Abstract
Deficiencies in mitochondrial protein production are associated with human disease and aging. Given the central role of transcription in gene expression, recent years have seen a renewed interest in understanding the molecular mechanisms controlling this process. In this review, we have focused on the mostly uncharacterized process of transcriptional termination. We review how several recent breakthroughs have provided insight into our understanding of the termination mechanism, the protein factors that mediate termination, and the functional relevance of different termination events. Furthermore, the identification of termination defects resulting from a number of mtDNA mutations has led to the suggestion that this could be a common mechanism influencing pathogenesis in a number of mitochondrial diseases, highlighting the importance of understanding the processes that regulate transcription in human mitochondria. We discuss how these recent findings set the stage for future studies on this important regulatory mechanism.
Keywords: mitochondrial transcription termination, MTERF, mitochondria, gene expression
1. Introduction
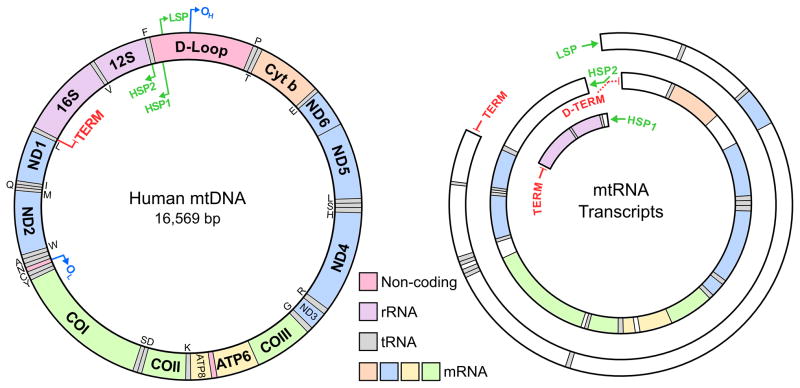
Mitochondria are involved in a vast array of cellular functions [1] but are most widely known for their role in generating ATP through oxidative phosphorylation [2]. This process is strictly dependent on expression of the mitochondrial genome, which encodes a small set of proteins that are essential for functionality of the electron-transport chain, as revealed by a number of human diseases linked to deficiencies in their synthesis or assembly (reviewed in [3–5]). The mitochondrial genome is encoded in a super-coiled double stranded circular DNA molecule of approximately 16.5 kilobases [6, 7]. Mammalian and other eukaryotic cells typically contain hundreds to thousands of mitochondrial DNA (mtDNA) genomes assembled into supramolecular structures called nucleoids [8–11]. In humans, each mtDNA genome contains 37 genes encoding the RNA components of the mitochondrial translation machinery: two ribosomal RNAs and 22 transfer RNAs, as well as mRNAs for 13 subunits of the respiratory chain [12] (see Figure 1). The two mtDNA strands have different buoyant densities in a cesium chloride gradient, which led to them being denoted heavy (H) and light (L) [13]. The 37 genes encoded in mtDNA are asymmetrically distributed, with the L-strand encoding only 8 tRNAs and one mRNA while the H-strand encodes the remaining two rRNAs, 14 tRNAs and 12 mRNAs [2]. In most, but not all cases, the mRNA and rRNA genes are flanked by at least one tRNA gene. Hence, excision of tRNA molecules is thought to be required for production of mature mRNA and rRNA molecules. This mode of RNA processing is known as the “tRNA punctuation model” [14]. In metazoa, the mitochondrial genome is extremely compact – there are no introns, and encoded polypeptide, tRNA and rRNA genes are smaller than their counterparts in the nucleus or in prokaryotes [15]. Furthermore, all of the genes are very closely spaced and little or no 5′ or 3′ flanking sequences exist on the mature mRNAs. The only sizeable non-coding region is the control region, or D-loop regulatory region, named after the triple-stranded structure or displacement loop that is formed by association of the nascent heavy strand in this region [2, 13, 16]. The roughly 1 kb long D-loop region contains two of the three promoters involved in transcription initiation (the L-strand promoter, LSP, and one of the two H-strand promoters, HSP1), as well as evolutionarily conserved regulatory sequences involved in DNA replication, D-loop formation, and presumably termination of transcription (see Figure 1) [17].
Figure 1. Schematic Representation of Human mtDNA: Genes, Transcripts, and cis-Acting Elements.
Transcription of the L-strand initiates from a single site, LSP, while transcription of the H-strand is initiated from two sites, HSP1 and HSP2. The MTERF1 protein binds to a site in the tRNALeu gene (TERM) and promotes termination of transcription initiated from LSP and HSP1. The putative termination site for HSP2 transcription (D-TERM) is also shown, although the termination factor acting at this site (if any) has not been identified. The tRNA genes encoded on each of the two strands are indicated with the standard one-letter symbols for amino acids. Abbreviations: COI, cytochrome c oxidase subunit I; COII, cytochrome c oxidase subunit II; COIII, cytochrome c oxidase subunit III; Cytb, cytochrome b; LSP, light-strand promoter; HSP, heavy-strand promoter; ND1–6, NADH dehydrogenase subunits 1–6; OH, origin of H-strand DNA replication; OL, origin of L-strand DNA replication.
2. Origins and initiation of mitochondrial transcription
The initial studies on mtDNA transcription were carried out about two decades ago, both in vitro and in vivo using cultured human and mouse cells. These studies characterized the mitochondrial RNAs in terms of their identity, structure and metabolic properties [18–22], and identified the cis-acting elements required for mitochondrial transcription [23–25]. Mapping studies of nascent transcripts and in vivo analysis of the kinetics of transcript synthesis suggested that transcription in human mitochondria starts at three different initiation points: one for the L-strand (LSP) and two for the H-strand (HSP1 and HSP2) [18, 19, 26].
The current model for transcription of the H-strand in humans involves two HSP promoters and two partially overlapping units. The first of these starts at the initiation site HSP1, which is located 19 bp upstream of the tRNAPhe gene (within the D-loop) and ends at the 3′ end of the 16S rRNA gene. The HSP1 transcription unit is responsible for the synthesis of the two ribosomal RNAs, tRNAPhe and tRNAVal (see Figure 1). The second transcription unit, operating with a frequency about 20 times lower [19], starts at the initiation site HSP2 which is located 2 bp upstream of the 5′ end of the 12S rRNA gene. Transcription from HSP2 produces a polycistronic RNA molecule covering almost the whole H-strand, encoding the mRNAs for the 12 H-strand encoded polypeptides and 12 tRNAs. Transcription of the L-strand begins at the LSP promoter, also located in the D-loop about 150 bp away from HSP, and produces only 8 tRNAs, one polypeptide mRNA and the H-strand replication primer. These primary transcripts are processed to produce the individual mRNA, rRNA, and tRNA molecules [14, 27, 28]. This transcription model explains how differential regulation of rRNA versus mRNA transcription could be accomplished through initiation of H-strand transcription at the two alternative sites [19]. Analysis of the effects of different ATP concentrations and intercalating drugs such as ethidium bromide on in organello transcription and the results of footprinting experiments have also provided support for this model of two H-strand transcription units [29–31]. However, it is important to note that this model has not yet been verified in vivo.
Mitochondria utilize unique enzyme systems responsible for mtDNA transcription (reviewed in [17, 32, 33]), which are nuclear-encoded, but distinct from those used in the nucleus. It is now more or less generally accepted that the core machinery involved in initiation of transcription consists of the mitochondrial polymerase (POLRMT) and two transcription factors, mitochondrial transcription factor A (TFAM) and mitochondrial transcription factor B2 (TFB2M) [34, 35]. POLRMT is a single-subunit RNA polymerase of the T-odd bacteriophage RNA polymerase family [36, 37]. However, unlike these other phage polymerases, which do not require interactions with transcription factors to initiate transcription, mammalian POLRMT requires the methyltransferase-related transcription factor, TFB2M, to initiate promoter-specific transcription from LSP and HSP1 in vitro [38, 39]. Initially, the TFB2M orthologue, TFB1M, was also thought to be essential for initiation, but recent studies indicate that only TFB2M is absolutely required for efficient, promoter-specific transcription in vitro [39]. Initiation also involves the high-mobility group (HMG) box DNA-binding protein, TFAM, which was the first mitochondrial transcription factor identified [40]. Human TFAM binds upstream of the LSP and HSP1 promoters [41, 42], and facilitates initiation in vitro by binding to TFB2M in a manner that requires its C-terminal tail [43] and possibly interactions with one of its two HMG box domains [42].
3. Termination of transcription by human MTERF1
Transcription initiated at the HSP1 promoter is preferentially terminated within the tRNALeu gene immediately downstream of the 16S rRNA gene [19, 44]. This site-specific termination event is mediated by a DNA-binding protein, mitochondrial transcription termination factor 1 (MTERF1), first identified by Attardi and colleagues [45], and later shown to be sufficient to mediate transcriptional termination in vitro [46]. Subsequent in vitro work has demonstrated that termination by MTERF1 is bidirectional [46, 47], and shows even higher efficiency when POLRMT proceeds in the direction of L-strand transcription [46].
