Summary
Autophagy is a cell biological process ubiquitous to all eukaryotic cells, often referred to as a catabolic, lysosomal degradative pathway. However, current studies in mammalian systems suggest that autophagy plays an unexpectedly broad biogenesis role in protein trafficking and secretion. Autophagy supports alternative trafficking pathways for delivery of integral membrane proteins to the plasma membrane and affects secretion, including the constitutive, regulated and unconventional secretion pathways. Autophagy-based unconventional secretion, termed here ‘autosecretion’, is one of the pathways enabling leaderless cytosolic proteins to exit the cell without entering the ER-to-Golgi secretory pathway. In this review, we discuss the emerging underlying mechanisms of how autophagy affects different facets of secretion. We also describe the physiological roles of autosecretory cargos that are often associated with inflammatory processes and also play a role in the formation of specialized tissues and in tissue remodeling, expanding the immediate sphere of influence of autophagy from the intracellular to the extracellular space.
Keywords: Autophagy, autosecretion, inflammasome, cystic fibrosis, IL-1, GRASP
Role of autophagy in biology exceeds degradation and catabolism
The sensu stricto autophagy (often referred to as macroautophagy) is a ubiquitous eukaryotic process dependent on evolutionarily conserved Atg (autophagy) factors and on formation of specialized internal membrane domains in the cytoplasm that form unique organelles called autophagosomes [1]. Autophagosomes capture various cytoplasmic cargo with different end purposes that typically fall into one of the following general categories: i) Quality control and removal of potentially harmful intracellular macromolecular aggregates and disused or defunct organelles (e.g. irreversibly depolarized or damaged mitochondria [2, 3]) often associated with aging and degenerative diseases [4, 5]; ii) Digestion of bulk cytoplasmic proteins to replenish amino acid pools and energy during starvation or growth factor withdrawal as well as to support energetic balance and supply anabolic precursors in cells driven by oncogenes to proliferate [6]; iii) Cooperation with the molecular machineries and organelles at the interface between cell survival and cell death [7]; iv) Regulatory and effector functions in innate and adaptive immunity and inflammation [8, 9].
Another emerging general function of autophagy, and the main topic of this review, is its role in protein secretion and trafficking of integral membrane proteins to the plasma membrane. The autophagy pathway and the Atg molecular machinery [1] are engaged in regulated secretion of the contents of granules or secondary lysosomes in specialized secretory cells that include protein [10–13] and potentially other [14] effector cargo. The secreted bioactive proteins and other products participate in tissue remodeling [11], inflammatory processes [10] and development of highly specialized aspects of certain tissues and organs [13, 15]. The organelles of this regulated secretion emerge from post-Golgi precursors directly or indirectly via complex trafficking pathways, and for the most part store biological mediators delivered from the ER-to-Golgi biosynthetic pathway. They mature and, in principle, fuse with the plasma membrane in response to physiological triggers. However, this is not the only aspect of conventional secretion that is affected by autophagy machinery. Recent evidence suggests that proteins exported via the constitutive branch of the ER-to-Golgi pathway are influenced by autophagy through a novel compartment named TASCC (TOR-autophagy spatial coupling compartment) [16]. Moreover, integral membrane proteins such as CFTR can be physiologically delivered to the plasma membrane via an unconventional trafficking pathway, bypassing the Golgi apparatus and vectored directly from the ER to the plasma membrane assisted by the autophagic machinery [17].
The role of autophagy in protein trafficking and secretion does not stop with the aspects of the conventional biosynthetic pathway utilized by integral membrane proteins and proteins that enter the ER lumen via signal peptides. Autophagy plays a role in extracellular release of a subset of leaderless cytosolic proteins, which cannot enter the ER but can nevertheless be secreted via a pathway dependent on the autophagic machinery [18–21]. In this review we give an update on the core autophagy pathway and provide an integrated view of the role of autophagy in protein secretion and trafficking.
Autophagy as a cell biological pathway
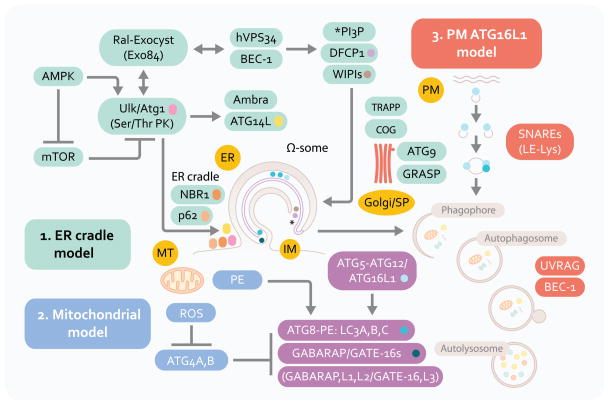
For a comprehensive list of autophagy factors in yeast, their equivalents in higher eukaryotes cells, and additional factors controlling autophagy in mammalian cells, the reader is referred to a recent comprehensive review [1]. In this section we cover aspects of autophagosome membrane biology that may be useful in understanding the connections of autophagic machinery with secretory manifestations discussed in this review. Autophagosomes are believed to emerge at least in part from the endoplasmic reticulum (ER) membranes [22, 23] via an ER cradle model, with the autophagic phagophore (isolation membrane) being formed from or in the vicinity of a phosphatidylinositol 3-phosphate (PI3P)-positive (albeit PI3P is atypical for the ER) and DFCP (double FYVE/PI3P-binding domain containing protein 1)-positive transient omegasome structures [24] (Figure 1, model 1). This likely occurs with participation of additional compartments and organelles supplying either assembly/signaling platforms, phospholipids, or membrane intermediates including those originating from the biosynthetic secretory pathway [25] [26–29], plasma membrane [30], and ER-mitochondria contact sites [31]. Several standalone alternatives or processes contributing to autophagosomes relative to the ER cradle have been proposed (Figure 1, model 2). In models highlighting mitochondria, this organelle has been postulated to be a direct source of autophagosomal membrane [31] as mitochondria at the very least may contribute a specific phospholipid, phosphatidylethanolamine (PE) as one site of its synthesis. PE is of significance, because one of the key autophagy factors, Atg8 (and multiple paralogs in mammals including LC3; Figure 1), is lipidated with PE at its C-terminus (when it is referred to as Atg8-PE; LC3-II in mammalian cells) thus permitting its association with membranes. More importantly, mitochondria are themselves prime substrates for autophagy and can induce autophagy via reactive oxygen species (ROS). ROS, through a redox-sensitive Cys residue in Atg4 (Cys81 in ATG4A and Cys78 in ATG4B), inactivate this protease [34] that otherwise reverses Atg8-PE modification required for autophagic membrane growth [33] and closure [32], with ROS thus favoring autophagosome expansion (Figure 1, model 2).
Fig. 1.
