Abstract
Megalencephaly-capillary malformation (MCAP) and megalencephaly-polymicrogyria-polydactyly-hydrocephalus (MPPH) syndromes are sporadic overgrowth disorders associated with markedly enlarged brain size and other recognizable features1-5. We performed exome sequencing in three families with MCAP or MPPH and confirmed our initial observations in exomes from 7 MCAP and 174 control individuals, as well as in 40 additional megalencephaly subjects using a combination of Sanger sequencing, restriction-enzyme assays, and targeted deep sequencing. We identified de novo germline or postzygotic mutations in three core components of the phosphatidylinositol-3-kinase (PI3K)/AKT pathway. These include two mutations of AKT3, one recurrent mutation of PIK3R2 in 11 unrelated MPPH families, and 15 mostly postzygotic mutations of PIK3CA in 23 MCAP and one MPPH patients. Our data highlight the central role of PI3K/AKT signaling in vascular, limb and brain development, and emphasize the power of massively parallel sequencing in a challenging context of phenotypic and genetic heterogeneity combined with postzygotic mosaicism.
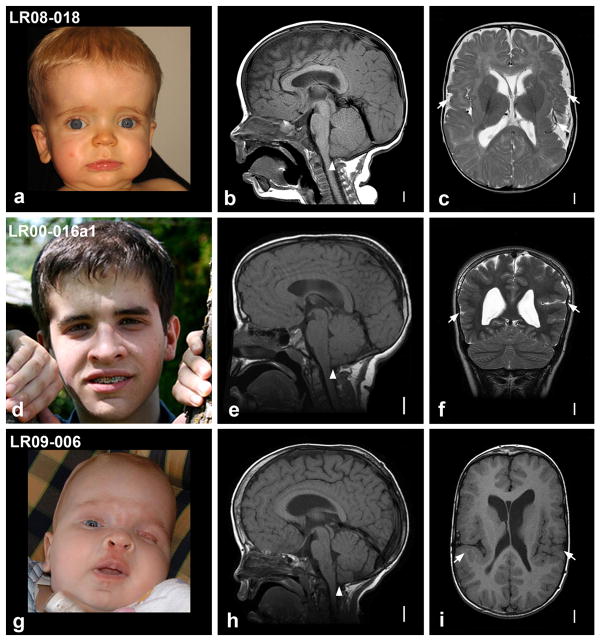
As described in our recent clinical analysis in 42 children with MCAP or MPPH4, the former consists of megalencephaly (or sometimes hemimegalencephaly), associated growth dysregulation with variable asymmetry, developmental vascular anomalies, distal limb malformations (syndactyly and polydactyly), variable cortical malformation, and a mild connective tissue dysplasia (Fig. 1 and Supplementary Fig. 1 and 2). The patchy skin vascular malformations and asymmetric overgrowth seen in MCAP meet criteria for the Klippel-Trenaunay subtype of vascular malformations and deregulated growth, which suggests that postzygotic mosaicism may be present in a subset of cases6,7. MPPH resembles MCAP but lacks vascular malformations and syndactyly4,8. We hypothesized that MCAP and MPPH result from mutations in the same pathway, and studied them together. Given the absence of recurrence of both syndromes in all reported families, we conducted exome sequencing in two parent-proband trios – one with MCAP (subject LR09-006, Fig. 1) – and one with clinical features overlapping between MCAP and MPPH (subject LR08-018, Fig. 1) – and searched for de novo mutations. We also performed exome sequencing in the oldest of three affected siblings with MPPH (subject LR00-016a1, Fig. 1) – the first known familial recurrence of this syndrome – assuming either autosomal recessive inheritance or germline mosaicism in one parent.
Figure 1.
Craniofacial appearance and magnetic resonance imaging (MRI) of the three index patients. Photos and brain MRI of patients LR08-018 (a–c), LR00-016a1 (d–f), and LR09-006 (g–i). Photos of patients LR08-018 (a), LR00-016a1 (d) and LR09-006 (g) were taken at 11 months, 15 years, and 5 months, respectively. Note the prominent forehead and apparent macrocephaly in all three patients (a, d, g), and a midline facial capillary malformation (or nevus flammeus) in LR09-006 (g). Midline sagittal brain images (b, e, h) show prominent forehead, increased cranium-to-face ratio and cerebellar tonsillar ectopia (arrowheads), while axial or coronal images (c, f, i) show bilateral perisylvian polymicrogyria (arrows). Scale bars correspond to 1 cm. Additional photos of patient LR09-006 and a clinical description of the three index patients are provided in the Supplementary Note. We obtained written consent to publish photographs of the patients.
We identified 247 to 254 rare protein-altering variants not previously seen in public variant databases or 112 other exomes per proband (Supplementary Table 1, Online Methods). Analysis of trio LR08-018 revealed a de novo mutation in AKT3 (p.Arg465Trp, Supplementary Table 2). Sanger sequencing of AKT3 in another 40 megalencephaly patients identified a second de novo mutation in LR11-354 (p.Asn229Ser, Table 1), which supports mutations in AKT3 as a rare cause of megalencephaly (P = 0.002, Online Methods). AKT3 encodes the brain-predominant isoform of AKT serine/threonine kinase, which is a major downstream mediator of PI3K signaling9,10, leading us to focus on genes in this pathway in other patients with megalencephaly.
Table 1.
Summary of AKT3, PIK3R2 and PIK3CA mutations identified in 37 megalencephaly families
| Group | Patient ID | Gene | Mutation coordinates (hg19) | cDNA change | Amino acid change | Inheritance | CpG site | COSMIC (samples)c | Grantham score | GERP score |
|---|---|---|---|---|---|---|---|---|---|---|
| Overlap | LR08-018a | AKT3 | chr1:243668598 G>A | c.1393C>T | p.Arg465Trp | De novo | Yes | No | 101 | 5.40 |
| MPPH | LR11-354 | AKT3 | chr1:243776983 T>C | c.686A>G | p.Asn229Ser | De novo | No | No | 46 | 5.43 |
|
| ||||||||||
| MPPH | LR00-016a1a,b | PIK3R2 | chr19:18273784 G>A | c.1117G>A | p.Gly373Arg | De novo | Yes | Nod | 125 | 3.69 |
| MPPH | LR00-016a2b | PIK3R2 | chr19:18273784 G>A | c.1117G>A | p.Gly373Arg | De novo | Yes | Nod | 125 | 3.69 |
| MPPH | LR00-016a3b | PIK3R2 | chr19:18273784 G>A | c.1117G>A | p.Gly373Arg | De novo | Yes | Nod | 125 | 3.69 |
| MPPH | LR01-164 | PIK3R2 | chr19:18273784 G>A | c.1117G>A | p.Gly373Arg | De novo | Yes | Nod | 125 | 3.69 |
| MPPH | LR04-069 | PIK3R2 | chr19:18273784 G>A | c.1117G>A | p.Gly373Arg | De novo | Yes | Nod | 125 | 3.69 |
| MPPH | LR04-032 | PIK3R2 | chr19:18273784 G>A | c.1117G>A | p.Gly373Arg | N/A | Yes | Nod | 125 | 3.69 |
| MPPH | LR04-373 | PIK3R2 | chr19:18273784 G>A | c.1117G>A | p.Gly373Arg | De novo | Yes | Nod | 125 | 3.69 |
| MPPH | LR08-263 | PIK3R2 | chr19:18273784 G>A | c.1117G>A | p.Gly373Arg | De novo | Yes | Nod | 125 | 3.69 |
| MPPH | LR08-422 | PIK3R2 | chr19:18273784 G>A | c.1117G>A | p.Gly373Arg | De novo | Yes | Nod | 125 | 3.69 |
| MPPH | LR04-181 | PIK3R2 | chr19:18273784 G>A | c.1117G>A | p.Gly373Arg | De novo | Yes | Nod | 125 | 3.69 |
| MPPH | LR11-021 | PIK3R2 | chr19:18273784 G>A | c.1117G>A | p.Gly373Arg | N/A | Yes | Nod | 125 | 3.69 |