In addition to binding the termination site in tRNALeu, MTERF1 may also bind an additional site in the HSP1 promoter region and stimulate transcription [48]. In a manner that bears some similarities with prokaryotic transcription, it has been proposed that simultaneous binding of MTERF1 to the HSP1 site and the canonical tRNALeu termination site causes a looping-out of the rDNA. This loop would allow recycling of POLRMT and other transcription components from the tRNALeu site to the HSP1 initiation site for efficiently starting another transcription cycle after termination occurs. This “ribomotor” model [49] is one way that the greater abundance of rRNAs compared to the downstream H-strand mRNAs can be explained [50], although differential stability of rRNA and mRNA species is thought to be a significant factor [50]. Moreover, this model would also explain the selective termination of transcription originating at HSP1 and suggests a mechanism for regulating the balance between termination and read-through. In addition, some evidence also suggests that MTERF1 may be involved in transcription initiation from the HSP2 [48]. The mechanistic basis for this remains unknown as does the precise protein components required for regulation at HSP2, which is the least studied of the three known human mtDNA promoters. Recently, Jacobs and colleagues showed that MTERF1 binds several sites in mtDNA in vivo and that altered MTERF1 protein levels affect mtDNA replication pausing at these sites [51], leading to a model in which MTERF1 may mediate transcription and replication passage on the same mtDNA molecule. It is important to note that the existence of such alternative MTERF1 binding sites is controversial, for example, as subsequent studies have not been able to reproduce binding of MTERF1 to the HSP [52]. Furthermore, in vitro experiments indicate that the specificity of MTERF1 seems to be exquisite for the tRNALeu site [53], implying that the affinity for other sites (if they exist) would be lower. This hypothesis is consistent with the striking degree of protection of the MTERF1 tRNALeu site observed in in vivo methylation [54] and DNaseI footprinting assays [55], as well as with ChIP studies that have predominately found MTERF1 bound to the tRNALeu site.
3.1 Structure of MTERF1
The x-ray crystallographic structure of human MTERF1 reveals that the 342-residue protein (mature form) has an all-α-helical structure that binds as a monomer to a 22-nucleotide termination sequence in the tRNALeu gene [53, 56]. The MTERF1 protein structure is modular, being configured around a motif of two α-helices and a 310 helix repeat known as the MTERF repeat. This MTERF repeat configuration was previously predicted by Roberti et al [57] and is similar in structure to other all-α-helical domains such as the HEAT domain [58], suggested to play a role in duplex DNA binding in DNA PK-cs [59], and the PUM/PUF domain, which is involved in RNA binding by PUMILIO proteins [60]. The 8 MTERF repeats found in MTERF1 constitute a helical fold that allows the protein to bind the major groove of duplex DNA containing the termination sequence. The extensive surface interactions observed between MTERF1 and the substrate DNA in the crystal structure indicate that the protein fold is likely dedicated to duplex DNA binding.
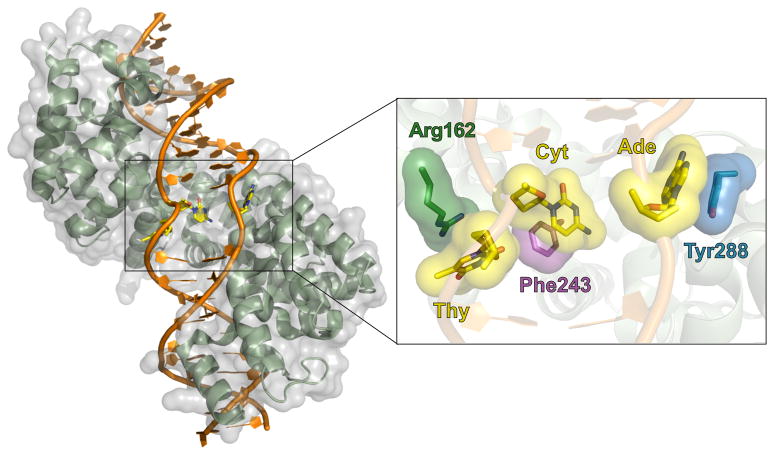
The MTERF1 crystal structure demonstrates that upon binding its target sequence, the protein alleviates the DNA duplex twist and promotes duplex melting and eversion of three nucleotide bases, leading to a novel and unique DNA binding mode (Figure 2). Although the mechanism by which it promotes this eversion, or base-flipping is not yet clear, MTERF1 stabilizes these three nucleotides in an extra-helical conformation through stacking interactions with three amino acid residues (Phe243, Tyr288 and Arg162) (see Figure 2 inset). We have demonstrated, using an MTERF1 triple mutant, that these three side chains are essential to maintaining the conformation observed in the crystal structure and although the MTERF1 triple mutant can still bind to the termination sequence, affinity for this termination site is significantly reduced. Moreover, the ability of the triple mutant MTERF1 to promote transcriptional termination in vitro is dramatically reduced.
Figure 2. The X-ray Crystallographic Structure of the Mitochondrial Transcription Termination Factor MTERF1 Bound to the tRNALeu Mitochondrial DNA Termination Sequence.
MTERF1 utilizes a unique DNA binding mode that results in duplex unwinding and eversion of three nucleotide bases. MTERF1 is shown in green with its molecular surface shown in transparent grey; the DNA termination site is shown in orange; everted bases are shown as yellow sticks. Inset: π-stacking interactions with the MTERF1 residues (dark green, purple, and blue) stabilize the three corresponding DNA bases (yellow) in an extrahelical position. These interactions are essential for termination activity.
With the exception of these stacking interactions and a few other important contacts (unpublished data), the majority of the interactions observed between MTERF1 and DNA involve the phosphate backbone of the substrate DNA sequence, and are therefore mostly electrostatic and non-specific. Sequence specificity appears to be determined in large part by a small number of key interactions between six arginine residues and guanine bases in the termination sequence. This type of major groove interaction is frequently seen in sequence-specific DNA binding proteins and is thought to be critical for MTERF function – we have shown that eliminating even a single one of these interactions can drastically affect both DNA binding and transcription termination [53]. Interestingly, this mechanism of sequence recognition implies that while the interaction between MTERF1 and its binding sequence involves contacts with 20 base pairs, only six of the 40 bases appear to be initially actively recognized by the protein. The extensive total number of protein-DNA interactions suggest that the entirety of the MTERF1 fold is involved in binding the termination sequence, but it is not immediately apparent how a single MTERF1 molecule could simultaneously bind both the HSP initiation and termination sites in the transcriptional loop model [48]. One possibility is that an additional molecule of MTERF1 or other factor(s) may mediate the association of these two sites and facilitate loop formation.
3.2 Model for termination by MTERF1
The MTERF1 crystal structure suggests a binding mechanism that involves establishment of site-specific interactions for sequence recognition followed by melting and unwinding of the DNA duplex. This unwinding would presumably destabilize base-pairing of the central nucleotides in the recognition sequence and facilitate subsequent base flipping, thereby stabilizing MTERF1 on the substrate DNA. We have demonstrated that stable MTERF1 binding and termination in vitro is dependent on base flipping, and the ability of MTERF1 to promote termination appears to be at least partially dependent on the strength of the cumulative interactions between MTERF1 and the termination sequence. This would suggest a model of transcription in which MTERF1 acts as a “roadblock,” preventing or interfering with transcriptional elongation. This model is consistent with the observation that MTERF1 terminates transcription bi-directionally and can arrest elongation by heterologous polymerases ([47] and our own unpublished results). However, in vitro termination by MTERF1 displays a distinct polarity, with MTERF1 being more efficient when terminating transcription originating from the light strand promoter than the heavy strand promoter [46, 53]. This polarity may be a result of the majority of protein-DNA interactions being established with the light strand [53] (ie, the strand transcribed from the LSP promoter) as well as the higher affinity of MTERF1 for this strand, as observed by Nam et al [61]. This suggests a mechanism in which MTERF1 might transiently bind to single-stranded DNA, although the measured affinity of MTERF1 for single-stranded DNA is extremely low ([61] and our own unpublished observations). Nevertheless, except for the asymmetrical distribution of interactions, no obvious structural feature provides an explanation for the observed polarity of termination. One possibility is that the observed orientation dependence of termination activity is due to the unique conformation of MTERF1 on DNA, although interactions between MTERF1 and mitochondrial POLRMT (or additional elongation factors) may also influence the polarity of termination events. Furthermore, it is not yet known whether other proteins can modulate termination polarity in vivo.
3.3 Significance of termination at tRNALeu
The strong polarity observed in termination assays in vitro combined with the lack of in vivo evidence supporting a role in HSP termination suggest that MTERF1-mediated HSP termination might only be a secondary role and that the main function of MTERF1 is termination of LSP transcription at the tRNALeu site. This hypothesis is supported by the fact that the light-strand does not encode any additional genes beyond tRNALeu. Recent observations in vivo have shown that manipulation of MTERF1 expression levels is strongly correlated with alterations in the relative amounts of antisense transcripts on both sides of the tRNALeu termination site [62]. These findings are consistent with the notion that termination at this site may be important for preventing the accumulation of antisense transcripts that would otherwise interfere with the assembly of the rRNAs into ribosomes. This idea would imply that MTERF1 function is, as originally thought, important for ribosome biogenesis, albeit by a different mechanism than first proposed.
