Abstract
PolyA (pA) tail binding proteins (PABPs) control mRNA polyadenylation, stability and translation. In a purified system, S. cerevisiae PABPs, Pab1p and Nab2p, are individually sufficient to provide normal pA tail length. However, it is unknown how this occurs in more complex environments. Here we find that the nuclear exosome subunit Rrp6p counteracts the in vitro and in vivo extension of mature pA tails by the non-canonical pA polymerase Trf4p. Moreover, PABP loading onto nascent pA tails is controlled by Rrp6p; while Pab1p is the major PABP, Nab2p only associates in the absence of Rrp6p. This is because Rrp6p can interact with Nab2p and displace it from pA tails, potentially leading to RNA turnover as evidenced for certain pre-mRNAs. We suggest that a nuclear mRNP surveillance step involves targeting of Rrp6p by Nab2p-bound pA-tailed RNPs and that pre-mRNA abundance is regulated at this level.
Introduction
To become functional, nascent eukaryotic mRNAs are processed in the nucleus. The pA tail, bound by PABPs, is a key attribute of this maturation and required for mRNA nuclear export and cytoplasmic translation (Eckmann et al., 2011; Kuhn and Wahle, 2004; Lemay et al., 2010a). Nuclear biogenesis of pA tails by the pre-mRNA 3′end processing machinery involves endonucleolytic cleavage of the nascent transcript followed by the 3′addition of adenosyl (A) residues. The length of mature eukaryotic pA tails is remarkably well defined, reaching ~70–80 nt during in vitro polyadenylation reactions in S. cerevisiae and ~250 nt in human cell extracts (Eckmann et al., 2011; Lemay et al., 2010b). Similar tail lengths are observed in vivo. Deviations from this norm can have dramatic consequences and modulation of pA tail length via its trimming or re-adenylation contributes in a major way to the post-transcriptional control of gene expression (Eckmann et al., 2011). However, while much is now known about the factors responsible for cleavage and pA tail synthesis, the mechanism determining final tail length is not well described. Most information derives from mammals where A-addition initiates in a distributive mode until the growing tail reaches a length sufficient for binding of the first nuclear PABP (PABPN1). This factor interacts directly with human PAP thereby promoting a switch to processive elongation (Eckmann et al., 2011). PABPN1 also helps terminate polyadenylation when the mature tail length is reached, presumably by forming a multimeric structure prohibitory to further PAP-mediated synthesis. Whether polyadenylation in S. cerevisiae goes through similar phases is not known, but there is so far no evidence for a direct interaction between PABPs and Pap1p.
S. cerevisiae contains two major PABPs, Pab1p and Nab2p, which are at steady state predominantly cytoplasmic and nuclear, respectively. In sequence, Pab1p is homologous to mammalian cytoplasmic PABP (PABPC), whereas Nab2p does not share any discernable homology to PABPN1. Pab1p functions in translation and also interacts with the Pan2p/Pan3p containing PAN deadenylase complex, an interaction suggested to be important for pA tail length control (Brown and Sachs, 1998). Despite its steady-state cytoplasmic localization, Pab1p shuttles between nucleus and cytoplasm and interacts with the strictly nuclear pre-mRNA cleavage factor IA (CFIA) complex (Amrani et al., 1997; Brune et al., 2005; Minvielle-Sebastia et al., 1997). Nab2p also shuttles and has been implicated in mRNA nuclear export via interactions with mRNA export factors Yra1p, Mex67p and the nuclear-pore associated protein Mlp1p (Fasken et al., 2008; Hector et al., 2002; Iglesias et al., 2010). Nab2p is exported with mRNA to the cytoplasm where it is thought to be exchanged for Pab1p (Lemay et al., 2010b). Unlike Pab1p, no direct interactions between Nab2p and 3′end processing factors have been reported.
Although both Nab2p and Pab1p carry out nuclear and cytoplasmic functions, the significance of their differential steady-state localizations in relation to pA tail production is elusive (Lemay et al., 2010b). S. cerevisiae strains carrying mutation of either pab1 or nab2 accumulate hyperadenylated mRNA in vivo. However, both Pab1p and Nab2p are individually capable of providing correct pA tail length control in in vitro assays using highly purified 3′end processing factors (Viphakone et al., 2008). This suggests that Pab1p and Nab2p serve overlapping nuclear functions, and indeed, Pab1p overexpression rescues the lethal phenotype of a NAB2 deletion (Hector et al., 2002). Yet, both proteins are essential, do not share significant sequence homology and interact individually with unrelated nuclear proteins. Thus, Nab2p and Pab1p are likely to also have specialized in vivo functions that are not recapitulated by reductionist approaches. It is therefore interesting to note that the S. pombe homolog of human PABPN1, Pab2p, is not ascribed any role in pA tail maturation. Instead, Pab2p reportedly targets select RNAs for trimming or complete degradation by the nuclear RNA exosome (Lemay et al., 2010a; St-Andre et al., 2010; Yamanaka et al., 2010). This echoes a reported functional link between Nab2p and the S. cerevisiae RNA exosome, allowing for the auto-regulation of NAB2 mRNA levels (Roth et al., 2005). In this case, binding of Nab2p to an A26 element in the 3′UTR of its own primary transcript is proposed to elicit Rrp6p-dependent degradation.
Rrp6p is restricted to the nucleus, and as an inhabitant of the nuclear exosome, its activity is stimulated by the TRAMP complex, which contains a non-canonical pA polymerase subunit, either Trf4p or Trf5p (Kadaba et al., 2004; LaCava et al., 2005; Vanacova et al., 2005; Wyers et al., 2005). TRAMP adds A-tails that can aid access of the exosome to its targets. This role of tails in priming RNAs for degradation is in striking contrast to the stabilizing function of Pap1p-produced, and PABP-bound, mature pA tails. A more global link between the S. cerevisiae nuclear exosome/TRAMP and mRNA polyadenylation is revealed via a quality control checkpoint activated upon blocks to mRNA nuclear export. Here Rrp6p is required for retention of mRNA, and together with TRAMP, involved in the degradation of transcripts as a result of their inefficient polyadenylation by Pap1p (Rougemaille et al., 2007; Saguez et al., 2008).
Here, we disclose an unexpected role for Rrp6p in controlling pA tail length and PABP-status in S. cerevisiae. Rrp6p interacts physically with Nab2p and can displace this protein from nuclear pA tails both in vitro and in vivo. A consequence of such interaction is pre-mRNA decay and we propose that nuclear mRNP biogenesis is not only monitored at the level of polyadenylation efficiency but also at the level of PABP association. This may create kinetic control of mRNP transport and provide regulatory potential.
Results
Rrp6p contributes to pA tail length control
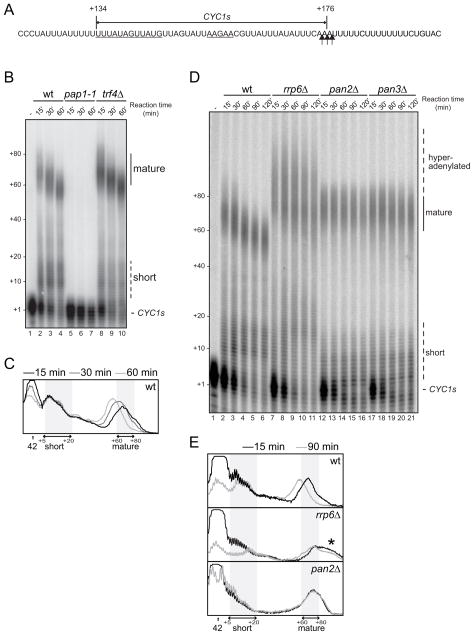
The polyadenylation phase of yeast mRNA 3′end processing can be studied separately in vitro using whole cell extract incubated with a ‘pre-cleaved’ substrate. We designed a short synthetic substrate (CYC1s) corresponding to the 42 nt upstream of the major pA-addition site of the CYC1 pre-mRNA (Figure 1A). When incubated in wild type (wt) yeast extract, 60–80 A’s were efficiently added to the 3′end of CYC1s (‘mature’, Figure 1B). These pA tails are produced by Pap1p as they were absent in extract containing a mutant form of the canonical pA polymerase Pap1p, but persisted in reactions using extract lacking Trf4p (Figure 1B). A similar tail length is also observed in reactions using longer substrates, and when measuring nascent tails in vivo. As observed with other substrates (Qu et al., 2009), a progressive pA tail shortening could be observed over the time course (Figure 1B, quantified in Figure 1C). Consistent with previous reports (Brown and Sachs, 1998; Qu et al., 2009), this can be attributed to deadenylation by the PAN complex (Figure 1D). We conclude that the CYC1s precursor recapitulates all major features of pA tail biogenesis.
