Abstract
Despite growing interest in multi-voxel pattern analysis (MVPA) methods for fMRI, a major problem remains—that of generating estimates in rapid event-related (ER) designs, where the BOLD responses of temporally adjacent events will overlap. While this problem has been investigated for methods that reduce each event to a single parameter per voxel (Mumford et al., 2012), most of these methods make strong parametric assumptions about the shape of the hemodynamic response, and require exact knowledge of the temporal profile of the underlying neural activity. A second class of methods uses multiple parameters per event (per voxel) to capture temporal information more faithfully. In addition to enabling a more accurate estimate of ER responses, this allows for the extension of the standard classification paradigm into the temporal domain (e.g., Mourão-Miranda et al., 2007). However, existing methods in this class were developed for use with block and slow ER data, and there has not yet been an exploration of how to adapt such methods to data collected using rapid ER designs. Here, we demonstrate that the use of multiple parameters preserves or improves classification accuracy, while additionally providing information on the evolution of class discrimination. Additionally, we explore an alternative to the method of Mourão-Miranda et al. tailored to use in rapid ER designs that yields equivalent classification accuracies, but is better at unmixing responses to temporally adjacent events. The current work paves the way for wider adoption of spatiotemporal classification analyses, and greater use of MVPA with rapid ER designs.
Keywords: Functional magnetic resonance imaging, Classification analysis, MVPA, Rapid event-related design
Introduction
Interest in multivariate methods for fMRI data analysis, and particularly multi-voxel pattern analysis (MVPA), has been growing since the start of the last decade, beginning with the work of Haxby and others (Haxby et al., 2001; Spiridon & Kanwisher, 2002). MVPA treats neural activity as occurring across ensembles of voxels, and attempts to find differences in the pattern of activity within these ensembles between events of different classes. More formally, MVPA assumes the BOLD response to every event can be represented as a point in a high-dimensional space. The vector defined by this point is referred to as the “observation vector.” In single-parameter approaches, the observation vector includes one value for each voxel in the region of interest (ROI). In multi-parameter approaches, the observation vector includes more than one value per voxel. Give a set of observation vectors, MVPA attempts to find a statistical machine that can separate observation vectors of one class from those of another. Many studies have investigated the various computational decisions involved in undertaking such an analysis (e.g. Cox & Savoy, 2003; Mitchell et al., 2004; Norman et al., 2006; Pereira et al., 2009).
The first step in applying MVPA is to choose a method for defining the observation vectors. In block designs, the most popular single-parameter method uses the mean BOLD activation over that block for each voxel. A popular multi-parameter method includes the BOLD response at all time points within a window containing the block. Because the observation vectors that result from this approach include both spatial (because the vector includes activity from multiple voxels) and temporal (because activity from multiple time points is included) information, this method of defining observation vectors is known as the "spatiotemporal" model (Mourão-Miranda et al., 2007). For slow ER designs, a common single-parameter method uses the GLM to model the BOLD response to each event using a canonical hemodynamic response function (HRF) convolved with a boxcar function. The single parameter per voxel is then the beta weight produced by fitting the GLM to the data from this voxel. For obvious reasons, this method is often referred to as "beta-series" analysis (Rissman et al., 2004). The spatiotemporal model for slow ER designs would be the same as in blocked designs, where the window would be large enough to capture the full BOLD response to the event (Mourão-Miranda et al., 2009).
Defining an observation vector for rapid ER designs is more difficult since the hemodynamic delay will cause the BOLD response from temporally proximate events to overlap.Mumford et al. (2012) proposed a single-parameter method that unmixes the BOLD response from overlapping event-specific activations more successfully than other existing single-parameter methods. The current article is the first to develop and test multi-parameter, spatiotemporal methods of defining observation vectors with rapid ER designs. There are two primary benefits to the methods we develop. First, the Mumford et al. approach assumes perfect knowledge of the HRF, that neural activation is exactly a boxcar function, and that the observed BOLD response is the convolution of the HRF and a boxcar. In contrast, the spatiotemporal method we develop makes none of those assumptions. The second benefit of the spatiotemporal model is it produces a weight for each time point that indicates how much the activation at that point contributed to classifying the trial types.
The first spatiotemporal method was first proposed by Mourão-Miranda (2007). A related method was proposed by Soon et al. (2008) and used a finite impulse response (FIR) function to extract average ER activity estimates for each of 10 functional runs in a free-choice response task, and demonstrated above-chance classification accuracy in certain brain regions, even at time points up to 10s before the self-reported decision to respond. Likewise, Harrison and Tong (2009) used what they call a “time-resolved decoding analysis” to show evidence for working memory influences in early visual areas, demonstrating above-chance classification accuracy for time points during a delay period in these areas. For all of these approaches, the researcher is still faced with the problem of extracting observations to be classified, which is a separate issue from how those observations are used to assay the temporal aspect of classification performance.
Although Chu et al. (2011) and Mourão-Miranda et al. (2007; 2009) found that block averages (i.e., a single parameter method) resulted in better classification accuracies than the spatiotemporal method, it isn't clear that this result would hold for rapid ER designs, since the analogous approach for rapid ER designs relies on an accurate model of the HRF and the neural activation. Additionally, as emphasized in Mourão-Miranda (2007), although the spatiotemporal model did not have better classification accuracy than block averages, the model gives insight to the dynamics of class discrimination. Lastly, software for carrying out the Mourão-Miranda approach already exists with recommendations for application to rapid ER designs (e.g., PyMVPA), yet has never been tested on these designs.
One important shortcoming of the method of Mourão-Miranda et al. is that it has no mechanism for unmixing the BOLD responses to adjacent events. What is needed is a GLM-based multi-parameter method that, unlike FIR, allows trial-by-trial estimation. This article proposes a novel approach that achieves this aim, i.e., it is spatiotemporal but also unmixes the BOLD responses to adjacent events. It achieves this by using the iterative approach to estimation employed by the best method of Mumford et al. (2012), but with a multi-parameter GLM rather than the parametric form used in that article.
This article has two goals: the first is to formally assess the performance of the estimation method of Mourão-Miranda et al. relative to established methods (in particular, the best method from Mumford et al., 2012) when applied to rapid ER data, and the second is to propose and test an alternative method that seeks to avoid some of the possible weaknesses of this approach (e.g., inability to unmix responses to temporally adjacent events). Additionally we have studied how activation estimate(s) for a single trial are impacted by the types of trials occurring before and after that trial. Specifically, we assess whether having the same versus different trial types in neighboring trials biases the activation estimate(s). Each of these methods is described in more detail below. The methods are applied to both simulated and real data, and classification accuracy based on the estimates generated using these methods is compared. As we will show, the novel method outperforms the other methods in all analyses, although with longer ISI structures, the difference between this method and that of Mourão-Miranda et al. diminishes.
