Abstract
Disruption of cellular processes affected by multiple genes and accumulation of numerous insults throughout life dictate the progression of age-related disorders, but their complex etiology is poorly understood. Postmitotic neurons, such as photoreceptor cells in the retina and epithelial cells in the adjacent retinal pigmented epithelium, are especially susceptible to cellular senescence, which contributes to age-related retinal degeneration (ARD). The multigenic and complex etiology of ARD in humans is reflected by the relative paucity of effective compounds for its early prevention and treatment. To understand the genetic differences that drive ARD pathogenesis, we studied A/J mice, which develop ARD more pronounced than that in other inbred mouse models. Although our investigation of consomic strains failed to identify a chromosome associated with the observed retinal deterioration, pathway analysis of RNA-Seq data from young mice prior to retinal pathological changes revealed that increased vulnerability to ARD in A/J mice was due to initially high levels of inflammatory factors and low levels of homeostatic neuroprotective factors. The genetic signatures of an uncompensated preinflammatory state and ARD progression identified here aid in understanding the susceptible genetic loci that underlie pathogenic mechanisms of age-associated disorders, including several human blinding diseases.
Introduction
Age-related disorders arise from failure of tissue maintenance and repair pathways that are accelerated by certain inherited and acquired factors (1). Long-lived nondividing cells, such as neurons, have markedly reduced tolerance to damage (2), and thus exhibit the most pronounced age-related changes. Neuronal cells in the retina are an especially attractive model system to study this phenomenon, owing to their accessibility and well-understood physiology. Rod and cone photoreceptors are retinal neuronal cells that initiate visual perception, a function requiring a competent neighboring retinal pigmented epithelium (RPE) for their normal operation (3). In postmitotic cells such as these photoreceptor and RPE cells, cellular senescence can ensue when shed oxidized photoreceptor outer segments (POS) are inadequately phagocytosed and digested by the RPE. This results in accumulation of damaged proteins, formation of toxic metabolic byproducts, inflammatory cell invasion, and cell death. RPE cells are the most affected because, in addition to processing POS, they serve as the conduit between photoreceptors and the choroidal blood supply for metabolite exchange (4). Acquisition of senescence-associated pathology stimulates cells to secrete various factors that contribute to tissue dysfunction. Conversely, adequate clearance of such cells can delay the onset of age-related tissue pathology (5).
Aging laboratory experimental animals are important models for studying age-related pathology, especially neurodegenerative disorders, which have been shown to be affected by their genetic backgrounds (6, 7). The A/J inbred mouse model, for instance, undergoes age-related retinal degeneration (ARD) much more rapidly than its albino (BALB/c) and pigmented C57BL/6J (B6) counterparts, presenting a natural model with which to study age-related pathological changes in the retina. Although different factors, such as those involved in inflammation and homeostatic processing originating from the photoreceptor and RPE layers, have been identified as possible markers of disease (8), the sets of genes that trigger progression to the chronic disease state are still poorly understood. A lack of methodology sufficiently powerful to reveal the complex interplay of genes that trigger and promote ARD progression has limited our understanding of disease pathogenesis. Studies to date have mostly relied on quantitative trait loci (QTL) (6) or chromosome-substituted strain (CSS) panels (9), but these methods cannot detect gene expression changes and SNPs on the global scale needed to identify distant interacting genetic factors.
To understand the molecular components that precipitate and perpetuate neurodegeneration associated with age-related pathology in the retina, we used A/J, BALB/c, and B6 inbred mouse models to focus on genetic factors contributing to ARD in a controlled environmental setting. With high-resolution imaging studies, we identified subtle pathological changes in the RPE in A/J mice, but not BALB/c or B6 mice, before phenotypic features of disease become apparent. We hypothesized that RNA-sequencing (RNA-Seq) (10) of young eye tissue from these 3 different genetic mouse backgrounds would allow us to detect global changes that trigger and drive this multigenic condition. RNA-Seq elucidated aberrant genetic pathways evident in A/J mice, which led us to propose that mice destined to develop ARD possess a retinal environment primed for inflammation. This condition is exacerbated with age as homeostatic proteins that already exhibit decreased expression, most notably those involved in the oxidative stress response, functionally decline. Moreover, identification of these targets revealed potentially susceptible genetic loci that may participate in pathogenic mechanisms responsible for age-associated retinal disorders, which holds particular relevance for human blinding diseases.
Results
Mice on the A/J genetic background undergo pronounced ARD.
Retinal cross-section images revealed a prominent decrease in outer nuclear layer (ONL) thickness attributable to photoreceptor loss as well as pathological changes in the RPE layer of 8- versus 1-month-old A/J mice (Figure 1A). Conversely, B6 mice displayed a negligible age-related decrease in ONL thickness and no other pathological changes (Figure 1A). Age-dependent ONL loss was observed globally in A/J mouse eyes (Figure 1B). Cone photoreceptor numbers also indicated a marked age-related decrease in A/J mice, whereas B6 mice exhibited only a slight decrease (Figure 1C). The decline in photoreceptor number in A/J mice at 8 months of age compromised visual function, as evidenced by attenuation of electroretinographic (ERG) responses (Figure 1D) and decreased levels of total retinoid content, especially the visual chromophore, 11-cis-retinal (Supplemental Figure 1; supplemental material available online with this article; doi: 10.1172/JCI64427DS1). More importantly, the pronounced aged retinal phenotype was independent of light-induced or pigmentary contributions (6). Similar to the above results obtained under normal lighting conditions, A/J mice reared in the dark also displayed a significant decline in ONL thickness at 8 months of age compared with 8-month-old B6 mice (Supplemental Figure 2). However, 8-month-old albino BALB/c mice exhibited only a 20% decline in ONL thickness and had a cone population similar to that of 8-month-old B6 mice (Supplemental Figure 3). To better elucidate factors that underlie the accelerated progression of this degenerative phenotype in A/J mice, but not BALB/c or B6 mice, we undertook a more in-depth retinal examination of mice with different genetic backgrounds.
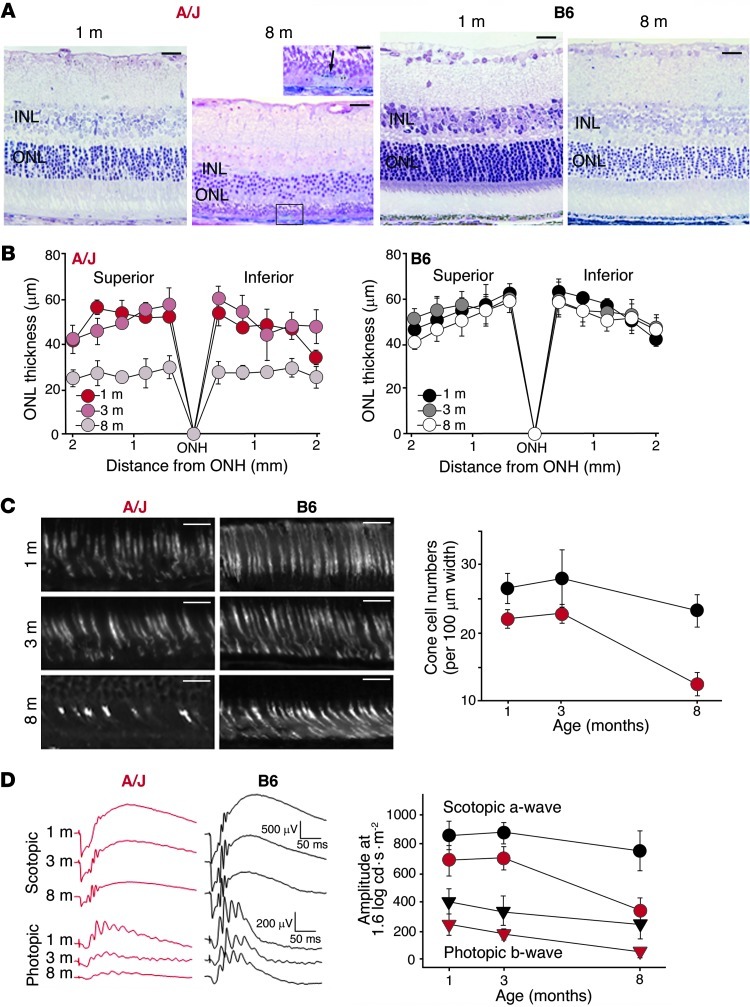
Figure 1. A/J mice display a pronounced age-dependent decline in vision.
(A) Light microscopy of A/J mouse retinas revealed a marked decrease in ONL and inner nuclear layer (INL) thickness in 8- versus 1-month-old animals, as well as pathological changes in the RPE layer at 8 months of age, as shown by the higher-magnification view of the boxed region (arrow, pyknotic cell). These changes were minimal in B6 mice. (B) ONL thickness, plotted as a function of distance from the optic nerve head (ONH), showed the most pronounced decline occurred between 3 and 8 months of age, a finding that was absent in B6 mice. (C) Cone cell sheaths were imaged by PNA staining at 1, 3, and 8 months of age. Average numbers of cone cells in both the superior and inferior retina in a 100-μm range located 500 μm from the optic nerve head were plotted (red, A/J; black, B6). A/J mice showed a marked decline between 3 and 8 months of age. (D) Representative ERG responses at 1.6 log cd•s•m–2, and functional a-wave and b-wave amplitudes, obtained from A/J (red) and B6 (black) mice at 1, 3, and 8 months of age. ERG responses were more attenuated with age in A/J versus B6 mice under both scotopic and photopic conditions (P < 0.001). Scale bars: 20 μm (A and C).