Figure 1. Rrp6p is required for normal pA tail length.
(A) The sequence of the CYC1s substrate used in this study is indicated by a double-arrowed line above the relevant CYC1 3′UTR sequence. Numbering is relative to the CYC1 ORF stop codon. Polyadenylation efficiency- (left) and positioning- (right), elements are underlined. Major cleavage sites are marked by vertical arrows.
(B) CYC1s in vitro polyadenylation time course in wt, pap1-1 and trf4Δ extracts. Positions of precursor (CYC1s), short and mature polyadenylated species are indicated to the right and pA tail lengths estimated from the migration of DNA size markers are shown to the left of the gel.
(C) Phosphorimager lane scans from wt reactions shown in (B) carried out for 15 min (black), 30 min (dark gray) or 60 min (light gray). The position of the precursor (42 nt) is indicated by an arrow. Areas covering short (5–20 nt) and mature (60–80 nt) pA tail regions are marked by double arrows and shaded.
(D) CYC1s in vitro polyadenylation time course in wt, rrp6Δ, pan2Δ and pan3Δ extracts. Image labeling as in (B). ‘Hyperadenylated’ denotes excess adenylation over the normal mature (wt) length.
(E) Lane scans of 15 min (light gray) and 90 min (dark gray) time points of wt (top), rrp6Δ (middle) and pan2Δ (bottom) reactions from (D). Image labeling as in (C). The region of hyperadenylation is indicated by an asterisk.
Curiously, these reactions also revealed the appearance of shorter 5–20 nt pA tails (‘short’, Figure 1B). Such pA tails have, to our knowledge, not previously been characterized in yeast. The appearance of short and mature pA tails is, however, reminiscent of the distributive and processive phases of polyadenylation reported for human reactions (Eckmann et al., 2011; Lemay et al., 2010b). Since the distributive addition of A’s by the nuclear exosome cofactor TRAMP creates Rrp6p-sensitive substrates and since Rrp6p influences the outcome of bona fide polyadenylation reactions under conditions where the reaction is slowed (Callahan and Butler, 2010; Milligan et al., 2005; Saguez et al., 2008), we asked how rrp6Δ extract would impact on CYC1s polyadenylation. Short tails were less abundant and distributed more broadly during the time course in rrp6Δ relative to wt extract (Figure 1D, 15 and 90 min time points quantified in Figure 1E). The relevance of this result for pA tail biogenesis will be described elsewhere (manuscript in preparation). More surprisingly, mature tails were also affected by Rrp6p removal and were considerably longer than those produced in wt extract (‘hyperadenylated’, Figure 1D and 1E). In contrast to reactions in pan2Δ and pan3Δ extracts, the CYC1s tails made in the rrp6Δ extract were elongated at the first 15 min time point and then slowly trimmed during prolonged incubation (Figure 1D and 1E), suggesting that the PAN complex is active on these hyperadenylated products. We conclude that the mode of pA tail length regulation by Rrp6p is different from the PAN-dependent pathway.
Trf4p targeting of mature CYC1s pA tails is revealed in the absence of Rrp6p
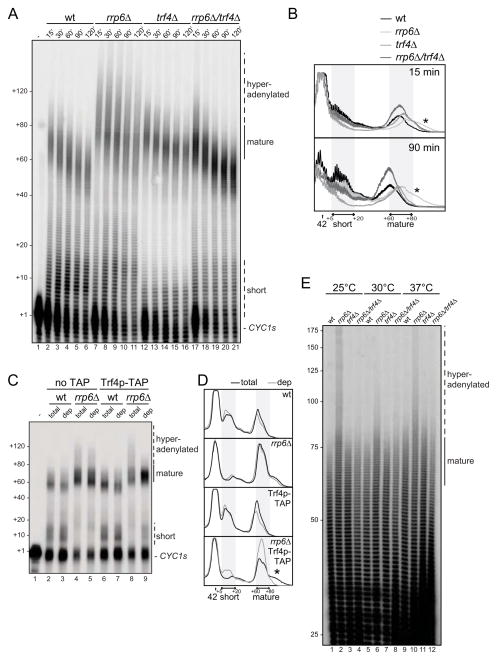
As Trf4p adenylation of exosome substrates often becomes detectable in rrp6Δ cells (Jensen and Moore, 2005), we tested whether hyperadenylation of mature CYC1s pA tails in rrp6Δ extract was mediated by Trf4p. Indeed, extract prepared from a rrp6Δ/trf4Δ double deletion strain did not produce the extended tails observed in the rrp6Δ background, but instead increased the signal intensity of mature tails by ~2 fold compared to wt extract (Figure 2A, 15 and 90 min time points quantified in Figure 2B). Comparable results were obtained when performing coupled cleavage-polyadenylation reactions on longer substrates derived from the 3′ends of CYC1, or two other RNAs, HSP104 and GAL7 (Figure S1A). Consistently, physical depletion (>90%) of TAP-tagged Trf4p from TRF4-TAP/rrp6Δ extract (Figure S1B) also eliminated most of the signal arising from hyperadenylated CYC1s species, and again the amount of mature tails increased relative to a reaction carried out in mock depleted wt extract (Figure 2C, quantified in Figure 2D). Importantly, Trf4p-TAP depletion did not alter the adenylation pattern of an otherwise wt extract. We therefore conclude that the hyperadenylation phenotype observed in rrp6Δ extracts derives from Trf4p extending otherwise mature pA tails produced by Pap1p. These extensions are presumably not observed in wt extract due to their rapid trimming back by Rrp6p.
Figure 2. In the absence of Rrp6p, Trf4p extends Pap1p-produced tails.
(A) CYC1s in vitro polyadenylation time course in wt, rrp6Δ, trf4Δ and rrp6Δ/trf4Δ extracts. Image labeling as in Figure 1D. A minor hyperadenylation phenotype is observed in trf4Δ extracts, which is likely due to lower levels of Pab1p in these extracts (data not shown), leading to decreased Pan2p/Pan3p deadenylation.
(B) Lane scans of the 15 min (top) and 90 min (bottom) reactions from wt (black), rrp6Δ (light gray), trf4Δ (medium gray) or rrp6Δ/trf4Δ (dark gray) extracts from (A). Image labeling as in Figure 1C and 1E.
(C) CYC1s in vitro polyadenylation reactions performed for 30 min in non-depleted (‘total’, lanes 6 and 8) or Trf4p-TAP depleted (‘dep’ lanes 7 and 9) TRF4-TAP and rrp6Δ/TRF4-TAP extracts. ‘Total’ and ‘dep’ versions of untagged extracts (lanes 2–5) were included as controls. Trf4p-TAP was depleted using magnetic beads coated with rabbit IgG (Figure S1A). Image labeling as in (A).
(D) Lane scans of reactions in (C) labeled as in (B). Total (dark colors) and depleted (light colors) lanes are overlaid in each panel.
(E) In vivo pA tail length measurements of total RNA purified from wt, rrp6Δ, trf4Δ or rrp6Δ/trf4Δ cells grown at 25°C, 30°C or temperature-shifted to 37°C for 60 min as indicated. pA tail lengths estimated from the migration of DNA size markers are shown to the left.
See also Figure S1.
As Trf4p/Rrp6p-dependent adenylation/trimming of RNA 3′ends has not previously been reported for mRNAs, it was important to examine whether such activities exist in vivo. To this end, we 3′end radiolabeled total RNA from wt, rrp6Δ, trf4Δ and rrp6Δ/trf4Δ strains and measured pA tail lengths globally by RNaseA and T1 digestion to leave only pA stretches. The size distribution of labeled tails with a maximum length of ~80 A’s in wt cells is consistent with the major part of this signal deriving from mRNA polyadenylated by Pap1p (Figure 2E). Like in the in vitro polyadenylation reactions, maximum tail length increased in rrp6Δ cells at all temperatures tested, and importantly, this extension was reversed in the rrp6Δ/trf4Δ double deletion background. The notion that mRNAs can be hyperadenylated in the absence of Rrp6p was confirmed by an alternative RT-PCR based method for the individual and highly expressed ENO2, PGK1 and RPL28 transcripts (Figure S1C). Although effects appear modest, they are significant as only a minor (nuclear) pool of each species contributes to the phenotype. Taken together, these data suggest that our in vitro results reflect events occurring in vivo. Moreover, they confirm that Trf4p can act on mature mRNA pA tails and that this is normally counteracted by Rrp6p.
