Outgoing Fellowship for Career Development.
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Citation for this article: J Clin Invest. 2012;122(8):2765–2768. doi:10.1172/JCI63549.
See the related article beginning on page 2793.
References
- 1.Guttman M, Rinn JL. Modular regulatory principles of large non-coding RNAs. Nature. 2012;482(7385):339–346. doi: 10.1038/nature10887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Salmena L, Poliseno L, Tay Y, Kats L, Pandolfi PP. A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell. 2011;146(3):353–358. doi: 10.1016/j.cell.2011.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bratkovic T, Rogelj B. Biology and applications of small nucleolar RNAs. Cell Mol Life Sci. 2011;68(23):3843–3851. doi: 10.1007/s00018-011-0762-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Matera AG, Terns RM, Terns MP. Non-coding RNAs: lessons from the small nuclear and small nucleolar RNAs. Nat Rev Mol Cell Biol. 2007;8(3):209–220. doi: 10.1038/nrm2124. [DOI] [PubMed] [Google Scholar]
- 5.Chu L, et al. Multiple myeloma–associated chromosomal translocation activates orphan snoRNA ACA11 to suppress oxidative stress. J Clin Invest. doi: 10.1172/JCI63051. doi:10.1172/JCI63051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Palumbo A, Anderson KN. Multiple myeloma. N Engl J Med. 2011;364(11):1046–1060. doi: 10.1056/NEJMra1011442. [DOI] [PubMed] [Google Scholar]
- 7.Chesi M, Nardini E, Lim RS, Smith KD, Kuehl WM, Bergsagel PL. The t(4;14) translocation in myeloma dysregulates both FGFR3 and a novel gene, MMSET, resulting in IgH/MMSET hybrid transcripts. Blood. 1998;92(9):3025–3034. [PubMed] [Google Scholar]
- 8.Hoeppner MP, White S, Jeffares DC, Poole AM. Evolutionarily stable association of intronic snoRNAs and microRNAs with their host genes. Genome Biol Evol. 2009;1:420–428. doi: 10.1093/gbe/evp045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136(2):215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Williams GT, Farzaneh F. Are snoRNAs and snoRNA host genes new players in cancer? Nat Rev Cancer. 2012;12(2):84–88. doi: 10.1038/nrc3195. [DOI] [PubMed] [Google Scholar]
- 11.Dong XY, et al. Implication of snoRNA U50 in human breast cancer. J Genet Genomics. 2009;36(8):447–454. doi: 10.1016/S1673-8527(08)60134-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Martens-Uzunova ES, et al. Diagnostic and prognostic signatures from the small non-coding RNA transcriptome in prostate cancer. Oncogene. 2012;31(8):978–991. doi: 10.1038/onc.2011.304. [DOI] [PubMed] [Google Scholar]
- 13.Smith CM, Steitz JA. Classification of gas5 as a multi-small-nucleolar-RNA (snoRNA) host gene and a member of the 5’-terminal oligopyrimidine gene family reveals common features of snoRNA host genes. Mol Cell Biol. 1998;18(12):6897–6909. doi: 10.1128/mcb.18.12.6897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kino T, Hurt DE, Ichijo T, Nader N, Chrousos GP. Noncoding RNA gas5 is a growth arrest- and starvation-associated repressor of the glucocorticoid receptor. Sci Signal. 2010;3(107):ra8. doi: 10.1126/scisignal.2000568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lauring J, et al. The multiple myeloma associated MMSET gene contributes to cellular adhesion, clonogenic growth, and tumorigenicity. Blood. 2008;111(2):856–864. doi: 10.1182/blood-2007-05-088674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Michel CI, et al. Small nucleolar RNAs U32a, U33, and U35a are critical mediators of metabolic stress. Cell Metab. 2011;14(1):33–44. doi: 10.1016/j.cmet.2011.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ender C, et al. A human snoRNA with microRNA-like functions. Mol Cell. 2008;32(4):519–528. doi: 10.1016/j.molcel.2008.10.017. [DOI] [PubMed] [Google Scholar]
- 18.Karreth FA, et al. In vivo identification of tumor- suppressive PTEN ceRNAs in an oncogenic BRAF-induced mouse model of melanoma. Cell. 2011;147(2):382–395. doi: 10.1016/j.cell.2011.09.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tay Y, et al. Coding-independent regulation of the tumor suppressor PTEN by competing endogenous mRNAs. Cell. 2011;147(2):344–357. doi: 10.1016/j.cell.2011.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cesana M, et al. A long noncoding RNA controls muscle differentiation by functioning as a competing endogenous RNA. Cell. 2011;147(2):358–369. doi: 10.1016/j.cell.2011.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sumazin P, et al. An extensive microRNA-mediated network of RNA-RNA interactions regulates established oncogenic pathways in glioblastoma. Cell. 2011;147(2):370–381. doi: 10.1016/j.cell.2011.09.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cazalla D, Yario T, Steitz JA. Down-regulation of a host microRNA by a Herpesvirus saimiri noncoding RNA. Science. 2010;328(5985):1563–1566. doi: 10.1126/science.1187197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Obad S, et al. Silencing of microRNA families by seed-targeting tiny LNAs. Nat Genet. 2011;43(4):371–378. doi: 10.1038/ng.786. [DOI] [PMC free article] [PubMed] [Google Scholar]