ARD in A/J mice is accompanied by inflammatory cell infiltration and RPE cell pathology.
In vivo scanning laser ophthalmoscopy (SLO) imaging of the fundus revealed a notable increase in autofluorescent spots in the outer retinal layers (where the photoreceptors and RPE are located) of 8- versus 1-month-old A/J mice, whereas no such age-related changes were observed in B6 mice (Figure 2A). Fundus autofluorescence levels did not correlate with amounts of age-related retinoid byproducts, such as N-retinyl-N-retinylidene-ethanolamine (A2E), in the eyes of these mice (Supplemental Figure 1B). These spots could, however, indicate immune cell activation and infiltration into the subretinal space (11). In vivo spectral domain–optical coherence tomography (SD-OCT) and plastic block sections indicated possible infiltrative cells in 8-month-old A/J mice, but not B6 mice (Figure 2B). As shown in Figure 2C, positive staining with the activated macrophage marker Iba-1 indicated that some of the cells were of inflammatory origin, whereas positive staining of other cells with the activated Müller cell marker glial fibrillary acidic protein (GFAP) indicated the stressed retinal environment of A/J mice, but not B6 mice, at 8 months of age.
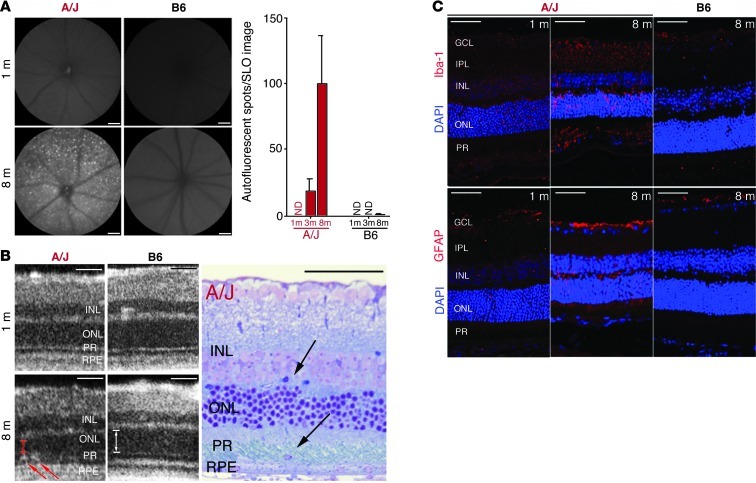
Figure 2. Increased retinal autofluorescence in A/J mice with age relates to inflammatory changes and immune cell infiltration.
(A) Representative SLO autofluorescent images of the outer retina (480 nm excitation; emission filter, 500–700 nm) of 1- and 8-month-old A/J and B6 mice. A/J mice exhibited increased autofluorescent spots with age, which were negligible in B6 mice (n > 3). ND, not detectable. (B) SD-OCT imaging of A/J and B6 mice at 1 and 8 months of age revealed infiltrating cells only in the subretinal space of 8-month-old A/J mice (left; red arrows) that were also observed in plastic block sections (right; black arrows). Thickness of ONL in OCT images is indicated by arrows to highlight the decline in A/J versus B6 mice at 8 months. (C) The cellular infiltration seen by OCT and in plastic sections was of inflammatory origin, as evidenced by increased Iba-1 staining (activated microglia cells) and increased GFAP staining (activated Müller glial cells and astrocytes) in retinas of 8- versus 1-month-old A/J mice. These age-related changes were absent from 8-month-old B6 mouse retina. GCL, ganglion cell layer; IPL, inner plexiform layer; PR, photoreceptor layer. Scale bars: 50 μm.
Age-dependent changes were further evaluated with high-resolution transmission electron microscopy (TEM) imaging of horizontal cross-sections of the RPE-photoreceptor interface. As early as 3 months of age, when no notable ultrastructural or functional decline was evident (Figure 1), A/J mice exhibited pathological changes in the RPE cell layer, as evidenced by swollen cells with abnormal intercellular inclusions resembling inadequately processed photoreceptor discs (Figure 3A). TEM imaging of retinal cross-sections showed abnormal processing of ingested photoreceptor discs by the RPE. These abnormalities were characterized by secondary pockets inside phagosomes (Figure 3, B and C) and abnormal membrane formation that exposed the contents of the discs to the RPE cell cytoplasm (Figure 3, D and E). In contrast, RPE cells imaged at 3 months of age in B6 mice revealed a smaller average cell width than A/J RPE cells with no pathological inclusions; furthermore, ingested discs appeared to be properly processed and transported (Figure 3, F–J). Age-dependent pathology of RPE cells was also assessed by 2-photon microscopy (TPM). There was a change in morphology and an increase in average RPE cell size in A/J mice from 1 to 8 months of age, whereas the size and morphology were well preserved with age in B6 mice (Figure 3K). The localization and function of major RPE-specific protein RPE65, which is critical for visual chromophore regeneration (12), was investigated to assess whether these pathologic changes impair RPE cell function. Immunohistochemical (IHC) staining revealed that RPE65 was homogeneously expressed in the RPE cell layer at 1 month of age, but then became heterogeneous by 8 months of age, in A/J mice (Figure 3L). This correlated with functional deterioration, as illustrated by a 75% depressed regeneration ratio of the visual chromophore, 11-cis-retinal, at 8 months of age in A/J mice (Figure 3M). This ratio was normal in A/J mice at 1 month of age (Supplemental Figure 1C). No significant changes in this ratio or RPE65 localization were evident in B6 mice.
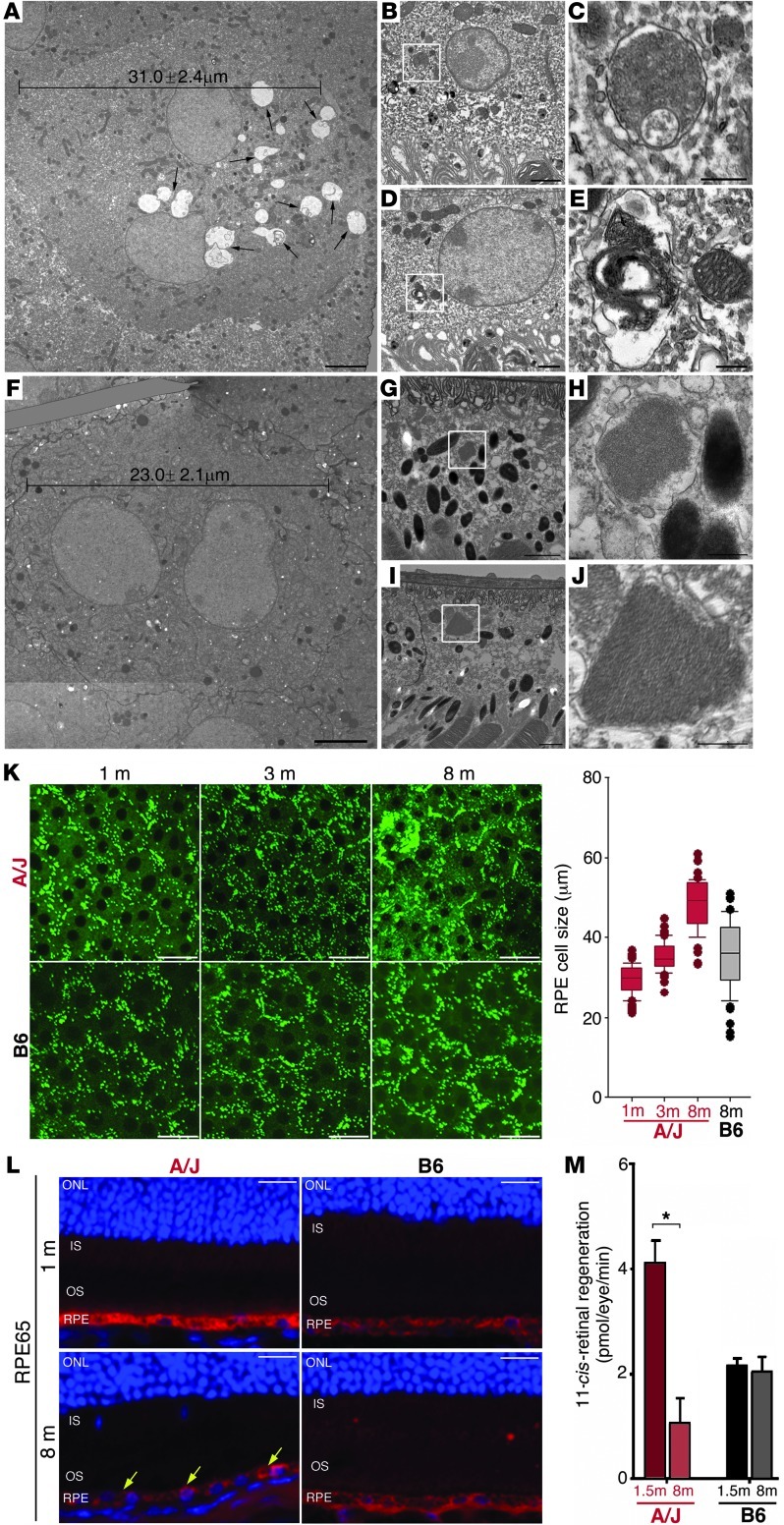
Figure 3. Substantial pathological changes are apparent in RPE cells of A/J mice before measurable visual decline.