3.4 Termination of HSP transcription at the distal site
Any discussion of mitochondrial termination is incomplete when only considering termination at the tRNALeu site. Evidently some degree of read-through must occur at the tRNALeu site so that HSP transcription can progress beyond this site and generate most of the mitochondrial mRNAs. How such read-through is regulated, and whether it is specific to transcription originating from HSP2 is in itself an interesting question for which no answer yet exists. In any case, read-through HSP transcription is thought to ultimately terminate at a distal site within the D-loop. The mouse HSP distal termination site was originally identified as a 22 bp region within the D-loop containing a conserved A/T rich sequence motif. Initial in vitro studies suggested that termination at this site is unidirectional and requires sequence-specific DNA binding proteins [63], but the mechanisms controlling distal termination in human mitochondria are far from being well understood. Since MTERF1 predominantly binds to the tRNALeu site, it is possible that distal termination may not depend on MTERF1, therefore raising the question of how termination is achieved at that site. Recent work by Wanrooij et al suggests that the mitochondrial polymerase, POLRMT, utilizes a mechanism similar to bacterial rho-independent termination to generate the RNA primers necessary for mitochondrial replication [64]. In T7 bacteriophages transcription termination occurs as a consequence of the polymerase interacting with spontaneously generated double-stranded RNA hairpins and falling off DNA due to the weak affinity of T7 RNA polymerase for double stranded structures [65]. In the case of mitochondrial POLRMT, Wanrooij et al have shown that stable G-quadruplex structures can form in the nascent RNA, suggesting that these structures then mediate transcription termination in a way reminiscent of RNA hairpins in rho-independent termination [64]. It is therefore possible that a similar mechanism is responsible for termination of transcription at the HSP distal site. Nevertheless, it cannot be excluded that MTERF1 might influence in termination at that site. As mentioned earlier, alternative MTERF1 binding sites have been described in the proximity of the distal site [51], and these sites will need to be further characterized in order to elucidate how termination is achieved and what role, if any, MTERF1 plays in the process. Finally, it is also possible that proteins other than MTERF1 might contribute to distal HSP termination. Preliminary experiments have identified several proteins that appear to bind at the mouse HSP distal termination region, including the Leucine-rich pentatricopeptide-repeat containing protein (LRPPRC) [66]. This protein is essential for the expression of mtDNA encoded respiratory chain subunits, and LRPPC deficient cells were reported to have a reduction in both oxygen consumption and expression of mRNA and tRNA [66, 67]. Recent in vivo work in an RNAi system further supports the concept that LRPPRC is involved in regulating expression of mitochondrial mRNAs [68]. Moreover, several homologues of MTERF1 have been identified in recent years, and like LRPPRC, they too have been implicated in the regulation of mitochondrial transcription and gene expression.
4. The MTERF family of proteins
MTERF1 is the founding member of a family of related proteins that are widely conserved throughout evolution (although notoriously absent from yeast) and that are defined by the presence of several copies of the MTERF motif. Three human MTERF1 paralogues, MTERF2-4, were identified in 2005 by Linder et al. and were all predicted to localize to mitochondria [69]. Three of these proteins, MTERF1-3, have since been demonstrated to influence the transcription process [52, 53, 70], while recent work suggests that MTERF4 may play a role in mitochondrial translation [71]. Interestingly, the structural similarity that MTERF1 shares with MTERF3 (rmsd of 2.7 Å over 218 C-α atoms), despite the latter being crystallized in the absence of substrate [72, 73], implies that MTERF proteins share a common fold and supports the idea that they have evolved to bind nucleic acids.
4.1 Mitochondrial transcription termination factor 2 (MTERF2)
MTERF2 (also known as mTERFL or mTERF.D3), was first characterized several years ago [74, 75]. Initial in vivo studies demonstrated that MTERF2 overexpression inhibits cell growth and that its expression is regulated in a reciprocal manner with that of MTERF1 [74]. Subsequent work has shown that MTERF2 is able to bind nonspecifically to DNA and is present in nucleoids [76]. Knocking out MTERF2 in mice is not lethal, but the mice exhibit respiratory defects when metabolically challenged with a high fat/low carbohydrate diet [77]. Under these conditions, the mice exhibit decreased steady-state levels of most mRNA and tRNA transcripts and reduced translation of several proteins. When fed a standard diet, however, the mice have normal levels of most mitochondrial RNA transcripts, with the exception of promoter-proximal tRNA species, which are increased, and promoter-distal tRNAs, which are decreased. Interestingly, it was observed that MTERF2 exhibited a ~10-fold preference for binding in the HSP promoter region that was confirmed in vivo by ChIP analysis [77]. These results may suggest a role for MTERF2 as a positive modulator of mitochondrial transcription, but the current evidence is unclear and the situation may be more complex. For instance, sequence-specific mtDNA binding by MTERF2 was not observed in cell culture experiments [78]. In addition, overexpression of MTERF2 in cells resulted in modest mtDNA copy number depletion (as well as an accumulation of specific replication intermediates), while MTERF2 knockout appears to cause an increase in mtDNA copy number. These findings may suggest an alternative or additional role for MTERF2 in regulating replication fork progression. While the effect of overexpressing MTERF2 on mtDNA copy number has since been confirmed by an independent study [79], further studies are needed to investigate the functions of MTERF2 in replication and transcription.
4.2 Mitochondrial termination factor 3 (MTERF3)
The x-ray crystallographic structure of MTERF3 (also known as mTERF.D1 or CGI-12) in the absence of DNA has been solved [72, 73] and shows striking similarity to the MTERF1 structure. Both proteins have nearly identical half-doughnut shapes consisting of MTERF motif repeats, suggesting that MTERF family members have similar folds and bind nucleic acids. However, there are also interesting differences, as only one of the five arginines necessary for sequence-specific DNA binding and only one of the three amino acids that stabilize base flipping are present in MTERF3 [73].
The in vivo function of this protein has recently been addressed through gene knockout studies in mice [52, 80]. Mammalian MTERF3 has been shown to localize to mitochondria, and while a global knockout of MTERF3 in mice is embryonic lethal, Park et al have analyzed a heart-specific knockout in detail [52]. Heart tissue from these mice exhibits an aberrant mitochondrial transcript profile leading to impaired respiration and subsequent demise. Specifically, both mRNA and tRNA transcripts proximal to the promoters are increased, whereas those more distal to the promoters are decreased. This imbalance in the steady-state level of promoter-proximal and promoter-distal tRNA species is reminiscent of the situation in the MTERF2 knockout mice. In addition, the authors observed that MTERF3 is able to bind mtDNA in the promoter region and that its immunodepletion from mitochondrial extracts leads to increased transcription. These findings led the authors to propose that increased transcription initiation in the absence of MTERF3 leads to collision of transcription complexes on opposite mtDNA strands and, hence, incomplete transcription of each strand. The Drosophila homolog of MTERF3, known as D-MTERF3, has been identified and it was recently reported that overexpression of this protein results in a modest decrease in mRNA levels [70]. Overall, the current evidence seems to point to a role for MTERF3 as a negative regulator of mitochondrial transcription. Interestingly, Jacobs and colleagues have also recently observed that MTERF3 overexpression has an inhibitory effect on mitochondrial replication, similar to that of MTERF2 [78].
4.3 Mitochondrial transcription termination factor 4 (MTERF4)
To date, MTERF4 (also known as mTERF.D2) has been the least studied MTERF family member but recent work has provided the first insight into the function of this protein in mitochondria. In vivo and in vitro studies have confirmed that MTERF4 localizes to mitochondria and demonstrated that this protein, like MTERF2 and −3, is essential to embryonic development in the mouse [71]. Furthermore, like MTERF3, a heart-specific conditional knockout of MTERF4 results in mitochondrial cardiomyopathy and respiratory chain deficiency. A substantial increase in the steady state levels of all mRNAs and rRNAs was observed (in some cases ~300% of normal levels). LSP transcripts encoding tRNAs were also increased for promoter-proximal genes, while those for promoter-distal genes were either unchanged or decreased. Despite the increased levels of transcripts, the MTERF4 knockout cells showed impaired translation and defective ribosome assembly, suggesting that the increase in de novo transcription is likely a response to the severe impairment in translation. MTERF4 is predicted to have a fold similar to MTERF1 and MTERF3, with positively charged surface areas for binding of nucleic acids [72]. Consistently, it was recently shown that MTERF4 binds the 16S rRNA and forms a stable stoichiometric complex with NSUN4, a mitochondrial rRNA methyltransferase, therefore targeting this protein to the large ribosomal subunit [71]. While this evidence points to a primary role in regulating translation, it is tempting to speculate that MTERF4 may also modulate mtDNA transcription, particularly in the context of the prokaryote-like organization of mitochondria, with no compartmentalization between transcription and translation, which may enable direct crosstalk between these two processes [81].
Overall, these new findings on MTERF family members underscore the importance of these proteins in regulating mitochondrial gene expression. The results strongly suggest that these proteins have evolved to bind nucleic acids, although with remarkable differences in their substrate specificity, being capable of specific and/or unspecific binding to ssDNA, dsDNA and RNA in the case of MTERF4. Furthermore, the different genetic experiments highlight their potential to serve multiple functional roles.
5. Termination of mitochondrial transcription in invertebrates
The MTERF protein family includes two invertebrate mitochondrial transcription termination factors, sea urchin mtDBP and Dros0ophila DmTTF. Both are involved in terminating transcription, and have also been implicated in transcription initiation and regulation of mtDNA replication. This multiplicity of functions appears to be a theme in the MTERF protein family, and our current understanding of these two invertebrate termination factors can provide insight into the roles of MTERF protein family members as well as the termination process.
