Abstract
A therapeutic strategy for treating cancer is to target and eradicate cancer stem cells (CSCs) without harming their normal stem cell counterparts. The success of this approach relies on identification of molecular pathways that selectively regulate CSC function. Using BCR-ABL-induced chronic myeloid leukemia (CML) as a disease model for CSCs, we show that BCR-ABL down-regulates the B lymphoid kinase (Blk) gene through c-Myc in leukemia stem cells (LSCs) in CML mice and that Blk functions as a tumor suppressor in LSCs but does not affect normal hematopoietic stem cells (HSCs) or hematopoiesis. Blk suppresses LSC function through a pathway involving an upstream regulator, Pax5, and a downstream effector, p27. Inhibition of this Blk pathway accelerates CML development, whereas increased activity of the Blk pathway delays CML development. Blk also suppresses human CML stem cells. Our results demonstrate the feasibility of selectively targeting LSCs, an approach that should be applicable to other cancers.
Cancer stem cells (CSCs) are required for cancer initiation in many hematologic malignancies and some solid tumors, and must be eradicated for cure 1–4. We identify genes essential for CSCs but not their normal stem cell counterparts. Previously we identified Alox5 as an important regulatory gene in leukemia stem cells (LSCs) but not normal hematopoietic stem cells (HSCs) 5, emphasizing the feasibility of this approach.
We used BCR-ABL-induced chronic myeloid leukemia (CML) as a stem cell disease model and identified LSCs for CML in mice 6. CML is a clonal HSC disorder associated with a reciprocal translocation between chromosomes 9 and 22 (t(9;22); also known as the Philadelphia chromosome), and chimeric BCR-ABL protein functions as a constitutively activated tyrosine kinase 7–9. Although BCR-ABL kinase inhibitors are highly effective in treating chronic phase CML patients 10–12, they do not efficiently kill LSCs 6,13,14. New therapeutic strategies are needed. LSCs share many properties with normal HSCs, such as self-renewal, pluripotency and signaling 2,15,16, it is important to develop therapies that specifically disturb the functions of LSCs.
In this study, we identify Blk, an Src family kinase, as a key regulator in CML LSCs. We show that Blk functions as a tumor suppressor in LSCs without affecting normal HSCs and mediates its inhibitory effect through a pathway involving an upstream regulator, Pax5, and a downstream effector, p27.
RESULTS
Blk has a tumor suppressor function in CML induction by BCR-ABL
LSCs in CML are insensitive to BCR-ABL inhibitors 6,13,14. Some genes are activated or inactivated by BCR-ABL in LSCs, but their expression is not affected by these inhibitors. Thus, expression of these genes is dependent on BCR-ABL protein but not its kinase activity. To identify this type of genes in LSCs, we compared gene expression between normal LSK cells (Lin−Sca-1+c-Kit+) and LSCs (BCR-ABL-expressing LSK) by DNA microarray as described previously5. We found that the Blk gene was down-regulated, and this down-regulation was not significantly reversed by imatinib treatment (Fig. 1a). Real-time RT-PCR confirmed the down-regulation of Blk by BCR-ABL and the inability of imatinib to restore Blk expression in LSCs (Fig. 1b). shRNA Knockdown of BCR-ABL restored Blk expression in leukemia cells (Supplementary Fig. 1a, b). Thus, BCR-ABL down-regulates Blk in a kinase activity-independent manner.