(A–J) TEM imaging of 3-month-old (A–E) A/J and (F–J) B6 retinas. (A) A/J RPE cells had an average width of 31.0 μm, with abnormal disc membrane accumulations (black arrows). (B) Ingested phagosomes in A/J mice showed (C) undigested accumulations. (D) Phagosomes were also disrupted, with (E) phagocytotic material exposed to the cytoplasm. (F) B6 RPE cells had an average width of 23.0 μm, with (G) uptake of discs that (H) were normally processed. (I) Ingested discs trafficked normally, with (J) membrane-enclosed disc elements clearly visible. Boxed regions in B, D, G, and I are shown at higher magnification in C, E, H, and J, respectively. (K) Ex vivo TPM imaging of A/J and B6 RPE cells at 1, 3, and 8 months of age. Dysmorphic features in 3-month-old A/J mice were exacerbated in 8-month-old A/J mice, but were absent in B6 mice. Boxes denote interquartile range, lines within boxes denote median, whiskers denote 10th and 90th percentiles, and symbols denote outliers. (L) RPE65 provided a localized uniform signal in 1-month-old A/J mouse retina, but a decreased and heterogeneously localized signal in 8-month-old A/J mouse retina (yellow arrows). No apparent age-related changes were noted in retinas of B6 mice. (M) Regeneration of 11-cis-retinal decreased 75% in A/J retinas by 8 months of age; no such change was noted in B6 mice. *P < 0.05. Scale bars: 5 μm (A and F); 1 μm (B, D, G, and I); 0.5 μm (C, E, H, and J); 20 μm (K and L).
BALB/c mice displayed a mild retinal degenerative phenotype compared with B6 mice, but not nearly as pronounced as the phenotype seen in A/J mice. Although SLO imaging revealed an increase in autofluorescent spots at 8 versus 1 month of age in BALB/c mice (Supplemental Figure 4A), only mild immune cell infiltration was detected (Supplemental Figure 4B), with no obvious pathological changes of the RPE cells (Supplemental Figure 4C), which suggests that BALB/c mice possessed important retinal-protective factors that curbed the inflammatory progression of ARD relative to A/J mice. These findings raised the question of what genetic differences among A/J, BALB/c, and B6 mice could initiate the progression of RPE pathologic changes that lead to a chronic inflammatory state in A/J mice. To address this, we used genetic tools such as CSSs and RNA-Seq technology to reveal multigenic contributions that precipitate and perpetuate this disease.
RNA-Seq reveals differential genetic background contributions to the transcriptome.
We used 22 CSSs of inbred mice, each carrying a single chromosome substituted from a donor strain (A/J) into a host background strain (B6) (Figure 4A), as a more powerful approach than QTL analysis to assess complex genetic traits. Unlike 8-month-old A/J mice, retinoid analysis and histological assessment of the CSS panel mice at 8 months of age revealed no significant phenotypic features of ARD (Figure 4, B–E). Moreover, the lack of significant differences in mice with substitution of chromosomes 3 or 7 further demonstrated that neither lighting conditions (13) nor pigmentary status (14) were major factors contributing to disease. Mislocalization of the centromeric portion of chromosome 1 (15) in the CSS panel was investigated, but no genetic variation or candidate genes from that region could account for the retinal phenotype observed. Thus, ARD in A/J mice is likely attributable to contributions from multiple chromosomes, which necessitates a more global genetic approach.
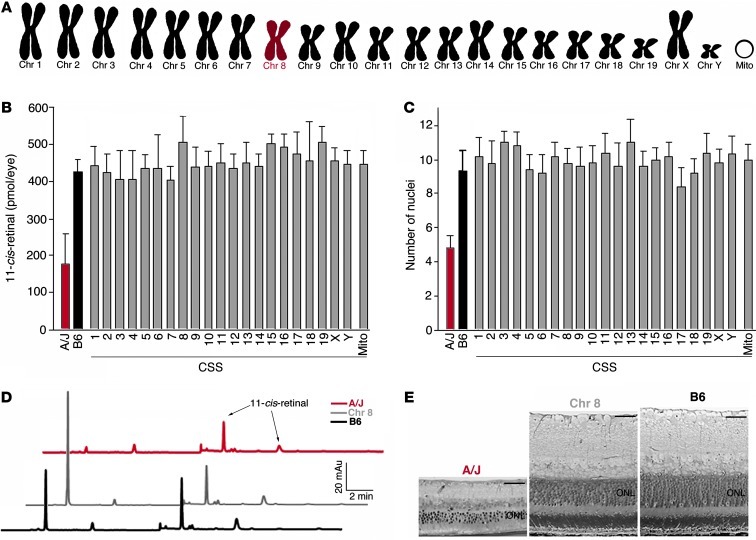
Figure 4. Genetic panel study reveals no significant phenotypic changes in B6 mice with single A/J chromosome substitutions.
A CSS panel was used in which single chromosomes (Chr) 1–19, X, Y, and mitochondrial DNA (Mito) from A/J mice were substituted into the B6 background. (A) An example of the genetic makeup of chromosome 8–substituted mice, which possess only chromosome 8 from A/J mice and all other chromosomes from B6 mice. No significant functional or structural changes were observed at 8 months when (B) 11-cis-retinal levels and (C) nuclei numbers in the ONL were compared in all CSSs relative to B6 mice. Representative (D) HPLC chromatograms and (E) plastic block sections of A/J, B6, and Chr 8 mice. Scale bars: 40 μm (E).
To investigate the putative multigenic etiology of ARD and identify more distant interacting genetic factors that contribute to its pathology, we carried out RNA-Seq of whole-eye tissues from A/J, BALB/c, and B6 mice. Importantly, 1-month-old mice from each genetic background were analyzed before obvious ARD-related phenotypic changes had occurred in order to identify genetic factors that could trigger this pathology. To obtain adequate statistical power, 3 biological replicates of ocular tissues from each strain were prepared for RNA-Seq. Replicate results from each strain had high correlation coefficients when plotted against each other (Supplemental Figure 5). A value of 1 fragment per kilobase per million reads (FPKM), equivalent to 1 transcript per cell (16), was used as the expression cutoff. Categorization of expressed transcripts — 12,606 in the A/J eye, 12,589 in the BALB/c eye, and 12,832 in the B6 eye — by GoTerm annotation did not reveal any significant differences between mouse strains at 1 month of age (Supplemental Table 1).
Performing 3 biological replicate RNA-Seq runs yielded an accurate determination of the differential expression profiles of the 3 different mouse strains. RNA-Seq results have been shown to correlate strongly with RT-PCR findings in the eye and retinal tissues (17, 18), and this was further demonstrated with a cohort of RT-PCR probes (Supplemental Figure 6, A and B). The RNA-Seq data were further validated for visual cycle proteins by immunoblotting (Supplemental Figure 6C). At least a 2-fold difference (P ≤ 0.05) was used as the cutoff to identify differentially expressed genes between mice with different genetic backgrounds. The RNA-Seq differential expression profile for A/J and B6 eyes showed that whereas most genes overlapped in expression, there were 332 genes that were differentially expressed between the 2 strains and required further analysis (Figure 5A). The single base resolution of RNA-Seq experiments also allowed SNP detection and characterization to complement the differential expression data: known SNPs in both Rpe65 (13) and Tyr (14) in A/J and BALB/c mice, but not B6 mice, are highlighted in Supplemental Figure 7, A–P. Moreover, the global scale of expression data from RNA-Seq allowed us to reexamine previous QTL studies in search of new insights. For example, in the QTL location on chromosome 10 proposed to account for the difference in cone photoreceptor numbers between A/J and B6 mice (19), we identified a 2-fold differential expression of only 1 gene, Enpp3 (encoding ectonucleotide pyrophosphate/phosphodiesterase 3). Interestingly, Enpp3 contained a SNP in the protein-coding sequence that leads to an amino acid residue change from Asn to Ser (Supplemental Figure 7, Q–V). SNP analysis of other differentially expressed genes, however, did not reveal any further molecular basis for the age-related phenotype in A/J mice (Supplemental Table 2). This prompted us to perform pathway analyses to define perturbations in cellular pathways across the differing genetic backgrounds in A/J, BALB/c, and B6 mice.