| MPPH | LR11-204 | PIK3R2 | chr19:18273784 G>A | c.1117G>A | p.Gly373Arg | De novo | Yes | Nod | 125 | 3.69 |
| MPPH | LR11-353 | PIK3R2 | chr19:18273784 G>A | c.1117G>A | p.Gly373Arg | N/A | Yes | Nod | 125 | 3.69 |
|
| ||||||||||
| MCAP | LR06-342 | PIK3CA | chr3:178916854 G>A | c.241 G>A | p.Glu81Lys | N/A | No | Yes (2) | 56 | 5.44 |
| MCAP | LR06-220 | PIK3CA | chr3:178916876 G>A | c.263G>A | p.Arg88Gln | De novo | Yes | Yes (26) | 43 | 5.44 |
| MCAP | LR11-068 | PIK3CA | chr3:178916876 G>A | c.263G>A | p.Arg88Gln | De novo | Yes | Yes (26) | 43 | 5.44 |
| MCAP | 07-0388a | PIK3CA | chr3:178922321 G>A | c.1090G>A | p.Gly364Arg | De novo | No | Yes (1) | 125 | 5.53 |
| MCAP | LR05-139 | PIK3CA | chr3:178922324 G>A | c.1093G>A | p.Glu365Lys | De novo | No | Yes (2) | 56 | 5.53 |
| MCAP | 44735a | PIK3CA | chr3:178922364 G>A | c.1133G>A | p.Cys378Tyr | De novo | No | No | 194 | 5.61 |
| MCAP | LR11-153 | PIK3CA | chr3:178928078 AGAA>A | c.1359_1361del | p.Glu453del | De novo | - | Yes (1) | - | - |
| MPPH | LR05-204 | PIK3CA | chr3:178928078 AGAA>A | c.1359_1361del | p.Glu453del | De novo | - | Yes (1) | - | - |
| MCAP | LR12-033 | PIK3CA | chr3:178936091 G>A | c.1633G>A | p.Glu545Lys | N/A | No | Yes (769) | 56 | 5.78 |
| MCAP | 162-001Pa | PIK3CA | chr3:178938934 G>A | c.2176G>A | p.Glu726Lys | De novo | No | Yes (2) | 56 | 5.67 |
| MCAP | LR08-261 | PIK3CA | chr3:178938934 G>A | c.2176G>A | p.Glu726Lys | N/A | No | Yes (2) | 56 | 5.67 |
| MCAP | LR06-333 | PIK3CA | chr3:178938934 G>A | c.2176G>A | p.Glu726Lys | N/A | No | Yes (2) | 56 | 5.67 |
| MCAP | LR09-006a | PIK3CA | chr3:178947865 G>A | c.2740G>A | p.Gly914Arg | De novo | No | No | 125 | 5.61 |
| MCAP | LR11-070 | PIK3CA | chr3:178947865 G>A | c.2740G>A | p.Gly914Arg | De novo | No | No | 125 | 5.61 |
| MCAP | LR06-341 | PIK3CA | chr3:178947865 G>A | c.2740G>A | p.Gly914Arg | De novo | No | No | 125 | 5.61 |
| MCAP | 11-0117a | PIK3CA | chr3:178947865 G>A | c.2740G>A | p.Gly914Arg | De novo | No | No | 125 | 5.61 |
| MCAP | LR11-212 | PIK3CA | chr3:178952007 A>G | c.3062A>G | p.Tyr1021Cys | De novo | No | Yes (17) | 194 | 6.08 |
| MCAP | LR11-069 | PIK3CA | chr3:178952018 A>G | c.3073A>G | p.Thr1025Ala | De novo | No | Yes (22) | 58 | 6.08 |
| MCAP | LR11-270 | PIK3CA | chr3:178952049 C>T | c.3104C>T | p.Ala1035Val | De novo | No | Yes (2) | 64 | 6.08 |
| MCAP | 115422a | PIK3CA | chr3:178952049 C>T | c.3104C>T | p.Ala1035Val | De novo | No | Yes (2) | 64 | 6.08 |
| MCAP | 86708a | PIK3CA | chr3:178952074 G>T | c.3129G>T | p.Met1043Ile | De novo | No | Yes (31) | 10 | 6.08 |
| MCAP | 121939a | PIK3CA | chr3:178952084 C>T | c.3139C>T | p. His1047Tyr | De novo | No | Yes (32) | 83 | 6.08 |
| MCAP | LR11-285 | PIK3CA | chr3:178952084 C>T | c.3139C>T | p. His1047Tyr | De novo | No | Yes (32) | 83 | 6.08 |
| MCAP | LR11-230 | PIK3CA | chr3:178952090 G>A | c.3145G>A | p.Gly1049Ser | De novo | No | Yes (10) | 56 | 6.08 |
N/A, parents not available. GERP, genomic evolutionary rate profiling.
Subjects analyzed by exome sequencing.
Affected siblings of family LR00-016.
Presence of the same amino acid change in the Catalogue of Somatic Mutations in Cancer (COSMIC, see URLs); numbers in parentheses indicate the number of samples reported in COSMIC. Gene accession numbers: NM_005465.4 (AKT3), NM_005027.2 (PIK3R2), and NM_006218.2 (PIK3CA). See Supplementary Table 4 for details on the levels of PIK3CA mutant alleles in different tissues as well as in 174 control exomes.
Although not reported in COSMIC, the PIK3R2 p.Gly373Arg was recently found in endometrial cancer (see Discussion for details). All mutations were absent from the Exome Variant Server, which consists of whole-exome sequencing data from ~5,400 subjects [NHLBI Exome Sequencing Project (ESP), Seattle, WA (see URLs, Date accessed: December, 2011)].
Analysis of LR00-016a1 revealed four genes consistent with autosomal recessive inheritance. However, filtering of variants using genome-wide single-nucleotide-polymorphism data from all three affected siblings excluded all four chromosomal loci (Supplementary Table 3, Online Methods). We next manually examined the list of 247 rare variants and identified a heterozygous mutation in PIK3R2 (p.Gly373Arg, Table 1), which encodes the p85β regulatory subunit of class IA PI3K11. Sanger sequencing confirmed the presence of the mutation in all three affected siblings and its absence in saliva and blood in both parents and the unaffected sister, demonstrating germline mosaicism in one parent. Sequencing of PIK3R2 in 40 megalencephaly patients identified the same nucleotide change in ten additional MPPH subjects, which proved to be de novo in all seven subjects with parental DNA available (Table 1). The mutation occurs at a CpG dinucleotide, which may explain its recurrence12.
We identified four candidate de novo variants in trio LR09-006, and confirmed a de novo substitution in the exon 3 splice donor consensus sequence of BDP1 (Supplementary Table 2). Screening of BPD1 in 12 additional subjects detected no other de novo mutations, thus failing to support a causative role for this sequence change. Given the clinical presentation of MCAP and the recent report of postzygotic mutations of AKT1 in Proteus syndrome13, we speculated that MCAP may result from low-level mosaic mutations missed by our standard pipeline. In parallel, we performed a second de novo analysis in this trio by including the raw variants that failed on our initial hard-filtering parameters (Online Methods). This approach revealed a missense change in PIK3CA (p.Gly914Arg, Supplementary Table 2), which encodes the p110α catalytic subunit of class IA PI3K14. This mutation was supported by 20/177 reads (11%) in the exome sequencing data, and confirmed as de novo and mosaic by Sanger sequencing and custom restriction-enzyme assay (Supplementary Fig. 3a, Online Methods). We then sequenced the coding exons of PIK3CA in 29 megalencephaly patients with no mutations in AKT3 or PIK3R2 and identified 14 additional PIK3CA mutations with mutant allele levels between 10 and 50% (Table 1, Supplementary Table 4).