Autophagy pathway: signaling systems and three proposed membrane sources in autophagosome formation. ER, endoplasmic reticulum. PM, plasma membrane. MT, mitochondria. Model 1: The central membranous structure, omegasome (Ω-some) is derived from the ER (ER cradle model), and is believed to be an early precursor of autophagic isolation membranes (IM) or phagophores. Phagophore crescents close to form double membrane autophagosomes that fuse with lysosomal intermediates to form the degradative organelles, autolysosomes. Model 2: Mitochondria may contribute membrane or phosphatidylinositol (PE) of relevance for LC3 (A, B and C; and other Atg8 paralogs, GABARAP and GATE-16) C-terminal lipidation into the LC3-II, autophagic membrane-associated form. Mitochondria may also be a source of reactive oxygen species that inactivate ATG4, an LC3 dilipidating enzyme. Mitochondria are also one of the major target substrates for autophagic elimination. Model 3: Plasma membrane Atg16L1-positive (initially LC3-negative) vesicles may contribute to autophagic membrane growth. Factors in the left upper corner represent upstream signaling systems (AMPK, mammalian Tor [mTor], Ral) controlling induction of autophagy in response to nutritional and cellular energetics signals. Beclin 1 (BEC-1) and class III phosphatidylinositol 3-kinase hVPS34 cooperate in control of phosphatidylinositol 3-phosphate (PI3P) structures that start with Ω-some, identifiable by the marker DFCP-1 for which a functional role in autophagy is yet to be established. NBR1 and p62 (also known as sequestosome 1) are autophagic adaptor proteins that capture cargo and interact with LC3; p62 is also present very early at the sites leading to omegasome formation, and is furthermore found in complexes with mTOR that sense amino acid starvation (not shown). Ambra and Atg14L are additional factors interacting with Beclin 1 complexes that are responsible for the early autophagosomal pathway. The lipid kinase hVPS34 interacts with UVRAG and additional factors (not shown) to control autophagosomal maturation into autolysosomes. Several systems and tethering systems along the different stages of the early secretory pathway and the Golgi apparatus (TRAPP, COG, GRASP) influence the formation, expansion (contributed by the only Atg integral membrane protein Atg9) and maturation of autophagosomal organelles.
Another alternative model that relies on plasma membrane as a source of autophagic precursors is based on a demonstration of homotypic fusion of ATG16L1+LC3− plasma membrane-derived profiles that fuse in a process dependent on late endosomal/lysosomal SNAREs (membrane fusion proteins such as Vamp7, Syntaxins 7 and 8 and Vti1b) to form autophagosomal membrane precursors (Figure 1, model 3). In yeast, the Golgi apparatus may contribute membranes to autophagosome expansion [25], and the regulatory/tethering complexes along different stations of the biosynthetic pathway -- including TRAPP (transport protein particle) [27, 28], the A lobe of COG (conserved oligomeric Golgi complex) [26], and exocyst (an octameric protein complex regulating vesicular trafficking, including targeting post-Golgi vesicles to the plasma membrane) [29] -- have been implicated in autophagosomal biogenesis or its control. Finally, precursor sites associated with mitochondria as Atg9-positive tubulo-vesicular clusters derived from the secretory pathway have been observed in yeast that can be sequentially promoted into an autophagosome-formation competent organelle [35] referred to in yeast as a pre-autophagosomal structure or phagophore assembly site (PAS). Atg9 is the only Atg membrane integral protein whose trafficking seems to be controlled by the sole known PI3P-binding Atg protein, Atg18 in yeast (in mammals represented by several orthologs termed WIPIs [WD-repeat proteins interacting with phosphoinosides]). In mammalian cells, it redistributes from its resting location in trans-Golgi network and late endosomes to peripheral LC3-positive profiles [36].
The processes of autophagosomal formation and autophagic flux are under the control of several systems [37] that have yet to be linked into a fully integrated model: (i) The first system represents the well-known protein conjugation systems (Atg5-Atg12/Atg16L and Atg7-Atg3-Atg8) culminating in the C-terminal lipidation of Atg8 with PE resulting in Atg8-PE, better known as LC3-II, the emblematic marker of mammalian autophagosomes [37]; (ii) The second of these systems is centered upon the Ser/Thr kinase Atg1 (mammalian Ulk1 and Ulk2) controlled by the upstream nutritional regulator mTOR (mammalian target of rapamycin) and, perhaps equally if not more importantly, directly by the cellular energy charge sensor and regulator AMPK (AMP-activated protein kinase) [38–41]. Ulk1 forms a complex with Atg13, Atg101 and FIP200, and although the full significance of this is not known at present, it is possible that these elements, with FIP200 being one of the likely key players, connect through yet to be delineated mechanisms with other parts of the autophagy initiation system including autophagic adapters (e.g. p62) and their cargo, the only known integral membrane autophagy protein -- Atg9L1-- and its recycling machinery (WIPIs/Atg18 and Atg2A/B) and the Atg conjugation system described above; (iii) The third system is centered on Atg6 (Beclin 1), which interacts with many proteins including the evolutionarily ancient Class III PI3K, Vps34 [42]. How precisely these three systems interact or cooperate to bring about autophagosome initiation, elongation and maturation is presently not known; only a small number of published hints exist (e.g. a finding that Ulk1 affects components such as Ambra thereby influencing the behavior of the Atg14L complex with Beclin 1 and hVPS34 [43], and the idea that exocyst as an effector of Ral may serve as an activating platform for both Ulk1 and Beclin 1-hVPS34 [29]).
In contrast to the paucity of details on the integration of the above systems, functions of the individual subsystems have been extensively studied. Although important details are still lacking, in sum, Ulk1/2 gathers signaling inputs coming from mTor (inhibitor of autophagy) and AMPK (inducer of autophagy) [39–41]. Both Ulk1/2 and one of the Beclin 1 interactors, Atg14L, mark the sites for initiation of autophagosome formation on the ER [37]. These structures are potentially but not necessarily equivalent to PAS in yeast, where this acronym refers to pre-autophagosomal structures or the phagophore assembly sites [37]. The Ulk1/2 and Atg14L puncta on the ER are formed in the vicinity of PI3P-positive structures referred to as omegasomes [24]. How the transition from Ulk1-driven processes to Beclin 1/PI3K-driven processes is made in the formation of autophagosomes is still a matter of conjecture. PI3P is believed to be generated by the Atg14L-Beclin 1-hVPS34 complex progressing through yet to be defined intermediates -- with participation of a number of proteins including WIPIs (Atg18 paralogs), Atg5-Atg12/Atg16L1, and Atg9 -- into LC3-II positive structures that elongate and wrap around their targets [37].
Significantly, the ‘PAS’ sites on the ER in mammalian cells seem to receive very early on at the Ulk1 recruitment sites [44] the autophagic adaptor proteins sequestosome 1/p62 and NBR1 (both referred to as sequestosome 1/p62-like receptors or SLRs), which will be discussed in other parts of this review as we explore links with innate immunity and inflammation. The p62 SLR is located at the very nidus of autophagosome formation in mammalian cells, even before their engagement in cargo recognition and capture [44]. Because SLRs have also been defined [45, 46] as complex inflammatory signaling platforms independent of their role as autophagic adaptors, this suggests potential for inflammatory signaling at the very initiation of the autophagic process. Additionally, p62/sequestosome 1 interacts with mTOR and plays a role in sensing nutritional signals [47]. How and why nutrition and inflammation are so closely wired through these systems remains to be fully understood. The answers may enlighten us about a subset of inflammatory conditions associated with metabolic state, and provide a deeper understanding of the core principles of regulatory and effector inputs and outputs of autophagy in mammalian systems.