5.1 Sea urchin mitochondrial transcription termination factor (mtDBP)
The mtDNA of the sea urchin Paracentrotus lividus encodes the same genes found in mammalian mtDNA, although the genetic organization is slightly different – the ribosomal RNA genes are separated by a 3.3 kb region that contains a cluster of 15 tRNA genes as well as genes for two polypeptides [82]. Like mammalian mtDNA, the sea urchin mtDNA also contains a non-coding D-loop region, although it is very short (~130 bp) and is located in the tRNA gene cluster downstream of the 12S rRNA gene. Mitochondrial transcription is thought to occur via multiple transcription units [83], but the precise initiation sites have not yet been defined. The sea urchin mitochondrial transcription termination factor, mtDBP, has been studied in detail by Cantatore and colleagues, and is known to bind with high affinity to two sites in the sea urchin mtDNA, one of which is located in the D-loop region, near the origin of replication [84]. The observation that mtDBP displays a significant homology with human MTERF1 led to the idea that it could possess a transcription termination activity [85], and initial in vitro experiments showed that indeed, mtDBP exhibits bidirectional transcription termination activity in the presence of human mitochondrial RNA polymerase, and unidirectional termination activity with bacteriophage polymerases [86]. More recently, an in vitro transcription assay using mtDBP and the endogenous sea urchin mitochondrial polymerase (mtRNAP) showed that termination of transcription occurred in a mtDBP-dependent manner when the enzyme approached the D-loop region binding site in the direction of L-strand transcription [87]. However, when the enzyme encountered the binding site in the opposite direction, termination occurred independently of mtDBP. This has led to the suggestion that, like human MTERF1, mtDBP acts as a polar termination factor, and that mitochondrial transcription termination in sea urchin may take place by two alternative modes [70]. In vitro experiments have also shown that mtDBP exhibits bidirectional contrahelicase activity [88], suggesting a role as a negative regulator of replication. Furthermore, Roberti et al have observed that H-strand read-through transcription and simultaneous sequence-dependent termination at the mtDBP binding site causes mtDBP to fall off the mtDNA, an event which may relieve helicase impairment and therefore allow mtDNA replication to resume [70]. Thus, it is likely that mtDBP may help regulate the balance between mtDNA transcription and replication in sea urchin mitochondria. Similarly, a role in mammalian mtDNA replication has also been suggested for MTERF1, based in part on the observation that the level of its expression in cultured human cells influences replication pausing in the vicinity of MTERF1 binding sites [89]. In a manner similar to that of mtDBP, it has been proposed that MTERF1 may facilitate the orderly passage of oppositely oriented transcription and replication complexes, helping to prevent collapse of the replication fork and subsequent generation of recombinogenic ends that could facilitate detrimental rearrangements in the mtDNA [89].
5.2 Drosophila transcription termination factor (DmTTF)
The Drosophila melanogaster mtDNA genome is larger than that of humans or sea urchin, mostly due to a large non-coding region of about 4.6 kbp that is A–T rich [90]. The genomic architecture is also markedly different from that of human and sea urchin genomes, with four gene clusters being equally distributed between the light and heavy strands. Initial mapping studies suggested that Drosophila mtDNA could be transcribed by multiple transcription units and several transcription termination sites were hypothesized [91]. In 2003, a Drosophila homologue of human MTERF1 and sea urchin mtDBP, known as DmTTF (for Drosophila mitochondrial transcription termination factor) was identified and characterized [92]. DmTTF binds two non-coding sequences located at the end gene blocks that are transcribed in opposite directions – one sequence between the tRNAGlu and tRNAPhe genes and another between tRNASer and a polypeptide gene. The locations of these DmTTF binding sequences coincide with two previously predicted transcription termination sites, which led to the suggestion that DmTTF may function as a transcription termination factor [91]. Subsequently, this function was confirmed in vitro by measuring the ability of DmTTF to arrest mitochondrial transcription in the presence of human POLRMT and a chimeric DNA template containing both the human HSP promoter and DmTTF binding site [93]. Like mtDBP, DmTTF acts in vitro to terminate transcription with directional properties depending on the polymerase used [92]. Although the mechanism of sequence recognition by DmTTF is not yet known, it is interesting to note that DmTTF binds to a sequence containing only A–T base pairs, indicating that sequence recognition is unlikely to involve arginine-guanine interactions as seen for MTERF1, further highlighting the flexibility of the protein-DNA interaction in different MTERF family members. Analyses of DmTTF function in cultured cells have shown that manipulation of its expression leads to complex effects on transcript levels [94], and whether or not DmTTF plays a role in replication (like its homologues MTERF1 and mtDBP) has not yet been determined.
6. Termination defects and implications for disease
Pathogenic mtDNA mutations were first reported in human patients more than two decades ago [95–97], and these important discoveries provided a genetic basis for the classification of mitochondrial disease. To date, a significant number of pathogenic mtDNA mutations have been identified and many of them have associated clinical phenotypes that have been characterized (reviewed in refs [98–100]). These mutations in the mtDNA often result in altered gene expression and therefore contribute to respiratory defects [98, 99, 101] and other mitochondrial pathologies that have been implicated in aging [102–104] as well as human disease. Most mtDNA mutations are thought to result in either defects in mitochondrial DNA maintenance or defects in mitochondrial translation. Such pathogenic defects in mitochondrial translation are frequently associated with mutations in tRNA genes, and interestingly, the tRNALeu gene contains the highest number of identified mutations of any mitochondrial tRNA gene. Not surprisingly, several of these mutations occur in the MTERF1 binding sequence. One such mutation, A3243G, is associated with mitochondrial myopathy, encephalomyopathy, lactic acidosis, and stroke-like episodes (MELAS), a syndrome which is characterized by lactic acidosis, episodic vomiting, seizures and recurrent cerebral insults resembling strokes that lead to hemiparesis, hemianopsia or cortical blindness [105]. The A3243G mutation, which has been shown to affect transcription in vitro [106], is the most common mtDNA mutation with a prevalence of more than 200 per 100,000 in a recent Australian study [107], and accounts for the majority (80%) of MELAS cases [108, 109]. Despite the observed in vitro termination defect, in vivo studies have shown that the A3243G mutation does not affect the balance between mitochondrial HSP transcripts upstream and downstream of the termination sequence [110]. Moreover, it has been shown that A3243G leads to defects in tRNA function, suggesting that these are the main cause of MELAS pathology rather than transcriptional alterations [111]. However, it is important to note that the transcript ratios measured in these in vivo studies only provide a measure of HSP termination, and therefore it is possible that defects in LSP termination might exist in A3243G carriers and contribute to MELAS pathogenesis. Furthermore, it has also been suggested that termination may be regulated in a tissue-specific manner in vivo [112], such that termination defects occur predominately in particular cell types that have not yet been studied in vivo.
After the MTERF1 structure was solved, revealing the basis for MTERF1 sequence specificity, transcription termination assays performed on seven other mutations within the termination sequence also revealed effects on transcriptional termination (see Table 1). Of these, two showed an effect that was significantly stronger than that observed with the A3243G mutation [53]. These mutations include the G3249A mutation that causes a variant of Kearns-Sayre syndrome [113] as well as the G3242A mutation, associated with an uncharacterized mitochondrial disorder [114]. Although tRNA mutations are generally thought to predominately affect the function of the mature tRNAs [115], these in vitro observations raise the possibility that the pathogenic effects of these mutations are also related to their effect on termination by MTERF1. Furthermore, given that MTERF1 appears to be needed for initiation at HSP2 and recycling of transcription at HSP1, it is possible that MTERF1 might play a role in promoter regulation, which could also be impacted by A3243G and other mtDNA mutations.
Table 1.
Pathogenic mtDNA mutations in the mitochondrial tRNALeu termination sequence
| mtDNA Mutation | Termination defect | Functional tRNA defect | Clinical disease |
|---|---|---|---|
| A3236G | + [53] | − | Sporadic bilateral optic neuropathy [119] |
| G3242A | ++ [53] | − | Uncharacterized mitochondrial myopathy [113] |
| A3243G | +++ [53, 105] | + [110, 120, 121] | MELAS (>80% of cases) [107, 108] |
| A3243T | ++ [53] | + [122] | Mitochondrial encephalopathy [123] |
| G3244A | ++ [53] | + [110] | Associated with MELAS, not fully characterized [113] |
| G3249A | +++ [53] | − | Variant of Kearns-Sayre syndrome [112] |
| T3250C | + [53] | + [124] | Mitochondrial myopathy [125] |
| A3251G | + [53] | − | Mitochondrial myopathy [126] |
| A3252G | − [53] | − | MELAS [127] |
In considering the effects of these mutations it is important to keep in mind that, in addition to potential effects in transcription, MTERF1 appears to be able to modulate replication and therefore affect mitochondrial DNA maintenance [89]. Moreover, patients harboring pathogenic mtDNA mutations often are heteroplasmic, i.e. have a mixture of wild-type and mutated mtDNA molecules [116]. Furthermore, the appearance and severity of clinical symptoms seems to correlate with the mutation load [117]. Interestingly, it has been reported that long-term culture of human cells with the A3243G mtDNA mutation results in an increase in the relative levels of A3243G mutant mtDNA [118, 119], suggesting the mutation confers a replicative advantage. With this in mind, it is tempting to speculate that in some cases, the primary deficiency caused by mutations in the tRNALeu MTERF1 binding site may be a defect in tRNA function, but the additional effect of these mutations on MTERF1 binding and/or function results in more rapid replication of the mutated genomes, thus facilitating an increase in the mutation load. This, in turn, would contribute to the appearance of clinical symptoms and/or an increase in their severity.