Nab2p is excluded from pA tails in a Rrp6p-dependent manner
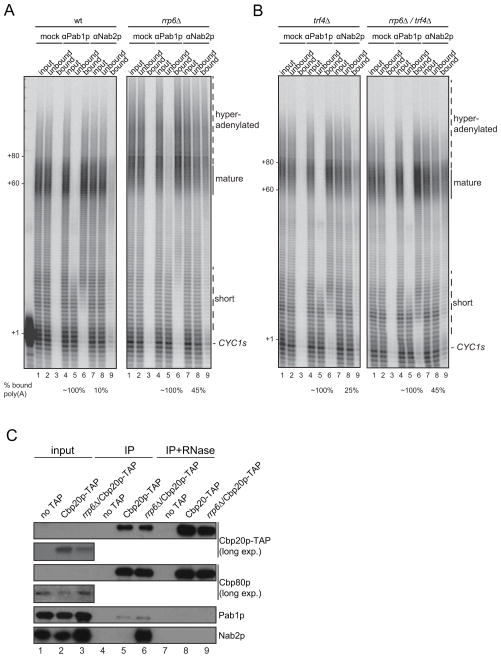
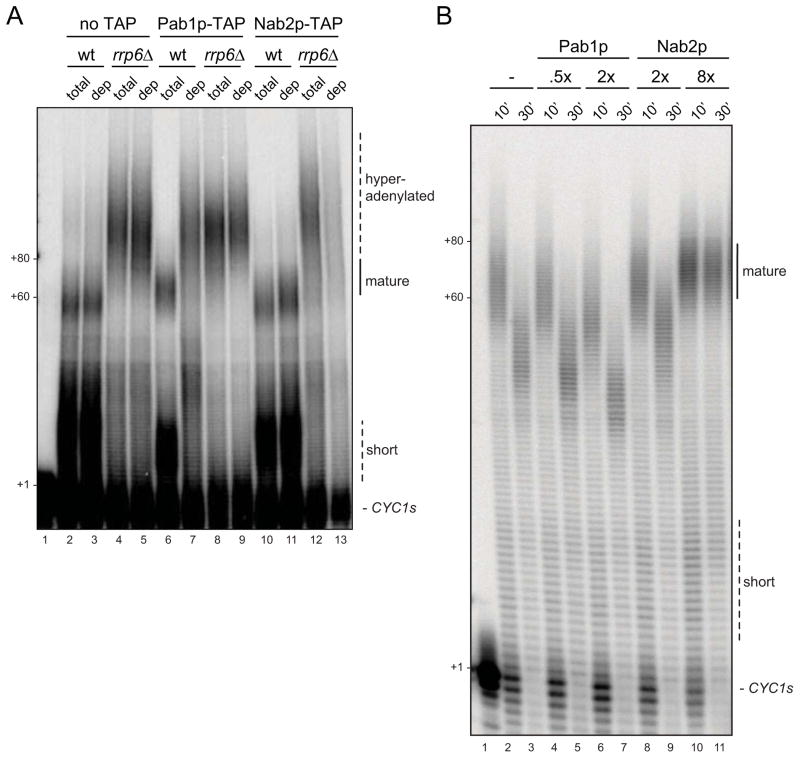
Our results suggest that mature pA tails are targeted by PAN, while Rrp6p serves to remove Trf4p-dependent extensions. As pA tail length is dictated by the RNP conformation provided by PABPs, we next examined how Pab1p and Nab2p impact CYC1s pA tail biogenesis and length control. To this end, we first measured PABP incorporation into pA tail RNPs by conducting immunoprecipitation (IP) assays using Pab1p or Nab2p antibodies on a polyadenylation reaction terminated after 30 min. Assays were carried out in wt or rrp6Δ extracts after which co-precipitated RNA and Pab1p/Nab2p proteins were analyzed by denaturing PAGE and western blotting analysis, respectively. IPs from wt extract revealed that mature-length and most short pA tails are bound by Pab1p, whereas Nab2p binding efficiency is ~10 fold lower (Figure 3A, left panel compare lanes 6 and 9). Interestingly, the Pab1p antibody pelleted all pA-containing material except for RNAs harboring tails shorter than ~10 A’s, which were instead found in the unbound fraction (Figure 3A, left panel compare lanes 4, 5 and 6). This is consistent with published data that Pab1p can only bind pA tails provided they have an adequate size of ~10–12 A’s (Sachs et al., 1987). In contrast to the situation for wt extract, 45% of total pA tail RNP produced in rrp6Δ extract could be precipitated by the Nab2p antibody, whereas the Pab1p antibody precipitated tails from this reaction at full efficiency (Figure 3A, right panel). Thus, these results surprisingly demonstrate that Nab2p is only incorporated into pA tail RNPs when Rrp6p is absent. However, in these conditions Nab2p incorporation is pronounced. In fact the measured 45% IP-efficiency is probably an underestimate as western blotting analysis of total, unbound and precipitated material demonstrated that despite the quantitative pull out of Pab1p and Nab2p by their respective antibodies only Pab1p was recovered at stoichiometric amounts in the bound fraction whereas some Nab2p protein was lost during washing (Figure S2). In any case, the data show that Pab1p and Nab2p can co-exist on a considerable number of the RNPs produced in rrp6Δ extracts, whereas pA tails produced in wt conditions are foremost bound by Pab1p. NAB2 mRNA is auto-regulated in an Rrp6p-dependent manner (Roth et al., 2005), resulting in slightly elevated (~1.5 fold) Nab2p levels in rrp6Δ compared to wt extracts (data not shown). However, increased Nab2p/pA tail association in the absence of Rrp6p is not simply explained by elevated Nab2p levels. This is because a similar result was obtained when employing an Rrp6p-insensitive Nab2p-TAP fusion as the sole source of Nab2p (data not shown).
Figure 3. Efficient Nab2p incorporation into pA tail RNP only occurs in the absence of Rrp6p.
(A) CYC1s in vitro polyadenylation reactions carried out for 30 min in wt or rrp6Δ extracts were terminated by the addition of excess dATP and subjected to IP by magnetic beads containing no (mock), α-Pab1p or α-Nab2p antibody. RNA was collected from input, unbound and bead-bound fractions and resolved by denaturing PAGE. Quantification of relative IP efficiencies of mature and hyperadenylated RNA species (see Experimental Procedures) are shown below the images labeled as in Figure 2A. The first lane (not numbered) was loaded with an equivalent amount of CYC1s precursor.
(B) Same as in (A) but using trf4Δ and rrp6Δ/trf4Δ extracts for polyadenylation reactions.
(C) Pab1p and Nab2p association with Cbp20p-bound RNPs in vivo. Cbp20p-TAP was purified using magnetic beads coated with rabbit IgG from wt or rrp6Δ cells and eluates were examined by western blotting analysis for their contents of Cbp20p-TAP, Cbp80p, Pab1p and Nab2p. ‘No TAP’ denotes a negative control strain carrying un-tagged Cbp20p. 0.5% of the total input (lanes 1–3) was loaded next to IP reactions carried out without (lanes 4–6) or with (lanes 7–9) addition of an RNase cocktail. Longer exposures of Cbp20p-TAP and Cbp80p blots are shown to reveal the presence of these proteins in input lanes.
See also Figure S2.
To test whether the Trf4p-specific adenylation of mature pA tails may be the feature that allows Nab2p to enter these RNPs, we repeated the Pab1p and Nab2p IP experiments on polyadenylation reactions performed in trf4Δ or rrp6Δ/trf4Δ extracts. As was observed for wt extract, Nab2p was less abundant on pA RNPs when the reaction was conducted in the Rrp6p-containing trf4Δ extract (Figure 3B, left panel). In rrp6Δ/trf4Δ extract, Nab2p and Pab1p associated with pA tails with efficiencies comparable to those observed for reactions in rrp6Δ extract (Figure 3B, right panel). We therefore conclude that Nab2p is perfectly capable of associating with mature-sized pA tails, but only if Rrp6p is absent. Interestingly though, even in the absence of Rrp6p, Nab2p does not associate with short tails (Figure 3B); i.e. binding of short products in the rrp6Δ/trf4Δ extract was restricted to Pab1p. This suggests a strong preference for recruitment of Pab1p during the early phase of polyadenylation, perhaps via a CFIA-Pab1p interaction (Amrani et al., 1997; Minvielle-Sebastia et al., 1997).