Methods
In this paper, we compare three methods for estimating the observation vectors evoked by separate events; two of these are existing methods and one is novel (shown schematically in Figure 1). The first method—included to allow comparison with the results of Mumford et al. (2012)—parametrically reduces the BOLD response elicited by each event to a single value, while the other two use multiple parameters to represent each event in a weakly-parametric fashion. The estimates returned by these latter two methods additionally provide temporal information. Although our application of these estimation methods will be for use in MVPA, any analysis requiring trial-by-trial estimates of ER activity in data collected using a rapid ER design could make use of our results—for instance, beta-series functional-connectivity analysis (Rissman et al., 2004), representational similarity analysis (Kriegeskorte et al., 2008), or kernel regression (Chu et al., 2011).
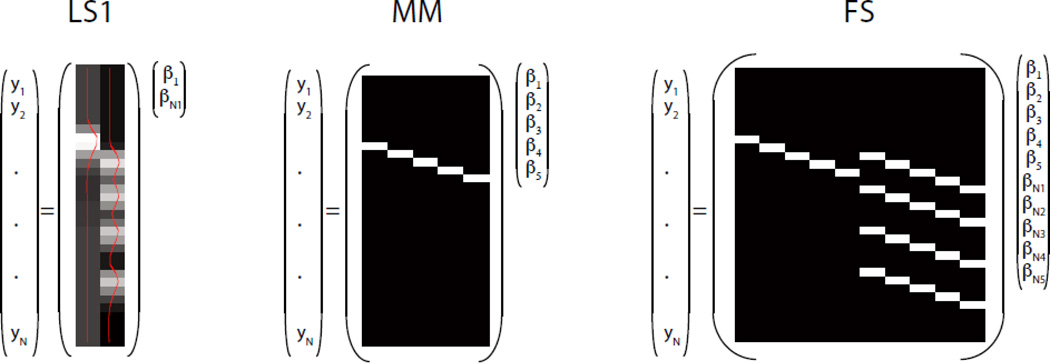
Figure 1.
Schematic representation of the design matrix for a single iteration of each of the three methods, shown with only a single class (and hence only one set of nuisance columns for FS); the parameters of interest are the βNs for LS1 and FS, and the βs for MM.
Estimation methods
The first method considered is the best-performing estimator (least squares–separate; LS-S) from Mumford et al. (2012). This is a single-parameter strongly parametric method that requires specifying a canonical HRF, which we will see has benefits but can also be restrictive in some situations. For each unique event, the LS-S method fits a new GLM with two predicted BOLD timecourses—one that reflects the expected BOLD response to the current event and another for the BOLD responses to all events except the current event. This returns two parameter estimates, one for the trial of interest and a nuisance parameter estimate representing the activation for all other trials. After fitting a separate model for every event in turn, the estimate for each unique event is taken as the regression weight for the predicted BOLD response when that event was the “current event” (left panel, Figure 1). We refer to this version of LS-S as LS1, as there is a single nuisance parameter for all trials. Additionally we considered a second model, referred to as LS2, which included two nuisance parameters. In this version, each GLM run included 3 regressors – one for the current event, one for all other events of the first type, and one for all other events of the second type (note that we only considered studies with two trial types)1.
The second method is a direct application of the method proposed in Mourão-Miranda et al. (2007; henceforth denoted as MM). This multi-parameter method treats as the observation vector for event i the raw BOLD values in the n TRs following onset of event i, with these temporal vectors concatenated across voxels to form the final spatiotemporal observation vector (described above). The middle panel of Figure 1 illustrates the GLM that would be used to extract the MM estimates for a single trial (like LS-S, the algorithm requires a new model for each trial). As discussed above, this method has been applied to data collected in block and slow ER designs, where there is no overlap between adjacent events, but never to data collected in a rapid ER design. However, the MM method forms the basis for an interesting reformulation of the standard classification approach introduced by Chu et al. (2011). This novel approach uses kernel regression to do multi-class classification; in the course of validating their method, they applied the MM method to ER data, where it returned accuracies ~15–20% lower than when it was applied to block-design data.
The MM method can be considered a multi-parameter extension of the Add6 method tested in Mumford et al. (2012), which populated the observation vector with the BOLD response six seconds after each event of interest. Specifically, the MM method simultaneously includes Add0, Add2, and so forth. One concern with this method is that the same BOLD data will appear unchanged in the observation vectors for different events. For example, suppose events 1, 2, and 3 were presented on TRs 1, 2, and 5. For any given voxel, the observation vector for the first event will include the BOLD values from the following TRs: [1,2,3,4,5,…], for the second event the observation vector will include TRs: [2,3,4,5,6,…]; and for the third event the vector will include TRs: [5,6,7,8,9,…]. Note that the BOLD response from the fifth TR appears unchanged in the estimates for all three events, thereby making them non-independent.
The final method we consider—which is presented here for the first time—aims to explicitly un-mix BOLD responses to temporally proximal events. Where the previous method was an extension of the Add6 method of Mumford et al. (2012), this method is more closely related to the above-discussed LS-S method from that study. However, rather than use the correlation-based model of Friston et al. (1995) to estimate events, this method uses what we call the finite BOLD response (FBR) model, which is closely related to the finite impulse response (FIR) model (Ollinger et al., 2001a, b; Serences, 2004). Whereas the FIR model (Dale & Buckner, 1997) treats the HRF as a series of finite impulses (which is then convolved with a boxcar), the FBR model treats the BOLD response as a series of finite impulses (and does not require convolution). In the FBR method, the parameter estimates are directly interpretable as the average unmixed BOLD response at each lag included in the model.
One limitation of the FBR method for trial-by-trial estimation is that it has many more parameters that need to be estimated for each event than the LS-S method. In particular, the researcher chooses a number n of TRs across which the BOLD response to each event is presumed to be nonzero, and this number is used to form a set of n columns in the design matrix. For the standard GLM approach, where events within a class are collapsed together, this means there need only be at least n × C TRs in order for the normal equations to be solvable (where C is the number of classes). However, for trial-by-trial estimation, there must be at least n × N TRs (where N is the number of unique events). In many rapid ER designs the number of TRs in the experiment will be less than this large number (i.e., less than n × N), making one-shot trial-by-trial estimation of observation vectors impossible with the FBR model.
In order to circumvent this limitation, it is necessary to use an iterative approach, as with the LS-S method. A simple iterative FBR method can be constructed in a similar way by using the FBR-based model, rather than the correlation-based model. Specifically, for each unique event, one set of FBR parameters models the current event and another set models all events except the current event. As shown in the right panel of Figure 1, the design matrix for a single iteration includes one set of columns representing all events except the current event, and an extra set of columns representing the current event. The estimated betas from these extra columns are taken as the estimate for the current event. By using this iterative approach, the constraint on the minimum number of TRs is reduced from n × N to n × 2. Here, we instead focus on the FBR analogue of LS2, i.e., we include two sets of nuisance columns in addition to the set of unique-event columns. We refer to this method as FS (FBR–separate).
Simulation study
The simulation procedures were identical to those used in Mumford et al. (2012). Briefly, there were 500 simulations in each of three (normally distributed noise variance: 0.82, 1.62, 32) × four (interstimulus interval, uniform over: 0–4s, 2–6s, 4–8s, 6–10s) conditions. Each simulation comprised three runs of data for a single voxel with 30 instances of each of two classes in each run. The two classes were modeled using a boxcar whose height was sampled at each presentation from a normal distribution with mean 5 (class 1) or 3 (class 2) and variance 0.52. The noise was AR(1) with Cor(zi,zj) = ρ|i – j|, where ρ = 0.12. On each iteration, the noise, along with event ordering, height, and timing were randomly generated.