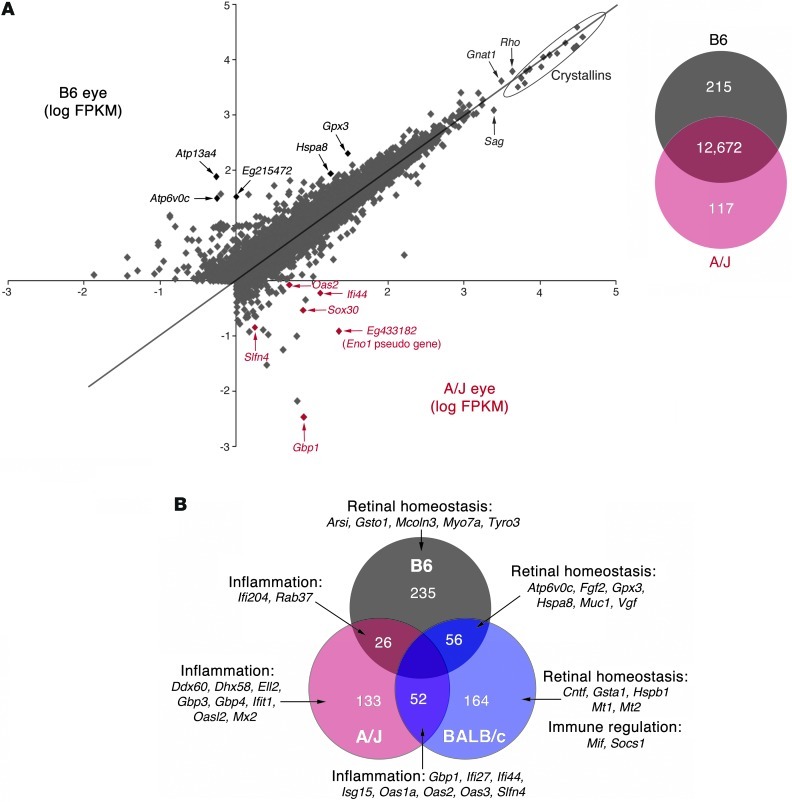
Figure 5. RNA-Seq of 3 individual biological replicates of 1-month-old A/J, BALB/c and B6 mouse eyes reveals differential transcriptome profiles.
(A) Left: Plot of log FPKM from A/J and B6 runs. The most highly expressed transcripts common to both A/J and B6 eyes were the lens crystallin genes (outlined) and rod photoreceptor genes such as Gnat1, Rho, and Sag. Most genes highly differentially expressed in the A/J eye relate to inflammation (red), whereas genes with the lowest differential expression in the A/J eye encode homeostatic proteins (black). Right: Whereas 12,672 genes had similar expression, 332 were differentially expressed by at least 2-fold (P ≤ 0.05) between A/J and B6 eyes. (B) Examination of all 3 mouse eye transcriptomes revealed those genes exclusively more highly expressed in a single mouse strain compared with the other 2 strains, and those sharing increased expression with respect to the third strain. For example, 235 genes are exclusively more highly expressed in the B6 eye; this strain shares 26 genes also highly expressed in the A/J eye and 56 also higher in the BALB/c eye. Importantly, a large cohort of inflammatory genes exhibits increased expression in A/J mice. Interestingly, although both A/J and BALB/c eyes share increased expression of inflammatory genes, only BALB/c exhibits a counteracting increased expression of retinal homeostatic and immune regulatory genes, either exclusively or shared with B6 eyes.
Pathway analysis highlights inflammatory priming coupled with impaired retinal homeostatic cellular pathways in 1-month-old A/J mice, before retinal pathology is evident.
RNA-Seq of whole eyes from 1-month-old mice of the 3 genetic backgrounds revealed genes that were exclusively expressed at a higher level by at least 2-fold (P ≤ 0.05) in each strain with respect to the other 2 strains, as well genes expressed at a higher level in 2 different strains compared with the third strain. The analysis identified 133, 164, and 235 genes with higher expression by at least 2-fold (P ≤ 0.05) exclusively in A/J, BALB/c, and B6 mice, respectively. We also found shared increased expression of 52 genes between A/J and BALB/c mice, 26 genes between A/J and B6 mice, and 56 genes between BALB/c and B6 mice (Figure 5B). Ingenuity pathway analysis of these genes revealed that A/J mice exhibited preferentially higher expression of a large cohort of inflammatory gene products, a subset of which was also shared with BALB/c mice. Genes with preferentially decreased expression in A/J compared with BALB/c and B6 mice included those encoding proteins in cellular pathways involved in maintaining normal retinal homeostasis and protecting against tissue damage. In contrast to A/J mice, BALB/c and B6 mice exhibited overlapping increased expression of a cohort of retinal homeostatic genes as well as exclusive strain-specific increased expression of such genes. Furthermore, there was selective increased expression of key immune-regulatory genes in BALB/c mice that could counteract the effects of inflammatory genes found to be elevated in common with A/J mice. Taken together, these network expression patterns present at 1 month of age in A/J mice indicated that the normal-appearing retina was primed for inflammation with an absence of key homeostatic and immune-protective molecules. This environment could preferentially trigger severe ARD in A/J mice.
Inflammatory priming in A/J retina is exacerbated with age.
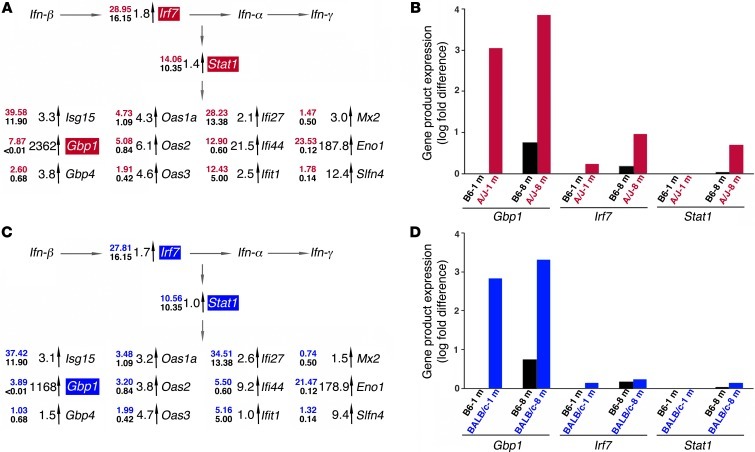
Ingenuity pathway analysis revealed an activated inflammatory priming pathway in young A/J and BALB/c mice centered on IFN signaling. This was evidenced by increased expression of inflammatory gene products such as IFN regulatory factor 7 (Irf7); the target of IFN-induced signaling, Stat1; and Stat1-inducible markers of inflammation, such as guanylate-binding protein 1 (Gbp1). Exhibiting a marked increase in young 1-month-old A/J mice (Figure 6A), expression of this entire network was further exacerbated with age, as evidenced by increased expression of Gbp1, Irf7, and Stat1 assessed by RT-PCR in 8-month-old A/J mice. There was minimal age-related elevation of expression detected in B6 mice over the same period (Figure 6B). This inflammatory network was also elevated in BALB/c versus B6 mice (Figure 6C), albeit at a lower level than in A/J mice. Moreover, BALB/c mice did not exhibit the marked age-related increase in Gbp1, Irf7, and Stat1 expression from 1 to 8 months of age (Figure 6D). This difference indicates that the inflammatory-primed network in A/J mice transitions to a chronic state of inflammation with age as a result of other factors, possibly involving RPE cells, which are key regulators of retinal inflammation.
Figure 6. Pathway analysis of RNA-Seq differential expression profiles reveals age-related inflammatory priming in eyes of A/J mice.
(A) Pathway analysis with Ingenuity software unveiled an aberrant inflammatory network in A/J mice characterized by priming of IFN at 1 month of age, as evidenced by increased activation of Irf7 and Stat1, coordinated increased expression of Stat1-induced secondary response genes, induction of positive regulatory loop genes in the inflammatory process, and expression of genes involved in immune cell activation. Numbers at left of each gene represent the RNA-Seq FPKM values from A/J (top; red) and B6 (bottom; black) eyes; fold differences are indicated next to the vertical arrows. (B) RT-PCR of Gbp1, Irf7, and Stat1 (shaded red in A) showed that this inflammatory priming network was preferentially exacerbated in older mice, with pronounced increases in gene product expression from 1 to 8 months of age in A/J relative to B6 mice. (C) BALB/c mice also exhibited features of inflammatory priming at 1 month of age when the same pathways were examined, but these changes were less pronounced than those in A/J mice. Numbers at left of each gene represent the RNA-Seq FPKM values from BALB/c (top; blue) and B6 (bottom; black) eyes; fold differences are indicated next to the vertical arrows. (D) RT-PCR of Gbp1, Irf7, and Stat1 (shaded blue in C) showed that in BALB/c mice, inflammatory priming was not exacerbated from 1 to 8 months of age, similar to findings in B6 mice.
Marginally expressed retinal homeostasis proteins exhibit abnormal RPE localization in A/J mice.