Standard variant calling in exomes from seven additional MCAP patients identified a mutation of PIK3CA supported by 68/250 reads (27%) in subject 44735 (p.Cys378Tyr, Table 1, Online Methods). This mutation showed variable levels of mosaicism depending on the tissue tested (Supplementary Fig. 3b). Manual inspection of the Sequence Alignment/Map files of the remaining six unsolved MCAP subjects using IGV15 revealed other candidate mosaic mutations in PIK3CA with mutant levels ranging from 2 to 15% of total reads. To differentiate between sequencing errors and putative mutations, we systematically searched for low-level mosaicism of PIK3CA in these six and 174 control exomes (Supplementary Table 5, Online Methods). Although data from both cohorts contained many sites with 1 or 2 variant reads (most expected to be sequencing errors), we found only 12 variant sites supported by 3 or more reads. A significantly higher frequency of such sites was found in the MCAP cohort. Using a threshold of 4 variant reads, we identified candidate mosaic mutations in five of six patients and none in control individuals (Supplementary Table 6). Sanger sequencing, a custom restriction-enzyme assay or both confirmed all five mutations (Supplementary Table 4). The only remaining unsolved MCAP subject (115422) had a p.Ala1035Val mutation supported by 3 of 185 reads (Supplementary Table 6) that was confirmed by Sanger sequencing and restriction-enzyme assay in DNA from saliva and buccal swab. We previously found and validated the same nucleotide substitution in four tissues from another MCAP subject (LR11-270, Supplementary Fig. 3c). Six other variant sites supported by 3 reads were identified in five individuals, including two sites in MCAP subject 162-001P in addition to the pathogenic mutation, neither of which was confirmed by Sanger sequencing in blood or saliva, and four sites in control individuals. This suggests that variant sites supported by 1–3 reads are mostly sequencing artifacts (Supplementary Table 6).
Given the limitations of Sanger sequencing for detecting low-level mosaic mutations, we performed targeted ultra-deep sequencing (>10,000-fold) of five mutation sites in 15 mutation-negative patients as well as in known mutation carriers and control individuals (Supplementary Table 7, Online Methods). This experiment confirmed all previously identified mutations and detected two additional low-level mosaic mutations missed by Sanger sequencing (Table 2). Both were confirmed by a second deep sequencing experiment and showed mutant levels of 1 to 8%.
Table 2.
PIK3CA mutations identified or confirmed by deep sequencing of five mutation sites
| Megalencephaly patients | Controls | |||||||
|---|---|---|---|---|---|---|---|---|
|
| ||||||||
| Patient ID | Source of DNA | Mutation Coordinates (hg19) | Amino acid change | Total readsa | Mutant readsa | Percent mutant allele | Mean mutant allele freqa,b (×10−4) | Max mutant allele freqa,c (×10−4) |
| 1. Confirmed PIK3CA mutations | ||||||||
| LR06-220 | Blood | chr3:178916876 G>A | p.Arg88Gln | 149,513 | 64,260 | 43 | 1.7 | 2.9 |
| 44735 | Blood | chr3:178922364 G>A | p.Cys378Tyr | 285,251 | 84,600 | 30 | 0.4 | 1.1 |
| LR08-261 | Blood | chr3:178938934 G>A | p.Glu726Lys | 293,540 | 35,865 | 12 | 0.5 | 0.7 |
| Buccal swab | chr3:178938934 G>A | p.Glu726Lys | 5,087 | 2,110 | 41 | 0.5 | 0.7 | |
| LR06-333 | LCL | chr3:178938934 G>A | p.Glu726Lys | 260,333 | 36,348 | 14 | 0.5 | 0.7 |
| Saliva | chr3:178938934 G>A | p.Glu726Lys | 125,336 | 19,195 | 15 | 0.5 | 0.7 | |
| LR09-006 | Blood | chr3:178947865 G>A | p.Gly914Arg | 392,036 | 61,427 | 16 | 19 | 23 |
| LR11-070 | LCL | chr3:178947865 G>A | p.Gly914Arg | 663,398 | 102,811 | 15 | 19 | 23 |
| Saliva | chr3:178947865 G>A | p.Gly914Arg | 327,763 | 56,313 | 17 | 19 | 23 | |
| LR06-341 | Blood | chr3:178947865 G>A | p.Gly914Arg | 728,792 | 112,818 | 15 | 19 | 23 |
| Saliva | chr3:178947865 G>A | p.Gly914Arg | 538,366 | 103,171 | 19 | 19 | 23 | |
| LR11-069 | LCL | chr3:178952018 A>G | p.Thr1025Ala | 49,105 | 11,856 | 24 | 18 | 19 |
| Saliva | chr3:178952018 A>G | p.Thr1025Ala | 12,126 | 1,440 | 12 | 18 | 19 | |
|
| ||||||||
| 2. New PIK3CA mutations | ||||||||
| LR06-342 | Saliva | chr3:178916854 G>A | p.Glu81Lys | 51,268 | 4,227 | 8 | 0.4 | 0.5 |
| LR11-068 | Blood | chr3:178916876 G>A | p.Arg88Gln | 117,487 | 2,921 | 2 | 1.7 | 2.9 |
|
| ||||||||
| 3. Validation of new PIK3CA mutations | ||||||||
| LR06-342 | LCL | chr3:178916854 G>A | p.Glu81Lys | 135,801 | 993 | 1 | 0.2 | 0.2 |
| Saliva | chr3:178916854 G>A | p.Glu81Lys | 82,864 | 2,783 | 3 | 0.2 | 0.2 | |
| LR11-068 | Blood | chr3:178916876 G>A | p.Arg88Gln | 282,771 | 3,378 | 1 | 0.2 | 0.3 |
| Saliva | chr3:178916876 G>A | p.Arg88Gln | 45,578 | 1,714 | 4 | 0.2 | 0.3 | |
Abbreviations: LCL: lymphoblastoid cell line.
Only bases with base quality 20 were considered. Controls section:
average frequency of the mutant allele in control individuals.
Highest mutant allele frequency found in control individuals.