Given the unusual and important roles of MTERF family members in transcription termination and initiation, as well as replication, more in-depth study of this interesting class of proteins is clearly warranted. A more detailed molecular understanding of how these proteins interact with the known transcription machinery, how they interact with each other, and how they are utilized differentially to control mtDNA expression in vivo will also provide insight into the pathogenesis of MELAS and other mitochondrial diseases.
HIGHLIGHTS.
Termination of transcription in mitochondria is not well characterized.
Human MTERF1 mediates termination at a major site in tRNALeu.
MTERF1 utilizes a unique binding mechanism to promote termination.
There are connections between transcription termination and mtDNA replication.
Other MTERF family members have been implicated in termination and replication.
Acknowledgments
The authors wish to thank the members of the Garcia-Diaz Lab for insightful discussions and suggestions. This work was supported by NIH R00 ES015421 to M.G.-D. and by the Medical Scientist Training Program NIH T32-GM008444 to K.E.G.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Kip E. Guja, Email: kip@pharm.stonybrook.edu.
Miguel Garcia-Diaz, Email: mgd@pharm.stonybrook.edu.
References
- 1.McBride HM, Neuspiel M, Wasiak S. Mitochondria: more than just a powerhouse. Current biology. 2006;16:551–560. doi: 10.1016/j.cub.2006.06.054. [DOI] [PubMed] [Google Scholar]
- 2.Attardi G, Schatz G. Biogenesis of mitochondria. Annu Rev Cell Biol. 1988;4:289–333. doi: 10.1146/annurev.cb.04.110188.001445. [DOI] [PubMed] [Google Scholar]
- 3.Wallace DC. Diseases of the mitochondrial DNA. Annual review of biochemistry. 1992;61:1175–1212. doi: 10.1146/annurev.bi.61.070192.005523. [DOI] [PubMed] [Google Scholar]
- 4.Wallace DC. Mitochondrial DNA mutations in disease and aging. Environmental and molecular mutagenesis. 2010;51:440–450. doi: 10.1002/em.20586. [DOI] [PubMed] [Google Scholar]
- 5.Shadel GS. Coupling the mitochondrial transcription machinery to human disease. Trends in genetics. 2004;20:513–519. doi: 10.1016/j.tig.2004.08.005. [DOI] [PubMed] [Google Scholar]
- 6.Anderson S, Bankier AT, Barrell BG, de Bruijn MH, Coulson AR, Drouin J, Eperon IC, Nierlich DP, Roe BA, Sanger F, Schreier PH, Smith AJ, Staden R, Young IG. Sequence and organization of the human mitochondrial genome. Nature. 1981;290:457–465. doi: 10.1038/290457a0. [DOI] [PubMed] [Google Scholar]
- 7.Andrews RM, Kubacka I, Chinnery PF, Lightowlers RN, Turnbull DM, Howell N. Reanalysis and revision of the Cambridge reference sequence for human mitochondrial DNA. Nature genetics. 1999;23:147. doi: 10.1038/13779. [DOI] [PubMed] [Google Scholar]
- 8.Miyakawa I, Sando N, Kawano S, Nakamura S, Kuroiwa T. Isolation of morphologically intact mitochondrial nucleoids from the yeast, Saccharomyces cerevisiae. J Cell Sci. 1987;88:431–439. doi: 10.1242/jcs.88.4.431. [DOI] [PubMed] [Google Scholar]
- 9.Kaufman BA, Newman SM, Hallberg RL, Slaughter CA, Perlman PS, Butow RA. In organello formaldehyde crosslinking of proteins to mtDNA: identification of bifunctional proteins. Proceedings of the National Academy of Sciences of the United States of America. 2000;97:7772–7777. doi: 10.1073/pnas.140063197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jacobs HT, Lehtinen SK, Spelbrink JN. No sex please, we’re mitochondria: a hypothesis on the somatic unit of inheritance of mammalian mtDNA. Bioessays. 2000;22:564–572. doi: 10.1002/(SICI)1521-1878(200006)22:6<564::AID-BIES9>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 11.Bogenhagen DF, Rousseau D, Burke S. The layered structure of human mitochondrial DNA nucleoids. The Journal of biological chemistry. 2008;283:3665–3675. doi: 10.1074/jbc.M708444200. [DOI] [PubMed] [Google Scholar]
- 12.Shuster RC, Rubenstein AJ, Wallace DC. Mitochondrial DNA in anucleate human blood cells. Biochemical and biophysical research communications. 1988;155:1360–1365. doi: 10.1016/s0006-291x(88)81291-9. [DOI] [PubMed] [Google Scholar]
- 13.Shadel GS, Clayton DA. Mitochondrial DNA maintenance in vertebrates. Annual review of biochemistry. 1997;66:409–435. doi: 10.1146/annurev.biochem.66.1.409. [DOI] [PubMed] [Google Scholar]
- 14.Ojala D, Montoya J, Attardi G. tRNA punctuation model of RNA processing in human mitochondria. Nature. 1981;290:470–474. doi: 10.1038/290470a0. [DOI] [PubMed] [Google Scholar]
- 15.Schneider A, Ebert D. Covariation of mitochondrial genome size with gene lengths: evidence for gene length reduction during mitochondrial evolution. Journal of molecular evolution. 2004;59:90–96. doi: 10.1007/s00239-004-2607-x. [DOI] [PubMed] [Google Scholar]
- 16.Fernández-Silva P, Enriquez JA, Montoya J. Replication and transcription of mammalian mitochondrial DNA. Experimental physiology. 2003;88:41–56. doi: 10.1113/eph8802514. [DOI] [PubMed] [Google Scholar]
- 17.Shadel GS. Expression and maintenance of mitochondrial DNA: new insights into human disease pathology. Am J Pathol. 2008;172:1445–1456. doi: 10.2353/ajpath.2008.071163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Montoya J, Christianson T, Levens D, Rabinowitz M, Attardi G. Identification of initiation sites for heavy-strand and light-strand transcription in human mitochondrial DNA. Proceedings of the National Academy of Sciences of the United States of America. 1982;79:7195–7199. doi: 10.1073/pnas.79.23.7195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Montoya J, Gaines GL, Attardi G. The pattern of transcription of the human mitochondrial rRNA genes reveals two overlapping transcription units. Cell. 1983;34:151–159. doi: 10.1016/0092-8674(83)90145-9. [DOI] [PubMed] [Google Scholar]
- 20.Walberg MW, Clayton DA. In vitro transcription of human mitochondrial DNA. Identification of specific light strand transcripts from the displacement loop region. The Journal of biological chemistry. 1983;258:1268–1275. [PubMed] [Google Scholar]
- 21.Chang DD, Clayton DA. Precise identification of individual promoters for transcription of each strand of human mitochondrial DNA. Cell. 1984;36:635–643. doi: 10.1016/0092-8674(84)90343-x. [DOI] [PubMed] [Google Scholar]
- 22.Yoza BK, Bogenhagen DF. Identification and in vitro capping of a primary transcript of human mitochondrial DNA. The Journal of biological chemistry. 1984;259:3909–3915. [PubMed] [Google Scholar]
- 23.Chang DD, Clayton DA. Precise assignment of the light-strand promoter of mouse mitochondrial DNA: a functional promoter consists of multiple upstream domains. Molecular and cellular biology. 1986;6:3253–3261. doi: 10.1128/mcb.6.9.3253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chang DD, Clayton DA. Precise assignment of the heavy-strand promoter of mouse mitochondrial DNA: cognate start sites are not required for transcriptional initiation. Molecular and cellular biology. 1986;6:3262–3267. doi: 10.1128/mcb.6.9.3262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fisher RP, Topper JN, Clayton DA. Promoter selection in human mitochondria involves binding of a transcription factor to orientation-independent upstream regulatory elements. Cell. 1987;50:247–258. doi: 10.1016/0092-8674(87)90220-0. [DOI] [PubMed] [Google Scholar]
- 26.Cantatore P, Attardi G. Mapping of nascent light and heavy strand transcripts on the physical map of HeLa cell mitochondrial DNA. Nucleic Acids Research. 1980;8:2605–2625. doi: 10.1093/nar/8.12.2605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Clayton DA. Replication and transcription of vertebrate mitochondrial DNA. Annu Rev Cell Biol. 1991;7:453–478. doi: 10.1146/annurev.cb.07.110191.002321. [DOI] [PubMed] [Google Scholar]
- 28.Montoya J, Ojala D, Attardi G. Distinctive features of the 5′-terminal sequences of the human mitochondrial mRNAs. Nature. 1981;290:465–470. doi: 10.1038/290465a0. [DOI] [PubMed] [Google Scholar]