Autophagy-based unconventional secretion (autosecretion)
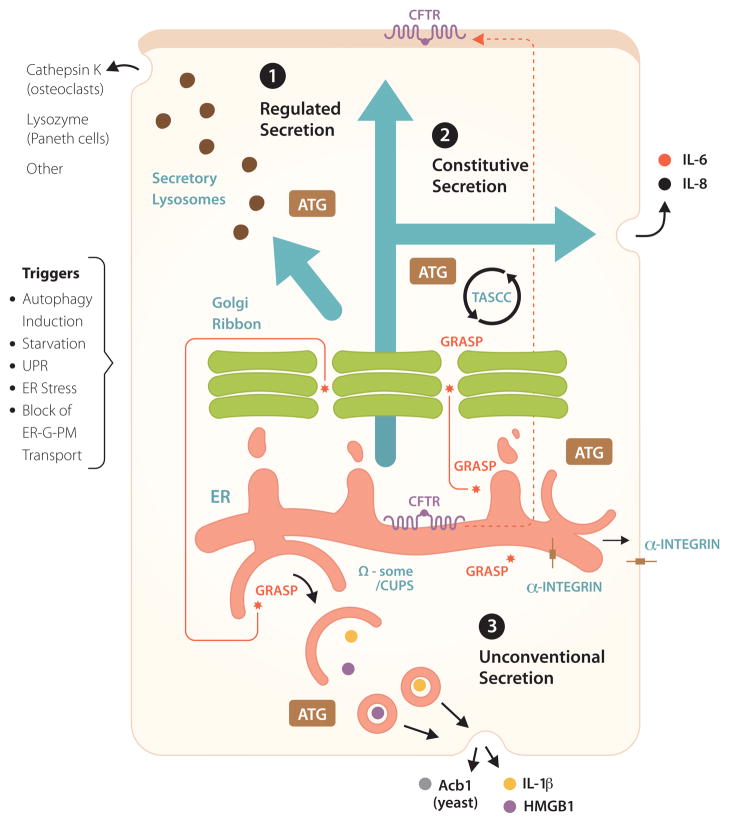
The role of autophagy in protein secretion and trafficking is a relatively recently recognized function of the autophagic machinery (Figure 2, pathways 1–3). One of the breakthroughs in this area was the realization that a subset of unconventionally secreted cytosolic proteins, which lack leader peptides and thus cannot enter the conventional ER-to-Golgi secretory pathway, depend on autophagy for extracellular export [18–21] (Figure 2, pathway 3).
Fig. 2.
Non-degradative roles of autophagy in conventional (constitutive and regulated) and unconventional secretion. 1. Regulated secretion: secretory lysosomes, granules and other organelles, partially derived from or affected by post-Golgi vesicles. ATG: symbolizes that Atg factors affect regulated secretion, delivering various biologically active cargo such as indicated. Other: includes non-proteinaceous cargo (e.g. ATP secreted from drug-treated cancer cells), provided that they are competent to undergo autophagy, with inflammatory consequences and clearance of transplanted tumors. 2. Autophagy affects constitutive secretion (e.g. IL-6, IL-8) via a compartment intermixed with autophagic organelles, called TASCC (TOR-autophagy spatial coupling compartment). 3. A subset of unconventional secretion processes depend on autophagy (autophagy-based unconventional secretion; ‘autosecretion’) for secretion of proinflammatory factors IL-1β and HMGB1 in mammalian cells and Acb1 in yeast. GRASP (note in Figure 1 that GRASP is normally localized to the Golgi and affects early stages of autophagy) is required for autophagy-based unconventional secretion (autosecretion). CUPS, a yeast structure implicated in autophagy-based unconventional secretion, may be equivalent to Ω-some in mammalian cells. In addition, autophagy plays a role in unconventional trafficking of the ER-form of CFTR (cystic fibrosis transmembrane conductance regulatory) to the apical aspect of the plasma membrane, bypassing the Golgi and rescuing function of mutant CFTR responsible for cystic fibrosis. GRASP plays a role in autophagy-dependent unconventional trafficking of CFTR and in unconventional trafficking of α-integrin to the basolateral plasma membrane in Drosophila (a role for autophagy has not been established as yet for α-integrin trafficking). Triggers: conditions (the list is not comprehensive) that contribute to induction of unconventional secretion or trafficking.
Unconventional secretion is a catch-all term for a range of diverse processes that can deliver different proteins localized in the cytosol to the extracellular environment or assist in trafficking of integral membrane proteins to the plasma membrane without having to pass through the Golgi en route to their final destination [48]. Thus, it is important to differentiate at the outset that not all unconventionally secreted proteins [48] are secreted by the same mechanism. In this context, we term autophagy-based unconventional secretion as ‘autosecretion’ to separate it from other, equally interesting but disparate processes of unconventional protein secretion.
A subset of unconventionally secreted proteins, such as Acb1 in yeast [18–20] and IL-1β, IL-18 and HMGB1 in mammalian cells [48] use, at least under some circumstances, autophagic organelles or their intermediates as a vehicle for extracellular export (Figure 2, pathway 3). The secretion of these leaderless proteins depends on Atg factors, and is distinguished by the participation of the somewhat enigmatic protein GRASP. GRASP (Golgi re-assembly and stacking protein), known as GrpA in Dictyostelium, Grh1 in yeast, dGRASP in Drosophila, and GRASP55 (GORASP2) and GRASP65 (GORASP1) in mammalian cells [49], was described first as a protein that helps organize the Golgi mini-stacks into Golgi ribbons but appears not to play a major role in conventional secretion through the Golgi apparatus [49]. It turns out, however, that GRASP is a specific factor specializing in unconventional secretion and trafficking; examples include AcbA/Acb1 in Dictyostelium [50] and yeast [18–20], IL-1β and IL-18 in mammalian cells [21], α-integrin in Drosophila [51], and CFTR in human epithelial cells [17]. Beyond GRASP, recent work has linked unconventional secretion and trafficking of certain proteins to autophagy [17–21]. Until recently, autophagosomes were believed to play a role primarily in digesting captured cargo. Thus, the capacity of autophagy to help secrete cytosolic proteins and perhaps other cargo represents a paradigm shift in how we view this process [18–21].
Autophagy-based unconventional secretion, omegasome and CUPS
The information regarding how autophagy promotes unconventional secretion comes, at present, from a handful of studies [17–21]]. The omegasome, acting as a cradle for generating nascent autophagosomes [24], or a potentially related structure in yeast termed CUPS (compartment for unconventional protein secretion) [19] (Figures 1 and 2) may represent the source of organelles or trafficking intermediates of the autosecretory pathway contributing to autophagy-based unconventional secretion.