Again, it was important to examine whether Rrp6p-dependent exclusion of Nab2p from pA-tailed RNA could be recapitulated in vivo. To focus on early events in pA tail biogenesis/metabolism, we took advantage of a previously published approach (Neil et al., 2009) to specifically IP nuclear RNPs by virtue of their association with Cbp20p of the nuclear cap-binding complex (CBC). Pull down of Cbp20p-TAP revealed a robust, and RNA-independent, association with Cbp80p of the CBC, an interaction which was equally efficient in wt and rrp6Δ environments (Figure 3C, compare lanes 5 and 6). Importantly, however, while Pab1p levels were unaffected in pA-RNPs purified from wt vs rrp6Δ cells, Nab2p levels dramatically increased upon RRP6 deletion. We note that the relatively low level of precipitated Pab1p is likely because this protein is very abundant and also interacts with cytoplasmic mRNAs. Both Pab1p and Nab2p association with Cbp20p-TAP was lost upon RNase treatment (Figure 3C, compare lanes 5 and 6 to 8 and 9), confirming that the assay monitors binding of Pab1p and Nab2p to nuclear RNPs and not direct protein-protein interactions. We conclude that Rrp6p also excludes Nab2p from pA-RNPs in vivo.
Rrp6p releases Nab2p from pA tails
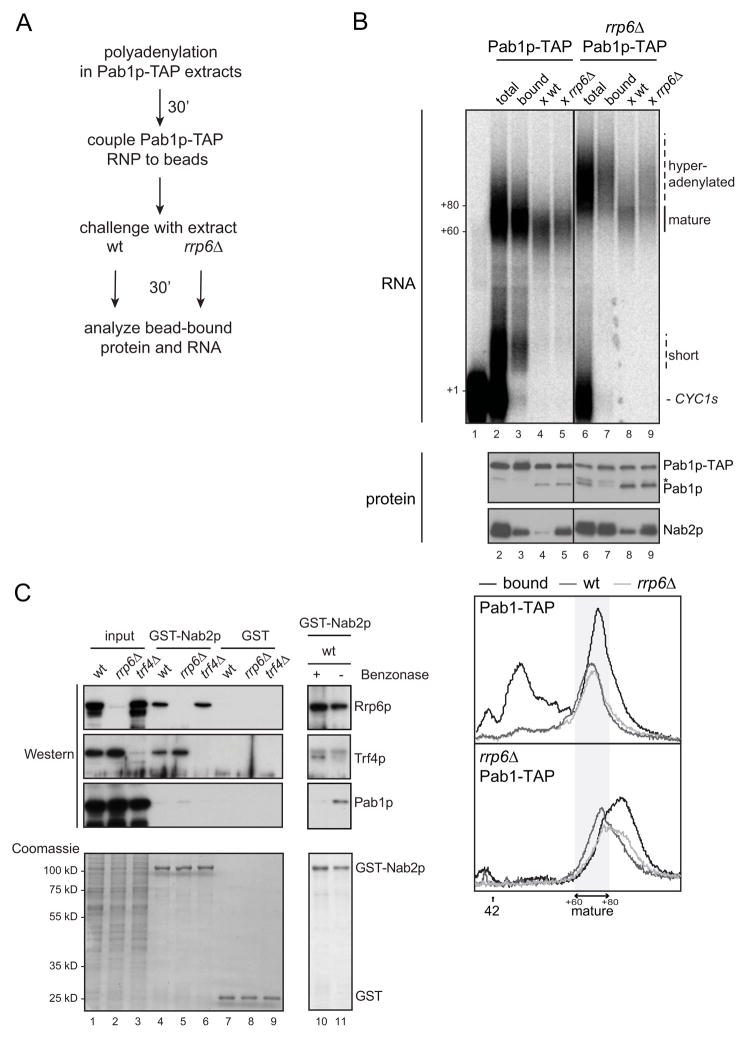
Why are Nab2p containing polyadenylated RNPs not detectable in the presence of Rrp6p? Does Rrp6p simply prevent Nab2p-tail association or can it remove Nab2p from pA RNPs? To address this question, we used a TAP-affinity column to isolate Pab1p-TAP bound CYC1s RNPs from a 30 min polyadenylation reaction performed in either PAB1-TAP or rrp6Δ/PAB1-TAP extracts. These RNPs were then treated with either wt or rrp6Δ extracts as outlined in Figure 4A. As expected, RNPs purified from the wt reaction primarily contained normal-length tails as well as more Pab1p-TAP relative to Nab2p as compared to the input extract (Figure 4B, compare lanes 2 and 3, ‘RNA’ and ‘protein’ panels, respectively). Conversely, RNPs precipitated from the rrp6Δ background harbored hyperadenylated tails and a higher Nab2p/Pab1p-TAP ratio (Figure 4B, compare lanes 6 and 7; and lanes 3 and 7, in ‘RNA’ and ‘protein’ panels, respectively). After the subsequent 30 min challenge of these wt or rrp6Δ pA-RNPs with either wt or rrp6Δ extract, tail lengths and Nab2p/Pab1p levels of bead-bound RNPs were assessed. Tails of RNPs purified from the wt background were trimmed to an equal extent during the 30 min chase period whether challenged with wt or rrp6Δ extract (Figure 4B ‘RNA’, lanes 4 and 5; quantified in bottom panels). In contrast, hyperadenylated tails contained in RNPs purified from the rrp6Δ reaction were only efficiently trimmed when exposed to wt extract (Figure 4B ‘RNA’, lanes 8 and 9; quantified in bottom panels). This underscores that Rrp6p specifically acts on these tail extensions. Interestingly, concomitant with tail trimming, pA tail-bound Nab2p was significantly reduced when the pA RNP was challenged with wt extract, but only to a small extent when exposed to rrp6Δ extract (Figure 4B ‘protein’, compare lanes 8 and 9). Moreover, the Nab2p residing in wt pA-RNPs was also selectively removed by wt extract (Figure 4B ‘protein’, compare lanes 4 and 5). These results demonstrate that Rrp6p can displace Nab2p from pA tails.
Figure 4. Rrp6p can displace pre-loaded Nab2p from pA tails.
(A) Schematic outline of the experiment analyzed in (B). See text for details.
(B) CYC1s polyadenylation reactions were carried out for 30 min in Pab1p-TAP or rrp6Δ/Pab1p-TAP extracts and then incubated with rabbit IgG Dynabeads for an additional 15 min to capture Pab1p-TAP (‘total’ lanes 2 and 6). Pab1p-TAP containing RNPs were then isolated (‘bound’, lanes 3 and 7) and aliquots of bead-bound material were incubated for 30 min in a chase reaction with either wt (lanes 4 and 8) or rrp6Δ (lanes 5 and 9) extracts. RNA isolated from the different steps was visualized by denaturing PAGE (‘RNA’) with image labeling as in Figure 1B, whereas the content of Pab1p-TAP and Nab2p in the samples was analyzed by western blotting analysis (‘protein’). The asterisk at the anti-Pab1p western image indicates a putative degradation fragment of Pab1p-TAP. Bottom: Scans of the indicated lanes as in Figure 1C and 1E.
(C) Pull-down of Nab2p interaction partners. GST-Nab2p or GST (negative control) was immobilized on Sepharose columns and incubated with wt, Δrrp6 or Δtrf4 cell extract as indicated. Columns were washed and eluates were probed by western blotting analysis for their presence of Rrp6p, Trf4p and Pab1p (top), and by Coomassie staining to control for equal elution of GST-Nab2p and GST between experiments (bottom). Similar fractions of input material (lanes 1–3), and eluate (lanes 4–9) were loaded. The GST-Nab2p IP of wt extract was carried out in parallel with (lane 10) or without (lane 11) addition of the nuclease Benzonase. Note that the RNA-mediated binding of Pab1p to GST-Nab2p is lost upon Benzonase addition, whereas the interactions of Rrp6p and Trf4p are not. An unspecific band migrating slightly lower than Trf4p in the Trf4p panel is marked with an asterisk.
See also Figure S3.
To characterize the nature of Rrp6p-dependent Nab2p displacement, we also analyzed its requirement for Rrp6p exonucleolytic activity. This was achieved in a similar experiment by including extract prepared from cells harboring the catalytically dead Rrp6p(D238A) mutant (Assenholt et al., 2008) as their only source of Rrp6p. Notably, rrp6-D238A extract also produced hyperadenylated CYC1s tails (Figure S3A). More importantly, this extract, like rrp6Δ extract, failed to efficiently trim hyperadenylated tails isolated from rrp6Δ extract and did not displace Nab2p (Figure S3B). Thus, displacement of Nab2p requires the exonuclease activity of Rrp6p, possible via its trimming of the pA tail.