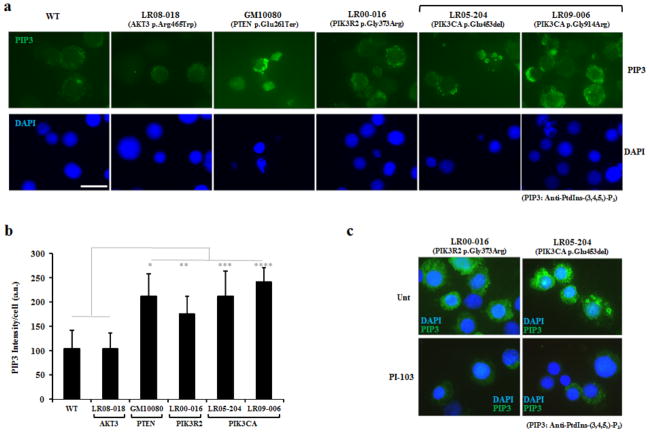
Class IA PI3Ks are heterodimeric enzymes that convert phosphatidylinositol-4,5-bisphosphate (PIP2) to phosphatidylinositol-4,5-trisphosphate (PIP3), a reaction reversed by PTEN phosphatase16. This reaction leads to activation of the PI3K-AKT-mTOR network, which can be monitored via phosphorylation levels of downstream targets such as S6 ribosomal protein and eukaryotic translation initiation factor 4E-binding protein 1 (4E-BP1)17. In order to assess the impact of AKT3, PIK3R2 and PIK3CA mutations on PI3K activity, we performed PIP3 immunostaining in lymphoblastoid cell lines derived from four megalencephaly mutation carriers and compared PIP3 levels to those in control and PTEN mutant cells (Fig. 2). Consistent with elevated PI3K activity – and similar to PTEN loss – all three PIK3R2 or PIK3CA mutant lines showed significantly increased PIP3 levels compared to control cells (Fig. 2b), as well as increased localization of active phosphoinositide-dependent kinase 1 (PDK1) to the cell membrane18 (Supplementary Fig. 4). Treatment with the PI3K inhibitor PI-103 resulted in decreased PIP3 levels in the PIK3R2 p.Gly373Arg and PIK3CA p.Glu453del mutant lines, confirming that these observations are PI3K-dependent (Fig. 2c). We found no evidence for increased PI3K activity in the AKT3 mutant line, consistent with a mutation affecting a downstream effector of PI3K. Western blot analysis showed elevated levels of phosphorylated S6 protein and 4E-BP1 in all mutant cell lines compared to controls (Supplementary Fig. 5). Although PI-103 treatment reduced S6 phosphorylation levels in control and mutant lines, the latter exhibited relative resistance to PI3K inhibition, consistent with elevated signaling through the pathway. Altogether, these observations support the conclusion that the megalencephaly-associated mutations result in increased PI3K activity and PI3K-mTOR signaling.
Figure 2.
PIP3 levels in lymphoblastoid cell lines derived from an unaffected control, a patient with Cowden disease (GM10080), and four megalencephaly patients. (a) Indirect immunofluorescence staining of PIP3 in exponentially growing lymphoblastoid cell lines using a mouse monoclonal anti-PIP3 antibody (Online Methods). Scale bar corresponds to 10μm. (b) Per-cell quantification of PIP3 levels based on anti-PIP3 signal intensity (a.u., arbitrary units). Levels of PIP3 signal in control cells (WT) are comparable to those of LR08-018 (AKT3 p.Arg465Trp). All other mutant cell lines show increased PIP3 signal compared to control cells. Elevated PIP3 signal is also evident in cells derived from the patient with Cowden disease (PTEN p.Glu261Ter), which served as a positive control. * Statistically significant difference compared to control cells (p<0.05 two-tailed t-test assuming unequal variance, n=30 to 50 cells per cell line). Error bars indicate standard deviation. (c) Levels of PIP3 in cell lines from LR00-016a1 (PIK3R2 p.Gly373Arg) and LR05-204 (PIK3CA p.Glu453del) can be reduced following treatment with the PI3K-inhibitor PI-103 (5μM for 16 hours). Scale bar corresponds to 10μm.
The PI3Ks are a highly conserved family of signaling enzymes that regulate a wide range of processes including cell growth, proliferation, survival, migration, metabolism, angiogenesis, apoptosis, tumorigenesis, and brain development16,17,19–21. As the predominant downstream effector of PI3K signaling, AKT kinases are involved in a wide range of human diseases and play a critical role in growth regulation10. Extensive mouse and human data have shown that loss- and gain-of-function mutations in AKT isoforms lead to opposite phenotypes13,22–25. Several reports have shown that heterozygous loss of AKT3 in human and homozygous loss in mouse cause microcephaly26–28, while a heterozygous missense mutation in Akt3 (p.Asp219Val) resulting in increased kinase activity was shown to cause seizures and brain overgrowth in mouse29. This phenotype resembles the clinical presentation of our two AKT3 mutation carriers (Supplementary Table 8). Further, another group recently reported that somatic activating mutations of AKT3 in brain cause hemimegalencephaly30. Many subjects in our series had asymmetric brain enlargement and several were diagnosed with hemimegalencephaly (Supplementary Fig. 2). These data, combined with our observations of elevated S6 and 4E-BP1 phosphorylation levels in a lymphoblastoid cell line harboring the AKT3 p.Arg465Trp mutation, strongly suggest a gain-of-function mechanism resulting in enhanced AKT activity for the two AKT3 mutations reported here.
Class IA PI3K dimers are composed of a p110 catalytic subunit and a p85 regulatory subunit, each with three isoforms encoded by three genes17. Mutations in five of these genes have been observed in many human cancers31–34. Our data show that mutations in the p85β (PIK3R2) regulatory and p110α (PIK3CA) catalytic subunits are a common cause of megalencephaly syndromes, albeit with a clear genotype-phenotype correlation as PIK3R2 and PIK3CA mutations are associated with MPPH (P = 3.3 × 10−6) and MCAP (P = 1.0 × 10−6), respectively (Supplementary Table 9, Online Methods). Both PIK3R1 and PIK3R2 have oncogenic potential, and mutations including the glycine-to-arginine substitution of PIK3R2 found in MPPH (p.Gly373Arg) and substitution of the homologous amino acid residue in PIK3R1 (p.Gly376Arg) have been found in cancer32. Available functional studies showed that several of these mutations disrupt the inactive conformation of the PI3K dimer and maintain the catalytic subunit in a high activity state32,35. Our observations in lymphoblastoid cells derived from patient LR00-016a1 show that the p.Gly373Arg mutation results in increased PI3K activity and elevated PI3K-mTOR signaling, further supporting this mechanism.
We identified 24 patients with PIK3CA mutations, and all but three (LR06-220, LR11-153, and LR11-230) demonstrated evidence of postzygotic mosaicism. These mutations were discovered and confirmed by independent experiments using four different methods and multiple, mostly uncultured tissues, thus ruling out the possibility of cell culture or technology artifacts. Similar to the study on Proteus syndrome13, we observed lower levels of mosaicism in blood compared to other tissues (Supplementary Table 4).
Somatic activating mutations in PIK3CA are frequently observed in several common human tumor types20. This may partly explain the mildly increased (~3%) incidence of cancer in MCAP and MPPH (Supplementary Note and Supplementary Table 10). Three mutational hot-spots (p.Glu542Lys, p.Glu545Lys and His1047Arg) account for eighty percent of all tumor-associated PIK3CA mutations and show the highest oncogenic activity20,33. Although 13 of 15 PIK3CA mutations found in our megalencephaly cohort are reported in the Catalogue of Somatic Mutations in Cancer (Table 1), only one megalencephaly patient carried one of these severe PIK3CA cancer mutation hotspots (LR12-033, Table 1). Interestingly, her brain phenotype appeared more severe than other patients (Supplementary Fig. 2). Among the MCAP-causing mutations previously associated with cancer, functional data reported for the p.Arg88Gln, p.Glu365Lys, p.Glu545Lys, p.Met1043Ile and p.His1047Tyr mutations demonstrate increased lipid kinase activity resulting in constitutive PI3K signaling36–38, consistent with our observations of increased PI3K activity and signaling in lymphoblastoid cell lines from MCAP patients carrying PIK3CA mutations.
Altogether, we identified germline or postzygotic mutations of AKT3, PIK3R2 or PIK3CA in 37 of 50 (74%) unrelated probands (Fig. 3, Supplementary Table 9). Mutations of all three genes led to pre- and postnatal overgrowth of brain and variably limbs. The brain overgrowth phenotype universally included significant megalencephaly and reduced extra-axial spaces, and frequently polymicrogyria, hydrocephalus, and cerebellar tonsillar ectopia including Chiari malformation (Supplementary Table 8). Patients with MCAP also presented striking – often asymmetric – overgrowth of many other tissues, especially vascular, subcutaneous, connective and lymphatic tissues. Considering the clinical complexity of these syndromes and the potential importance of our findings for clinical care, we have summarized our provisional recommendations for clinical management in the Supplementary Note.