There are indications that autophagy, autosecretion and the conventional secretory pathway converge upon interrelated domains on the ER, specifically the secretory aspect of ER oriented towards the Golgi known as the ER-Golgi-intermediate compartment or vesicular-tubular clusters. In autophagy-based unconventional secretion this starts with an omegasome-like structure termed CUPS [19], forming in the vicinity of the ER exit sites marked by Sec13 but not fully overlapping with Sec13. It is well known that organizers of the ER exit sites (Sec12 and Sec16) and COPII components (Sec23 and Sec24), but not the other parts of COPII (Sec13 and Sec31), are important for autophagosome formation [52]. Participation of the regulators of the early secretory pathway in autophagy is also seen in mammalian cells. This is exemplified by the roles of Sar1 [53], a GTPase affected by the Sec12 GEF, and other regulators of the early ER-to-Golgi secretory pathway in omegasome and autophagosome formation [54]. It appears likely that CUPS in yeast and omegasomes in mammalian cells are related, but this remains to be established. In contrast, there are some discordances regarding which of the factors play a role in CUPS versus PAS formation in yeast; for example, Sec23 of COPII is required for autophagy [52] but its mutation does not interrupt unconventional secretion of Acb1, whereas Sec12 matters for autophagy but appears to be dispensable for CUPS formation [19, 52]. Whether these differences indicate subcompartmentalization of omegasome domains destined for degradative autophagy versus autosecretion remains to be determined.
Starvation induces both CUPS [19] and omegasome [24] formation. Although there may be subtle molecular and physiological differences as noted above, yeast CUPS [19] is strikingly similar by morphology and localization to the omegasome structures associated with autophagosome formation in mammals [24]. The salient features are that both structures are induced by starvation, are PI3P-positive and are associated with Atg factors. CUPS is distinct from the Cvt structure (marked by Ape1) and is not induced by rapamycin [19]. Of note, the original description of the omegasome and its role in autophagy along with its PI3P positivity did not report use of rapamycin and was based solely on starvation-dependent induction of autophagy [24]. Like the omegasome, which is a very early structure preceding the typical LC3/Atg8-positive autophagosomes [1], CUPS forms in a process independent of Atg8 and Atg9 [19]. Upon starvation of yeast cells, CUPS structures (one to three per cell, as monitored with Grh1-GFP) form in the vicinity of Sec13-labeled transitional ER, sites known to be positive for Grh1 [55]. Among other distinguishing features of CUPS are the presence of PI3P, Atg8, and Atg9; thus, these structures resemble PAS [19]. CUPS also contains Vps23 [19], known in mammalian systems as TSG101, the significance of which is not known given that other ESCRT (endosomal sorting complex required for transport) proteins were not found in CUPS. Vps23 is a member of the ESCRT-I complex usually associated with multivesicular endosomal sorting but playing other cellular roles. Although Vps23 is required for Acb1 unconventional secretion in yeast [20], deletion of Vps23 did not affect Grh+ CUPS formation, suggesting that Vps23 may be a part of the downstream sorting effector functions of CUPS. Among other ESCRT members tested, Vps27 (ortholog of the mammalian Hrs) and several other members of ESCRT complexes 0, I, II and III, when the corresponding mutant yeast strains were tested, had only negligible or minor kinetic effects on CUPS and thus do not equal the role of Vps23. Perhaps of further significance is that VPS23/TSG101 in its sorting function can bind to ubiquitinated cargo. This has yet to be explored in CUPS function and in terms of putative adaptors selecting and delivering cargo destined for autophagy-based unconventional secretion.
Surprisingly, deletion of Atg1 in yeast did not inhibit translocation of the yeast GRASP (Grh1) to CUPS, thus dissociating Atg1 action from localization effects on Grh1. A similar absence of effects was seen with Atg8 and Atg18 mutants [19]. Thus, it is not known which molecular events regulated by potential upstream factors lead to redistribution of the yeast GRASP ortholog to CUPS upon starvation. GRASP nevertheless plays a role in autophagosome formation in mammalian cells [21]. In yeast, the GRASP equivalent is important for Atg9 relocalization following starvation [19] in addition to Vps23 localization to CUPS. The effect of the yeast GRASP ortholog Grh1 on the sole integral membrane autophagy factor (Atg9) is in keeping with the reported role of the mammalian GRASPs (GRAPS55) in controlling autophagy initiation [21].
Dual role of autophagy in unconventional secretion of alarmins and control of inflammasome activation
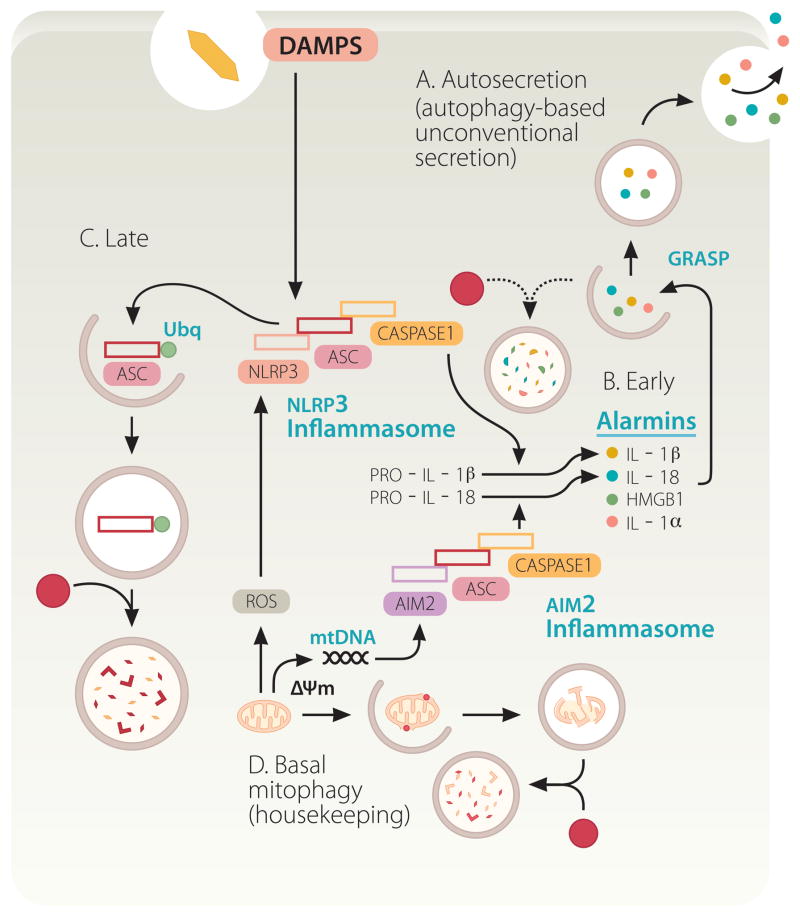
It may have appeared as a foregone conclusion that IL-1β, a leaderless cytosolic protein secreted form the cell via membranous organelles, would be a substrate for autophagy-based unconventional secretion, at least according to some predictions [18, 20, 56]. However, this issue has turned out to be far more complicated (Fig. 3A–D). A number of converging reports [21, 57–60] studying the effects of autophagy on IL-1β from the immunological perspective, have unequivocally indicated that autophagy plays a negative role in IL-1β activation. This is in keeping with the initial report that Atg16L1 loss increases IL-1β production in mice [61], and is precisely the opposite from the cell biology-based predictions for a positive role of autophagy in IL-1β secretion [18, 20, 56]. Autophagy inhibits IL-1β activation indirectly, by lowering the endogenous sources of inflammasome activation [57, 58] (Figure 3D). It may also act directly via a yet to be fully demonstrated autophagic degradation of inflammasome components [59, 60] (Figure 3C). The inflammasome is a protein complex consisting of at least three components: an NLR (Nod-like receptor) protein such as NLRP3, adaptor protein ASC, and caspase-1 (Figure 3). Upon activation by agonists, inflammasome processes pro-IL-1β (along with several other pro-proteins) into a mature, biologically active IL-1β, ready for secretion. Autophagy suppresses inflammasome activation by removing depolarized or otherwise defunct mitochondria that provide endogenous sources of inflammasome agonists [57, 58]. When unkempt mitochondria accumulate in cells defective for autophagy, they become a source of endogenous irritants (ROS, mitochondrial DNA) that act as inflammasome activators (Figure 3D). This is consistent with the archetypal role for autophagy in keeping the cellular interior clean of potentially harmful macromolecular aggregates and dysfunctional organelles. Thus, in the absence of basal autophagy, endogenous factors lead to inflammasome activation and increased IL-1β processing and represent sources of sterile inflammation.