In order to test the flexibility of each model with respect to its ability to deal with mis-specification of the HRF, four additional data sets were simulated, which were identical to the first except that the HRF used to generate the data was lagged by 0.5–2.0s; the boxcar heights, jitter, and AR(1) noise all were constant between the two, so that on each iteration, any differences in accuracy can be directly attributed to the mis-specification of the HRF, allowing us to use paired comparisons across lags in assessing the impact of HRF mis-specification.
The classification procedure for the simulated data is likewise identical to Mumford et al. (2012). On each iteration, two runs were held out to fit a logistic regression model that was used to generate predicted labels for the third run, which were compared with the true labels to calculate the classification accuracy. Of course, for the multi-parameter methods, the true label was regressed on n parameters when fitting the logistic regression, whereas for the single-parameter methods, there was a single regressor (both models also include an intercept term). As in Mumford et al. (2012), we also performed an additional correlation analysis that correlated the estimated and true trial-by-trial responses. However, the pattern of results was identical to that observed for accuracy, so these results are not reported.
Finally, because the major distinction between MM and FS is the latter’s ability to unmix responses to temporally adjacent events, we sought to formally test the impact of temporal adjacency on classifier performance. To this end, after performing the classification above, we performed the following analysis, only for the simulations with an unshifted HRF in the high SNR, low ISI case, which gives the greatest opportunity to see differences among the methods. First, define the margin of event i as
where ŷi is the output of the logistic regression model. Note that because ŷi ranges between 0 and 1, margini takes the value of 0 in the case of a perfect misprediction and the value of 1 if the prediction is perfect. Thus, unlike ŷi, margini is a measure of accuracy. We modeled the effect of neighboring trials on margin through a multiple linear regression, where the dependent variable was margin. Define the onset of event i as time 0. Next, define 18 lags centered around 0, where the first lag variable—lag 1—includes the interval (−18s, −16s], the interval covered by lag 2 is (−16s, −14s], and lag 18 includes the interval (16s, 18s]. Next define the dummy variables
and
where I is the indicator function [i.e., I(true) = 1 and I(false) = 0]. This defines a total of 36 regressors—18 XSj and 18 XDj. Thus, the linear regression is
(Note that due to jittering, the design matrix is not rank deficient.) Each parameter estimate then reflects the adjusted change in margin due to the lag and trial type. Positive betas imply a bias toward correct classification and negative betas imply a bias toward incorrect classification.
Real data: data description
The real data come from a study on the neural substrates of category learning (unpublished). Data were collected on the UC Santa Barbara Siemens TIM Trio 3T scanner with a standard 12-channel coil using a T2*-weighted whole-brain echoplanar (EPI) sequence (2s repetition time (TR), 30ms echo time (TE), 90° flip angle). Each volume consisted of 33 slices acquired parallel to the AC-PC line (interleaved acquisition; 3-mm slice thickness, 64 × 64 matrix). Subjects were scanned in 6 runs spanning approximately 450s each. During each run, participants had to respond to sinusoidal grating patterns that belonged to one of four categories on which the subject had been pretrained. The stimulus and response both occurred in the same TR, i.e., the motor response occurred within 2s of the onset of the stimulus. For all estimation measures, time ‘0’ corresponds to the onset of this stimulus. Although there were four responses, two were left-hand responses and two were right-hand responses. This left-right distinction is the one we will train the classifier to make in order to assess the estimation methods. This study used a partial trials design with 100 stimulus TRs per function run. Each stimulus TR was preceded by a cue TR with probability 0.5 and followed by a feedback TR with probability 0.75. The distribution of intervals between for any two adjacent stimulus-response events (regardless of class, left or right) was [0, 2, 4] s with probabilities [0.125, 0.5, 0.375], respectively.
Fifteen healthy normal subjects were scanned performing this task. All trials were included, irrespective of the accuracy of the response emitted by the participant. Prior to estimating the observations for classification, the data were pre-processed using FEAT v5.98, part of FSL (Smith et al., 2004). The following preprocessing steps were applied: motion correction using the MCFLIRT tool; non-brain removal using the BET brain extraction tool; grand-mean intensity normalization of the entire 4D dataset by a single multiplicative factor; and highpass temporal filtering (Gaussian-weighted least-squares straight line fitting, with sigma=25.0s).
Finally, prior to entry into classification, feature selection was applied using an orthogonal selection method to isolate voxels that might be task-involved. In particular, the voxels included as features for subject M were selected based on a top-level GLM analysis including all subjects except M, calculated using FSL’s FLAME 1 analysis. For every subject, we took the 1000 voxels with the largest statistic values from each direct contrast between our two events for a total of 2000 voxels. Because the features included in the training and test sets were selected without reference to the data comprising either set, this procedure avoids any “peeking” bias that might inflate the classification accuracy.
Real data: data analysis
All of the methods discussed above were applied to the real data to generate estimates for use in MVPA. For each method, the estimates from each functional run were generated independently for every subject. Prior to generating the estimates, the data from each voxel for a given block were demeaned, and then the data for all voxels within the run were normalized to have unit variance across all voxels and time points (i.e., there were no constraints placed on the variance across TRs within a voxel or across voxels for a given TR). The classifier was a linear SVM2 (implemented using Matlab’s Bioinformatics Toolbox); an initial double cross-validation (CV) was used to find the optimal value of C for each fold of the primary CV (see Appendix). Additionally, by default, the dimensions were each normalized to have zero mean and unit variance—note that this operation is distinct from the pre-estimation normalization, which operates on the time series, whereas this normalization operates on the estimates. As with the simulation analysis, the space in which the SVM operated for the multi-parameter methods was larger by a factor of n than that LS1 and LS2.
In order to assess the significance of the classification results against the null hypothesis of no class–response associations, a permutation analysis was conducted (Golland & Fischl, 2003). Across each of 500 iterations, a leave-one-run-out cross-validation was run with each of the methods, with the vector of labels for all events in the training set randomly shuffled. Within each iteration, the same shuffle is used across all subjects, folds, and methods. This allows us to use these permutation results to test for differences between methods, in addition to testing each method individually against chance. Moreover—because researchers might be interested in class-specific accuracy as well as overall accuracy—for both the true analysis and the permutation analysis, accuracy was computed separately for the two classes in addition to the standard across-class average accuracy.
As with the simulated data, we are also interested in the impact of proximal events on classification performance. However, for the real data results, there are several changes in how we assess this relationship from the simulation analysis. First, because we are using SVM rather than logistic regression, the margin is not bounded between 0 and 1 and can no longer be interpreted as a probability. For this reason, we conducted a repeated measures logistic regression analysis where we predicted the classification accuracy of each event (0 or 1), rather than the classifier margin. Second, in addition to the independent variables we had before (indicators of the presence of an event of the same or opposite class at each of the preceding and subsequent 9 TRs), the model also included a nuisance variable indicating class. Finally, the intercept was allowed to vary randomly over runs nested within subjects. As before, if a method’s classification accuracy is impacted by overlap in the responses to temporally-adjacent events, this analysis will demonstrate the change in accuracy caused by these adjacent events.