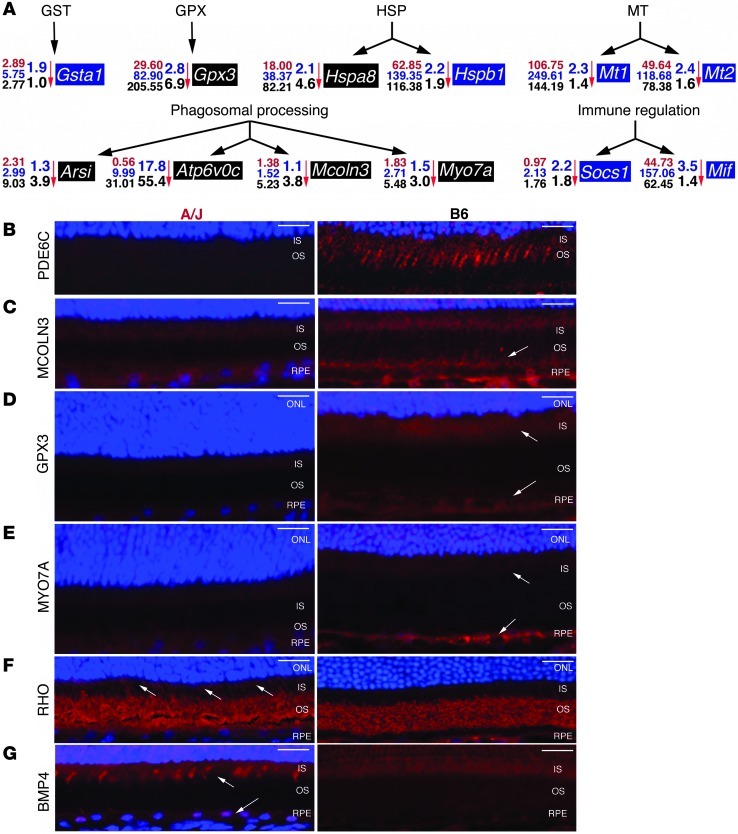
Numerous genes with preferentially decreased expression in A/J compared with BALB/c and/or B6 mouse eyes are involved in homeostatic maintenance of the retinal environment. These genes encode diverse groups of proteins, such as those in the glutathione S-transferase (GST), glutathione peroxidase (GPX), heat shock protein (HSP), and metallothionein (MT) families (Figure 7A). Expression of genes potentially involved in phagosomal processing, such as Arsi (20), Atp6v0c (21), and Mcoln3 (22), and key genes in immune regulation, such as Socs1 (23), were also decreased in A/J mouse eyes (Figure 7A). IHC staining revealed that expression levels in mouse retinas correlated strongly with transcript abundance identified by RNA-Seq in mouse eyes. Examination of the cone marker phosphodiesterase-6c (PDE6C) illustrated the increased cone numbers in B6 compared with A/J mice (Figure 7B). Because many of the highlighted genes play important roles in RPE processing and response to oxidative stress (24), the localization of markers like mucolipin 3 (MCOLN3) and GPX3 were examined. Both displayed dim signals in A/J RPE, but much stronger signals in B6 RPE (Figure 7, C and D). The MCOLN3 staining distribution also suggested that this protein extends to RPE cell processes that interact with the photoreceptor layer. IHC staining of myosin VIIa (MYO7A), a well-known component of the photoreceptor cilium involved in RPE-mediated protein movement and photoreceptor phagocytosis (25, 26), provided only weak signals in the A/J inner segment and RPE cell layer, whereas staining was more pronounced in both these layers of the B6 retina (Figure 7E). Attenuated expression of MYO7A can contribute to mislocalization of key photoreceptor and RPE proteins, such as rhodopsin (RHO) and RPE65. This was evidenced by a RHO signal in the inner segments of A/J retinas that was absent in B6 retinas (Figure 7F) and the heterogeneous RPE65 signal in RPE cells of A/J mice, but not B6 mice, at 8 months of age (Figure 3L). Moreover, staining retinas for ROS with the marker dihydroethidium revealed an enhanced signal in 8-month-old A/J retinas before light exposure that became even more pronounced after this procedure. Only a background signal was apparent in BALB/c and B6 retinas (Supplemental Figure 8). Furthermore, staining for bone morphogenic protein 4 (BMP4), a marker for oxidative stress–induced RPE senescence (27), produced a more enhanced signal in A/J RPE and inner segments of cone photoreceptors than in B6 retina at 1 month of age (Figure 7G).
Figure 7. Homeostatic processing genes with decreased expression in A/J mice display protein expression in RPE and photoreceptor compartments of the retina.
(A) Expression of genes of the GST, GPX, HSP, and MT families, as well as genes involved in phagosomal processing and in immune regulation, exhibited markedly decreased expression in A/J compared with both BALB/c and B6 mice. Numbers at left of each gene represent the RNA-Seq FPKM values from A/J (red), BALB/c (blue), and B6 (black) eyes; fold difference in A/J versus BALB/c (top; blue) and A/J versus B6 (bottom; black) is indicated next to each vertical arrow. Blue shading denotes genes with greater expression in BALB/c; black shading denotes genes with greater expression in B6. (B–G) IHC of 1-month-old A/J and B6 retinas was done with (B) PDE6C, cone cell marker; (C) MCOLN3, involved in lysosomal degradation; (D) GPX3, involved in ROS detoxification; (E) MYO7A, involved in protein trafficking; (F) RHO, visual pigment in rod photoreceptors; and (G) BMP4, marker of RPE cell senescence. MCOLN3, GPX3, and MYO7A staining was much more pronounced in B6 RPE (arrows in C–E). The aberrant nature of the A/J RPE cell was evidenced by mislocalization of RHO, as evidenced by signals in both the outer and inner segment (OS and IS, respectively; arrows in F) and increased expression of BMP4 in inner segment and RPE (arrows in G) in A/J mice. Scale bars: 20 μm.
Discussion
Phenotypic variability based on genetic background differences observed in mice (28, 29) can provide models for complex human conditions and explain the variable susceptibility to certain conditions among different individuals with the same allelic variant of disease. With no reliable markers to date for ARD before it becomes clinically evident, genetic studies have involved only affected individuals, making it difficult to identify factors that trigger this condition. Compared with BALB/c and B6 mice, A/J mice in this study displayed accelerated retinal degeneration stemming from differences in their natural genetic background. Declining visual function with age, associated with worsening RPE and photoreceptor cell pathology, was documented in these mice. Given that the retina is fully developed after 1 month in mice, we used a comprehensive set of approaches to identify genetic factors in 1-month-old mice that led to severe ARD at 8 months in A/J mice, but not BALB/c or B6 mice.
Identifying global gene variations associated with complex diseases of multigenic origin can be difficult. The present studies highlighted that CSS panels, although capable of revealing some complex disease associations (15, 30), lacked the breadth to identify distant interacting genetic factors characteristic of a complex disease like ARD. RNA-Seq allows for more complete documentation of genetic changes relevant to complex disease etiology (31). This is especially true for age-related neurodegenerative conditions, as illustrated by the recent discovery of 2 novel genetic loci associated with Alzheimer disease by high-throughput sequencing methodology (32). The challenge of RNA-Seq technology lies in the large-scale translation of genetic variations into knowledge about the molecular pathogenesis of complex diseases. By studying cellular pathways encoded by these genes as integrated systems, we can begin to understand their derived phenotypes (33) and identify genes that critically contribute to complex disease traits. In this study, RNA-Seq of young mouse eye tissue across different genetic backgrounds was performed before substantial pathology was present. This analysis revealed global gene expression changes that identified factors apparently predisposing A/J mice to a severe ARD phenotype, as well as factors in BALB/c and B6 mice that may prevent this pathology. Complementing morphologic assessment of ARD with deep sequencing technology allowed us to identify early signatures of disease in inbred mice with different genetic backgrounds.
RPE cells are critical for the maintenance of the blood/retina barrier and retinal neurons and must possess mechanisms that protect the retina against oxidative stress generated by exposure to light and high oxygen tension. Compared with BALB/c and B6 mice, A/J mice displayed decreased expression of key gene families needed by RPE cells to mitigate such stress. Most notable were those families involved in glutathione-mediated oxidative stress protection. Abundant in the retina, the GPX family encodes a critical group of retinal detoxification enzymes. Only Gpx3 expression from this family was markedly decreased in A/J mice, and GPX3 largely localized to RPE cells. Inadequate activity of antioxidant enzymes like GPX in other organ systems has been implicated in the progression to chronic inflammatory pathology (34), and polymorphisms of Gpx3 have been implicated in human phenotypes of ARD (35, 36). This finding extends to other protein families, such as the HSPs, of which both Hspa8 and Hspb1 showed decreased expression in A/J mice. In addition to chaperone-mediated activity (37), HSPs combat oxidative stress by increasing levels of glutathione and modulating the redox status of cells (38). Decreasing levels of glutathione and associated proteins with age (39, 40), coupled with constitutively decreased expression of these important homeostatic gene families in young A/J mice compared with BALB/c and B6 mice, could explain the accelerated ARD phenotype found in A/J mice. Pathologic changes seen in A/J RPE, characterized by cell swelling and buildup of undigested disc elements, have also been reported in albino and pigmented rats deprived of selenium (41, 42), a critical cofactor for activation of antioxidant proteins such as GPX3. Inadequate protection against oxidative stress was also accompanied by accelerated loss of photoreceptors, especially in the central retina of these rats, which suggests that oxidative damage to RPE cells may initiate the detrimental process and that photoreceptor loss is a secondary effect. An inadequate oxidative stress response could thus produce wide-ranging adverse effects in the retina. One detrimental age-related effect — namely, cellular senescence — was evidenced by increased BMP4 expression in A/J mice as young as 1 month of age, and this could lead to secretion of various factors that fuel chronic inflammation (43).