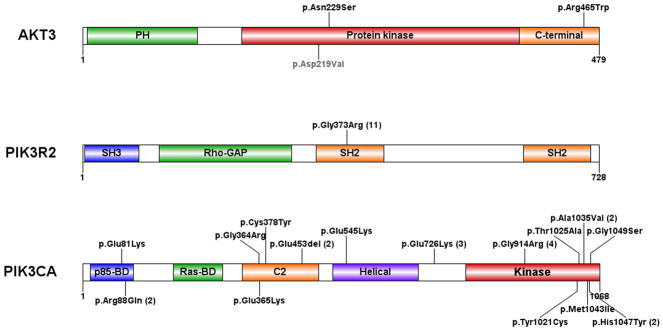
Figure 3.
Distribution of mutations in AKT3, PIK3R2, and PIK3CA. The activating Akt3 mutation in mouse is indicated in grey (p.Asp219Val). For recurrent mutations, the number of occurrences is indicated in parentheses. PH: pleckstrin homology domain; C-terminal: carboxyl-terminal domain; SH2 and SH3: Src-homology-2 and -3 domains; Rho-GAP: Rho GTPase-activating protein domain; p85-BD and RAS-BD: p85- and RAS-binding domains; C2: protein-kinase-C-homology-2 domain. The PIK3CA mutations affect a total of 15 residues, mainly localized in the p85-binding, C2, and catalytic lipid kinase domains.
Although we cannot rule out the possibility of low-level mosaic mutations undetectable by Sanger sequencing, none of the 13 unexplained patients had mutations in PTEN or the four other subunits of class IA PI3K (Online Methods). Further, some unexplained patients may carry mosaic PIK3CA mutations undetectable by Sanger sequencing in the tissues tested. Our data emphasize the challenges of detecting low-level mosaic mutations, but nonetheless highlight the power of massively parallel sequencing for discovering postzygotic mutations genome-wide.
MPPH and MCAP share clinical features with Proteus syndrome and a spectrum of overgrowth disorders resulting from loss-of-function mutations in PTEN, including Bannayan–Riley–Ruvalcaba syndrome, Cowden disease and autism with severe megalencephaly39–41. Our results extend the list of overgrowth syndromes associated with aberrant PI3K/AKT signaling and demonstrate that dysfunction of this pathway causes a constellation of brain, vascular and limb malformations. Finally, our findings combined with the development of PI3K inhibitors for human disease42,43 may open the door to new therapeutic opportunities for megalencephaly and other developmental disorders.
METHODS
Study subjects
The study included 52 affected individuals from 50 unrelated families. Informed consent was obtained from all subjects (see Supplementary Note). Patients were diagnosed as having MCAP or MPPH using our published diagnostic criteria4. Genomic DNA was extracted from different tissues using standard procedures. When parental DNA samples were available, paternity-maternity testing was performed by genotyping six highly polymorphic short-tandem repeats. All exome capture and sequencing experiments were performed using whole-blood genomic DNA.
Exome sequencing of the three index families
Exome capture and sequencing of trio LR08-018 was performed at the PerkinElmer DNA Sequencing Service. Libraries were generated using the 50 megabases (Mb) SureSelect human exome kit (Agilent) and sequenced on a HiSeq 2000 (Illumina) according to the manufacturer’s recommendations for paired-end 100-bp reads. Exome sequencing of trio LR09-006 and subject LR00-016a1 were carried-out at the University of Washington Genome Sciences Genomic Resource Center using the SeqCap EZ Exome Library v2.0 capture kit (Roche) combined with sequencing of paired-end 76-bp reads on a Genome Analyzer IIx (Illumina). Reads were aligned to the human reference genome (hg19) as previously described44.
Single-nucleotide variants and small indels were identified using the GATK Unified Genotyper45 and a variant quality score ≥ 10, and annotated using SeattleSeq SNP annotation (see URLs). Variants were then filtered using standard hard-filtering parameters45. Specifically, only variants with a quality score ≥ 30, sequencing depth ≥ 4, quality/depth ratio ≥ 5, length of homopolymer run ≤ 5.0, and allelic balance ≤ 0.80 were considered for downstream analysis. We also used exome sequencing data from 112 individuals sequenced at the University of Washington Genome Sciences or PerkinElmer DNA Sequencing Service. These included 95 unrelated control individuals from the University of Washington NIEHS Exome Project (see URLs) and 17 in-house exomes consisting of healthy individuals and patients with unrelated phenotypes.
Family-based analysis
For each index patient (LR08-018, LR09-006, and LR00-016a1), we generated a list of “rare variants” by focusing on protein-altering and splice-site DNA changes absent from dbSNP build 132, the 1000 Genomes project46, and 112 control exomes. As described previously44, we systematically identified potential de novo mutational events from the list of rare variants generated for subjects LR08-018 and LR09-006 by focusing on variants supported by ≥ 2 reads in the proband and absent in both parents (< 2 variant reads) at base-pair positions covered by ≥ 4 reads in the entire trio. Candidate de novo mutations were manually inspected using the Integrative Genomics Viewer (IGV)15. This approach was unsuccessful for MCAP trio LR09-006, so we relaxed our criteria to include all variants that failed on the initial hard-filtering parameters used for variant calling, and generated a second list of rare variants and candidate de novo events using the aforementioned criteria.
Genomic DNA samples from LR00-016a1 and both affected siblings were genotyped using the InfiniumII HumanHap610 Quad BeadChip array (Illumina) at the Center for Applied Genomics at Children’s Hospital of Philadelphia prior to the current study. We assumed autosomal recessive inheritance and pulled genes with either homozygous or two heterozygous rare variants. We then used the genome-wide single-nucleotide polymorphism (SNP) data obtained on all three affected siblings to further reduce the number of candidate genes. For a dominant mode of inheritance, we manually examined candidate heterozygous variants from the list of rare variants generated for LR00-016a1.
Additional MCAP patients and control exomes
Exome capture and sequencing from seven unrelated MCAP patients was performed at the McGill University and Genome Quebec Innovation Centre (Montreal, Canada) using the 50 Mb SureSelect human exome kit (Agilent) and sequencing of 100-bp paired-end reads on Illumina Hiseq. We generated over 15 Gb of sequence per individual such that approximately 90% of the coding bases of the genome defined by the consensus coding sequence project were covered by at least 20 reads. Reads were aligned to hg19 with BWA47, and duplicate reads were marked using Picard (see URLs) and excluded from downstream analysis. For each sample, single-nucleotide variants and indels were called using SAMtools pileup and varFilter48 with the base alignment quality adjustment disabled, and were then quality filtered using a threshold of at least 20% of reads supporting the variant call. Due to low-level mosaicism in most MCAP patients, this initial analysis missed all but one mutation of PIK3CA, which were later found using other methods. The study also included exome sequencing data from 174 individuals sequenced at the University of Washington Genome Sciences using the SeqCap EZ Exome Library v2.0 (Roche) and Illumina sequencing platform. These 174 individuals consist of unrelated healthy individuals of European descent that are parents of children with sporadic autism.
To detect low-level mosaicism, we systematically analyzed the 3,287 coding and splice-site bases of PIK3CA in exomes from the remaining six unsolved MCAP patients and 174 unrelated control individuals. Standard variant calling49 in the patient and control datasets identified 36 heterozygous variants supported by 41 to 66% of reads, and one homozygous change found in 99% of reads. These sites were considered as germline variants and excluded from downstream analysis. To detect candidate mosaic changes, we first pulled all sites with at least one read not matching the reference sequence using the SAMtools (v0.1.7) pileup48. Raw variant sites were then run through a custom pipeline to identify candidate mosaic changes supported by at least 1, 2, 3 or 4 variant reads using a base quality threshold of 20 and at least 1% of reads supporting the variant.