Results
Simulation study
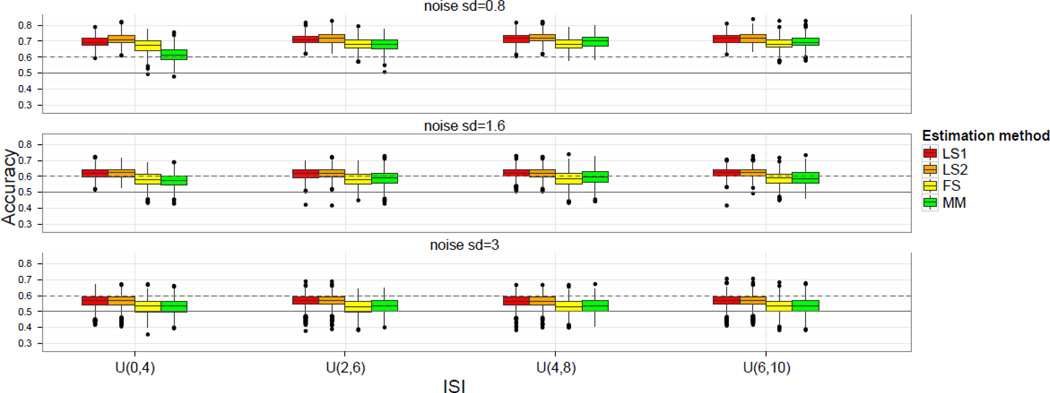
Classification accuracies for all the methods discussed above are shown in Figure 2. Rows of Figure 2 correspond to different levels of signal-to-noise ratio (SNR), whereas columns correspond to different ISI structures. For each combination of values, a boxplot shows the accuracy of all four methods. In general, the single-parameter methods slightly outperform the multi-parameter methods. Among the multi-parameter methods, the FS and MM methods perform similarly, with the FS method showing an advantage under conditions of low noise and short ISI, and MM perhaps showing a much smaller advantage with low noise and long ISI3. The addition of a second nuisance column (i.e., LS2 vs. LS1) has a negligible effect on accuracy except in the case of low noise and short ISI, where it evinces an accuracy advantage of ~2%. Under these same conditions, the disadvantage of the multi-parameter methods relative to the single-parameter methods is particularly clear: MM is ~10% lower than LS1, and FS suffers a drop of ~5% accuracy from the appropriate single-parameter comparison (i.e., LS2).
Figure 2.
Classification accuracy results for the simulated data for six methods. Signal-to-noise ratio decreases across rows, while ISI increases across columns. The solid horizontal line indicates chance (50%), while the dashed line indicates accuracy significantly better than chance according to the binomial distribution.
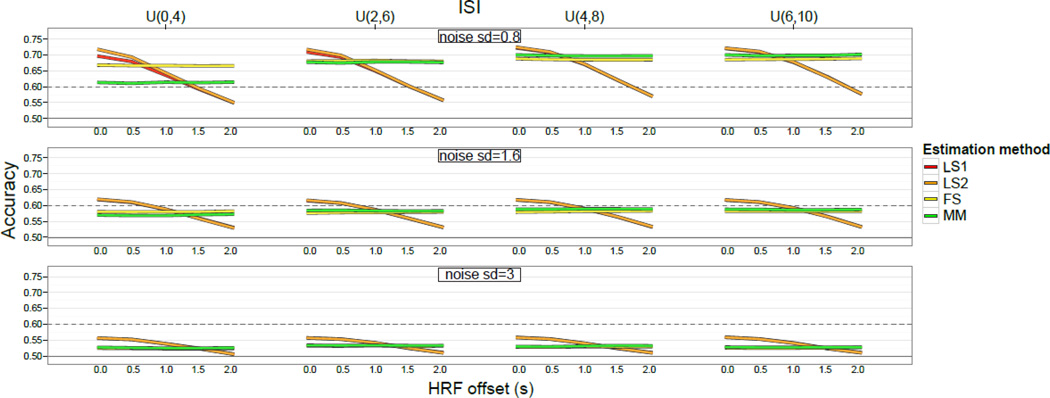
Recall that both LS1 and LS2 require specifying an HRF, and that the HRF assumed by these methods was the same as the HRF used to generate the simulated data. This gives these methods a distinct advantage when tested against these data. As described in the Methods section above, to examine this issue more closely, we generated additional simulated data sets that were identical except that they used an HRF that was temporally shifted by 0.5–2.0s. Thus, when applied to these new data, LS1 and LS2 use a mis-specified HRF (i.e., relative to the HRF used to generate the data). The classification accuracy on these data sets is shown in Figure 3 for each method. Note that for all the multi-parameter methods, the change in accuracy is negligible. However, for LS1 and LS2, there is a noticeable drop in accuracy, particularly with relatively high SNR. The qualitative difference between the multi- and single-parameter methods here is due not to the difference in number of parameters per se, but rather to the fact that LS1 and LS2 are strictly parametric, whereas the multi-parameter methods are nonparametric, or at least much more weakly parametric. This differs from the situation in Mourão-Miranda et al. (2007), where the best-performing single-parameter method (i.e., averaging across a subset of TRs within a block) was also essentially non-parametric4.
Figure 3.
Decrease in accuracy caused by using a shifted HRF to generate simulated data. Each line runs from 0 s shift to 2 s shift in steps of 0.5 s. The solid horizontal line indicates chance (50%), while the dashed line indicates accuracy significantly better than chance according to the binomial distribution.