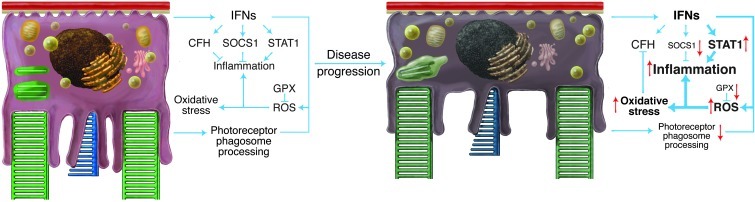
The RPE is a central site of immune regulation in the retina (44). Moreover, constant stress and cellular senescence can exacerbate immune responses that lead to chronic inflammation, which results in disease progression because of changes in homeostatic set points (45). Although A/J mice have natural deficiencies of complement (C5a) and NOD (Naip5) genes involved in susceptibility to bacterial/fungal infection, recombinant congenic strains (46, 47) have shown that these genes are controlled by additional factors and have little effect on the inflammatory-priming pathways highlighted in this study. In young A/J mice (and to a lesser degree BALB/c mice, but not B6 mice), RNA-Seq revealed an inflammatory-primed network characterized by increased expression of IFN gene products, such as Stat1 and its downstream effectors (23). The inflammatory-primed state identified in A/J mice may not be limited to the retina, as it also likely contributes to the enhanced A/J inflammatory response in experimental allergic asthma (48). Moreover, the graded retinal inflammatory changes we saw from A/J to BALB/c to B6 mice are consistent with studies of allergic airway inflammation in the lungs of these strains (49). These prior studies indicate that A/J mice are most susceptible to inflammation in tissues of high oxygen tension (retina and lungs), further implicating an inadequate oxidative stress response as a driving force of late stages of inflammatory disease progression. In the present study, the inflammatory-primed state of the retina in A/J mice was compounded by markedly lower levels of immune-regulatory enzymes. Compared with BALB/c mice, A/J mice exhibited decreased expression of genes such as Tyro3 and its downstream effector Socs1, which serve to control this inflammatory response (50). Moreover, inadequate function of enzymes like GPX3 would lead to increased oxidative stress and hydrogen peroxide production, which reduce the expression and function of complement factor H (51), a key complement-regulatory enzyme implicated in disease pathogenesis (52). Thus, the inflammatory-primed state of the retina in young A/J mice reflects a parainflammatory state (53) that, accentuated by increased oxidative stress, transitions to chronic inflammation and cellular dysfunction with increasing age (Figure 8), just as observed in diseased patients (54). Priming of RPE cells can translate to secondary effects on critical cellular processes such as phagocytosis and digestion of POS, which are increasingly affected in A/J mice with age. Another secondary effect is the production of anti-retinal antibodies to α-crystallin, GFAP, and α-enolase found in sera of ARD patients (55). Preferential activation of Stat1 has been detected in several autoimmune/inflammatory conditions (23), and this increased significantly and exclusively in A/J mice with age. We also noted increased GFAP expression with age, and α-enolase was elevated at the transcript level in A/J mice as early as 1 month of age. RPE/retina barrier function is most detrimentally affected when both oxidative stress and immune activation are induced (56), which suggests that these additive effects of aging in A/J mice could potentially lead to autoimmune responses. Ocular immune privilege is normally responsible for the impaired tolerance to retinal antigens (57), but exposure to such antigens upon age-related barrier compromise could further contribute to disease progression.
Figure 8. Inadequate protection by the RPE from stress drives the retina from an inflammatory-primed state to a chronic disease state.
In a normal homeostatic state (left), there is a delicate balance between stress and the resulting tissue response. Stress caused by POS accumulation in the RPE from daily ingestion of oxidized photoreceptor discs could result in oxidative damage and inflammation unless modulated by a network of enzymes such as GPX. Moreover, activation of inflammatory factors like STAT1 in response to such RPE cell stress is controlled by regulatory factors such as SOCS1 and complement factor H (CFH). In contrast, low or declining levels of protein expression from these complex sets of interconnected gene networks (right) result in inadequate stress protection and inflammatory changes that cause chronic retinal degeneration. Thus, decreased expression of homeostatic genes like Gpx foster increased oxidative stress and can reduce other protective factors like Cfh. This is compounded by decreased expression of immune regulatory factors that can exacerbate inflammation and drive disease progression. Subtle genetic differences therefore can have profound effects on the predisposition to and pathogenesis of ARD.
Our preliminary RNA-Seq analysis of whole eye tissues from both nocturnal (Long-Evans rat) and diurnal (Nile rat) rodent species at a young age revealed gene signatures most closely related to B6 mice and clearly distinct from A/J mice (Supplemental Table 3). This observation emphasizes that our present findings may extend beyond model mouse species and provide an additional frameworks for understanding etiologic factors contributing to complex age-associated diseases in higher organisms. This is especially relevant to human blinding conditions associated with aging, such as age-related macular degeneration (AMD), the leading cause of blindness in the industrial world and now considered one of the major blinding diseases worldwide (58). The complex etiology of AMD is reflected by the relative paucity of effective compounds for its early prevention and treatment, with the main risk factor being increasing age. Complex trait analysis of genome-wide association studies — having identified the vast majority of variations in noncoding regions (59) and the recent identification of large intervening noncoding RNAs implicated in controlling a diverse set of biological processes (60, 61), including differentiation of the murine retina (62), a discovery accelerated by application of next-generation sequencing technology (63) — could serve as the next frontier to facilitate understanding of complex disease phenotypes.
In conclusion, we used 3 inbred background strains of mice with differing susceptibilities to ARD and used RNA-Seq to identify genes encoding components of cellular pathways that contribute to this blinding condition at a young age, before phenotypic changes are apparent. We identified relatively high expression of proinflammatory factors coupled with low expression of homeostatic protective factors, such as those involved in oxidative stress response, and localized them to the A/J mouse retina, most notably to the RPE. This combination made A/J mice especially vulnerable to rapidly progressive ARD compared with BALB/c and B6 mice. A/J mice reared in the dark still developed ARD, which indicates that the pathogenic mechanisms described here differ from those involving excessive accumulation of the all-trans-retinal chromophore (64). These results, in conjunction with imaging techniques that demonstrated elevated level of ROS and changes in phagocytotic processing in the RPE, allow a more comprehensive understanding of a complex neuronal degeneration. With advances in phenotypic and genotypic characterization technology, the methodologies outlined herein represent a powerful paradigm to unveil cellular changes that trigger and drive progression of complex neurodegenerative diseases extending beyond the eye.
Methods
Materials.
See Supplemental Methods for details and sources of antibodies and probe sets.
Animals.
A/J, BALB/c, and B6 inbred mice and A/J CSS mice were obtained from The Jackson Laboratory. Long-Evans rats were purchased from Harlan Laboratories. Nile rats were obtained from the laboratory of L. Smale (Michigan State University, Lansing, Michigan, USA). Mice and rats were housed in the animal facility at the School of Medicine, Case Western Reserve University (CWRU), where they were maintained on a standard chow diet in a 12-hour light (∼10 lux), 12-hour dark cycle.
Histology and IHC.
Histological and IHC procedures were carried out as previously described (65). IHC sections were viewed with a Zeiss LSM 510 inverted Laser Scan Confocal Microscope.
ERGs.
All ERG procedures were performed according to previously published methods (65).
Retinoid and A2E analyses.
All experimental procedures related to extraction, derivatization, and separation of retinoids from dissected mouse eyes were carried out as described previously (65). Quantification of A2E by HPLC was achieved by comparison with known concentrations of pure synthetic A2E standards (66).
Ultra-high-resolution SD-OCT and SLO imaging.
Both ultra-high-resolution SD-OCT (Bioptigen) and SLO (HRAII; Heidelberg Engineering) were used for in vivo imaging of mouse retinas as previously described (11).
Library preparation and Illumina RNA-Seq runs.
Mouse eye tissue libraries were prepared as previously described (18). Pooled total RNA samples from 5 eyes were used for each whole-eye library preparation. 3 separate biological replicate libraries were made from mice with A/J, BALB/c, or B6 backgrounds. Each murine eye library was run on 1 lane of the Genome Analyzer IIx in the CWRU Genomics Core Facility using 54-, 76-, or 79-bp single-end sequencing. The number of mapped single reads was 19.9 × 106, 22.5 × 106, and 21.7 × 106 from the 3 different A/J eye libraries; 41.2 × 106, 38.9 × 106, and 27.4 × 106 from the 3 different BALB/c eye libraries; and 16.8 × 106, 17.7 × 106, and 38.2 × 106 from the 3 different B6 eye libraries. Data were processed and aligned with the University of California, Santa Cruz, mouse genome assembly and transcript annotation (mm9) and processed as previously described to calculate FPKM statistics by gene, transcript, and exon (18). The output for CASAVA was visualized with the GenomeStudio RNA Sequencing Module (Illumina), which allowed comparisons between samples. The processed and raw fastq files were deposited in GEO (accession no. GSE38359).
Pathway generation and analysis.
RNA-Seq data were analyzed with Ingenuity pathway analysis software (Ingenuity Systems). Average FPKM values from RNA-Seq runs of A/J, BALB/c, and B6 mouse eyes were uploaded along with gene identifiers and statistical P value calculations. Core analysis of a set of 11,339 mapped genes was done to identify perturbed molecular networks. Pathway generation of differentially expressed gene was also done by hand with Ingenuity, Pubmed database, and known retinal localization as guides.
Real-time RT-PCR.
2 μg isolated total retinal RNA from 3 pooled A/J, BALB/c, or B6 eye samples was converted to cDNA with the High Capacity RNA-to-cDNA kit (Applied Biosystems). All real-time experiments were done in triplicate with an ABI Step-One Plus qRT-PCR machine (Applied BioSystems). Fold changes were calculated based on differences in Ct between A/J or BALB/c and B6 samples after normalization to 18S rRNA.
TEM.
Mouse eye cups were prepared as previously described (18). Blocks were cut with a microtome, and ultrathin sections (0.07 μm) were stained with uranyl acetate and adsorbed onto carbon grids. A Tecnai T12 EM (FEI) operating at 120 kV with a LaB6 filament was used for final imaging.
TPM.
