Few measurements in biochemistry are as fundamental as mass. Whereas the mass of a macromolecule is often inferred by its migration through polymer gels, direct measure may only be determined by mass spectrometry. Most scientists' expectation of mass spectrometry is that of determining the mass of an unknown. In the postgenomic era, this has resulted in a fundamental shift in our ability to approach biological investigation. Consider the problem of elucidating the identity of a protein resolved on a two-dimensional polyacrylamide gel. What was once a dissertation project is now a service in which protein is extracted from the gel and proteolysed, and the fragment masses determined by mass spectrometry. Given the specificity of the protease and the organism's genome, the identity of the protein is determined by a database search (1). Historically, most mass spectrometry is conducted by first ionizing molecules from harsh conditions, e.g., desiccated in the case of matrix assisted laser desorption ionization, or heated and acidified aqueous/organic mixtures in the case of electrospray ionization. It is, however, possible to ionize biomolecules from aqueous buffers at neutral pH and room temperature. This is done with some difficulty using conventional electrospray ionization, and with relative ease using nano-electrospray ionization (2, 3). As a result, a great many biochemists are finding novel applications of mass spectrometry in instances where the mass is already known (4).
In this issue of PNAS, Fändrich et al. (5) use mass spectrometry to examine the oligomeric structure of MtGimC from M. thermoautotrophicum. MtGimC is an archael homologue of the eukaryotic chaperone prefoldin (or GimC; ref. 5), the role of which in S. cerevisiae is to escort newly synthesized actin and tubulin to a cytosolic chaperone, CCT (6). It has been suggested that the role of the archael homologue is that of a general chaperone of newly synthesized polypeptides (7). The work of Fändrich et al. focuses on the supramolecular assembly of MtGimC, its stability and kinetics. Whereas prefoldin is a heteromeric complex of six polypeptides, MtGimC is composed of only two subunits termed α and β. Ionization of this heteromeric complex from native conditions using a minimum of electric potential to drive the ions into the mass analyzer gives rise to a single distribution of charge states corresponding to a mass with only a single possible stoichiometry: 4 β and 2 α.
This approach to stoichiometry determination has two significant advantages. First, the mass accuracy under these conditions is better than one part per thousand. As the substituents' masses are known, this trivially allows unequivocal determination of stoichiometry. Second, this approach does not require separation of free precursors from the complex, thus allowing direct measurement without risk of solution dissociation contributing to the observation.
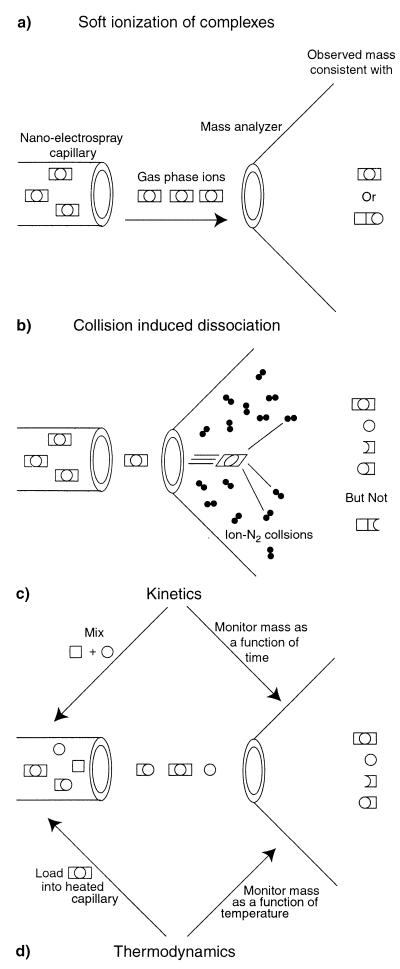
To understand these applications, it is important to understand the basis of nano-electrospray ionization. An overview of the procedure, in brief, begins when an analyte solution (≈1 μl at ≈10 μM) is placed in a metal coated glass capillary (Fig. 1). The tip of the capillary has an inner diameter of about 1 μm and is held at a high potential difference (1–2 kV) with respect to the orifice of the mass analyzer. As a result, there is a local separation of charges at the tip. The repulsion of these charges overwhelms the surface tension and gives rise to a jet of charged droplets. These droplets have an initial volume on the order of 10−11 μl and flow at about 10 nl/min. Cycles of evaporation followed by droplet fission ultimately result in desorption of the macromolecular ion. During most of the 100 μs over which this process takes place, the macromolecule is still in solution. Once ions are formed, they are analyzed by using a variety of techniques (e.g., time-of-flight) all of which give rise to a mass/charge (m/z) spectrum. Thus, a pure protein sample will give a spectrum composed of a series of peaks, each corresponding to the same mass, but different numbers of charges on the protein. Of particular relevance to the biochemist is that a few μl of sample may be analyzed for hours while requiring as little as 1–2 seconds for a spectrum to be taken.
Figure 1.