Finally, we investigated whether the ability of Rrp6p to displace Nab2p could be based on a physical interaction between these factors. To this end Nab2p was expressed in E. coli as a GST-fusion protein, coupled to glutathione-beads and incubated with yeast extracts prepared from wt, rrp6Δ or trf4Δ cells. Subsequent western blotting analysis revealed that both Rrp6p and Trf4p could be retrieved from extracts by GST-Nab2p, but not by the GST control (Figure 4C). Both Rrp6p and Trf4p interactions with GST-Nab2p were RNA-independent (Figure 4C, compare lanes 10 and 11), and interestingly, binding of both factors did also occur when the other was genetically depleted. Thus, Rrp6p and Trf4p can interact with Nab2p independent of each other.
Nab2p is dispensable for pA tail biogenesis in yeast extract
The above results imply that Pab1p and Nab2p have very different roles in pA tail formation, and that it may be possible to synthesize mature tails in the absence of Nab2p. To address this idea directly, we depleted either Pab1p-TAP or Nab2p-TAP proteins from wt or rrp6Δ extracts and performed CYC1s polyadenylation reactions. Depletions were efficient and reduced levels of Pab1p-TAP and Nab2p-TAP to ~90% and ~95%, respectively (Figure S4A and S4B). Removal of Pab1p from wt extracts caused hyperadenylation of mature pA tails (Figure 5A, compare lanes 6 and 7), whereas Pab1p-TAP depletion from rrp6Δ extract resulted in little change in the CYC1s pA tail profile (Figure 5A, compare lanes 8 and 9). Importantly, hyperadenylation upon Pab1p-TAP depletion was also observed when employing rrp6Δ/trf4Δ extracts (Figure S4C). Excess adenylation is therefore not due to Trf4p and the phenotype is distinct from the one observed in rrp6Δ extracts. Moreover, pA tails are considerably longer than in pan2Δ or pan3Δ mutant extracts (Figure 1D), and pA tail length is rescued by adding back recombinant Pab1p (Figure S4D). Consistent with previous reports, we therefore suggest that Pab1p-depletion not only prevents PAN-dependent trimming, but also causes a failure of Pap1p in terminating at the mature pA tail length (Brown and Sachs, 1998; Dheur et al., 2005). In contrast, Nab2p-TAP depletion had no effect on CYC1s pA tail length in the wt extract, but instead led to decreased processing efficiency in the rrp6Δ context (Figure 5A, compare lanes 10 and 11, and 12 and 13). These results are consistent with our Pab1p and Nab2p pA tail binding profiles and suggest that Pab1p is the major PABP determinant for the biogenesis of pA RNP; at least in the context of yeast extract (see Discussion). Nab2p, on the other hand, only binds, and functionally contributes, to polyadenylation reactions in an rrp6Δ context. This argues for an alternative role of Nab2p in pA tail biogenesis/metabolism.
Figure 5. Nab2p is not required for normal pA tail biogenesis.
(A) CYC1s in vitro polyadenylation reactions carried out for 30 min in wt or rrp6Δ extracts before (total) or after (dep) no TAP-, Pab1p-TAP- or Nab2p-TAP-depletion as indicated. Pab1p-TAP and Nab2p-TAP proteins were depleted using rabbit IgG antibody. Image labeling as in Figure 1B.
(B) CYC1s in vitro polyadenylation reactions carried out for 10 or 30 min in wt extracts supplemented with buffer (lanes 2 and 3), 0.5-fold (lanes 4 and 5) or 2-fold (lanes 6 and 7) excess recombinant Pab1; or 2-fold (lanes 8 and 9) or 8-fold (lanes 10 and 11) excess recombinant Nab2 as indicated.
See also Figure S4.
One likely consequence of Nab2p binding to pA tails is a delay in PAN trimming. Indeed, careful inspection of pA tail profiles suggested slightly decreased deadenylation kinetics in rrp6Δ compared to wt extracts (e.g. Figure 1D and 1E). To further corroborate this, we produced and purified recombinant Pab1p and Nab2p (Figure S4E and S4F), added either factor to a wt polyadenylation reaction and compared tail lengths at the 10 and 30 min time points. Addition of recombinant Pab1p resulted in a concentration-dependent tail shortening at both time points (Figure 5B, lanes 2–6). Notably, excess Pab1p did not cause tail shortening in pan2Δ extracts (data not shown), underscoring published data that Pab1p stimulates PAN-dependent deadenylation (Brown and Sachs, 1998; Dheur et al., 2005). In contrast, 2-fold excess recombinant Nab2p caused a delay in tail shortening at the 30 min time point and completely abolished PAN deadenylation when added in 8-fold excess over Nab2p levels contained in the extract (Figure 5B, lanes 8–11). These findings demonstrate yet another differential behavior of Pab1p- vs Nab2p-bound pA tails. Moreover, we speculate that the somewhat unexpected low levels of pA-tailed material in Nab2p-depleted rrp6Δ extract (Figure 5A, lane 13) may be due to unchallenged Pab1p/PAN trimming.
Nab2p and Rrp6p partake in pre-mRNA turnover
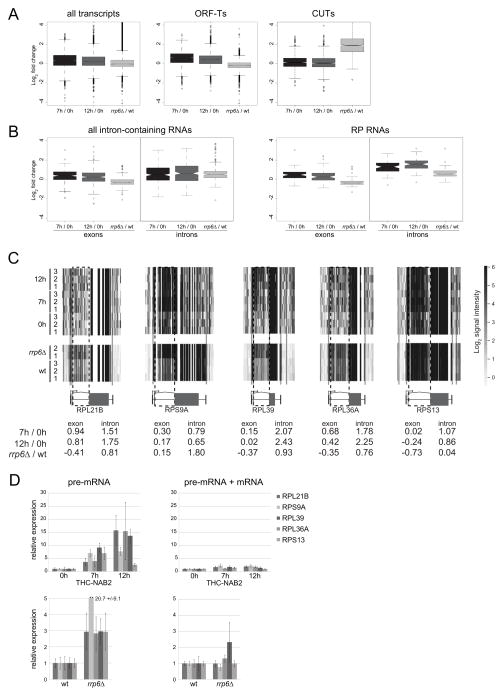
The close ties between Nab2p and Rrp6p reported here, taken together with the established role of Rrp6p as a nuclear RNA decay factor, suggested a role for Nab2p as a co-factor for Rrp6p-directed RNA turnover. To address this prediction, we utilized commercially available cells containing the NAB2 gene under a tetracycline-repressible promoter and harvested triplicate sets of RNA samples for tiling array analysis before (‘0h’) and after 7h (‘7h’) or 12h (‘12h’) of transcriptional repression. These repression conditions resulted in Nab2p levels of around 20% and below 5% of ‘0h’ levels, respectively (Figure S5A). Nab2p-depletion led to a small, but significant, overall increase in transcript levels (Figure 6A, left panel, median log2-fold change: 0.43 and 0.30 for 7h/0h and 12h/0h ratios, respectively), which is mostly due to a minor global upregulation of mRNAs (ORF-Ts) (Figure 6A, compare left panel and middle panel, data not shown). This effect was not observed when comparing tiling array analysis of total RNA harvested from rrp6Δ and wt cells; and details of this result will be described elsewhere. Conversely, the strongly Rrp6p-sensitve cryptic unstable transcripts (CUTs) were only marginally upregulated upon Nab2p-depletion and actually below the transcriptome-wide average (median log2-fold change: 0.10 and 0.06 for 7h/0h and 12h/0h ratios, respectively; Figure 6A, right panel). Thus, it appears that Nab2p affects certain RNAs Rrp6p-independently and Rrp6p-mediated decay does not generally require Nab2p. We focused on identifying transcripts affected by Nab2p-dependent recruitment of Rrp6p and therefore compared features responding both to Nab2p-depletion and Rrp6p-deletion. Rrp6p is implicated in pre-mRNA turnover (Bousquet-Antonelli et al., 2000). Nab2p is directly associated with the pre-mRNA retention factor Mlp1p (Fasken et al., 2008) and the S. pombe nuclear PABP, Pab2p, has recently been implied in the degradation of pre-mRNA (Lemieux et al., 2011). We therefore inspected intron-containing genes separately and observed increased intron signals from numerous genes upon Nab2p-depletion (Figure 6B left panels, compare ‘exons’ vs. ‘introns’ signals). This was particularly evident for 72 detectable introns derived from ribosomal protein-encoding (RP) genes. Of these, 47 were more than 2-fold upregulated and 64 were increased 1.5-fold at both the 7h and 12h time points (Figure 6B right panels, compare ‘exons’ vs. ‘introns’ signals; Figure 6C and Table S1). A more modest, but still very significant, upregulation of intronic signal was also apparent when comparing ‘rrp6Δ’ to ‘wt’ samples; 10 and 35 introns were more than 2-fold and 1.5-fold upregulated, respectively (Table S1). Six of these 10 most upregulated introns were also more than 2-fold increased in the Nab2p-depleted samples (for individual examples, see Figure 6C). Importantly, levels of exonic regions of these pre-mRNAs did generally not change compared to array-averages and mRNAs lacking introns. In contrast, probes spanning intron-exon junctions detected increases similar to intronic probes, arguing that elevated levels of pre-mRNAs, and not of excised introns, is the basis for the observed differences (Figure S6B). This notion was confirmed for five pre-mRNAs by quantitative RT-PCR using amplicons spanning exon-intron junctions (Figure 6D). Taken together, these data strongly suggest that Nab2p, in addition to its proposed functions in mRNA export and pA tail length control, also contributes to lowering pre-mRNA levels, at least in part by recruiting Rrp6p.