Excitation TPM was performed with a Leica TCS SP2 scanning head (Leica) attached to a DM IRBE2 inverted microscope stand with Leica LCS 3-dimensional software. Laser pulses from a mode-locked Ti:Sapphire laser (Chameleon-XR) were focused on the tissue sample by an HCX PL APO ×40 oil immersion objective lens (NA, 1.25; Leica). Imaging and sample conditions were as described previously (67).
In vivo detection of ROS.
ROS detection in vivo was performed as previously published (68). The ROS probe dihydroethidium (Invitrogen), at a dose of 20 mg/kg body weight, was administered to mice via intraperitoneal injection 30 minutes prior to light exposure. Eye cups — obtained after removing the cornea, lens, and vitreous body from enucleated eye globes 3 hours after light illumination — were fixed in 4% paraformaldehyde. Cryosections were prepared from fixed eye cups and cut at 12-μm thickness for microscopic assessment of ROS signal fluorescence in the retina.
Statistics.
Experimental results were analyzed by 1-way ANOVA. A P value of 0.05 or less was considered statistically significant. Data are presented graphically in figures as mean ± SD.
Study approval.
All animal procedures and experiments were performed in accordance with US animal protection laws, were approved by the CWRU Animal Care Committees, and conformed to the recommendations of both the American Veterinary Medical Association Panel on Euthanasia and the Association of Research for Vision and Ophthalmology.
Supplementary Material
Acknowledgments
We thank Leslie T. Webster Jr., Johannes von Lintig, Michael E. Maguire, Clifford V. Harding, George Dubyak, Timothy S. Kern (CWRU), Thaddeus Dryja (Novartis), and Alexander Moise (University of Kansas, Lawrence, Kansas, USA) for valuable comments about the manuscript. We thank Mark Adams, Akiko Maeda, Yu Chen, Brian Kevany, Hisashi Fujioka, Neil Molyneaux, Simone Edelheit, Lindsay Perusek, Melissa Matosky, Satsumi Roos, Midori Hitomi, and Anne E. Baskin (CWRU) for technical assistance. This research was supported by NIH grants EY008061, EY019478, K08EY019880, and P30 EY11373; by NCRR grant RR12305; by the Research to Prevent Blindness Foundation; by Foundation Fighting Blindness; by Fight for Sight; and by the Ohio Lions Eye Research Foundation. D. Mustafi was supported in part by CWRU Medical Scientist Training Program Grant T32GM007250 and by Visual Sciences Training Grant T32EY007157 from the NIH. K. Palczewski is the John H. Hord Professor of Pharmacology.
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Citation for this article: J Clin Invest. 2012;122(8):2989–3001. doi:10.1172/JCI64427.
References
- 1.Kirkwood TBL. Understanding the odd science of aging. Cell. 2005;120(4):437–447. doi: 10.1016/j.cell.2005.01.027. [DOI] [PubMed] [Google Scholar]
- 2.Medzhitov R, Schneider DS, Soares MP. Disease tolerance as a defense strategy. Science. 2012;335(6071):936–941. doi: 10.1126/science.1214935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Luo DG, Xue T, Yau KW. How vision begins: An odyssey. Proc Natl Acad Sci U S A. 2008;105(29):9855–9862. doi: 10.1073/pnas.0708405105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Strauss O. The retinal pigment epithelium in visual function. Physiol Rev. 2005;85(3):845–881. doi: 10.1152/physrev.00021.2004. [DOI] [PubMed] [Google Scholar]
- 5.Baker DJ, et al. Clearance of p16(Ink4a)-positive senescent cells delays ageing-associated disorders. Nature. 2011;479(7372):232–237. doi: 10.1038/nature10600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Danciger M, et al. Quantitative genetics of age-related retinal degeneration: a second F1 intercross between the A/J and C57BL/6 strains. Mol Vis. 2007;13:79–85. [PMC free article] [PubMed] [Google Scholar]
- 7.Pierce EA, Liu Q, Saveliev A. The severity of retinal degeneration in Rp1h gene-targeted mice is dependent on genetic background. Invest Ophthalmol Vis Sci. 2009;50(4):1566–1574. doi: 10.1167/iovs.08-2776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Swaroop A, Chew EY, Rickman CB, Abecasis GR. Unraveling a multifactorial late-onset disease: from genetic susceptibility to disease mechanisms for age-related macular degeneration. Annu Rev Genomics Hum Genet. 2009;10:19–43. doi: 10.1146/annurev.genom.9.081307.164350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nadeau JH, Singer JB, Matin A, Lander ES. Analysing complex genetic traits with chromosome substitution strains. Nat Genet. 2000;24(3):221–225. doi: 10.1038/73427. [DOI] [PubMed] [Google Scholar]
- 10.Wang Z, Gerstein M, Snyder M. RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet. 2009;10(1):57–63. doi: 10.1038/nrg2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Maeda A, et al. Palmitoylation stabilizes unliganded rod opsin. Proc Natl Acad Sci U S A. 2010;107(18):8428–8433. doi: 10.1073/pnas.1000640107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kiser PD, Palczewski K. Membrane-binding and enzymatic properties of RPE65. Prog Retin Eye Res. 2010;29(5):428–442. doi: 10.1016/j.preteyeres.2010.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wenzel A, Reme CE, Williams TP, Hafezi F, Grimm C. The Rpe65 Leu450Met variation increases retinal resistance against light-induced degeneration by slowing rhodopsin regeneration. J Neurosci. 2001;21(1):53–58. doi: 10.1523/JNEUROSCI.21-01-00053.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yokoyama T, Silversides DW, Waymire KG, Kwon BS, Takeuchi T, Overbeek PA. Conserved cysteine to serine mutation in tyrosinase is responsible for the classical albino mutation in laboratory mice. Nucleic Acids Res. 1990;18(24):7293–7298. doi: 10.1093/nar/18.24.7293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Singer JB, et al. Genetic dissection of complex traits with chromosome substitution strains of mice. Science. 2004;304(5669):445–448. doi: 10.1126/science.1093139. [DOI] [PubMed] [Google Scholar]
- 16.Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods. 2008;5(7):621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- 17.Brooks MJ, Rajasimha HK, Roger JE, Swaroop A. Next-generation sequencing facilitates quantitative analysis of wild-type and Nrl(–/–) retinal transcriptomes. Mol Vis. 2011;17:3034–3054. [PMC free article] [PubMed] [Google Scholar]
- 18.Mustafi D, et al. Defective photoreceptor phagocytosis in a mouse model of enhanced S-cone syndrome causes progressive retinal degeneration. FASEB J. 2011;25(9):3157–3176. doi: 10.1096/fj.11-186767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Whitney IE, Raven MA, Lu L, Williams RW, Reese BE. A QTL on chromosome 10 modulates cone photoreceptor number in the mouse retina. Invest Ophthalmol Vis Sci. 2011;52(6):3228–3236. doi: 10.1167/iovs.10-6693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Oshikawa M, Usami R, Kato S. Characterization of the arylsulfatase I (ARSI) gene preferentially expressed in the human retinal pigment epithelium cell line ARPE-19. Mol Vis. 2009;15:482–494. [PMC free article] [PubMed] [Google Scholar]
- 21.Peri F, Nusslein-Volhard C. Live imaging of neuronal degradation by microglia reveals a role for v0-ATPase a1 in phagosomal fusion in vivo. Cell. 2008;133(5):916–927. doi: 10.1016/j.cell.2008.04.037. [DOI] [PubMed] [Google Scholar]
- 22.Lelouvier B, Puertollano R. Mucolipin-3 regulates luminal calcium, acidification, and membrane fusion in the endosomal pathway. J Biol Chem. 2011;286(11):9826–9832. doi: 10.1074/jbc.M110.169185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hu XY, Chakravarty SD, Ivashkiv LB. Regulation of interferon and Toll-like receptor signaling during macrophage activation by opposing feedforward and feedback inhibition mechanisms. Immunol Rev. 2008;226:41–56. doi: 10.1111/j.1600-065X.2008.00707.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Newsome DA, Miceli MV, Liles MR, Tate DJ, Oliver PD. Antioxidants in the retinal-pigment epithelium. Prog Retin Eye Res. 1994;13(I):101–123. doi: 10.1016/1350-9462(94)90006-X. [DOI] [Google Scholar]
- 25.Lopes VS, et al. The Usher 1B protein, MYO7A, is required for normal localization and function of the visual retinoid cycle enzyme, RPE65. Hum Mol Genet. 2011;20(13):2560–2570. doi: 10.1093/hmg/ddr155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu XR, Udovichenko IP, Brown SDM, Steel KP, Williams DS. Myosin VIIa participates in opsin transport through the photoreceptor cilium. J Neurosci. 1999;19(15):6267–6274. doi: 10.1523/JNEUROSCI.19-15-06267.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhu DH, Wu J, Spee C, Ryan SJ, Hinton DR. BMP4 mediates oxidative stress-induced retinal pigment epithelial cell senescence and is overexpressed in age-related macular degeneration. J Biol Chem. 2009;284(14):9529–9539. doi: 10.1074/jbc.M809393200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Keane TM, et al. Mouse genomic variation and its effect on phenotypes and gene regulation. Nature. 2011;477(7364):289–294. doi: 10.1038/nature10413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jelcick AS, et al. Genetic variations strongly influence phenotypic outcome in the mouse retina. PloS One. 2011;6(7):e21858. doi: 10.1371/journal.pone.0021858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Malek RL, et al. Physiogenomic resources for rat models of heart, lung and blood disorders. Nat Genet. 2006;38(2):234–239. doi: 10.1038/ng1693. [DOI] [PubMed] [Google Scholar]