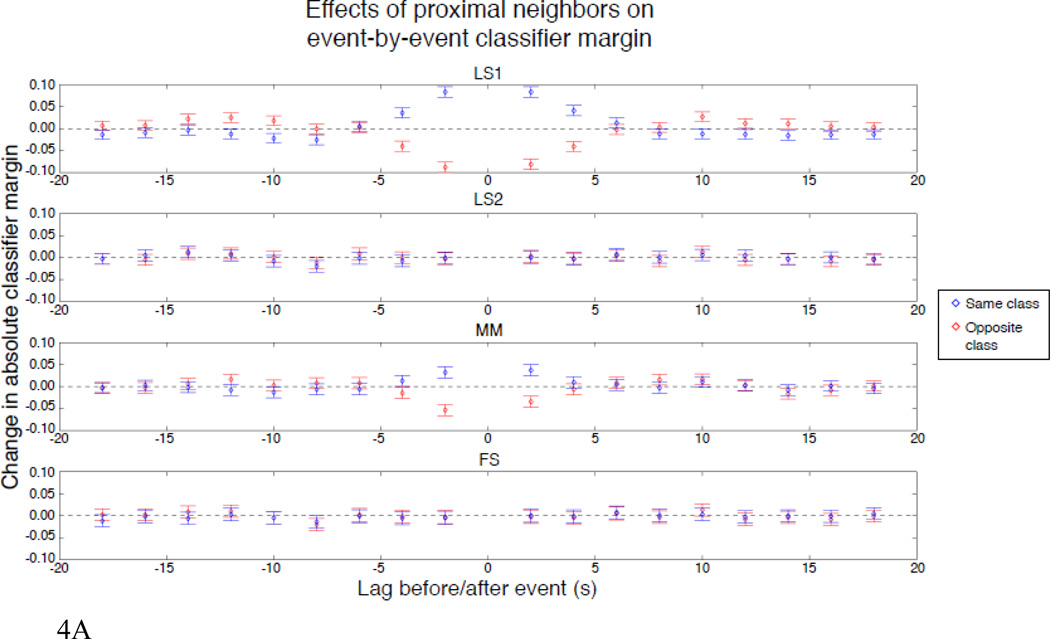
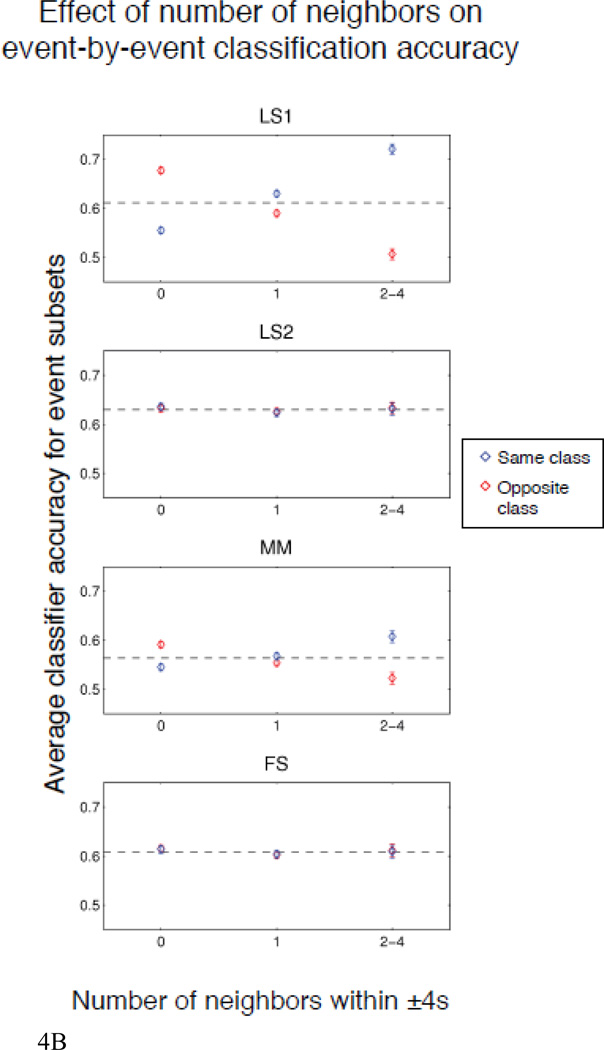
The fact that the difference between MM and FS is largest for the shortest ISI distribution and decreases as the distribution shifts to longer ISIs is expected since MM does not unmix the BOLD responses to nearby events. For this reason, the MM activation estimates will not be as accurate when the ISIs are short. The FS approach, on the other hand, specifically unmixes the observed BOLD response into the component BOLD responses to each event. We explicitly tested the nature of the bias that occurs because of the failure of MM to unmix by asking whether the presence of neighbors of the same or opposite class caused any consistent change in the classifier’s absolute margin (the degree to which the classifier makes a perfect prediction). We repeated the simulation with high SNR, low ISI, and unshifted HRF 50 times, i.e., 9000 events— the same number as there was across all subjects in the real data analysis. We then constructed a separate model that predicted the classifier’s margin—generated using estimates from each of the methods—as a function of the presence of same- or opposite-class neighbors at each of the 9 preceding and subsequent TRs. The beta estimates for this analysis represent the change in classifier margin caused by a neighbor of the same or opposite class at the given lag, holding everything else constant. The results, given as the change in margin (above or below baseline, i.e. higher values correspond to better performance) for all lag-class pairs (i.e., the parameters of this model), are shown in Figure 4a. Another way to view this effect is to examine the average classification accuracy as a function of the number of neighbors within ±4s for the same and opposite classes, as shown in Figure 4b5.
Figure 4.
a. Beta weights (±95% confidence intervals) showing the influence of neighboring events for 50 simulated runs with 0–4s ISI and noise standard deviation = 0.8. The ordinate shows change in classifier margin (y-axis of each frame) as a function of the presence of a neighbor at each 2s lag ±18s (x-axis), and whether that neighbor was of the same (blue) or opposite (red) class.
b. Average classifier accuracy (± 95% confidence interval) for each method as a function of number of same- or opposite-class neighbors within ±4s, for 50 simulated runs with 0–4s ISI and noise standard deviation = 0.8.
As expected, FS is immune to neighbors of either the same or opposite class, whereas the performance of MM is affected by these neighbors, and this effect is most severe when there are at least two events in the preceding or subsequent two TRs. LS1 is even more strongly affected by neighbors, which may initially be surprising, because this method was designed to unmix adjacent events. However, it is easy to explain: in the situation where there are two classes with distinct responses, LS1 allows only a single nuisance regressor, which will take a value somewhere between the two true responses. Therefore, it is systematically overestimating the response to one class, and underestimating the response to the other. When the unique event we are trying to estimate has many other events of a single class nearby, the predicted timecourse in the vicinity of the unique event will be systematically too low or too high (i.e., the residuals will not be zero-mean in this local neighborhood), which will cause the regressor for the unique event to be larger or smaller than it ought to be in order to accommodate. Because LS2 includes one nuisance regressor per class, it avoids this problem, so neighbor influences should be minimized when using LS2 (except to the degree that the choice of an incorrect HRF introduces systematic bias).
Real data
The classification accuracy results for the real data are given in Table 1 for each of the methods considered in this analysis for the baseline case of unrestricted feature selection and using a linear SVM classifier. To test that classifier accuracy was above chance for each of these results, we compared the true average accuracy for each class separately and across classes to the distribution of permuted average accuracies. The number of instances of a permuted average accuracy falling above the observed average accuracy for a method serves as an empirical estimate of the p-value of that accuracy result. This analysis revealed that all class-separate and class-averaged accuracies obtained using each of the estimation methods were significantly higher than chance (see Table 1).
Table 1.
Mean accuracy for each method. Null accuracies come from permutation analysis. All observed accuracies are significantly different than chance, p < 0.001, two-tailed (uncorrected for multiple comparisons).
| Method | Accuracy (event 1) | Accuracy (event 2) | Accuracy (average) | |||
|---|---|---|---|---|---|---|
| Observed | Null | Observed | Null | Observed | Null | |
| LS1 | 73.9 | 51.4 | 72.5 | 48.7 | 73.2 | 50.1 |
| LS2 | 76.1 | 51.5 | 74.0 | 48.6 | 75.1 | 50.0 |
| MM | 83.5 | 53.0 | 81.8 | 47.1 | 82.6 | 50.1 |
| FS | 83.0 | 53.0 | 81.9 | 47.1 | 82.5 | 50.1 |
In order to compare methods, difference scores were computed between every reasonable pairwise combination of methods, and the above procedure was repeated: the mean of each of these difference score distributions was compared against the distribution of permuted mean differences, giving empirical p-values for each of the five pairwise comparisons that we considered (Table 2). This analysis revealed that, relative to chance performance, the difference between LS1 and LS2 was significant for one of the classes and for the average accuracies, indicating an advantage with the addition of the second nuisance column. Additionally, both multi-parameter methods yielded significantly higher class-separate and class-average classification accuracies than their single-parameter referents. There was no significant difference in classification accuracy between the two multi-parameter methods.