Figure 6. Nab2p and Rrp6p regulate pre-mRNA levels.
(A) and (B) Boxplots (see Supplemental Experimental Procedures) of global expression changes of indicated transcript classes identified by tiling microarray analysis. Individual plots depict log2-fold expression changes comparing total RNA from THC-NAB2 cells depleted for 7h (black) or 12h (dark gray) relative to non-depleted (0h) samples, as well as total RNA from rrp6Δ relative to wt (light gray) cells. Note that Nab2p-depletion data cDNA was obtained from total RNA, whereas pA-enriched RNA was used for wt and rrp6Δ samples. Hence, signal values for most transcripts are lower for THC-NAB2 than for rrp6Δ samples. ‘All transcripts’ include exons and introns of protein-encoding transcripts as well as non-coding transcripts. Panel ‘ORF-T’ (top, middle panel) includes only exons of protein-encoding transcripts (ORF-Ts). RP: Ribosomal protein.
(C) Screenshots of tiling array tracks for five RP genes, displaying normalized probe signal intensities of individual probes (gray-scaled as shown on the right) of three independent biological replicates of THC-NAB2 12h, 7h and 0h samples as well as duplicates of rrp6Δ and wt samples (see text for details). Position of introns and transcription directions are indicated by dashed boxes and arrows, respectively. Vertical lines indicate refer to transcript boundaries. Calculated log2-fold changes for displayed exonic and intronic RNA are shown at the bottom.
(D) Validation of results displayed in (C) using oligo(dT)-directed qPCR with amplicons spanning exon-intron junctions (‘pre-mRNA’, left panels) or amplicons entirely within exon2 (‘mRNA+pre-mRNA’, right panels). Expression values depicted are normalized to the intron-less mRNA PMA1 and to the 0h time point for the THC-NAB2 strain (top panel) or to the wt control in the bottom panel. Error bars are from triplicate PCRs of THC-NAB2 culture 1 (similar results were confirmed for culture 2 and 3, data not shown) and for untreated wt and rrp6Δ cultures.
Discussion
An emerging theme in mRNA biology is the somewhat paradoxical concept that the processing and assembly of mRNA into mRNP is often coupled to degradative activities, a relationship suggested to be key in establishing efficient quality control during mRNP biogenesis (Houseley and Tollervey, 2009). For example, it has long been known that S. cerevisiae Pab1p recruits 3′-5′ exonucleolytic activity in the form of the PAN deadenylase complex (Brown and Sachs, 1998), the exact in vivo role of which is not clear. In this paper, we demonstrate that this circumstance extends to Rrp6p interacting with the other major S. cerevisiae PABP, Nab2p.
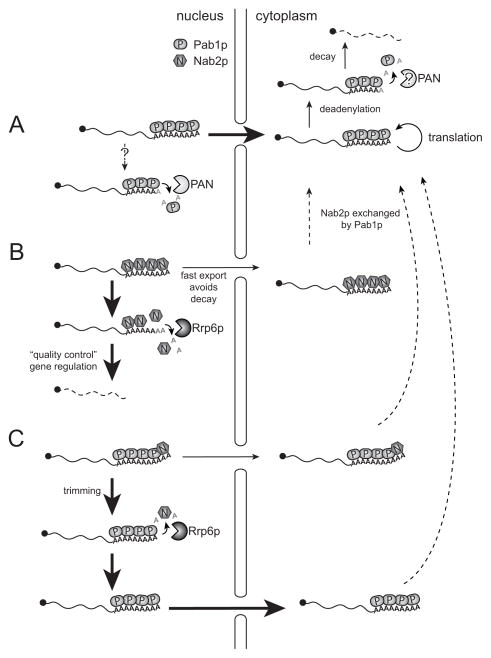
Our work shows that in the absence of Rrp6p, otherwise mature pA tails are extended in vitro and in vivo by Trf4p, the PAP subunit of the nuclear TRAMP complex. Absence of Rrp6p also allows Nab2p binding to the growing pA tail both in vitro and in vivo. This extra Nab2p can be removed if mRNPs are challenged with extract containing Rrp6p. We also found evidence for a physical interaction between Nab2p and Rrp6p, suggesting that Nab2p provides a critical platform for Rrp6p recruitment to pA tails. As the ensuing displacement of Nab2p requires Rrp6p to be exonucleolytically active, we propose that pA tail deadenylation eventually leads Nab2p to detach. Based on our findings, we propose that Rrp6p can impact mRNP fate at the level of PABP/pA tail association in the nucleus (Figure 7).
Figure 7. Models of functional significance of Nab2p-Rrp6p targeting.
Nuclear pA-tails are either loaded with Pab1p (A), Nab2p (B) or a mixture of the two (C).
(A) In wt cells, the majority of mRNA pA tails are Pab1p-bound and destined for nuclear export and cytoplasmic translation. Pab1p recruits the PAN deadenylase complex, but the location in which this interaction is functionally significant is presently unclear (indicated by question marks). The influence of Pab1p and Nab2p on the activity of the major deadenylase activity from the Ccr4/NOT complex (not shown) is not known.
(B) Nab2p-bound tails may escape nuclear targeting by Rrp6p by rapid export. In the cytoplasm, Nab2p is exchanged for Pab1p. If export is delayed, Rrp6p is recruited for complete mRNA destruction. This pathway is involved in quality control and gene expression regulation.
(C) Pab1p-decorated nuclear pA tails harboring Nab2p at their 3′ends can target Rrp6p for trimming and pA tail length control. Symbols: Pab1p (oval), Nab2p (hexagon), PAN complex (light pacman symbol), Rrp6p (dark pacman symbol).
See text for further details.
What might be the purpose of an Rrp6p-dependent equilibrium between Nab2p addition and removal from nuclear pA tails? Our observation that mature pA tails are still produced upon Nab2p depletion, whereas removal of Pab1p results in aberrantly long pA tails (Figure 5A), positions Pab1p as the main determinant for creation of a normal pA tail, seemingly offering the most direct route to final mRNP maturation, nuclear export and cytoplasmic translation (Figure 7A). Nonetheless, both PABPs are likely capable of contacting export competent mRNP as e.g. suggested by the interaction of Nab2p with mRNA export factors Yra1p and Mex67p (Fasken et al., 2008; Hector et al., 2002; Iglesias et al., 2010). However, Rrp6p recruitment by Nab2p may also present an opportunity to elicit nuclear RNA decay (Figure 7B) or trimming (Figure 7C). An extended nuclear exposure due to mRNP disability has been suggested to represent a kinetic quality control check point (Libri, 2010; Lemieux et al., 2011), and the Nab2p-Rrp6p link could act as such at the level of PABP/pA tail configuration. In fact, an mRNP proofreading step in S. cerevisiae occurs at the nuclear periphery (Tutucci and Stutz, 2011). The incorporation of Nab2p with mRNA export factors into mRNP at this site would therefore provide an opportunity for the destruction of incapacitated or otherwise slowly maturing mRNP (Figure 7B). Our observed stabilization of pre-mRNAs upon Nab2p-depletion is consistent with this idea as these transcripts are restricted to cell nuclei and known to be targeted by nuclear decay factors, including Rrp6p (Bousquet-Antonelli et al., 2000; Domenico Libri personal communication). Since we observe more and stronger effects on pre-mRNA accumulation by depleting Nab2p than deleting Rrp6p, it is possible that Nab2p also operates via alternative decay pathways. We consider it unlikely, however, that higher pre-mRNA levels arise from inefficient splicing as corresponding mRNA levels are not decreased by Nab2p-depletion and because Nab2p has no ascribed role in this process. Instead, we note that nuclear retention of splicing-deficient mRNA is mediated by the Nab2p-interacting Mlp1/2p proteins (Galy et al., 2004). Nab2p may therefore be a surveillance factor providing a direct pathway for targeting Mlp-anchored nuclear-retained pre-mRNA for turnover. Such an activity could also generally serve to remove excess ‘overproduced’ pre-mRNA from normally growing cells, providing a reservoir of transcript for rapid post-transcriptional regulation of gene expression in case of changing conditions and as reported for several links between RNP factors and degradation enzymes (Figure 7B; Muhlemann and Jensen, 2012).