- 31.Pickrell JK, et al. Understanding mechanisms underlying human gene expression variation with RNA sequencing. Nature. 2010;464(7289):768–772. doi: 10.1038/nature08872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Harold D, et al. Genome-wide association study identifies variants at CLU and PICALM associated with Alzheimer’s disease. Nat Genet. 2009;41(10):1088–1093. doi: 10.1038/ng.440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Slonim DK. From patterns to pathways: gene expression data analysis comes of age. Nat Genet. 2002;32(suppl):502–508. doi: 10.1038/ng1033. [DOI] [PubMed] [Google Scholar]
- 34.Chu FF, Esworthy RS, Doroshow JH. Role of Se-dependent glutathoine peroxidases in gastrointestinal inflammation and cancer. Free Radic Biol Med. 2004;36(12):1481–1495. doi: 10.1016/j.freeradbiomed.2004.04.010. [DOI] [PubMed] [Google Scholar]
- 35.Conley YP, Gorin MB, Mah TS, Weeks DE, Ferrell RE. Glutathione peroxidase 3: Identification of promoter region polymorphisms and association with age-related maculopathy. Am J Hum Genet. 1999;65:A104. doi: 10.1086/302467. [DOI] [Google Scholar]
- 36.Weeks DE, et al. A full genome scan for age-related maculopathy. Hum Mol Genet. 2000;9(9):1329–1349. doi: 10.1093/hmg/9.9.1329. [DOI] [PubMed] [Google Scholar]
- 37.Morimoto RI. Proteotoxic stress and inducible chaperone networks in neurodegenerative disease and aging. Genes Dev. 2008;22(11):1427–1438. doi: 10.1101/gad.1657108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mehlen P, KretzRemy C, Preville X, Arrigo AP. Human hsp27, Drosophila hsp27 and human alpha B-crystallin expression-mediated increase in glutathione is essential for the protective activity of these proteins against TNF alpha-induced cell death. EMBO J. 1996;15(11):2695–2706. [PMC free article] [PubMed] [Google Scholar]
- 39.Sternberg P, et al. Glutathione in human plasma: Decline in association with aging, age-related macular degeneration, and diabetes. Free Radic Biol Med. 1998;24(5):699–704. doi: 10.1016/S0891-5849(97)00286-4. [DOI] [PubMed] [Google Scholar]
- 40.Castorina C, Campisi A, Digiacomo C, Sorrenti V, Russo A, Vanella A. Lipid peroxidation and antioxidant enzymatic systems in rat retina as a function of age. Neurochem Res. 1992;17(6):599–604. doi: 10.1007/BF00968789. [DOI] [PubMed] [Google Scholar]
- 41.Katz ML, Parker KR, Handelman GJ, Bramel TL, Dratz EA. Effects of antioxidant nutrient deficiency on the retina and retinal pigment epithelium of albino rats: a light and electron microscopic study. Exp Eye Res. 1982;34(3):339–369. doi: 10.1016/0014-4835(82)90082-3. [DOI] [PubMed] [Google Scholar]
- 42.Katz ML, Stone WL, Dratz EA. Fluorescent pigment accumulation in retinal pigment epithelium of antioxidant-deficient rats. Invest Ophthalmol Vis Sci. 1978;17(11):1049–1058. [PubMed] [Google Scholar]
- 43.Freund A, Orjalo AV, Desprez PY, Campisi J. Inflammatory networks during cellular senescence: causes and consequences. Trends Mol Med. 2010;16(5):238–246. doi: 10.1016/j.molmed.2010.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Detrick B, Hooks JJ. Immune regulation in the retina. Immunol Res. 2010;47(1–3):153–161. doi: 10.1007/s12026-009-8146-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Medzhitov R. Origin and physiological roles of inflammation. Nature. 2008;454(7203):428–435. doi: 10.1038/nature07201. [DOI] [PubMed] [Google Scholar]
- 46.Fortier A, Faucher SP, Diallo K, Gros P. Global cellular changes induced by Legionella pneumophila infection of bone marrow-derived macrophages. Immunobiology. 2011;216(12):1274–1285. doi: 10.1016/j.imbio.2011.06.008. [DOI] [PubMed] [Google Scholar]
- 47.Tuite A, Elias M, Picard S, Mullick A, Gros P. Genetic control of suceptibility to Candida albicans in susceptible A/J and resistant C57BL/6J mice. Genes Immun. 2005;6(8):672–682. doi: 10.1038/sj.gene.6364254. [DOI] [PubMed] [Google Scholar]
- 48.Karp CL, et al. Identification of complement factor 5 as a susceptibility locus for experimental allergic asthma. Nat Immunol. 2000;1(3):221–226. doi: 10.1038/79759. [DOI] [PubMed] [Google Scholar]
- 49.Singh B, Shinagawa K, Taube C, Gelfand EW, Pabst R. Strain-specific differences in perivascular inflammation in lungs in two murine models of allergic airway inflammation. Clin Exp Immunol. 2005;141(2):223–229. doi: 10.1111/j.1365-2249.2005.02841.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rothlin CV, Ghosh S, Zuniga EI, Oldstone MBA, Lemke G. TAM receptors are pleiotropic inhibitors of the innate immune response. Cell. 2007;131(6):1124–1136. doi: 10.1016/j.cell.2007.10.034. [DOI] [PubMed] [Google Scholar]
- 51.Wu ZH, Lauer TW, Sick A, Hackett SF, Campochiaro PA. Oxidative stress modulates complement factor H expression in retinal Pigmented epithelial cells by acetylation of FOXO3. J Biol Chem. 2007;282(31):22414–22425. doi: 10.1074/jbc.M702321200. [DOI] [PubMed] [Google Scholar]
- 52.Klein RJ, et al. Complement factor H polymorphism in age-related macular degeneration. Science. 2005;308(5720):385–389. doi: 10.1126/science.1109557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Xu HP, Chen M, Forrester JV. Para-inflammation in the aging retina. Prog Retin Eye Res. 2009;28(5):348–368. doi: 10.1016/j.preteyeres.2009.06.001. [DOI] [PubMed] [Google Scholar]
- 54.Hollyfield JG, et al. Oxidative damage-induced inflammation initiates age-related macular degeneration. Nat Med. 2008;14(2):194–198. doi: 10.1038/nm1709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Joachim SC, Bruns K, Lackner KJ, Pfeiffer N, Grus FH. Analysis of IgG antibody patterns against retinal antigens and antibodies to alpha-crystallin, GFAP, and alpha-enolase in sera of patients with “wet” age-related macular degeneration. Graefes Arch Clin Exp Ophthalmol. 2007;245(5):619–626. doi: 10.1007/s00417-006-0429-9. [DOI] [PubMed] [Google Scholar]
- 56.Thurman JM, et al. Oxidative stress renders retinal pigment epithelial cells susceptible to complement-mediated injury. J Biol Chem. 2009;284(25):16939–16947. doi: 10.1074/jbc.M808166200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Caspi RR. Ocular autoimmunity: the price of privilege? Immunol Rev. 2006;213:23–35. doi: 10.1111/j.1600-065X.2006.00439.x. [DOI] [PubMed] [Google Scholar]
- 58.Bird AC. Therapeutic targets in age-related macular disease. J Clin Invest. 2010;120(9):3033–3041. doi: 10.1172/JCI42437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Manolio TA, Brooks LD, Collins FS. A HapMap harvest of insights into the genetics of common disease. J Clin Invest. 2008;118(5):1590–1605. doi: 10.1172/JCI34772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Khalil AM, et al. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc Natl Acad Sci U S A. 2009;106(28):11667–11672. doi: 10.1073/pnas.0904715106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Guttman M, et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature. 2009;458(7235):223–227. doi: 10.1038/nature07672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Young TL, Matsuda T, Cepko CL. The noncoding RNA Taurine upregulated gene 1 is required for differentiation of the murine retina. Curr Biol. 2005;15(6):501–512. doi: 10.1016/j.cub.2005.02.027. [DOI] [PubMed] [Google Scholar]
- 63.Guttman M, et al. Ab initio reconstruction of cell type-specific transcriptomes in mouse reveals the conserved multi-exonic structure of lincRNAs. Nat Biotechnol. 2010;28(5):503–510. doi: 10.1038/nbt.1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Maeda A, et al. Primary amines protect against retinal degeneration in mouse models of retinopathies. Nat Chem Biol. 2012;8(2):170–178. doi: 10.1038/nchembio.759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Maeda A, et al. Role of photoreceptor-specific retinol dehydrogenase in the retinoid cycle in vivo. J Biol Chem. 2005;280(19):18822–18832. doi: 10.1074/jbc.M501757200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Maeda A, Maeda T, Sun W, Zhang H, Baehr W, Palczewski K. Redundant and unique roles of retinol dehydrogenases in the mouse retina. Proc Natl Acad Sci U S A. 2007;104(49):19565–19570. doi: 10.1073/pnas.0707477104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Imanishi Y, Gerke V, Palczewski K. Retinosomes: new insights into intracellular managing of hydrophobic substances in lipid bodies. J Cell Biol. 2004;166(4):447–453. doi: 10.1083/jcb.200405110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Chen Y, et al. Mechanism of all-trans-retinal toxicity with implications for Stargardt disease and age-related macular degeneration. J Biol Chem. 2012;287(7):5059–5069. doi: 10.1074/jbc.M111.315432. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.