Table 2.
Paired comparisons between methods, giving mean differences in percent accuracy and two-tailed p-values (uncorrected for multiple comparisons) for the unmasked data using a linear SVM. p-values derived empirically from permutation analysis—because of the finite number of permutations used, p-values are capped at minimum value of 0.001.
| Paired methods | Difference (event 1) |
p-value | Difference (event 2) |
p-value | Difference (average) |
p-value |
|---|---|---|---|---|---|---|
| LS2 – LS1 | 2.1 | 0.016 | 1.4 | 0.100 | 1.8 | 0.008 |
| MM – LS1 | 9.7 | 0.001 | 9.4 | 0.001 | 9.5 | 0.001 |
| FS – LS2 | 6.8 | 0.001 | 7.7 | 0.001 | 7.3 | 0.001 |
| MM – FS | 0.8 | 0.352 | 0.2 | 0.756 | 0.5 | 0.324 |
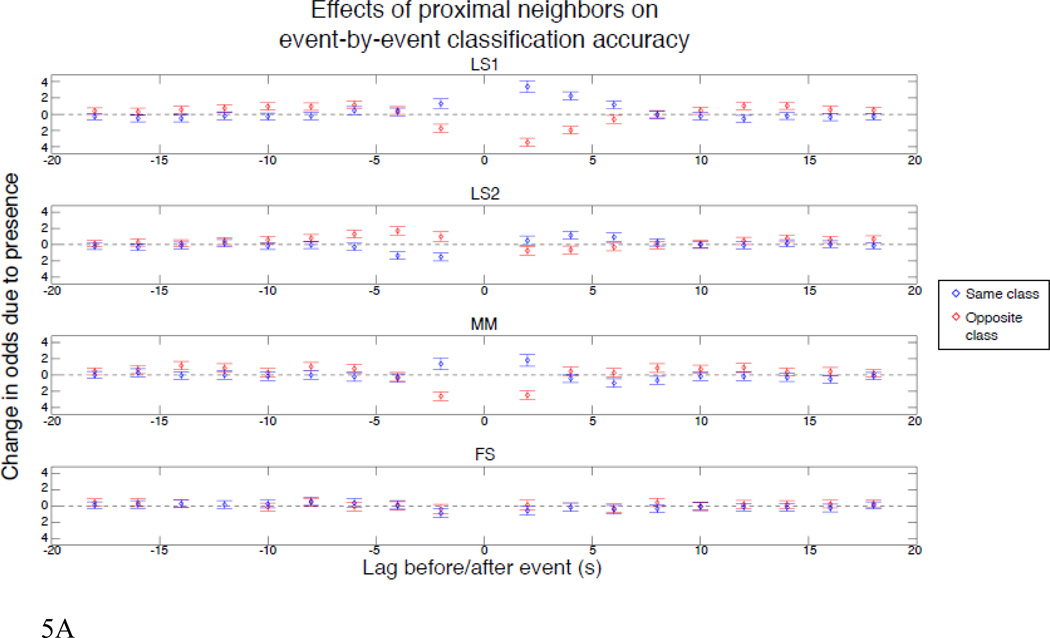
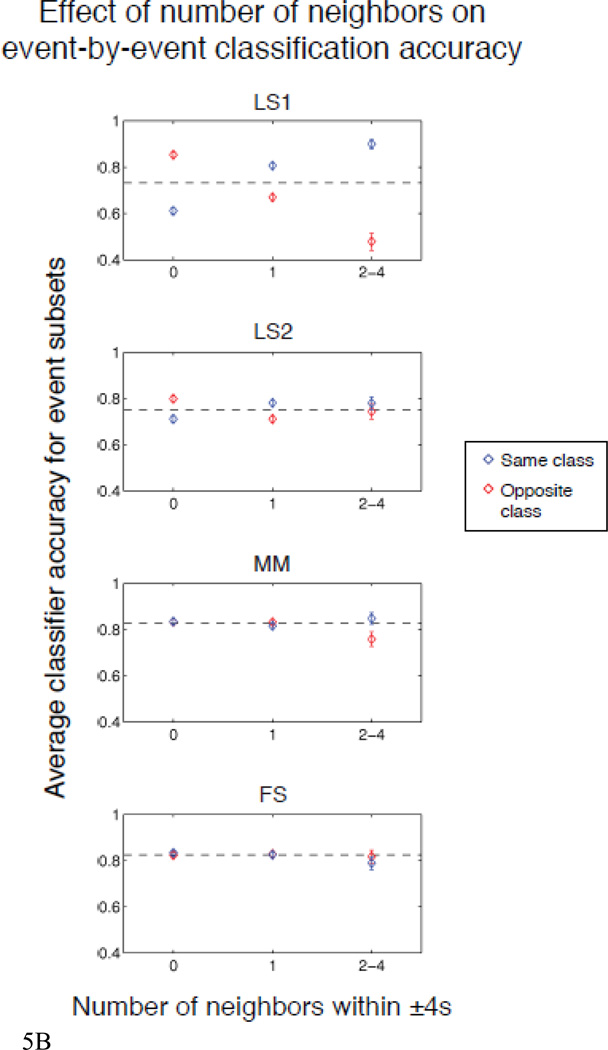
In addition to examining the accuracy across all events, we tested the effect of same- and opposite-class neighbors on event-by-event accuracy. To do this, we used a logistic regression model to predict the event-by-event accuracies (i.e., 1 or 0) as a function of the presence or absence of neighbors of the same or opposite class at each of the preceding and subsequent 9 TRs. The parameter estimates give the log of the ratio of the odds of correct/incorrect classifications for presence versus absence. In other words, exponentiating any parameter estimate reveals how much more likely—or less likely—correct classification is than incorrect classification given presence versus absence for that estimate’s type and lag. A parameter estimate of 0 indicates that the corresponding neighbor has no impact on accuracy, i.e., the probability correct is the same given presence or absence. Positive parameter estimates reflect a beneficial effect of the presence of neighbors, and vice versa for negative parameter estimates. The results of this analysis are given in Figure 5a, which shows the parameter estimates for each method. To demonstrate the direct impact of the neighbor effects on classification accuracy that this analysis implies, Fig. 5b shows the average classification accuracy for each method as a function of the number of same- or opposite-class neighbors within ±4s6. As with the simulated data, LS2 shows a large advantage over LS1 and FS shows a small advantage over MM.