Nano-electrospray ionization of a hypothetical trimeric heterocomplex composed of two square and one round subunits. (a) Soft ionization results in a single mass which unequivocally determines the stoichiometry of the complex. Supramolecular assembly cannot, however, be determined. (b) By increasing the electric potential in the source of the mass spectrometer, the energy of collisions between the complex and neutral residual gas causes the complex to fall apart. The identity of the subsequently measured subcomplexes reveals that the assembly is square-circle-square. (c) The assembly of complexes can be determined in real time by mixing the subcomponents and loading the reaction into the capillary needle of the mass spectrometer. Unique to mass spectrometry, the separate rates of assembly may be measured. (d) Fändrich et al. have now gone further by developing a temperature controller for the nanospray capillary which allows thermal melts to be conducted on the sample. The products of heat-induced dissociation can be measured directly. As the subcomplexes are separately detected, denaturation curves can be assigned explicitly to particular subcomponents.
The first observations that ionization of this type was sufficiently gentle to analyze conformation came from the observation that the charge state distribution of a protein is sensitive to the solution from which it is ionized (8). Proteins ionized from denaturing solution conditions give rise to many more charge states than proteins ionized from native conditions. Furthermore, the most abundant charge state resulting from native state ionization tends to be at higher m/z. The explanation for this is that unfolded proteins adopt extended conformations, which have a greater capacity to stabilize charge.
Deuterium labeling at labile positions, principally the amide NH, is a powerful tool for analyzing residue-specific structure formation and stability. Deuterium at these locations are readily exchanged back to hydrogen from water unless the conformation of the protein excludes solvent and/or the amide has a hydrogen bonding partner (9). The ability of investigators to perform these experiments indicates that nothing extreme is happening to the protein in the 100 μs during which ionization takes place. In ion cyclotron resonance mass spectrometry, strong magnetic and electric fields may be used to trap molecules after ionization. In recent experiments, ions formed from troponin C were trapped and exposed to D2O gas. The mass of the molecules increases on exchange with D2O in the gas phase. Remarkably, these profiles show a dependence on the solution conditions from which the ions were made (10). Although it is impossible to establish that gas phase ions are native-like, gaseous protein ions clearly have discrete and interconvertable conformations (11).
Lastly and most recently, it has been possible to ionize and measure the mass of intact noncovalently associated macromolecular complexes (4). This has several important advantages over other methods. Firstly, it does not require separation of complex from precursor. Methods such as size exclusion chromatography rely on dissociation rates to be slow compared with the time of analysis. Second, material requirements are exceedingly small. Volumes on the order of 1 μl at concentrations of the order of 10 μM are readily analyzed. These properties, when combined with the accuracy and resolution of mass spectrometry, have resulted in a variety of fascinating applications. For example, the coordination of metal by the 7 zinc metallothionein MT-2 (12). This investigation permitted identification of cooperative formation of a 4 zinc center before formation of a 3 zinc center. In a second recent example, the stoichiometry of the PA28 regulatory complex of the eukaryotic proteasome has been investigated by electrospray ionization (ESI)-MS (13). This complex is formed from two homologous (≈50% identity) subunits, α and β, and is particularly interesting because a homomeric complex may be formed from isolated α. Coexpression of the two subunits in Escherichia coli gave rise to a complex whose mass spectrum is dominated by a heterocomplex consisting of 3 α and 4 β subunits.
These observations strongly suggest that the process of ionization does not perturb protein conformation. This is an observation no less alarming than the observation that enzymes may be transferred to organic solvent and maintain structure and activity (14). Furthermore, maintenance of protein structure in organic solvent depends on strict elimination of water (15). Given the recent evidence that water may catalyze secondary structure interconversion (16, 17), it is possible that gas phase protein ions maintain a native-like structure as a result of being kinetically trapped.
Protein ions collide with residual neutral gas molecules in the source of a mass spectrometer (Fig. 1b). The energy of these collisions may be increased simply by raising one of the potential differences used to guide the ions through the mass analyzer to the detector. Such collisions permit the relatively weak noncovalent associations to be broken. In a number of systems of known structure [e.g., the ribosome (18)], partial disassembly within the mass spectrometer yields subcomplexes which reveal the superstucture of the intact complex. In Fig. 1a, the mass of a hypothetical trimeric complex is consistent with two alternative assemblies. On collision-induced dissociation within the mass spectrometer (Fig. 1b), subcomplexes are seen that only correspond to one of the two possibilities. Confidence has now been established in these approaches, permitting Fändrich et al. to describe a structure consistent with a central dimer of α subunits surrounded by 4 β subunits which interact with the α dimer, but not each other.
The pathway of assembly can also be measured (Fig. 1c). Reactants are mixed and placed in the capillary of a mass spectrometer by using settings which correspond to those used for measuring the intact complex. Kinetics may be measured by monitoring the disappearance of the signal of the precursor (19) and/or appearance of the complex. Intermediates of such a reaction may be inferred either indirectly from a disparity in these two rates, or directly from observation of the intermediates themselves. By using this approach, Fändrich et al. note that there is no appearance of intermediate forms in the assembly of MtGimC. This suggests either that formation of MtGimC is highly cooperative, or that subunits are not sufficiently stable to be observed. As they had already been able to detect subcomplexes by using collisions in the mass spectrometer, they are able to infer that their observations are a reflection of cooperative formation of the complex.