- 29.Gaines G, Attardi G. Highly efficient RNA-synthesizing system that uses isolated human mitochondria: new initiation events and in vivo-like processing patterns. Molecular and cellular biology. 1984;4:1605–1617. doi: 10.1128/mcb.4.8.1605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gaines G, Rossi C, Attardi G. Markedly different ATP requirements for rRNA synthesis and mtDNA light strand transcription versus mRNA synthesis in isolated human mitochondria. The Journal of biological chemistry. 1987;262:1907–1915. [PubMed] [Google Scholar]
- 31.Micol V, Fernandez-Silva P, Attardi G. Functional analysis of in vivo and in organello footprinting of HeLa cell mitochondrial DNA in relationship to ATP and ethidium bromide effects on transcription. The Journal of biological chemistry. 1997;272:18896–18904. doi: 10.1074/jbc.272.30.18896. [DOI] [PubMed] [Google Scholar]
- 32.Bonawitz ND, Clayton DA, Shadel GS. Initiation and beyond: multiple functions of the human mitochondrial transcription machinery. Molecular cell. 2006;24:813–825. doi: 10.1016/j.molcel.2006.11.024. [DOI] [PubMed] [Google Scholar]
- 33.Falkenberg M, Larsson NG, Gustafsson CM. DNA Replication and Transcription in Mammalian Mitochondria. Annual review of biochemistry. 2007;76:679–699. doi: 10.1146/annurev.biochem.76.060305.152028. [DOI] [PubMed] [Google Scholar]
- 34.Falkenberg M, Gaspari M, Rantanen A, Trifunovic A, Larsson NG, Gustafsson CM. Mitochondrial transcription factors B1 and B2 activate transcription of human mtDNA. Nature genetics. 2002;31:289–294. doi: 10.1038/ng909. [DOI] [PubMed] [Google Scholar]
- 35.Gaspari M, Falkenberg M, Gustafsson CM. The mitochondrial RNA polymerase contributes critically to promoter specificity in mammalian cells. The EMBO journal. 2004;23:4606–4614. doi: 10.1038/sj.emboj.7600465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Masters BS, Stohl LL, Clayton DA. Yeast mitochondrial RNA polymerase is homologous to those encoded by bacteriophages T3 and T7. Cell. 1987;51:89–99. doi: 10.1016/0092-8674(87)90013-4. [DOI] [PubMed] [Google Scholar]
- 37.Tiranti V, Savoia A, Forti F, D’Apolito MF, Centra M, Rocchi M, Zeviani M. Identification of the gene encoding the human mitochondrial RNA polymerase (h-mtRPOL) by cyberscreening of the Expressed Sequence Tags database. Human molecular genetics. 1997;6:615–625. doi: 10.1093/hmg/6.4.615. [DOI] [PubMed] [Google Scholar]
- 38.Litonin D, Sologub M, Shi Y, Savkina M, Anikin M, Falkenberg M, Gustafsson CM, Temiakov D. Human mitochondrial transcription revisited: only TFAM and TFB2M are required for transcription of the mitochondrial genes in vitro. The Journal of biological chemistry. 2010;285:18129–18133. doi: 10.1074/jbc.C110.128918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shutt TE, Lodeiro MF, Cotney J, Cameron CE, Shadel GS. Core human mitochondrial transcription apparatus is a regulated two-component system in vitro. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:12133–12138. doi: 10.1073/pnas.0910581107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fisher RP, Clayton DA. A transcription factor required for promoter recognition by human mitochondrial RNA polymerase. Accurate initiation at the heavy- and light-strand promoters dissected and reconstituted in vitro. The Journal of biological chemistry. 1985;260:11330–11338. [PubMed] [Google Scholar]
- 41.Kaufman BA, Durisic N, Mativetsky JM, Costantino S, Hancock MA, Grutter P, Shoubridge EA. The mitochondrial transcription factor TFAM coordinates the assembly of multiple DNA molecules into nucleoid-like structures. Mol Biol Cell. 2007;18:3225–3236. doi: 10.1091/mbc.E07-05-0404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gangelhoff TA, Mungalachetty PS, Nix JC, Churchill ME. Structural analysis and DNA binding of the HMG domains of the human mitochondrial transcription factor A. Nucleic Acids Research. 2009;37:3153–3164. doi: 10.1093/nar/gkp157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.McCulloch V, Shadel GS. Human mitochondrial transcription factor B1 interacts with the C-terminal activation region of h-mtTFA and stimulates transcription independently of its RNA methyltransferase activity. Molecular and cellular biology. 2003;23:5816–5824. doi: 10.1128/MCB.23.16.5816-5824.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Martínez-Azorín F. The mitochondrial ribomotor hypothesis. IUBMB life. 2005;57:27–30. doi: 10.1080/15216540500088755. [DOI] [PubMed] [Google Scholar]
- 45.Kruse B, Narasimhan N, Attardi G. Termination of transcription in human mitochondria: identification and purification of a DNA binding protein factor that promotes termination. Cell. 1989;58:391–397. doi: 10.1016/0092-8674(89)90853-2. [DOI] [PubMed] [Google Scholar]
- 46.Asin-Cayuela J, Schwend T, Farge G, Gustafsson CM. The human mitochondrial transcription termination factor (mTERF) is fully active in vitro in the non-phosphorylated form. Journal of Biological Chemistry. 2005;280:25499–25505. doi: 10.1074/jbc.M501145200. [DOI] [PubMed] [Google Scholar]
- 47.Shang J, Clayton DA. Human mitochondrial transcription termination exhibits RNA polymerase independence and biased bipolarity in vitro. The Journal of biological chemistry. 1994;269:29112–29120. [PubMed] [Google Scholar]
- 48.Martin M, Cho J, Cesare AJ, Griffith JD, Attardi G. Termination factor-mediated DNA loop between termination and initiation sites drives mitochondrial rRNA synthesis. Cell. 2005;123:1227–1240. doi: 10.1016/j.cell.2005.09.040. [DOI] [PubMed] [Google Scholar]
- 49.Martinez-Azorin F. The mitochondrial ribomotor hypothesis. IUBMB life. 2005;57:27–30. doi: 10.1080/15216540500088755. [DOI] [PubMed] [Google Scholar]
- 50.Gelfand R, Attardi G. Synthesis and turnover of mitochondrial ribonucleic acid in HeLa cells: the mature ribosomal and messenger ribonucleic acid species are metabolically unstable. Molecular and cellular biology. 1981;1:497–511. doi: 10.1128/mcb.1.6.497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hyvärinen AK, Pohjoismäki JLO, Reyes A, Wanrooij S, Yasukawa T, Karhunen PJ, Spelbrink JN, Holt IJ, Jacobs HT. The mitochondrial transcription termination factor mTERF modulates replication pausing in human mitochondrial DNA. Nucleic Acids Res. 2007;35:6458–6474. doi: 10.1093/nar/gkm676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Park CB, Asin-Cayuela J, Cámara Y, Shi Y, Pellegrini M, Gaspari M, Wibom R, Hultenby K, Erdjument-Bromage H, Tempst P, Falkenberg M, Gustafsson CM, Larsson NG. MTERF3 is a negative regulator of mammalian mtDNA transcription. Cell. 2007;130:273–285. doi: 10.1016/j.cell.2007.05.046. [DOI] [PubMed] [Google Scholar]
- 53.Yakubovskaya E, Mejia E, Byrnes J, Hambardjieva E, Garcia-Diaz M. Helix unwinding and base flipping enable human MTERF1 to terminate mitochondrial transcription. Cell. 2010;141:982–993. doi: 10.1016/j.cell.2010.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rebelo AP, Williams SL, Moraes CT. In vivo methylation of mtDNA reveals the dynamics of protein-mtDNA interactions. Nucleic Acids Res. 2009;37:6701–6715. doi: 10.1093/nar/gkp727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mercer TR, Neph S, Dinger ME, Crawford J, Smith MA, Shearwood AM, Haugen E, Bracken CP, Rackham O, Stamatoyannopoulos JA, Filipovska A, Mattick JS. The human mitochondrial transcriptome. Cell. 2011;146:645–658. doi: 10.1016/j.cell.2011.06.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jiménez-Menéndez N, Fernández-Millán P, Rubio-Cosials A, Arnan C, Montoya J, Jacobs HT, Bernadó P, Miquel C, Usón I, Solà M. Human mitochondrial mTERF wraps around DNA through a left-handed superhelical tandem repeat. Nature structural & molecular biology. 2010;17:891–893. doi: 10.1038/nsmb.1859. [DOI] [PubMed] [Google Scholar]
- 57.Roberti M, Bruni F, Loguercio Polosa P, Manzari C, Gadaleta MN, Cantatore P. MTERF3, the most conserved member of the mTERF-family, is a modular factor involved in mitochondrial protein synthesis. Biochimica et Biophysica Acta. 2006;1757:1199–1206. doi: 10.1016/j.bbabio.2006.04.026. [DOI] [PubMed] [Google Scholar]
- 58.Groves MR, Barford D. Topological characteristics of helical repeat proteins. Current opinion in structural biology. 1999;9:383–389. doi: 10.1016/s0959-440x(99)80052-9. [DOI] [PubMed] [Google Scholar]