Sanger sequencing
We amplified the coding exons of AKT3 (NM_005465.4 and NM_181690.1), BDP1 (NM_018429.2), PIK3R1 (NM_181523.1, NM_181524.1 and NM_181504.2), PIK3R2 (NM_005027.2), PIK3R3 (AK302049.1 and EU832531.1), PIK3CA (NM_006218.2), PIK3CB (NM_006219.1), PIK3CD (NM_005026.3 and U57843.1), and PTEN (NM_000314.4) using custom intronic primers and standard polymerase-chain-reaction (PCR) protocols (primer sequences available on request) combined with Sanger sequencing. Amplicons were sequenced at the University of Washington High-Throughput Genomics Unit and Seattle BioMed Sequencing Core Facility. All mutations were tested in at least two independent amplification and sequencing reactions in the proband and available relatives. Sequence traces were analyzed using Mutation Surveyor v3.97 (SoftGenetics). For all mutations, nucleotide-level conservation and impact of amino-acid substitutions were assessed using GERP50 and Grantham matrix scores51, respectively.
Restriction-enzyme and genotyping assays
Six mutation sites (p.Glu453del, p.Glu726Lys, p.Gly914Arg, p.Ala1035Val, p.Met1043Ile, and p. His1047Tyr) were confirmed by PCR using HotStarTaq Plus DNA polymerase (Qiagen), and primer pairs including a carboxyfluorescein (FAM)-labeled primer and an unlabeled primer with a GTTTCTT sequence on the 5′ end52. Amplicons were digested using restriction enzymes (New England BioLabs) cutting either the wild-type or the mutant allele, depending on the mutation examined (Supplementary Table 11). Fragments were detected on the ABI 3730XL DNA Analyzer (Applied Biosystems) and results were analyzed using GenMarker v1.90 software (SoftGenetics). The fraction of mutant allele was calculated as follows: mutant peak area/(wild-type peak area + mutant peak area).
The p.Glu726Lys PIK3CA mutation is located in a 7.2 kb segmental duplication (chr3:178932477-178939690) sharing 98% identity with a region on chromosome 22 (chr22:17049390-17056254). To rule out the possibility of non-specific amplification, we amplified a 12.2 kb fragment using the TaKaRa LA Taq and primers flanking the segmental duplication (primer sequences are listed in Supplementary Table 11). We then used the long-range PCR product as a template for our restriction-enzyme assay.
The p.Glu453del PIK3CA mutation was confirmed by standard PCR using a forward FAM-labeled primer and an unlabeled reverse primer to amplify 238 and 235 bp fragments corresponding to the wild-type and mutant alleles, respectively.
Deep sequencing of mutation sites
We performed targeted ultra-deep sequencing of five mutation sites in PIK3CA in a series of MCAP and MPPH patients and unaffected parents using chimeric oligonucleotides containing Illumina adapter sequences combined with locus specific primers. Reverse oligonucleotides also contained 12 unique 8-mer barcodes for multiplexing of up to 12 samples per lane (Supplementary Table 12). Amplification was carried out in a MiniOpticon Real-time PCR system (Bio-Rad) using the iProof High-Fidelity Master Mix (Bio-Rad), 50 ng of genomic DNA, and SYBR Green. Samples were removed from the PCR machine before fluorescence began to plateau. Amplicons were purified using Ampure beads (Agencourt) and directly sequenced on an Illumina HiSeq 2000 using 100 bp paired-end reads. Sequencing reads were aligned to hg19 using BWA47. We used the SAMtools (v0.1.7) pileup48 combined with custom scripts to pull variant sites supported by at least 1% of reads with a base quality score threshold of 20.
The two newly identified mutations (p.Glu81Lys and p.Arg88Gln in subjects LR06-342 and LR11-068, respectively) were validated by an independent ultra-deep sequencing experiment including two DNA sources per affected individual and DNA samples from parents of LR11-068. All six DNA samples were amplified and sequenced twice using 8-mer barcodes.
Lymphoblastoid cell lines
Epstein-Barr virus immortalized lymphoblastoid cell lines were established from peripheral blood samples of four megalencephaly patients (LR00-016a1, LR08-018, LR05-204, and LR09-006), one unaffected individual, and one patient with Cowden disease (GM10080). Lymphoblastoid cell lines were cultured in RPMI medium with 15% fetal calf serum (FCS), L-Gln and antibiotics (Pen-Strep) at 5% CO2.
Antibodies and PI3K inhibitor
Anti-phosphatidylinositol-3,4,5-trisphosphate (PIP3) monoclonal antibody was from Caltag-Medsystems Ltd (UK). The following antibodies were obtained from Cell Signalling Technology: PDK1-pSer241, S6-pSer240/244, 4E-BP-pThr37/46, and β-tubulin. Conjugated secondary antibodies, anti-mouse IgG-FITC (whole molecule), and anti-rabbit IgG-Cy3 F(ab′)2 fragment were from SIGMA-ALDRICH (UK). We used the PI3K inhibitor InSolution™ PI-103 (Calbiochem) from Merck-Millipore.
Indirect immunofluorescence
Exponentially growing lymphoblastoid cell lines were pelleted, swollen in 75mM KCL (10 minutes), immobilized onto poly-lysine coated slides by cytospinning (CytoSpin. Shandon), permeabilized (0.1% triton X-100 in 5% BSA/PBS for 2 minutes) and blocked in 5% BSA/PBS (10 minutes), prior to sequential incubation with primary and secondary antibodies. Slides were counterstained with DAPI and preserved in anti-fade mounting media (Vectashield). Slides were analyzed using the Zeiss AxioPlan platform. Images were captured using SimplePCI software and the same exposure time for each sample. Images (TIFF format) were then used to quantify signal (RGB) using ImageJ software.
SDS-page and Western Blot
Whole-cell extracts were prepared by lysing the cells in urea buffer (9M urea, 50mM Tris-HCl at pH 7.5 and 10mM 2-mercaptoethanol), followed by 15sec sonication, 30% amplitude using a micro-tip (SIGMA-Aldrich). The supernatant was quantified by Bradford Assay. Extracts were resolved by SDS-PAGE, semi-dry blotted onto polyvinylidene fluoride (PVDF) membranes for blocking and antibody incubation.
Statistical analysis
The likelihood of observing a second de novo mutation in AKT3 was calculated with the following: P = 1.28 × 10−8 × 1,540 × 98; with 1.28 × 10−8 as the estimated haploid substitution rate53, 1,540 as the number of coding and splice-site bases of AKT3 (NM_005465.4 and NM_181690.1), and 98 as the number of chromosomes tested (49 unrelated patients). We used two-tailed Fisher’s exact tests for genotype-phenotype correlations.
Supplementary Material
Acknowledgments
We wish to thank all of the children and families in this study, their referring physicians, and the M-CM Network (see URLs) for their help with this project over many years. We thank the members of the Northwest Genomics Center and McGill University and Genome Quebec Innovation Centre for their excellent technical assistance. We also thank the FORGE (Finding of Rare Disease Genes) Canada Consortium, especially Janet Marcadier (Clinical Coordinator) for her contribution to the infrastructure.