Figure 5.
a. Exponentiated logistic regression weights (±95% confidence intervals) showing the influence of neighboring events for real data. The regression predicts event-by-event accuracy of an SVM classifier as a function of the presence of a neighbor at each 2s lag ±18s (x-axis), and whether that neighbor was of the same (blue) or opposite (red) class. The ordinate (log scaled for visualization) gives the change in odds ratio. For example, a value of 2 above zero corresponds to a doubling in p(correct)/p(incorrect) with presence as compared to absence, while a value of 2 below zero corresponds to a doubling in p(incorrect)/p(correct) for presence versus absence.
b. Average SVM classifier accuracy (± 95% confidence interval) for each method as a function of number of same- (blue) or opposite- (red) class neighbors within ±4s.
Discussion
Mourão-Miranda and colleagues (2007) showed that the addition of temporal information resulted in identical classification performance to the best single-parameter method they considered, and added the benefit of showing the temporal evolution of class-discriminating information across the brain. Relative to many of the methods that are commonly applied to rapid ER data (e.g., Misaki et al., 2010; Mumford et al., 2012), their method also has the advantage of being more flexible, due to its extra parameters and non-parametric form. However, the MM method was developed for data collected using a block design, and although it has since been applied to data collected using slow ER designs, the goal of this article was to extend the method to data collected in rapid ER designs. Although direct application of MM to rapid ER data is gaining traction, there has never been a systematic assessment of how it compares to single-parameter methods, or to other potential multi-parameter methods.
Our results show that, although the classification accuracy of the multi-parameter methods was slightly lower than that of the single-parameter methods on the simulated data, this disadvantage only occurred when we used the same HRF during estimation that was used to create the simulated data. The single-parameter methods considered in this article are parametric, whereas the spatiotemporal methods are all nonparametric. Any parametric statistical technique performs best when its parametric assumptions are valid. Of course, with real data, it is unlikely that a user would be able to specify the HRF with no error7, and therefore we should not necessarily expect the single-parameter methods to show the same advantage when applied to real data. In fact, the superiority of the single-parameter methods disappeared in our real data analysis. For a data set comprising 15 subjects with 6 functional runs each in a task of category learning, both MM and FS outperformed LS1 and LS2 in spite of having to overcome the curse of dimensionality in a much higher-dimensional space.
The advantage of FS over MM is likely to depend on the event timing structure of the experiment: when the shortest ISI is longer than the time it takes for the hemodynamic response to return to baseline, MM will be able to perfectly capture the BOLD response to each event. Similarly, the difference between FS and LS-S will increase as the event timing becomes more uncertain (or equivalently, as the model used for the HRF becomes less well-specified). Each of these effects is demonstrated in our simulated data results. Moreover, in the real data analysis, we show that FS—but not MM, LS1 or LS2—successfully unmixes the responses to temporally adjacent events. For MM, this is because MM lacks any mechanism for unmixing adjacent events; for LS1, it is because of the systematic over- and under-estimation of the two classes by use of a single nuisance regressor; and for LS2, it is likely because the parametric shape of the assumed response is systematically biased relative to the true shape. Therefore, in any design where the responses to subsequent events may overlap, these latter methods will result in contaminated estimates for each event, and therefore in more variable classification performance. Note that even though classification accuracy actually improves in the latter methods as more events of the same class are proximal, this should be considered a liability of the methods rather than an asset, because it still reflects a failure to estimate event-by-event activity in an unbiased way. On the other hand, FS does not exhibit this failing, but instead demonstrates an ability to reliably capture the veridical event-by-event responses in a way that may be useful to researchers applying techniques extending beyond simply maximizing classification accuracy in MVPA.
Spatiotemporal methods are an obvious choice even for researchers not interested in classification performance per se, because their classification weight vectors identify when, as well as where, class-discriminating information arises (see Mourão-Miranda et al., 2007). These methods incur little extra cost relative to the single-parameter methods, yet they offer several potential advantages. The most obvious, of course, is a possible improvement in accuracy. More significant, though, is that the spatiotemporal methods open the temporal domain to analysis, and thus raise the hope of addressing questions that are beyond the scope of current pattern classification methods. Regardless of the aims of the researcher—spatiotemporal analyses, pattern information assessment as indexed by classification accuracy, or other analysis techniques that rely on trial-by-trial activity estimates—FS makes an appealing choice when the data are collected in a rapid ER design. While future work remains in determining the effects of, e.g., varying degrees of averaging or other classifier types, it is our hope that the current work encourages wider adoption of multivariate and trial-specific methods by researchers who have previously regarded these areas as inaccessible because they use rapid ER designs.
Acknowledgements
This research was supported in part by the U.S. Army Research Office through the Institute for Collaborative Biotechnologies under grant W911NF-07-1-0072 and by Award Number P01NS044393 from the National Institute of Neurological Disorders and Stroke. The real data were collected with support from the Institute for Collaborative Biotechnologies through grant W911NF-09-0001 from the U.S. Army Research Office.
Appendix
Double cross-validation
In order to tune the hyper-parameters associated with the classifier we used in this paper, we performed a double cross-validation as follows:
For a given fold within the primary cross-validation, select a functional run to be held out as the test run. The remaining five runs comprise the training set.
Within the test set, select a run to be held out as the cross-validation test run; the remaining four runs now comprise the cross-validation training set.
Loop through candidate values of C, choosing exponents of 10 from −6 to 6 in steps of size 0.5.
Train the classifier with the current value of the hyper-parameter on the cross-validation training set, and observe the error on the cross-validation test run, defined as −1 times the phi coefficient (Matthews correlation) of the predicted and actual labels.
Repeat steps 2)–4), holding out each other run in the original training set as the cross-validation test run, and using the remaining runs as the cross-validation training set.
For the given fold of the primary cross-validation, the optimal hyper-parameter is the one that returns the lowest error across all folds of the double cross-validation. This hyper-parameter is used to train and test the classifier with the original primary cross-validation training and test sets—note that the value was chosen independent of its impact on classification performance for the primary cross-validation test set.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Regardless of whether one or two sets of nuisance columns is used, the iterative nature of LS-S requires that the events in the training and test sets be estimated in separate models in order to maintain independence. This condition is met in both our real and simulated analyses, both of which use leave-one-run-out cross-validation, where the observation vectors for each block are estimated independently.
Because a major advantage of the spatiotemporal methods lies in the interpretation of the SVM weight vector, we decided against using a nonlinear kernel, which requires much more effort to interpret (e.g., Rasmussen et al., 2010).
It is unsurprising that, as the ISI structure moves toward longer ISIs, the MM method should be increasingly similar to FBR. In the limit of perfect separation between successive events, the two are very similar, and MM will be able to perfectly extract the BOLD response to each event without having to resort to any averaging, which might in fact give MM a slight advantage over FBR.
This would be akin to combining parameters in the multi-parameter methods prior to classification, as was demonstrated for the kNN analysis. The classifier no longer has direct access to temporal information, but estimation was still non-parametric.
Note that these are unconditioned counts, i.e., the two types are anticorrelated: the mean number of opposite-class neighbors within ±4s given 0, 1, or ≥2 same-class neighbors in the same interval is 1.03, 0.67, and 0.37, respectively. Likewise, the mean number of same-class neighbors given 0, 1, or ≥2 opposite class neighbors is 1.00, 0.67, and 0.31.