- 59.Sibanda BL, Chirgadze DY, Blundell TL. Crystal structure of DNA-PKcs reveals a large open-ring cradle comprised of HEAT repeats. Nature. 2010;463:118–121. doi: 10.1038/nature08648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lu G, Dolgner SJ, Hall TM. Understanding and engineering RNA sequence specificity of PUF proteins. Current opinion in structural biology. 2009;19:110–115. doi: 10.1016/j.sbi.2008.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Nam SC, Kang C. DNA light-strand preferential recognition of human mitochondria transcription termination factor mTERF. Journal of biochemistry and molecular biology. 2005;38:690–694. doi: 10.5483/bmbrep.2005.38.6.690. [DOI] [PubMed] [Google Scholar]
- 62.Hyvärinen AK, Kumanto MK, Marjavaara SK, Jacobs HT. Effects on mitochondrial transcription of manipulating mTERF protein levels in cultured human HEK293 cells. BMC molecular biology. 2010;11:72. doi: 10.1186/1471-2199-11-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Camasamudram V, Fang JK, Avadhani NG. Transcription termination at the mouse mitochondrial H-strand promoter distal site requires an A/T rich sequence motif and sequence specific DNA binding proteins. Eur J Biochem. 2003;270:1128–1140. doi: 10.1046/j.1432-1033.2003.03461.x. [DOI] [PubMed] [Google Scholar]
- 64.Wanrooij PH, Uhler JP, Simonsson T, Falkenberg M, Gustafsson CM. G-quadruplex structures in RNA stimulate mitochondrial transcription termination and primer formation. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:16072–16077. doi: 10.1073/pnas.1006026107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kochetkov SN, Rusakova EE, Tunitskaya VL. Recent studies of T7 RNA polymerase mechanism. FEBS letters. 1998;440:264–267. doi: 10.1016/s0014-5793(98)01484-7. [DOI] [PubMed] [Google Scholar]
- 66.Sondheimer N, Fang JK, Polyak E, Falk MJ, Avadhani NG. Leucine-rich pentatricopeptide-repeat containing protein regulates mitochondrial transcription. Biochemistry. 2010;49:7467–7473. doi: 10.1021/bi1008479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sasarman F, Brunel-Guitton C, Antonicka H, Wai T, Shoubridge EA. LRPPRC and SLIRP interact in a ribonucleoprotein complex that regulates posttranscriptional gene expression in mitochondria. Mol Biol Cell. 2010;21:1315–1323. doi: 10.1091/mbc.E10-01-0047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Gohil VM, Nilsson R, Belcher-Timme CA, Luo B, Root DE, Mootha VK. Mitochondrial and nuclear genomic responses to loss of LRPPRC expression. The Journal of biological chemistry. 2010;285:13742–13747. doi: 10.1074/jbc.M109.098400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Linder T, Park CB, Asin-Cayuela J, Pellegrini M, Larsson NG, Falkenberg M, Samuelsson T, Gustafsson CM. A family of putative transcription termination factors shared amongst metazoans and plants. Current genetics. 2005;48:265–269. doi: 10.1007/s00294-005-0022-5. [DOI] [PubMed] [Google Scholar]
- 70.Roberti M, Polosa PL, Bruni F, Manzari C, Deceglie S, Gadaleta MN, Cantatore P. The MTERF family proteins: mitochondrial transcription regulators and beyond. Biochimica et Biophysica Acta. 2009;1787:303–311. doi: 10.1016/j.bbabio.2009.01.013. [DOI] [PubMed] [Google Scholar]
- 71.Cámara Y, Asin-Cayuela J, Park CB, Metodiev MD, Shi Y, Kukat C, Habermann B, Wibom R, Hultenby K, Franz T, Erdjument-Bromage H, Tempst P, Hällberg BM, Gustafsson CM, Larsson NG. MTERF4 regulates translation by targeting the methyltransferase NSUN4 to the mammalian mitochondrial ribosome. Cell metabolism. 2011;13:527–539. doi: 10.1016/j.cmet.2011.04.002. [DOI] [PubMed] [Google Scholar]
- 72.Spåhr H, Samuelsson T, Hällberg BM, Gustafsson CM. Structure of mitochondrial transcription termination factor 3 reveals a novel nucleic acid-binding domain. Biochemical and biophysical research communications. 2010;397:386–390. doi: 10.1016/j.bbrc.2010.04.130. [DOI] [PubMed] [Google Scholar]
- 73.Kim D-U, Cho S-G, Kim K-L, Cho H-S. Human MTERF3 crystal structure forms a left-handed superhelix. Molecules and cells. 2011 doi: 10.1007/s10059-011-0264-7. [DOI] [PubMed] [Google Scholar]
- 74.Chen Y, Zhou G, Yu M, He Y, Tang W, Lai J, He J, Liu W, Tan D. Cloning and functional analysis of human mTERFL encoding a novel mitochondrial transcription termination factor-like protein. Biochemical and biophysical research communications. 2005;337:1112–1118. doi: 10.1016/j.bbrc.2005.09.164. [DOI] [PubMed] [Google Scholar]
- 75.Luca CC, Moraes CT. Functional analysis of mouse mTERF.D3, a novel mitochondrial transcription termination-like factor. Mitochondrion. 2006;6:15–15. [Google Scholar]
- 76.Pellegrini M, Asin-Cayuela J, Erdjument-Bromage H, Tempst P, Larsson NG, Gustafsson CM. MTERF2 is a nucleoid component in mammalian mitochondria. Biochimica et Biophysica Acta. 2009;1787:296–302. doi: 10.1016/j.bbabio.2009.01.018. [DOI] [PubMed] [Google Scholar]
- 77.Wenz T, Luca C, Torraco A, Moraes CT. mTERF2 regulates oxidative phosphorylation by modulating mtDNA transcription. Cell metabolism. 2009;9:499–511. doi: 10.1016/j.cmet.2009.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 78.Hyvarinen AK, Pohjoismaki JL, Holt IJ, Jacobs HT. Overexpression of MTERFD1 or MTERFD3 impairs the completion of mitochondrial DNA replication. Molecular biology reports. 2011;38:1321–1328. doi: 10.1007/s11033-010-0233-9. [DOI] [PubMed] [Google Scholar]
- 79.Huang W, Yu M, Jiao Y, Ma J, Ma M, Wang Z, Wu H, Tan D. Mitochondrial transcription termination factor 2 binds to entire mitochondrial DNA and negatively regulates mitochondrial gene expression. Acta Biochim Biophys Sin (Shanghai) 2011;43:472–479. doi: 10.1093/abbs/gmr035. [DOI] [PubMed] [Google Scholar]
- 80.Andersson DC, Fauconnier J, Park CB, Zhang S-J, Thireau J, Ivarsson N, Larsson N-G, Westerblad H. Enhanced cardiomyocyte Ca2+ cycling precedes terminal AV-block in mitochondrial cardiomyopathy Mterf3 KO mice. Antioxidants & redox signaling. 2011 doi: 10.1089/ars.2011.3915. [DOI] [PubMed] [Google Scholar]
- 81.Wang Z, Cotney J, Shadel GS. Human mitochondrial ribosomal protein MRPL12 interacts directly with mitochondrial RNA polymerase to modulate mitochondrial gene expression. The Journal of biological chemistry. 2007;282:12610–12618. doi: 10.1074/jbc.M700461200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Cantatore P, Roberti M, Rainaldi G, Gadaleta MN, Saccone C. The complete nucleotide sequence, gene organization, and genetic code of the mitochondrial genome of Paracentrotus lividus. The Journal of biological chemistry. 1989;264:10965–10975. [PubMed] [Google Scholar]
- 83.Cantatore P, Roberti M, Loguercio Polosa P, Mustich A, Gadaleta MN. Mapping and characterization of Paracentrotus lividus mitochondrial transcripts: multiple and overlapping transcription units. Current genetics. 1990;17:235–245. doi: 10.1007/BF00312615. [DOI] [PubMed] [Google Scholar]
- 84.Roberti M, Mustich A, Gadaleta MN, Cantatore P. Identification of two homologous mitochondrial DNA sequences, which bind strongly and specifically to a mitochondrial protein of Paracentrotus lividus. Nucleic Acids Research. 1991;19:6249–6254. doi: 10.1093/nar/19.22.6249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Loguercio Polosa P, Roberti M, Musicco C, Gadaleta MN, Quagliariello E, Cantatore P. Cloning and characterisation of mtDBP, a DNA-binding protein which binds two distinct regions of sea urchin mitochondrial DNA. Nucleic Acids Res. 1999;27:1890–1899. doi: 10.1093/nar/27.8.1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Fernandez-Silva P, Polosa PL, Roberti M, Di Ponzio B, Gadaleta MN, Montoya J, Cantatore P. Sea urchin mtDBP is a two-faced transcription termination factor with a biased polarity depending on the RNA polymerase. Nucleic Acids Res. 2001;29:4736–4743. doi: 10.1093/nar/29.22.4736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Polosa PL, Deceglie S, Falkenberg M, Roberti M, Di Ponzio B, Gadaleta MN, Cantatore P. Cloning of the sea urchin mitochondrial RNA polymerase and reconstitution of the transcription termination system. Nucleic Acids Res. 2007;35:2413–2427. doi: 10.1093/nar/gkm159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Polosa PL, Deceglie S, Roberti M, Gadaleta MN, Cantatore P. Contrahelicase activity of the mitochondrial transcription termination factor mtDBP. Nucleic Acids Res. 2005;33:3812–3820. doi: 10.1093/nar/gki693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Hyvarinen AK, Pohjoismaki JL, Reyes A, Wanrooij S, Yasukawa T, Karhunen PJ, Spelbrink JN, Holt IJ, Jacobs HT. The mitochondrial transcription termination factor mTERF modulates replication pausing in human mitochondrial DNA. Nucleic Acids Research. 2007;35:6458–6474. doi: 10.1093/nar/gkm676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Lewis DL, Farr CL, Kaguni LS. Drosophila melanogaster mitochondrial DNA: completion of the nucleotide sequence and evolutionary comparisons. Insect Mol Biol. 1995;4:263–278. doi: 10.1111/j.1365-2583.1995.tb00032.x. [DOI] [PubMed] [Google Scholar]