This work was funded by the US National Institutes of Health under NINDS grant NS058721 (to W.B.D.), NICHD grant HD36657 and NIGMS grant 5-T32-GM08243 (to J.M.G. Jr.), and the Government of Canada (to FORGE) through Genome Canada, the Canadian Institutes of Health Research (CIHR), and the Ontario Genomics Institute (OGI-049). Additional funding was provided to FORGE by Genome Quebec and Genome British Columbia. J.-B.R. is supported by a Banting Postdoctoral Fellowship from the CIHR. K.M.B. is supported by a Clinical Investigatorship Award from the CIHR Institute of Genetics. M.O.D. laboratory is funded by Cancer Research UK (CR-UK), Medical Research Council (UK) and Leukaemia Lymphoma Research (UK). M.O.D. is a Senior CR-UK Research Fellow.
We would like to thank the Simons Foundation Autism Research Initiative (SFARI) for providing control exome data (grant 191889 to J.S). We also thank the NIEHS Environmental Genome Project (contract No.HHSN273200800010C) and the NHLBI GO Exome Sequencing Project and its ongoing studies – the Lung GO (HL-102923), Broad GO (HL-102925), Seattle GO (HL-102926), Heart GO (HL-103010) and WHI (HL-102924) Sequencing Projects – for providing exome variant calls for comparison.
Footnotes
URLs
M-CM Network, http://www.m-cm.net/
Catalogue of Somatic Mutations in Cancer, http://www.sanger.ac.uk/genetics/CGP/cosmic/
NHLBI Exome Sequencing Project, http://evs.gs.washington.edu/EVS/
SeattleSeq Annotation 131, http://snp.gs.washington.edu/SeattleSeqAnnotation131/
NIEHS Exome Project; http://evs.gs.washington.edu/niehsExome/
Picard, http://picard.sourceforge.net/
DATA ACCESS
The exome sequencing data from individuals of the three index families has been deposited in dbGaP under accession number phs000455.v1.p1.
COMPETING FINANCIAL INTERESTS
The authors report no competing financial interests.
AUTHOR CONTRIBUTIONS
J.-B.R., G.M.M., K.M.B. and W.B.D. designed the study. J.-B.R., B.J.O. and J.S.-O. designed and performed the genetics experiments. M.B., T.W., C.T.S. and T.R.W. contributed to the genetics experiments. J.-B.R., J.A.S. and B.J.O. performed the bioinformatics experiments. D.A. performed the experiments in lymphoblastoid cell lines. G.M.M., R.L.C., K.W.G., S.M.N., B.A., C.M.A., L.A., O.C., C.C., B.A.D., A.M.I., J.L.L., A.E.L., G.M.S.M., W.S.M., J.D.R., A.K.S., T.L.-S., G.U., R.W., B.Z., J.M.G.Jr., K.M.B. and W.B.D. recruited and evaluated the study subjects. H.E.B., N.A.K. and C.L.B. provided administrative support and recruited the study subjects. J.M., D.E.B., M.O.D., J.S., K.M.B. and W.B.D supervised the study. J.-B.R., G.M.M. and W.B.D. wrote the paper.
References
- 1.Clayton-Smith J, et al. Macrocephaly with cutis marmorata, haemangioma and syndactyly--a distinctive overgrowth syndrome. Clin Dysmorphol. 1997;6:291–302. doi: 10.1097/00019605-199710000-00001. [DOI] [PubMed] [Google Scholar]
- 2.Mirzaa G, et al. Megalencephaly and perisylvian polymicrogyria with postaxial polydactyly and hydrocephalus: a rare brain malformation syndrome associated with mental retardation and seizures. Neuropediatrics. 2004;35:353–9. doi: 10.1055/s-2004-830497. [DOI] [PubMed] [Google Scholar]
- 3.Moore CA, et al. Macrocephaly-cutis marmorata telangiectatica congenita: a distinct disorder with developmental delay and connective tissue abnormalities. Am J Med Genet. 1997;70:67–73. [PubMed] [Google Scholar]
- 4.Mirzaa GM, et al. Megalencephaly-capillary malformation (MCAP) and megalencephaly-polydactyly-polymicrogyria-hydrocephalus (MPPH) syndromes: two closely related disorders of brain overgrowth and abnormal brain and body morphogenesis. Am J Med Genet A. 2012;158A:269–91. doi: 10.1002/ajmg.a.34402. [DOI] [PubMed] [Google Scholar]
- 5.Conway RL, et al. Neuroimaging findings in macrocephaly-capillary malformation: a longitudinal study of 17 patients. Am J Med Genet A. 2007;143A:2981–3008. doi: 10.1002/ajmg.a.32040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Oduber CE, et al. A proposal for classification of entities combining vascular malformations and deregulated growth. Eur J Med Genet. 2011;54:262–71. doi: 10.1016/j.ejmg.2011.02.007. [DOI] [PubMed] [Google Scholar]
- 7.Happle R. Lethal genes surviving by mosaicism: a possible explanation for sporadic birth defects involving the skin. J Am Acad Dermatol. 1987;16:899–906. doi: 10.1016/s0190-9622(87)80249-9. [DOI] [PubMed] [Google Scholar]
- 8.Gripp KW, et al. Significant overlap and possible identity of macrocephaly capillary malformation and megalencephaly polymicrogyria-polydactyly hydrocephalus syndromes. Am J Med Genet A. 2009;149A:868–76. doi: 10.1002/ajmg.a.32732. [DOI] [PubMed] [Google Scholar]
- 9.Brodbeck D, Cron P, Hemmings BA. A human protein kinase Bgamma with regulatory phosphorylation sites in the activation loop and in the C-terminal hydrophobic domain. J Biol Chem. 1999;274:9133–6. doi: 10.1074/jbc.274.14.9133. [DOI] [PubMed] [Google Scholar]
- 10.Hers I, Vincent EE, Tavare JM. Akt signalling in health and disease. Cell Signal. 2011;23:1515–27. doi: 10.1016/j.cellsig.2011.05.004. [DOI] [PubMed] [Google Scholar]
- 11.Otsu M, et al. Characterization of two 85 kd proteins that associate with receptor tyrosine kinases, middle-T/pp60c-src complexes, and PI3-kinase. Cell. 1991;65:91–104. doi: 10.1016/0092-8674(91)90411-q. [DOI] [PubMed] [Google Scholar]
- 12.Scarano E, Iaccarino M, Grippo P, Parisi E. The heterogeneity of thymine methyl group origin in DNA pyrimidine isostichs of developing sea urchin embryos. Proc Natl Acad Sci U S A. 1967;57:1394–400. doi: 10.1073/pnas.57.5.1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lindhurst MJ, et al. A mosaic activating mutation in AKT1 associated with the Proteus syndrome. N Engl J Med. 2011;365:611–9. doi: 10.1056/NEJMoa1104017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Volinia S, et al. Molecular cloning, cDNA sequence, and chromosomal localization of the human phosphatidylinositol 3-kinase p110 alpha (PIK3CA) gene. Genomics. 1994;24:472–7. doi: 10.1006/geno.1994.1655. [DOI] [PubMed] [Google Scholar]
- 15.Robinson JT, et al. Integrative genomics viewer. Nat Biotechnol. 2011;29:24–6. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vanhaesebroeck B, et al. Synthesis and function of 3-phosphorylated inositol lipids. Annu Rev Biochem. 2001;70:535–602. doi: 10.1146/annurev.biochem.70.1.535. [DOI] [PubMed] [Google Scholar]
- 17.Engelman JA, Luo J, Cantley LC. The evolution of phosphatidylinositol 3-kinases as regulators of growth and metabolism. Nat Rev Genet. 2006;7:606–19. doi: 10.1038/nrg1879. [DOI] [PubMed] [Google Scholar]
- 18.Mora A, Komander D, van Aalten DM, Alessi DR. PDK1, the master regulator of AGC kinase signal transduction. Semin Cell Dev Biol. 2004;15:161–70. doi: 10.1016/j.semcdb.2003.12.022. [DOI] [PubMed] [Google Scholar]