Fig. 3.
Autophagy plays a dual role in pro-inflammatory processes based on inflammasome activation and autosecretion. Autophagy suppresses basal levels of inflammasome activation. Inflammasomes (composed of an NLR, in the illustrated case NLRP3, or AIM2, ASC, and caspase 1) are multi-protein platforms assembled and activated in response to damage/danger associated molecular patterns (DAMPS), resulting in caspase-1 activation and proteolytic processing of cytosolic pro-forms of cytokines such as IL-1β, a cardinal mediator of infection-associated and sterile inflammation. Autophagy keeps the sources of endogenous agonists low by removing depolarized (ΔΨm) or unkempt mitochondria (sources of ROS and mitochondrial DNA [mtDNA]). However, autophagy also enables the cytosolic IL-1β (and other alarmins such as HMGB1; see Figure 2) to be delivered to the outside of the cell via autosecretion. Finally, autophagy may downregulate inflammasomes by digesting its components. A, B. Autosecretion (autophagy-based unconventional secretion; see Figure 2 and text for explanations) enables secretion of cytosolic proteins such as IL-1β and HMGB1 per the illustrated process controlled by Atg factors and GRASP. Autosecretion occurs early in the process of stimulation. C. Autosecretion is likely shut down by deactivation, or possibly by digestion as in the illustration, as a way of downregulating the system following its physiologically useful period of activation. D. In resting cells, the effect of autosecretion is masked by the dominant tonic negative role of autophagy in inhibiting basal levels of inflammasome activation.
Autophagy does play a role in unconventional secretion of IL-1β [21]. This is, however, difficult to dissect away from the effects of basal autophagy in keeping the cellular interiors clean and basal levels of inflammasome activation down [21, 57–59]. When autophagy is induced by starvation, early on in the process this leads to enhanced secretion of IL-1β provided that inflammasome is activated with conventional inflammasome agonists such as silica, nigericin, β-amyloid fibrils, and alum [21] (Figure 3B, Early stages). This effect wanes with time and the negative regulation of inflammasome by autophagy becomes dominant at later time points [60] (Figure 3C, Late stages). In summary, autophagy negatively controls inflammasome activation and positively controls IL-1β secretion, with the net result being the product of these two opposing actions. Induced autophagy promotes IL-1β secretion and secretion of other cytosolic inflammasome substrates such as IL-18, and additional alarmins such as HMGB1 (High-mobility group protein B1) [21]. These acute effects can be best detected very early afterstimulation [21].
Autophagy and GRASP in unconventional trafficking of proteins to plasma membrane
The role of GRASP and autophagy in vectorial transport of proteins is not limited to autosecretion of leaderless cytosolic proteins but also involves unconventional trafficking of integral membrane proteins to the polarized domains of plasma membrane, bypassing the conventional ER-Golgi-plasma membrane pathway. Recently, ΔF508 CFTR (cystic fibrosis transmembrane conductance regulator), the most common form of mutant CFTR protein causing cystic fibrosis, was shown to utilize an ATG5- and ATG7-dependent, as well as GRASP55-dependent, unconventional transport to the plasma membrane (Figure 2) [17]. Blocking the conventional ER-to-Golgi secretory pathway (using dominant negative expression constructs of Sar1, Arf1, and Syntaxin 5) unexpectedly enhanced plasma membrane expression of the ER form of both ΔF508 and wild type CFTR (core glycosylated, band B) (Figure 2). Examination of the role of GRASPs, predominantly GRASP55, indicated that GRASP myristoylation (and thus membrane association) was needed, but more importantly that phosphorylation of the GRASP55 residue Ser-441 and association of the C-terminal signal of CFTR with the first PDZ (PSD95, Dlg1 and zonula occludens-1 anchoring) domain (PDZ1) of GRASP55 was essential for the unconventional trafficking of CFTR to the plasma membrane [17].
The unconventionally transported CFTR that reached the plasma membrane was biologically functional and CFTR was in the correct – apical – domain of the plasma membrane [17]. It displayed functional channel activity and prevented the typical malnourished, growth retardation phenotype in transgenic mice homozygous for the ΔF508 CFTR (but not in the mice null for CFTR, thus establishing CFTR specificity) [17]. The fidelity of the GRASP-dependent unconventional trafficking is also illustrated in the delivery of a Drosophila integrin to the basolateral plasma membrane domain where this integrin performs its physiological function [51]. Thus, albeit surprisingly, the unconventional trafficking to the plasma membrane retains polarization specificity and should be viewed not as a random process but as a potentially high fidelity pathway that has evolved as a default or as a salvage trafficking process under certain circumstances.
Of significance in the context of the reported unconventional CFTR trafficking [17] is that this trafficking was induced by ER stress signaling and the unfolded protein response (UPR). Depletion of ER Ca2+ stores with thapsigargin resulted in activation of the GRASP-dependent (either GRASP55 or GRASP65, depending on the dominant form expressed in the cell line tested) unconventional trafficking of the core-glycosylated CFTR. The signaling leading to these events depended specifically on the Ser/Thr kinase IRE1 but not on PERK and ATF6, all three being examined in the CFTR study [17] as the key signal transducers of UPR. The role of IRE1 was outside of its most commonly appreciated endonuclease function of activating XBP1 (X-box binding protein 1 transcriptional factor) during ER stress, because TRAF2 (TNF-receptor associated factor 2)-dependent activation of JNK (c-Jun N-terminal kinase/stress-activated protein kinase) [62] appeared to play a role [17]. Furthermore, GRASP55 was found to be phosphorylated following ER stress (or upon the ER-to-Golgi transport blockade) and a specific residue, Ser-441, was required for activation of the unconventional trafficking pathway delivering ΔF508 CFTR to the plasma membrane. The exact links (i.e. the identity of the kinase phosphorylating Ser-441 on GRASP55) between IRE1 signaling and GRASP55 phosphorylation remain to be established; however, overexpression of the wild type, phosphorylatable GRASP55 required IRE1 for its ability to induce unconventional trafficking of ΔF508 CFTR to the plasma membrane [17]. Thus, UPR, IRE1 and GRASP55 are sequentially connected in this pathway that also depends on autophagy factors [17].