- 91.Berthier F, Renaud M, Alziari S, Durand R. RNA mapping on Drosophila mitochondrial DNA: precursors and template strands. Nucleic Acids Research. 1986;14:4519–4533. doi: 10.1093/nar/14.11.4519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Roberti M, Polosa PL, Bruni F, Musicco C, Gadaleta MN, Cantatore P. DmTTF, a novel mitochondrial transcription termination factor that recognises two sequences of Drosophila melanogaster mitochondrial DNA. Nucleic Acids Res. 2003;31:1597–1604. doi: 10.1093/nar/gkg272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Roberti M, Fernandez-Silva P, Polosa PL, Fernandez-Vizarra E, Bruni F, Deceglie S, Montoya J, Gadaleta MN, Cantatore P. In vitro transcription termination activity of the Drosophila mitochondrial DNA-binding protein DmTTF. Biochemical and biophysical research communications. 2005;331:357–362. doi: 10.1016/j.bbrc.2005.03.173. [DOI] [PubMed] [Google Scholar]
- 94.Roberti M, Bruni F, Polosa PL, Gadaleta MN, Cantatore P. The Drosophila termination factor DmTTF regulates in vivo mitochondrial transcription. Nucleic Acids Research. 2006;34:2109–2116. doi: 10.1093/nar/gkl181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Holt IJ, Harding AE, Morgan-Hughes JA. Deletions of muscle mitochondrial DNA in patients with mitochondrial myopathies. Nature. 1988;331:717–719. doi: 10.1038/331717a0. [DOI] [PubMed] [Google Scholar]
- 96.Wallace DC, Singh G, Lott MT, Hodge JA, Schurr TG, Lezza AM, Elsas LJ, 2nd, Nikoskelainen EK. Mitochondrial DNA mutation associated with Leber’s hereditary optic neuropathy. Science. 1988;242:1427–1430. doi: 10.1126/science.3201231. [DOI] [PubMed] [Google Scholar]
- 97.Zeviani M, Moraes CT, DiMauro S, Nakase H, Bonilla E, Schon EA, Rowland LP. Deletions of mitochondrial DNA in Kearns-Sayre syndrome. Neurology. 1988;38:1339–1346. doi: 10.1212/wnl.38.9.1339. [DOI] [PubMed] [Google Scholar]
- 98.Larsson NG, Clayton DA. Molecular genetic aspects of human mitochondrial disorders. Annual review of genetics. 1995;29:151–178. doi: 10.1146/annurev.ge.29.120195.001055. [DOI] [PubMed] [Google Scholar]
- 99.Munnich A, Rustin P. Clinical spectrum and diagnosis of mitochondrial disorders. Am J Med Genet. 2001;106:4–17. doi: 10.1002/ajmg.1391. [DOI] [PubMed] [Google Scholar]
- 100.Zeviani M. Mitochondrial disorders. Suppl Clin Neurophysiol. 2004;57:304–312. [PubMed] [Google Scholar]
- 101.Zeviani M, Di Donato S. Mitochondrial disorders. Brain : a journal of neurology. 2004;127:2153–2172. doi: 10.1093/brain/awh259. [DOI] [PubMed] [Google Scholar]
- 102.Krishnan KJ, Greaves LC, Reeve AK, Turnbull D. The ageing mitochondrial genome. Nucleic Acids Research. 2007;35:7399–7405. doi: 10.1093/nar/gkm635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Krishnan KJ, Greaves LC, Reeve AK, Turnbull DM. Mitochondrial DNA mutations and aging. Ann N Y Acad Sci. 2007;1100:227–240. doi: 10.1196/annals.1395.024. [DOI] [PubMed] [Google Scholar]
- 104.Larsson N-G. Somatic Mitochondrial DNA Mutations in Mammalian Aging. Annual review of biochemistry. 2010;79:683–706. doi: 10.1146/annurev-biochem-060408-093701. [DOI] [PubMed] [Google Scholar]
- 105.Pavlakis SG, Phillips PC, DiMauro S, De Vivo DC, Rowland LP. Mitochondrial myopathy, encephalopathy, lactic acidosis, and strokelike episodes: a distinctive clinical syndrome. Ann Neurol. 1984;16:481–488. doi: 10.1002/ana.410160409. [DOI] [PubMed] [Google Scholar]
- 106.Hess JF, Parisi MA, Bennett JL, Clayton DA. Impairment of mitochondrial transcription termination by a point mutation associated with the MELAS subgroup of mitochondrial encephalomyopathies. Nature. 1991;351:236–239. doi: 10.1038/351236a0. [DOI] [PubMed] [Google Scholar]
- 107.Manwaring N, Jones MM, Wang JJ, Rochtchina E, Howard C, Mitchell P, Sue CM. Population prevalence of the MELAS A3243G mutation. Mitochondrion. 2007;7:230–233. doi: 10.1016/j.mito.2006.12.004. [DOI] [PubMed] [Google Scholar]
- 108.Goto Y, Nonaka I, Horai S. A mutation in the tRNA(Leu)(UUR) gene associated with the MELAS subgroup of mitochondrial encephalomyopathies. Nature. 1990;348:651–653. doi: 10.1038/348651a0. [DOI] [PubMed] [Google Scholar]
- 109.Kobayashi Y, Momoi MY, Tominaga K, Momoi T, Nihei K, Yanagisawa M, Kagawa Y, Ohta S. A point mutation in the mitochondrial tRNA(Leu)(UUR) gene in MELAS (mitochondrial myopathy, encephalopathy, lactic acidosis and stroke-like episodes) Biochemical and biophysical research communications. 1990;173:816–822. doi: 10.1016/s0006-291x(05)80860-5. [DOI] [PubMed] [Google Scholar]
- 110.Chomyn A, Martinuzzi A, Yoneda M, Daga A, Hurko O, Johns D, Lai ST, Nonaka I, Angelini C, Attardi G. MELAS mutation in mtDNA binding site for transcription termination factor causes defects in protein synthesis and in respiration but no change in levels of upstream and downstream mature transcripts. Proc Natl Acad Sci U S A. 1992;89:4221–4225. doi: 10.1073/pnas.89.10.4221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Kirino Y, Yasukawa T, Ohta S, Akira S, Ishihara K, Watanabe K, Suzuki T. Codon-specific translational defect caused by a wobble modification deficiency in mutant tRNA from a human mitochondrial disease. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:15070–15075. doi: 10.1073/pnas.0405173101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Shutt TE, Shadel GS. A compendium of human mitochondrial gene expression machinery with links to disease. Environmental and molecular mutagenesis. 2010;51:360–379. doi: 10.1002/em.20571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Seneca S, Verhelst H, De Meirleir L, Meire F, Ceuterick-De Groote C, Lissens W, Van Coster R. A new mitochondrial point mutation in the transfer RNA(Leu) gene in a patient with a clinical phenotype resembling Kearns-Sayre syndrome. Arch Neurol. 2001;58:1113–1118. doi: 10.1001/archneur.58.7.1113. [DOI] [PubMed] [Google Scholar]
- 114.Mimaki M, Hatakeyama H, Ichiyama T, Isumi H, Furukawa S, Akasaka M, Kamei A, Komaki H, Nishino I, Nonaka I, Goto Y. Different effects of novel mtDNA G3242A and G3244A base changes adjacent to a common A3243G mutation in patients with mitochondrial disorders. Mitochondrion. 2009;9:115–122. doi: 10.1016/j.mito.2009.01.005. [DOI] [PubMed] [Google Scholar]
- 115.Sasarman F, Antonicka H, Shoubridge EA. The A3243G tRNALeu(UUR) MELAS mutation causes amino acid misincorporation and a combined respiratory chain assembly defect partially suppressed by overexpression of EFTu and EFG2. Human molecular genetics. 2008;17:3697–3707. doi: 10.1093/hmg/ddn265. [DOI] [PubMed] [Google Scholar]
- 116.Wallace DC. Mitochondrial diseases in man and mouse. Science. 1999;283:1482–1488. doi: 10.1126/science.283.5407.1482. [DOI] [PubMed] [Google Scholar]
- 117.Brown MD, Lott MT. Emery and Rimoin’s Principles and Practice of Medical Genetics. Churchill Livingstone; London: 1996. [Google Scholar]
- 118.Yoneda M, Chomyn A, Martinuzzi A, Hurko O, Attardi G. Marked replicative advantage of human mtDNA carrying a point mutation that causes the MELAS encephalomyopathy. Proc Natl Acad Sci U S A. 1992;89:11164–11168. doi: 10.1073/pnas.89.23.11164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Shoubridge EA. Segregation of mitochondrial DNAs carrying a pathogenic point mutation (tRNA(leu3243)) in cybrid cells. Biochemical and biophysical research communications. 1995;213:189–195. doi: 10.1006/bbrc.1995.2115. [DOI] [PubMed] [Google Scholar]