- 19.Katso R, et al. Cellular function of phosphoinositide 3-kinases: implications for development, homeostasis, and cancer. Annu Rev Cell Dev Biol. 2001;17:615–75. doi: 10.1146/annurev.cellbio.17.1.615. [DOI] [PubMed] [Google Scholar]
- 20.Samuels Y, Ericson K. Oncogenic PI3K and its role in cancer. Curr Opin Oncol. 2006;18:77–82. doi: 10.1097/01.cco.0000198021.99347.b9. [DOI] [PubMed] [Google Scholar]
- 21.Sansal I, Sellers WR. The biology and clinical relevance of the PTEN tumor suppressor pathway. J Clin Oncol. 2004;22:2954–63. doi: 10.1200/JCO.2004.02.141. [DOI] [PubMed] [Google Scholar]
- 22.Cho H, et al. Insulin resistance and a diabetes mellitus-like syndrome in mice lacking the protein kinase Akt2 (PKB beta) Science. 2001;292:1728–31. doi: 10.1126/science.292.5522.1728. [DOI] [PubMed] [Google Scholar]
- 23.Cho H, Thorvaldsen JL, Chu Q, Feng F, Birnbaum MJ. Akt1/PKBalpha is required for normal growth but dispensable for maintenance of glucose homeostasis in mice. J Biol Chem. 2001;276:38349–52. doi: 10.1074/jbc.C100462200. [DOI] [PubMed] [Google Scholar]
- 24.George S, et al. A family with severe insulin resistance and diabetes due to a mutation in AKT2. Science. 2004;304:1325–8. doi: 10.1126/science.1096706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hussain K, et al. An activating mutation of AKT2 and human hypoglycemia. Science. 2011;334:474. doi: 10.1126/science.1210878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ballif BC, et al. High-resolution array CGH defines critical regions and candidate genes for microcephaly, abnormalities of the corpus callosum, and seizure phenotypes in patients with microdeletions of 1q43q44. Hum Genet. 2012;131:145–56. doi: 10.1007/s00439-011-1073-y. [DOI] [PubMed] [Google Scholar]
- 27.Boland E, et al. Mapping of deletion and translocation breakpoints in 1q44 implicates the serine/threonine kinase AKT3 in postnatal microcephaly and agenesis of the corpus callosum. Am J Hum Genet. 2007;81:292–303. doi: 10.1086/519999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tschopp O, et al. Essential role of protein kinase B gamma (PKB gamma/Akt3) in postnatal brain development but not in glucose homeostasis. Development. 2005;132:2943–54. doi: 10.1242/dev.01864. [DOI] [PubMed] [Google Scholar]
- 29.Tokuda S, et al. A novel Akt3 mutation associated with enhanced kinase activity and seizure susceptibility in mice. Hum Mol Genet. 2011;20:988–99. doi: 10.1093/hmg/ddq544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Poduri A, et al. Somatic Activation of AKT3 Causes Hemispheric Developmental Brain Malformations. Neuron. 2012;74:41–8. doi: 10.1016/j.neuron.2012.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455:1061–8. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cheung LW, et al. High Frequency of PIK3R1 and PIK3R2 Mutations in Endometrial Cancer Elucidates a Novel Mechanism for Regulation of PTEN Protein Stability. Cancer Discov. 2011;1:170–185. doi: 10.1158/2159-8290.CD-11-0039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Samuels Y, et al. High frequency of mutations of the PIK3CA gene in human cancers. Science. 2004;304:554. doi: 10.1126/science.1096502. [DOI] [PubMed] [Google Scholar]
- 34.Thomas RK, et al. High-throughput oncogene mutation profiling in human cancer. Nat Genet. 2007;39:347–51. doi: 10.1038/ng1975. [DOI] [PubMed] [Google Scholar]
- 35.Jaiswal BS, et al. Somatic mutations in p85alpha promote tumorigenesis through class IA PI3K activation. Cancer Cell. 2009;16:463–74. doi: 10.1016/j.ccr.2009.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gymnopoulos M, Elsliger MA, Vogt PK. Rare cancer-specific mutations in PIK3CA show gain of function. Proc Natl Acad Sci U S A. 2007;104:5569–74. doi: 10.1073/pnas.0701005104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ikenoue T, et al. Functional analysis of PIK3CA gene mutations in human colorectal cancer. Cancer Res. 2005;65:4562–7. doi: 10.1158/0008-5472.CAN-04-4114. [DOI] [PubMed] [Google Scholar]
- 38.Oda K, et al. PIK3CA cooperates with other phosphatidylinositol 3′-kinase pathway mutations to effect oncogenic transformation. Cancer Res. 2008;68:8127–36. doi: 10.1158/0008-5472.CAN-08-0755. [DOI] [PubMed] [Google Scholar]
- 39.Butler MG, et al. Subset of individuals with autism spectrum disorders and extreme macrocephaly associated with germline PTEN tumour suppressor gene mutations. J Med Genet. 2005;42:318–21. doi: 10.1136/jmg.2004.024646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liaw D, et al. Germline mutations of the PTEN gene in Cowden disease, an inherited breast and thyroid cancer syndrome. Nat Genet. 1997;16:64–7. doi: 10.1038/ng0597-64. [DOI] [PubMed] [Google Scholar]
- 41.Marsh DJ, et al. Mutation spectrum and genotype-phenotype analyses in Cowden disease and Bannayan-Zonana syndrome, two hamartoma syndromes with germline PTEN mutation. Hum Mol Genet. 1998;7:507–15. doi: 10.1093/hmg/7.3.507. [DOI] [PubMed] [Google Scholar]
- 42.Kong D, Yamori T. Phosphatidylinositol 3-kinase inhibitors: promising drug candidates for cancer therapy. Cancer Sci. 2008;99:1734–40. doi: 10.1111/j.1349-7006.2008.00891.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Marone R, Cmiljanovic V, Giese B, Wymann MP. Targeting phosphoinositide 3-kinase: moving towards therapy. Biochim Biophys Acta. 2008;1784:159–85. doi: 10.1016/j.bbapap.2007.10.003. [DOI] [PubMed] [Google Scholar]
- 44.Riviere JB, et al. De novo mutations in the actin genes ACTB and ACTG1 cause Baraitser-Winter syndrome. Nat Genet. 2012;44:440–4. S1–2. doi: 10.1038/ng.1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.DePristo MA, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 2011;43:491–8. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–73. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–60. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li H, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–9. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.O’Roak BJ, et al. Exome sequencing in sporadic autism spectrum disorders identifies severe de novo mutations. Nat Genet. 2011;43:585–9. doi: 10.1038/ng.835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cooper GM, et al. Single-nucleotide evolutionary constraint scores highlight disease-causing mutations. Nat Methods. 2010;7:250–1. doi: 10.1038/nmeth0410-250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Grantham R. Amino acid difference formula to help explain protein evolution. Science. 1974;185:862–4. doi: 10.1126/science.185.4154.862. [DOI] [PubMed] [Google Scholar]
- 52.Brownstein MJ, Carpten JD, Smith JR. Modulation of non-templated nucleotide addition by Taq DNA polymerase: primer modifications that facilitate genotyping. Biotechniques. 1996;20:1004–6. 1008–10. doi: 10.2144/96206st01. [DOI] [PubMed] [Google Scholar]
- 53.Lynch M. Rate, molecular spectrum, and consequences of human mutation. Proc Natl Acad Sci U S A. 2010;107:961–8. doi: 10.1073/pnas.0912629107. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.