It is notable that isolated β subunits of MtGimC are unfolded at the normal growth temperature of 65°C, whereas α are folded. Fändrich et al. have developed a separate temperature controller for the capillary of the electrospray source (Fig. 1d) enabling them to conduct thermal melts of the complex with direct detection of dissociated products. This permitted them to directly measure the dissociation of the complex as well as make observations of the dissociation products. As a result, they were able to clearly observe that the β subunits remain a component of the complex even at temperatures above the melting transition for isolated β subunits.
There are a number of mechanistic questions critical to understanding how MTGimC might work. Which subunits bind unfolded peptides? What oligomeric state of MtGimC binds unfolded peptides. Do MtGimC ligands have a consensus sequence? Does the binding of unfolded peptides affect assembly rates and or the stability of intact complex and subcomplexes. These are intriguing questions with many alternative answers. The alternatives, however, have distinct masses. The ability of ESI-MS to monitor assembly in real time, the recent addition of thermal control of the sample, and its unique ability to simultaneously measure multiple species makes mass spectrometry ideal for these investigations.
Acknowledgments
This contribution was funded by the National Institute of Diabetes, Digestive and Kidney Diseases of the National Institutes of Health and by the Pew Scholars Program in the Biomedical Sciences.
Footnotes
See companion article on page 14151.
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.011526498.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.011526498
References
- 1.Yates J R., III Electrophoresis. 1998;19:893–900. doi: 10.1002/elps.1150190604. [DOI] [PubMed] [Google Scholar]
- 2.Kebarle P. J Mass Spectrom. 2000;35:804–817. doi: 10.1002/1096-9888(200007)35:7<804::AID-JMS22>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 3.Cole R B. J Mass Spectrom. 2000;35:763–772. doi: 10.1002/1096-9888(200007)35:7<763::AID-JMS16>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- 4.Miranker A D. Curr Opin Struct Biol. 2000;10:601–606. doi: 10.1016/s0959-440x(00)00137-8. [DOI] [PubMed] [Google Scholar]
- 5.Fändrich M, Tito M A, Leroux M R, Rostom A A, Hartl F U, Dobson C M, Robinson C V. Proc Natl Acad Sci USA. 2000;97:14151–14155. doi: 10.1073/pnas.240326597. . (First Published November 21, 2000; 10.1073/pnas.240326597) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vainberg I E, Lewis S A, Rommelaere H, Ampe C, Vandekerckhove J, Klein H L, Cowan N J. Cell. 1998;93:863–873. doi: 10.1016/s0092-8674(00)81446-4. [DOI] [PubMed] [Google Scholar]
- 7.Leroux M R, Fandrich M, Klunker D, Siegers K, Lupas A N, Brown J R, Schiebel E, Dobson C M, Hartl F U. EMBO J. 1999;18:6730–6743. doi: 10.1093/emboj/18.23.6730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chowdhury S K, Katta V, Chait B T. J Am Chem Soc. 1990;112:9012–9013. [Google Scholar]
- 9.Miranker A, Robinson C V, Radford S E, Dobson C M. FASEB J. 1996;10:93–101. doi: 10.1096/fasebj.10.1.8566553. [DOI] [PubMed] [Google Scholar]
- 10.Wang F, Freitas M, Marshall A, Sykes B. Int J Mass Spectrom. 1999;192:319–325. [Google Scholar]
- 11.McLafferty F, Guan Z, Haupts U, Wood T, Kelleher N. J Am Chem Soc. 1998;120:4732–4740. [Google Scholar]
- 12.Gehrig P M, You C, Dallinger R, Gruber C, Brouwer M, Kagi J H, Hunziker P E. Protein Sci. 2000;9:395–402. doi: 10.1110/ps.9.2.395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang Z, Krutchinsky A, Endicott S, Realini C, Rechsteiner M, Standing K G. Biochemistry. 1999;38:5651–5658. doi: 10.1021/bi990056+. [DOI] [PubMed] [Google Scholar]
- 14.Fitzpatrick P A, Steinmetz A C, Ringe D, Klibanov A M. Proc Natl Acad Sci USA. 1993;90:8653–8657. doi: 10.1073/pnas.90.18.8653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Griebenow K, Klibanov A M. J Am Chem Soc. 1996;118:11695–11700. [Google Scholar]
- 16.Barron L D, Hecht L, Wilson G. Biochemistry. 1997;36:13143–13147. doi: 10.1021/bi971323j. [DOI] [PubMed] [Google Scholar]
- 17.Xu F, Cross T A. Proc Natl Acad Sci USA. 1999;96:9057–9061. doi: 10.1073/pnas.96.16.9057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Benjamin D R, Robinson C V, Hendrick J P, Hartl F U, Dobson C M. Proc Natl Acad Sci USA. 1998;95:7391–7395. doi: 10.1073/pnas.95.13.7391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Larson J L, Ko E, Miranker A D. Protein Sci. 2000;9:427–431. doi: 10.1110/ps.9.2.427. [DOI] [PMC free article] [PubMed] [Google Scholar]