Removal of excess primary transcript is also evident during the Nab2p/Rrp6p-dependent autoregulation of NAB2 mRNA levels (Roth et al., 2005). Here NAB2 mRNA 3′end formation occurs in a non-conventional manner that may allow enough time for Nab2p-Rrp6p targeting to occur. In addition, this type of 3′end formation likely disfavors the involvement of Pab1p, further sensitizing the transcript to Nab2p-directed exosomal-decay. A potentially similar mechanistic principle was recently described for Pab2p/Rrp6p-mediated RNA control in the fission yeast S. pombe. Here, modulation of pre-mRNA stability during changing growth conditions regulates levels of many intron-containing genes (Lemieux et al., 2011) and also serves to ensure the selective elimination of an estimated >20 meiotic mRNAs during vegetative growth (Lemay et al., 2010a; St-Andre et al., 2010; Yamanaka et al., 2010).
Given the huge regulatory potential, it is important to consider to which extent and under which circumstances Pab1p and Nab2p bind nuclear pA tails. At first glance, Nab2p association appears to be favored because of its higher steady-state concentration in the nucleus and the similar binding affinities for pA sequence (Hector et al., 2002). However, Pab1p interacts with the polyadenylation co-factor CFIA, which may facilitate Pab1p hand off to the growing tail until a mature length is reached (Amrani et al., 1997; Minvielle-Sebastia et al., 1997). Thereafter, polyadenylation may continue in a distributive manner, using PAP as described for the mammalian reaction (Eckmann et al., 2011), or Trf4p as implied by this paper. Such distributively added tails may attract Nab2p and consequent recruitment of Rrp6p would provide pA tail length control (Figure 7C). Alternatively, Nab2p binding at the end of the pA tail could protect Pab1p-bound tails from PAN trimming in the nucleus. Our results show that Pab1b binding is strongly favored in wt cell nuclei while Nab2p binding is very transient due to Rrp6p-mediated removal. The fact that overexpression, or nuclear targeting, of Pab1p can rescue growth of nab2Δ cells also favors the interpretation that Nab2p is not critically required for pA tail formation (Hector et al., 2002). However, it also implies that nuclear Pab1p may only be present in limiting amounts. Conversely, the otherwise lethality of a pab1Δ deletion strain can be rescued by mutation of cytoplasmic factors (Boeck et al., 1998; Bonnerot et al., 2000; Wyers et al., 2000). While such strains grow very slowly, it does argue that Nab2p alone is also capable of providing at least some nuclear pA tail production and export. We therefore suggest that the mutual exclusive targeting of Rrp6p by Nab2p-bound tails and PAN by Pab1p-bound tails is decisive for the fate of polyadenylated RNAs and thereby provides an option for differential gene regulation and quality control during early mRNA production in the nucleus. A careful characterization of pA tail conformation will be an important goal for future investigation and should uncover further specific messages and situations where the pathways described above are employed.
Experimental Procedures
Details of the individual protocols are presented in Supplemental Experimental Procedures.
In vitro processing reactions
Processing extracts and reactions were carried out essentially as described in (Saguez et al., 2008). In brief, processing extracts were prepared by grinding of yeast cells in liquid nitrogen, ultracentrifugation and concentration by ammonium sulfate precipitation. Processing reactions were carried out by incubating a 5′ end radio-labeled synthetic RNA template (CYC1s) with processing extracts and appropriate buffers containing ATP at 30°C. Reaction products were separated on polyacrylamide/urea gels and analyzed by phosphorimaging. Details of IP, depletion and challenge of processing reactions are described in Supplemental Experimental Procedures.
Tiling microarray analysis
Total RNA was extracted from exponentially growing THC-NAB2 controls (0h) or cells treated for 7h and 12h with 50 μg/ml doxycycline and amplified by RT using a mix of random hexamers and oligo(dT) primers. For wt and rrp6Δ cells, a similar RT reaction was performed on pA-enriched RNA. cDNAs were fragmented, labeled and hybridized to Affymetrix tiling arrays (PN520055). Expression data are deposited in the ArrayExpress archive.
Accession numbers
ArrayExpress E-MTAB-981 (THC-NAB2 samples) and E-MTAB-1089 (wt and rrp6Δ data).
Supplementary Material
Highlights.
Nab2p/Rrp6p pathway degrades pre-mRNA
Rrp6p displaces Nab2p from nuclear pA tails
Pab1p associates predominantly with pA tails
Rrp6p counteracts Trf4p-dependent extension of pA tails
Acknowledgments
We thank Alain Jacquier, Domenico Libri, Iain Mattaj, Ditlev Brodersen, Alan Sachs, Lionel Minvielle-Sebastia and Stepanka Vanacova for their generous gifts of expression constructs, yeast strains and antibodies. Members of the CM and THJ laboratories are thanked for stimulating discussions and Ditlev Brodersen, Domenico Libri, Eckhard Jankowsky and Francoise Stutz are acknowledged for critical reading of the manuscript. The work was supported by the Danish National Research Foundation (THJ), the Danish Council for Independent Research: Natural Sciences (THJ), the NOVO Nordisk Foundation (THJ), an EMBO postdoctoral Fellowship (VP) and the NIH (LMS and CM (grant GM041752)).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Amrani N, Minet M, Le Gouar M, Lacroute F, Wyers F. Yeast Pab1 interacts with Rna15 and participates in the control of the poly(A) tail length in vitro. Mol Cell Biol. 1997;17:3694–3701. doi: 10.1128/mcb.17.7.3694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Assenholt J, Mouaikel J, Andersen KR, Brodersen DE, Libri D, Jensen TH. Exonucleolysis is required for nuclear mRNA quality control in yeast THO mutants. Rna. 2008;14:2305–2313. doi: 10.1261/rna.1108008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boeck R, Lapeyre B, Brown CE, Sachs AB. Capped mRNA degradation intermediates accumulate in the yeast spb8-2 mutant. Mol Cell Biol. 1998;18:5062–5072. doi: 10.1128/mcb.18.9.5062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonnerot C, Boeck R, Lapeyre B. The two proteins Pat1p (Mrt1p) and Spb8p interact in vivo, are required for mRNA decay, and are functionally linked to Pab1p. Mol Cell Biol. 2000;20:5939–5946. doi: 10.1128/mcb.20.16.5939-5946.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bousquet-Antonelli C, Presutti C, Tollervey D. Identification of a regulated pathway for nuclear pre-mRNA turnover. Cell. 2000;102:765–775. doi: 10.1016/s0092-8674(00)00065-9. [DOI] [PubMed] [Google Scholar]
- Brown CE, Sachs AB. Poly(A) tail length control in Saccharomyces cerevisiae occurs by message-specific deadenylation. Mol Cell Biol. 1998;18:6548–6559. doi: 10.1128/mcb.18.11.6548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brune C, Munchel SE, Fischer N, Podtelejnikov AV, Weis K. Yeast poly(A)-binding protein Pab1 shuttles between the nucleus and the cytoplasm and functions in mRNA export. Rna. 2005;11:517–531. doi: 10.1261/rna.7291205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callahan KP, Butler JS. TRAMP complex enhances RNA degradation by the nuclear exosome component Rrp6. J Biol Chem. 2010;285:3540–3547. doi: 10.1074/jbc.M109.058396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dheur S, Nykamp KR, Viphakone N, Swanson MS, Minvielle-Sebastia L. Yeast mRNA Poly(A) tail length control can be reconstituted in vitro in the absence of Pab1p-dependent Poly(A) nuclease activity. J Biol Chem. 2005;280:24532–24538. doi: 10.1074/jbc.M504720200. [DOI] [PubMed] [Google Scholar]