Autophagy and constitutive secretion
A recent report [16] uncovered another intersection between autophagy and the constitutive secretory pathway (Figure 2, pathway 2). In this case, the authors described the existence of a specialized compartment, TASCC (TASCC (TOR-autophagy spatial coupling compartment), interfacing with autophagic degradation and indirectly favoring production of a subset of secreted proteins that utilize conventional (ER-to-Golgi-to-plasma membrane) secretion. TASCC is physically recognizable in senescent cells as a cluster of lysosomal-like organelles in a perinuclear region juxtaposed to the Golgi; it is formed during mutant Ras upregulation-related senescence [16]. TASCC positioning is nocodazol-insensitive but is perturbed by brefeldin A and thus dependent on the maintenance of the conventional secretory organelles. TASCC is largely composed of degradative organelles with features of late/mature phagosomal compartments based on being p62-positive, LAMP-positive, variably LC3-positive, and, importantly in functional terms, mTOR-positive. This compartment appears to play a role in favoring biogenesis and secretion of a subset of conventional secretory cargo. One interpretation of how this system may work is that it focuses cellular secretion on a few key proteins while degrading bulk proteins via autophagy. In this model, autophagosomes capture and deliver cellular proteins, slated to be trimmed down in TASCC where the targeted proteins are degraded, while simultaneously enabling the TASCC-resident mTOR (activated by amino acids released from autolysosomes) to selectively promote translation and secretion of a limited number of favored proteins. This was shown in the case of enhanced secretion of IL-6 by senescent cells (Figure 2). However, there were indications in the same study that this process (even if morphologically not as discernible) may be in use in differentiated cells in tissues to streamline or tighten their repertoire of secreted proteins.
Autophagy and regulated secretion
The effects of autophagy on secretion do not end with the constitutive conventional secretory pathway and unconventional secretion. Several studies have linked defects in autophagy or Atg genes to alterations in regulated secretion (Figure 2, pathway 1) delivering the contents stored in secretory granules or lysosomes [10–12]. A forerunner of the role of autophagy in the above events is the report of enhanced Atg-dependent fusion between phagosomes and lysosomes in the process of microbial ingestion by macrophages [63]. Further examples of events at the plasma membrane include secretion of lysozyme from Paneth cells implicated in Crohn’s disease [10], secretion under certain conditions of ATP (acting as an extracellular signaling molecule in its immunological role of an alarmin) by autophagy-competent cancer cells [14], cathepsin K secretion in the ruffled border by osteoclasts during bone resorption [11], and degranulation of LC3-positive secretory granules in mast cells with consequences for anaphylaxis [12]. Autophagy is also necessary for secretion by vestibular epithelial cells of otoconins, the glycoproteins required for normal development of organic calcium carbonate crystals (otoconia) essential for equilibrioception in the middle ear [13]. Autophagy also has been implicated in the maturation of melanosomes [15]. Melanosomes are specialized lysosome-related organelles with components derived from the Golgi that are subject to complex trafficking via plasma membrane through sorting endosomes. Atg5 and LC3 colocalize with pre-melanosomes marked by PMEL17 [15], the amyloid-like fibrils providing structural foundation to pre-melanosomes, which mature to become laden with the pigments eumelanin or pheomelanins for eventual transfer from melanocytes to recipient cells such as keratinocytes.
Autophagy directly or indirectly affects secretory processes in pancreatic β-cells of the islets of Langerhans [64, 65]. In islets, autophagy affects, most likely indirectly, the status of intracellular organelles such as mitochondria and ER, which are needed for glucose-stimulated insulin secretion via cytosolic Ca2+ transients [65] and for β-cell mass increase as part of normal glucose tolerance adaptations under conditions (e.g. free fatty acids) promoting peripheral insulin resistance and diabetes [64, 65].
How autophagy or Atg factors affect regulated secretion is not known. This could have roots in the common aspects of the secretory pathway -- post-Golgi carriers and fusion properties of the plasma membrane domains -- but may also entail unique and highly specialized maturation steps of secretory organelles and maintenance of their integrity and quality by autophagy. Among the proposed mechanisms are also the effects of autophagy on clathrin-associated adaptor AP3 complex levels (expression and stability) in vestibular cells [13] (of note is that melanosome biogenesis in melanocytes also depends on AP3 for tyrosinase delivery) and organization of polarized plasma membrane domains (e.g. ruffled border within the actin ring in osteoclasts). The latter case of specialized fusion acceptor sites may provide homing addresses for the regulated secretory lysosomes and their cargo [11]. In this context, it is possible that lipidated LC3-II or other Atg8 paralogs may be required to mark the acceptor membranes (e.g. ruffled border [11]) destined for fusion with lysosomes, a phenomenon applicable to other cellular membrane domains. For example, these relationships are akin to lysosomal fusion with plasma membrane-derived intracellular compartments such as pathogen-harboring phagosomes in professional phagocytic cells [63] or in epithelial cells during entosis, a process whereby detached epithelial cells can invade neighboring cells where they either survive in entotic vacuoles or die through ATG-dependent fusion with lysosomes [66]. Although this at first may seem an unexpected function of autophagic machinery and LC3, it falls neatly within the function of LC3/Atg8 as a tethering/fusion device [67, 68], and recapitulates the successive steps of the classical autodigestive autophagy pathway because the conventional LC3-positive autophagosomes complete their function by being targeted for fusion with lysosomal organelles.