Same caveat applies as for Fig. 4b: the mean number of opposite-class neighbors within ±4s given 0, 1, or ≥2 same-class neighbors in the same interval is 0.94, 0.51, and 0.11, respectively. Likewise, the mean number of same-class neighbors given 0, 1, or ≥2 opposite class neighbors is 0.94, 0.50, and 0.11.
Of course, it would be possible to try various forms of the HRF, but this introduces problems with multiple comparisons, requiring additional cross-validation to control the false-positive rate, and still imposes the constraint that the HRF be identical throughout the brain—both of which are avoided by more weakly-parametric methods.
References
- Ashby FG. Statistical analysis of fMRI data. Cambridge, MA: MIT Press; 2011. [Google Scholar]
- Chu C, Mourão-Miranda J, Chiu Y-C, Kriegeskorte N, Tan G, Ashburner J. Utilizing temporal information in fMRI decoding: Classifier using kernel regression methods. NeuroImage. 2011;58:560–571. doi: 10.1016/j.neuroimage.2011.06.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox DD, Savoy RL. Functional magnetic resonance imaging (fMRI) ”brain reading”: Detecting and classifying distributed patterns of fMRI activity in human visual cortex. NeuroImage. 2003;19:261–270. doi: 10.1016/s1053-8119(03)00049-1. [DOI] [PubMed] [Google Scholar]
- Dale AM, Buckner RL. Selective averaging of rapidly presented individual trials using fMRI. Human Brain Mapping. 1997;5:329–340. doi: 10.1002/(SICI)1097-0193(1997)5:5<329::AID-HBM1>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- Friston KJ, Holmes AP, Worsley KJ, Poline J-P, Frith CD, Frackowiak RSJ. Statistical parametric maps in functional imaging: A general linear approach. Human Brain Mapping. 1995;2:189–210. [Google Scholar]
- Golland P, Fischl B. Permutation tests for classification: Toward statistical significance in image-based studies. Information Processing in Medical Imaging. 2003;2732:330–341. doi: 10.1007/978-3-540-45087-0_28. [DOI] [PubMed] [Google Scholar]
- Harrison FA, Tong F. Decoding reveals the contents of visual working memory in early visual areas. Nature. 2009;458:632–635. doi: 10.1038/nature07832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henson R, Friston K. Convolution models for fMRI. In: Friston K, Ashburner J, Kiebel S, Nichols T, Penny W, editors. Statistical parametric mapping: The analysis of functional brain images. Amsterdam: Elsevier; 2007. pp. 178–192. [Google Scholar]
- Haxby JV, Gobbini MI, Furey ML, Ishai A, Schouten JL, Pietrini P. Distributed and overlapping representations of faces and objects in ventral temporal cortex. Science. 2001;293:2425–2430. doi: 10.1126/science.1063736. [DOI] [PubMed] [Google Scholar]
- Kriegeskorte N, Mur M, Bandettini PA. Representational similarity analysis – Connecting the branches of systems neuroscience. Frontiers in Systems Neuroscience. 2008;2(4):1–28. doi: 10.3389/neuro.06.004.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misaki M, Kim Y, Bandettini PA, Kriegeskorte N. Comparison of multivariate classifiers and response normalizations for pattern-information fMRI. NeuroImage. 2010;53:103–118. doi: 10.1016/j.neuroimage.2010.05.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell TM, Hutchinson R, Niculescu RS, Pereira F, Wang X, Just M, Newman S. Learning to decode cognitive states from brain images. Machine Learning. 2004;57:145–175. [Google Scholar]
- Mourão-Miranda J, Friston KJ, Brammer M. Dynamic discrimination analysis: A spatial–temporal SVM. NeuroImage. 2007;36:88–99. doi: 10.1016/j.neuroimage.2007.02.020. [DOI] [PubMed] [Google Scholar]
- Mourão-Miranda J, Ecker C, Sato JR, Brammer M. Dynamic changes in the mental rotation network revealed by pattern recognition analysis of fMRI data. Journal of Cognitive Neuroscience. 2009;21:890–904. doi: 10.1162/jocn.2009.21078. [DOI] [PubMed] [Google Scholar]
- Mumford JA, Turner BO, Ashby FG, Poldrack RA. Deconvolving BOLD activation in event-related designs for multivoxel pattern classification analyses. NeuroImage. 2012;59:2636–2343. doi: 10.1016/j.neuroimage.2011.08.076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norman KA, Polyn SM, Detre GJ, Haxby JV. Beyond mind-reading: Multi-voxel pattern analysis of fMRI data. Trends in Cognitive Sciences. 2006;10:424–430. doi: 10.1016/j.tics.2006.07.005. [DOI] [PubMed] [Google Scholar]
- Ollinger JM, Shulman GL, Corbetta M. Separating processes within a trial in event-related functional MRI: I. The method. NeuroImage. 2001a;13:210–217. doi: 10.1006/nimg.2000.0710. [DOI] [PubMed] [Google Scholar]
- Ollinger JM, Corbetta M, Shulman GL. Separating processes within a trial in event-related functional MRI: II. Analysis. NeuroImage. 2001b;13:218–229. doi: 10.1006/nimg.2000.0711. [DOI] [PubMed] [Google Scholar]
- Pereira F, Mitchell TM, Botvinick M. Machine learning classifiers and fMRI: A tutorial overview. NeuroImage. 2009;45:S199–S209. doi: 10.1016/j.neuroimage.2008.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen PM, Madsen KH, Lund TE, Hansen LK. Visualization of nonlinear kernel models in neuroimaging by sensitivity maps. NeuroImage. 2010;55:1120–1131. doi: 10.1016/j.neuroimage.2010.12.035. [DOI] [PubMed] [Google Scholar]
- Rissman J, Gazzaley A, D’Esposito M. Measuring functional connectivity during distinct stages of a cognitive task. NeuroImage. 2004;23:752–763. doi: 10.1016/j.neuroimage.2004.06.035. [DOI] [PubMed] [Google Scholar]
- Serences JT. A comparison of methods for characterizing the event-related BOLD timeseries in rapid fMRI. NeuroImage. 2004;21:1690–1700. doi: 10.1016/j.neuroimage.2003.12.021. [DOI] [PubMed] [Google Scholar]
- Smith SM, Jenkinson M, Woolrich MW, Beckmann CF, Behrens TEJ, Johansen-Berg H, Bannister PR, De Luca M, Drobnjak I, Flitney DE, Niazy R, Saunders J, Vickers J, Zhang Y, De Stefano N, Brady JM, Matthews PM. Advances in functional and structural MR image analysis and implementation as FSL. NeuroImage. 2004;23:S208–S219. doi: 10.1016/j.neuroimage.2004.07.051. [DOI] [PubMed] [Google Scholar]
- Soon CS, Brass M, Heinze HJ, Haynes JD. Unconscious determinants of free decisions in the human brain. Nat. Neurosci. 2008;11:543–545. doi: 10.1038/nn.2112. [DOI] [PubMed] [Google Scholar]
- Spiridon M, Kanwisher N. How distributed is visual category information in human occipito-temporal cortex? An fMRI study. Neuron. 2002;35:1157–1165. doi: 10.1016/s0896-6273(02)00877-2. [DOI] [PubMed] [Google Scholar]