- Eckmann CR, Rammelt C, Wahle E. Control of poly(A) tail length. Wiley Interdisciplinary Reviews: RNA. 2011;2:348–361. doi: 10.1002/wrna.56. [DOI] [PubMed] [Google Scholar]
- Fasken MB, Stewart M, Corbett AH. Functional significance of the interaction between the mRNA-binding protein, Nab2, and the nuclear pore-associated protein, Mlp1, in mRNA export. J Biol Chem. 2008;283:27130–27143. doi: 10.1074/jbc.M803649200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galy V, Gadal O, Fromont-Racine M, Romano A, Jacquier A, Nehrbass U. Nuclear retention of unspliced mRNAs in yeast is mediated by perinuclear Mlp1. Cell. 2004;116:63–73. doi: 10.1016/s0092-8674(03)01026-2. [DOI] [PubMed] [Google Scholar]
- Hector RE, Nykamp KR, Dheur S, Anderson JT, Non PJ, Urbinati CR, Wilson SM, Minvielle-Sebastia L, Swanson MS. Dual requirement for yeast hnRNP Nab2p in mRNA poly(A) tail length control and nuclear export. The EMBO journal. 2002;21:1800–1810. doi: 10.1093/emboj/21.7.1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houseley J, Tollervey D. The many pathways of RNA degradation. Cell. 2009;136:763–776. doi: 10.1016/j.cell.2009.01.019. [DOI] [PubMed] [Google Scholar]
- Iglesias N, Tutucci E, Gwizdek C, Vinciguerra P, Von Dach E, Corbett AH, Dargemont C, Stutz F. Ubiquitin-mediated mRNP dynamics and surveillance prior to budding yeast mRNA export. Genes Dev. 2010;24:1927–1938. doi: 10.1101/gad.583310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen TH, Moore C. Reviving the exosome. Cell. 2005;121:660–662. doi: 10.1016/j.cell.2005.05.018. [DOI] [PubMed] [Google Scholar]
- Kadaba S, Krueger A, Trice T, Krecic AM, Hinnebusch AG, Anderson J. Nuclear surveillance and degradation of hypomodified initiator tRNAMet in S. cerevisiae. Genes Dev. 2004;18:1227–1240. doi: 10.1101/gad.1183804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn U, Gundel M, Knoth A, Kerwitz Y, Rudel S, Wahle E. Poly(A) tail length is controlled by the nuclear poly(A)-binding protein regulating the interaction between poly(A) polymerase and the cleavage and polyadenylation specificity factor. J Biol Chem. 2009;284:22803–22814. doi: 10.1074/jbc.M109.018226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaCava J, Houseley J, Saveanu C, Petfalski E, Thompson E, Jacquier A, Tollervey D. RNA degradation by the exosome is promoted by a nuclear polyadenylation complex. Cell. 2005;121:713–724. doi: 10.1016/j.cell.2005.04.029. [DOI] [PubMed] [Google Scholar]
- Lemay JF, D’Amours A, Lemieux C, Lackner DH, St-Sauveur VG, Bahler J, Bachand F. The nuclear poly(A)-binding protein interacts with the exosome to promote synthesis of noncoding small nucleolar RNAs. Mol Cell. 2010a;37:34–45. doi: 10.1016/j.molcel.2009.12.019. [DOI] [PubMed] [Google Scholar]
- Lemay JF, Lemieux C, St-Andre O, Bachand F. Crossing the borders: poly(A)-binding proteins working on both sides of the fence. RNA Biol. 2010b;7:291–295. doi: 10.4161/rna.7.3.11649. [DOI] [PubMed] [Google Scholar]
- Lemieux C, Marguerat S, Lafontaine J, Barbezier N, Bahler J, Bachand F. A Pre-mRNA degradation pathway that selectively targets intron-containing genes requires the nuclear poly(A)-binding protein. Mol Cell. 2011;44:108–119. doi: 10.1016/j.molcel.2011.06.035. [DOI] [PubMed] [Google Scholar]
- Libri D. Nuclear poly(a)-binding proteins and nuclear degradation: take the mRNA and run? Mol Cell. 2010;37:3–5. doi: 10.1016/j.molcel.2009.12.029. [DOI] [PubMed] [Google Scholar]
- Milligan L, Torchet C, Allmang C, Shipman T, Tollervey D. A nuclear surveillance pathway for mRNAs with defective polyadenylation. Mol Cell Biol. 2005;25:9996–10004. doi: 10.1128/MCB.25.22.9996-10004.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minvielle-Sebastia L, Preker PJ, Wiederkehr T, Strahm Y, Keller W. The major yeast poly(A)-binding protein is associated with cleavage factor IA and functions in premessenger RNA 3′-end formation. Proceedings of the National Academy of Sciences of the United States of America. 1997;94:7897–7902. doi: 10.1073/pnas.94.15.7897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muhlemann O, Jensen TH. mRNP quality control goes regulatory. Trends in genetics: TIG. 2012;28:70–77. doi: 10.1016/j.tig.2011.11.001. [DOI] [PubMed] [Google Scholar]
- Neil H, Malabat C, d’Aubenton-Carafa Y, Xu Z, Steinmetz LM, Jacquier A. Widespread bidirectional promoters are the major source of cryptic transcripts in yeast. Nature. 2009;457:1038–1042. doi: 10.1038/nature07747. [DOI] [PubMed] [Google Scholar]
- Qu X, Lykke-Andersen S, Nasser T, Saguez C, Bertrand E, Jensen TH, Moore C. Assembly of an export-competent mRNP is needed for efficient release of the 3′-end processing complex after polyadenylation. Mol Cell Biol. 2009;29:5327–5338. doi: 10.1128/MCB.00468-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth KM, Wolf MK, Rossi M, Butler JS. The nuclear exosome contributes to autogenous control of NAB2 mRNA levels. Mol Cell Biol. 2005;25:1577–1585. doi: 10.1128/MCB.25.5.1577-1585.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rougemaille M, Gudipati RK, Olesen JR, Thomsen R, Seraphin B, Libri D, Jensen TH. Dissecting mechanisms of nuclear mRNA surveillance in THO/sub2 complex mutants. The EMBO journal. 2007;26:2317–2326. doi: 10.1038/sj.emboj.7601669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saguez C, Schmid M, Olesen JR, Ghazy MA, Qu X, Poulsen MB, Nasser T, Moore C, Jensen TH. Nuclear mRNA surveillance in THO/sub2 mutants is triggered by inefficient polyadenylation. Mol Cell. 2008;31:91–103. doi: 10.1016/j.molcel.2008.04.030. [DOI] [PubMed] [Google Scholar]
- St-Andre O, Lemieux C, Perreault A, Lackner DH, Bahler J, Bachand F. Negative regulation of meiotic gene expression by the nuclear poly(a)-binding protein in fission yeast. J Biol Chem. 2010;285:27859–27868. doi: 10.1074/jbc.M110.150748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tutucci E, Stutz F. Keeping mRNPs in check during assembly and nuclear export. Nature reviews Molecular cell biology. 2011;12:377–384. doi: 10.1038/nrm3119. [DOI] [PubMed] [Google Scholar]
- Vanacova S, Wolf J, Martin G, Blank D, Dettwiler S, Friedlein A, Langen H, Keith G, Keller W. A new yeast poly(A) polymerase complex involved in RNA quality control. PLoS biology. 2005;3:e189. doi: 10.1371/journal.pbio.0030189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viphakone N, Voisinet-Hakil F, Minvielle-Sebastia L. Molecular dissection of mRNA poly(A) tail length control in yeast. Nucleic Acids Res. 2008;36:2418–2433. doi: 10.1093/nar/gkn080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyers F, Minet M, Dufour ME, Vo LT, Lacroute F. Deletion of the PAT1 gene affects translation initiation and suppresses a PAB1 gene deletion in yeast. Mol Cell Biol. 2000;20:3538–3549. doi: 10.1128/mcb.20.10.3538-3549.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyers F, Rougemaille M, Badis G, Rousselle JC, Dufour ME, Boulay J, Regnault B, Devaux F, Namane A, Seraphin B, et al. Cryptic pol II transcripts are degraded by a nuclear quality control pathway involving a new poly(A) polymerase. Cell. 2005;121:725–737. doi: 10.1016/j.cell.2005.04.030. [DOI] [PubMed] [Google Scholar]
- Yamanaka S, Yamashita A, Harigaya Y, Iwata R, Yamamoto M. Importance of polyadenylation in the selective elimination of meiotic mRNAs in growing S. pombe cells. The EMBO journal. 2010;29:2173–2181. doi: 10.1038/emboj.2010.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.