Concluding remarks
The newly uncovered numerous intersections of autophagy with biosynthetic processes such as conventional and unconventional secretion of biologically active cargo and trafficking of integral membrane proteins expand the range of physiological roles of autophagy. Several specialized tissues and organs are influenced by autophagic machinery in this biogenesis role. Bone remodeling by osteoclasts [11], melanosome maturation in skin melanocytes and retinal pigment epithelial cells [15], and secretion of inflammatory mediators [12, 14, 16, 21] are potentially the forerunners of an expanding impact of autophagy in this area. With these developments, the domain of autophagy influence expands to the extracellular space both in complex metazoan tissues and in the environments surrounding unicellular eukaryotes such as yeast [18–20]. Complex syndromes, sometimes with strong inflammatory components such as Crohn’s disease [10, 61, 69] and cystic fibrosis [17], appear to be impacted by this newly appreciated role of autophagy in addition to its more conventional manifestations. One wonders what is next, as surprises in the breadth and depth of the impact of autophagy in biology seem endless.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Mizushima N, et al. The role of atg proteins in autophagosome formation. Annual review of cell and developmental biology. 2011;27:107–132. doi: 10.1146/annurev-cellbio-092910-154005. [DOI] [PubMed] [Google Scholar]
- 2.Liu L, et al. Mitochondrial outer-membrane protein FUNDC1 mediates hypoxia-induced mitophagy in mammalian cells. Nat Cell Biol. 2012;14:177–185. doi: 10.1038/ncb2422. [DOI] [PubMed] [Google Scholar]
- 3.Youle RJ, Narendra DP. Mechanisms of mitophagy. Nat Rev Mol Cell Biol. 2011;12:9–14. doi: 10.1038/nrm3028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Green DR, et al. Mitochondria and the autophagy-inflammation-cell death axis in organismal aging. Science. 2011;333:1109–1112. doi: 10.1126/science.1201940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rubinsztein DC, et al. Autophagy and aging. Cell. 2011;146:682–695. doi: 10.1016/j.cell.2011.07.030. [DOI] [PubMed] [Google Scholar]
- 6.Rabinowitz JD, White E. Autophagy and metabolism. Science. 2010;330:1344–1348. doi: 10.1126/science.1193497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rubinstein AD, et al. The autophagy protein atg12 associates with antiapoptotic bcl-2 family members to promote mitochondrial apoptosis. Mol Cell. 2011;44:698–709. doi: 10.1016/j.molcel.2011.10.014. [DOI] [PubMed] [Google Scholar]
- 8.Levine B, et al. Autophagy in immunity and inflammation. Nature. 2011;469:323–335. doi: 10.1038/nature09782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Deretic V. Autophagy as an innate immunity paradigm: expanding the scope and repertoire of pattern recognition receptors. Curr Opin Immunol. 2012;24:21–31. doi: 10.1016/j.coi.2011.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cadwell K, et al. A key role for autophagy and the autophagy gene Atg16l1 in mouse and human intestinal Paneth cells. Nature. 2008;456:259–263. doi: 10.1038/nature07416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.DeSelm CJ, et al. Autophagy proteins regulate the secretory component of osteoclastic bone resorption. Dev Cell. 2011;21:966–974. doi: 10.1016/j.devcel.2011.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ushio H, et al. Crucial role for autophagy in degranulation of mast cells. J Allergy Clin Immunol. 2011;127:1267–1276. e1266. doi: 10.1016/j.jaci.2010.12.1078. [DOI] [PubMed] [Google Scholar]
- 13.Marino G, et al. Autophagy is essential for mouse sense of balance. J Clin Invest. 2010;120:2331–2344. doi: 10.1172/JCI42601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Michaud M, et al. Autophagy-dependent anticancer immune responses induced by chemotherapeutic agents in mice. Science. 2011;334:1573–1577. doi: 10.1126/science.1208347. [DOI] [PubMed] [Google Scholar]
- 15.Ganesan AK, et al. Genome-wide siRNA-based functional genomics of pigmentation identifies novel genes and pathways that impact melanogenesis in human cells. PLoS Genet. 2008;4:e1000298. doi: 10.1371/journal.pgen.1000298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Narita M, et al. Spatial coupling of mTOR and autophagy augments secretory phenotypes. Science. 2011;332:966–970. doi: 10.1126/science.1205407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gee HY, et al. Rescue of DeltaF508-CFTR Trafficking via a GRASP-Dependent Unconventional Secretion Pathway. Cell. 2011;146:746–760. doi: 10.1016/j.cell.2011.07.021. [DOI] [PubMed] [Google Scholar]
- 18.Manjithaya R, et al. Unconventional secretion of Pichia pastoris Acb1 is dependent on GRASP protein, peroxisomal functions, and autophagosome formation. J Cell Biol. 2010;188:537–546. doi: 10.1083/jcb.200911149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bruns C, et al. Biogenesis of a novel compartment for autophagosome-mediated unconventional protein secretion. J Cell Biol. 2011;195:979–992. doi: 10.1083/jcb.201106098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Duran JM, et al. Unconventional secretion of Acb1 is mediated by autophagosomes. J Cell Biol. 2010;188:527–536. doi: 10.1083/jcb.200911154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dupont N, et al. Autophagy-based unconventional secretory pathway for extracellular delivery of IL-1beta. The EMBO journal. 2011;30:4701–4711. doi: 10.1038/emboj.2011.398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hayashi-Nishino M, et al. A subdomain of the endoplasmic reticulum forms a cradle for autophagosome formation. Nat Cell Biol. 2009;11:1433–1437. doi: 10.1038/ncb1991. [DOI] [PubMed] [Google Scholar]
- 23.Yla-Anttila P, et al. 3D tomography reveals connections between the phagophore and endoplasmic reticulum. Autophagy. 2009;5:1180–1185. doi: 10.4161/auto.5.8.10274. [DOI] [PubMed] [Google Scholar]
- 24.Axe EL, et al. Autophagosome formation from membrane compartments enriched in phosphatidylinositol 3-phosphate and dynamically connected to the endoplasmic reticulum. J Cell Biol. 2008;182:685–701. doi: 10.1083/jcb.200803137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.van der Vaart A, et al. Exit from the golgi is required for the expansion of the autophagosomal phagophore in yeast Saccharomyces cerevisiae. Mol Biol Cell. 2010;21:2270–2284. doi: 10.1091/mbc.E09-04-0345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yen WL, et al. The conserved oligomeric Golgi complex is involved in double-membrane vesicle formation during autophagy. J Cell Biol. 2010;188:101–114. doi: 10.1083/jcb.200904075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Meiling-Wesse K, et al. Trs85 (Gsg1), a component of the TRAPP complexes, is required for the organization of the preautophagosomal structure during selective autophagy via the Cvt pathway. J Biol Chem. 2005;280:33669–33678. doi: 10.1074/jbc.M501701200. [DOI] [PubMed] [Google Scholar]
- 28.Lynch-Day MA, et al. Trs85 directs a Ypt1 GEF, TRAPPIII, to the phagophore to promote autophagy. Proc Natl Acad Sci U S A. 2010;107:7811–7816. doi: 10.1073/pnas.1000063107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bodemann BO, et al. RalB and the Exocyst Mediate the Cellular Starvation Response by Direct Activation of Autophagosome Assembly. Cell. 2011;144:253–267. doi: 10.1016/j.cell.2010.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Moreau K, et al. Autophagosome precursor maturation requires homotypic fusion. Cell. 2011;146:303–317. doi: 10.1016/j.cell.2011.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hailey DW, et al. Mitochondria supply membranes for autophagosome biogenesis during starvation. Cell. 2010;141:656–667. doi: 10.1016/j.cell.2010.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Weidberg H, et al. LC3 and GATE-16/GABARAP subfamilies are both essential yet act differently in autophagosome biogenesis. The EMBO journal. 2010;29:1792–1802. doi: 10.1038/emboj.2010.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fujita N, et al. An Atg4B mutant hampers the lipidation of LC3 paralogues and causes defects in autophagosome closure. Mol Biol Cell. 2008;19:4651–4659. doi: 10.1091/mbc.E08-03-0312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Scherz-Shouval R, et al. Reactive oxygen species are essential for autophagy and specifically regulate the activity of Atg4. Embo J. 2007;26:1749–1760. doi: 10.1038/sj.emboj.7601623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mari M, et al. An Atg9-containing compartment that functions in the early steps of autophagosome biogenesis. J Cell Biol. 2010;190:1005–1022. doi: 10.1083/jcb.200912089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Young AR, et al. Starvation and ULK1-dependent cycling of mammalian Atg9 between the TGN and endosomes. J Cell Sci. 2006;119:3888–3900. doi: 10.1242/jcs.03172. [DOI] [PubMed] [Google Scholar]
- 37.Mizushima N, Levine B. Autophagy in mammalian development and differentiation. Nat Cell Biol. 2010;12:823–830. doi: 10.1038/ncb0910-823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chan EY, et al. Kinase-inactivated ULK proteins inhibit autophagy via their conserved C-terminal domains using an Atg13-independent mechanism. Mol Cell Biol. 2009;29:157–171. doi: 10.1128/MCB.01082-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Egan DF, et al. Phosphorylation of ULK1 (hATG1) by AMP-activated protein kinase connects energy sensing to mitophagy. Science. 2011;331:456–461. doi: 10.1126/science.1196371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shang L, et al. Nutrient starvation elicits an acute autophagic response mediated by Ulk1 dephosphorylation and its subsequent dissociation from AMPK. Proc Natl Acad Sci U S A. 2011;108:4788–4793. doi: 10.1073/pnas.1100844108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kim J, et al. AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nat Cell Biol. 2011;13:132–141. doi: 10.1038/ncb2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.He C, Levine B. The Beclin 1 interactome. Current opinion in cell biology. 2010;22:140–149. doi: 10.1016/j.ceb.2010.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Di Bartolomeo S, et al. The dynamic interaction of AMBRA1 with the dynein motor complex regulates mammalian autophagy. J Cell Biol. 2010;191:155–168. doi: 10.1083/jcb.201002100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Itakura E, Mizushima N. p62 Targeting to the autophagosome formation site requires self-oligomerization but not LC3 binding. J Cell Biol. 2011;192:17–27. doi: 10.1083/jcb.201009067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yang JQ, et al. NBR1 is a new PB1 signalling adapter in Th2 differentiation and allergic airway inflammation in vivo. The EMBO journal. 2010;29:3421–3433. doi: 10.1038/emboj.2010.214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Moscat J, Diaz-Meco MT. p62 at the crossroads of autophagy, apoptosis, and cancer. Cell. 2009;137:1001–1004. doi: 10.1016/j.cell.2009.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Duran A, et al. p62 is a key regulator of nutrient sensing in the mTORC1 pathway. Mol Cell. 2011;44:134–146. doi: 10.1016/j.molcel.2011.06.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nickel W, Rabouille C. Mechanisms of regulated unconventional protein secretion. Nat Rev Mol Cell Biol. 2009;10:148–155. doi: 10.1038/nrm2617. [DOI] [PubMed] [Google Scholar]
- 49.Giuliani F, et al. Unconventional secretion: a stress on GRASP. Current opinion in cell biology. 2011;23:498–504. doi: 10.1016/j.ceb.2011.04.005. [DOI] [PubMed] [Google Scholar]
- 50.Kinseth MA, et al. The Golgi-associated protein GRASP is required for unconventional protein secretion during development. Cell. 2007;130:524–534. doi: 10.1016/j.cell.2007.06.029. [DOI] [PubMed] [Google Scholar]
- 51.Schotman H, et al. dGRASP-mediated noncanonical integrin secretion is required for Drosophila epithelial remodeling. Dev Cell. 2008;14:171–182. doi: 10.1016/j.devcel.2007.12.006. [DOI] [PubMed] [Google Scholar]
- 52.Ishihara N, et al. Autophagosome requires specific early Sec proteins for its formation and NSF/SNARE for vacuolar fusion. Mol Biol Cell. 2001;12:3690–3702. doi: 10.1091/mbc.12.11.3690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zoppino FC, et al. Autophagosome formation depends on the small GTPase Rab1 and functional ER exit sites. Traffic. 2010;11:1246–1261. doi: 10.1111/j.1600-0854.2010.01086.x. [DOI] [PubMed] [Google Scholar]
- 54.Winslow AR, et al. alpha-Synuclein impairs macroautophagy: implications for Parkinson’s disease. J Cell Biol. 2010;190:1023–1037. doi: 10.1083/jcb.201003122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Levi SK, et al. The yeast GRASP Grh1 colocalizes with COPII and is dispensable for organizing the secretory pathway. Traffic. 2010;11:1168–1179. doi: 10.1111/j.1600-0854.2010.01089.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pfeffer SR. Unconventional secretion by autophagosome exocytosis. J Cell Biol. 2010;188:451–452. doi: 10.1083/jcb.201001121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhou R, et al. A role for mitochondria in NLRP3 inflammasome activation. Nature. 2011;469:221–225. doi: 10.1038/nature09663. [DOI] [PubMed] [Google Scholar]
- 58.Nakahira K, et al. Autophagy proteins regulate innate immune responses by inhibiting the release of mitochondrial DNA mediated by the NALP3 inflammasome. Nature immunology. 2011;12:222–230. doi: 10.1038/ni.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Harris J, et al. Autophagy controls IL-1{beta} secretion by targeting pro-IL-1{beta} for degradation. J Biol Chem. 2011 doi: 10.1074/jbc.M110.202911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Shi CS, et al. Activation of autophagy by inflammatory signals limits IL-1beta production by targeting ubiquitinated inflammasomes for destruction. Nature immunology. 2012 doi: 10.1038/ni.2215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Saitoh T, et al. Loss of the autophagy protein Atg16L1 enhances endotoxin-induced IL-1beta production. Nature. 2008;456:264–268. doi: 10.1038/nature07383. [DOI] [PubMed] [Google Scholar]
- 62.Urano F, et al. Coupling of stress in the ER to activation of JNK protein kinases by transmembrane protein kinase IRE1. Science. 2000;287:664–666. doi: 10.1126/science.287.5453.664. [DOI] [PubMed] [Google Scholar]
- 63.Sanjuan MA, et al. Toll-like receptor signalling in macrophages links the autophagy pathway to phagocytosis. Nature. 2007;450:1253–1257. doi: 10.1038/nature06421. [DOI] [PubMed] [Google Scholar]
- 64.Ebato C, et al. Autophagy is important in islet homeostasis and compensatory increase of beta cell mass in response to high-fat diet. Cell Metab. 2008;8:325–332. doi: 10.1016/j.cmet.2008.08.009. [DOI] [PubMed] [Google Scholar]
- 65.Jung HS, et al. Loss of autophagy diminishes pancreatic beta cell mass and function with resultant hyperglycemia. Cell Metab. 2008;8:318–324. doi: 10.1016/j.cmet.2008.08.013. [DOI] [PubMed] [Google Scholar]
- 66.Florey O, et al. Autophagy machinery mediates macroendocytic processing and entotic cell death by targeting single membranes. Nat Cell Biol. 2011;13:1335–1343. doi: 10.1038/ncb2363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Nakatogawa H, et al. Atg8, a ubiquitin-like protein required for autophagosome formation, mediates membrane tethering and hemifusion. Cell. 2007;130:165–178. doi: 10.1016/j.cell.2007.05.021. [DOI] [PubMed] [Google Scholar]
- 68.Weidberg H, et al. LC3 and GATE-16 N termini mediate membrane fusion processes required for autophagosome biogenesis. Dev Cell. 2011;20:444–454. doi: 10.1016/j.devcel.2011.02.006. [DOI] [PubMed] [Google Scholar]
- 69.Cadwell K, et al. Virus-plus-susceptibility gene interaction determines Crohn’s disease gene Atg16L1 phenotypes in intestine. Cell. 2010;141:1135–1145. doi: 10.1016/j.cell.2010.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]