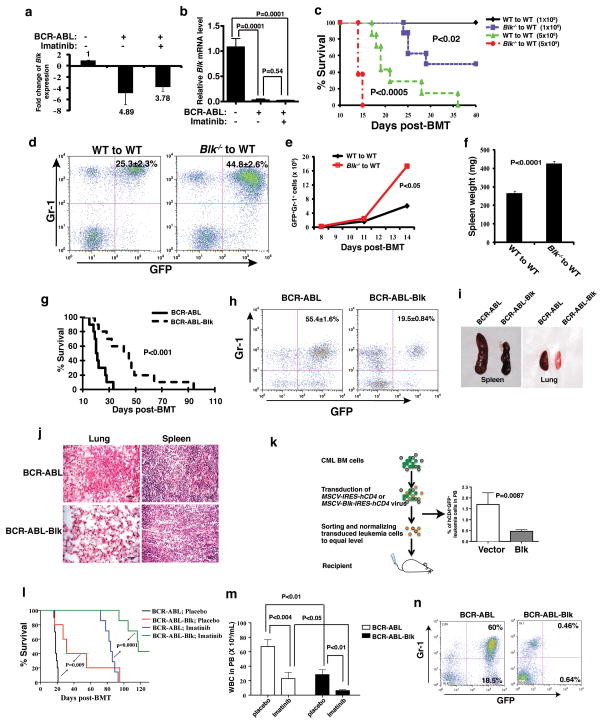
Figure 1. Blk suppresses CML induction by BCR-ABL.
(a) Microarray analysis Blk expression in LSCs of CML mice and upon imatinib treatment. Mean ± s.e.m. (b) Real-time RT-PCR analysis of Blk expression in LSCs of CML mice compared to GFP vector-transduced normal stem cells. Mean ± s.e.m. (c) Kaplan-Meier survival curves for recipients of BCR-ABL-transduced bone marrow cells from WT (n=7) or Blk−/− (n=8) donor mice. (d) The percentage of GFP+Gr-1+ cells in peripheral blood at day 11 after transplantation. (P<0.002). (e) Total number of GFP+Gr-1+ cells in peripheral blood at days 8, 11, 14 after transplantation. (P<0.05). (f) Spleen weight at day 11 after transplantation. Mean ± s.e.m. (g) Kaplan-Meier survival curves for recipients of BCR-ABL (n=10) or BCR-ABL-Blk (n=10) transduced bone marrow cells. (h) The percentage of GFP+Gr-1+ cells in peripheral blood at day 15 after transplantation. (P<0.001). (i) Gross appearance of the lungs and spleens at day 15 after transplantation. (j) Photomicrographs of haematoxylin and eosin-stained lung and spleen sections (Scale bar = 100μm). (k) Leukemia cell growth in recipients transplanted with equal numbers of GFP+hCD4+ cells. Mean ± s.e.m. (l) Kaplan-Meier survival curves for recipients of BCR-ABL (n=7) or BCR-ABL-Blk (n=7) transduced bone marrow cells treated with a placebo or imatinib (P=0.0001). (m) Total number of white blood cells in peripheral blood at 1 week after the treatment with a placebo or imatinib. Results are given as mean ± s.e.m. (n) FACS analysis showing the percentage of GFP+ leukemia cells at 8 weeks after imatinib treatment.
The expression results raised the possibility that Blk suppresses CML development. We first studied the role of Blk in CML development using Blk homozygous knockout (Blk−/−) mice (Supplementary Fig. 2a). Wild type (WT) or Blk−/− donor bone marrow cells in the C57BL/6 (B6) background were used to induce CML. Fig. 1c shows that recipients of BCR-ABL-transduced bone marrow cells from 5-FU-treated Blk−/− donor mice developed CML significantly faster than did recipients of BCR-ABL-transduced WT bone marrow cells. The accelerated disease phenotype correlated with a higher percentage and number of myeloid leukemia cells (GFP+Gr-1+) in peripheral blood (Fig. 1d, e) and more severe infiltration of leukemia cells in the spleen (Fig. 1f). The lack of Blk did not affect BCR-ABL retroviral transduction efficiency (Supplementary Fig. 2b) or homing of normal (Supplementary Fig. 2c) and BCR-ABL-transduced (Supplementary Fig. 2d) cells to bone marrow after transplantation. Reversely, we overexpressed Blk in donor bone marrow cells by transducing the cells with retrovirus expressing both BCR-ABL and Blk (Supplementary Fig. 3), and survival of CML mice increased (Fig. 1g), correlating with a lower percentage of myeloid leukemia cells in peripheral blood (Fig. 1h) and decreased infiltration of leukemic cells in the spleen and lung (Fig. 1i, j). To determine whether Blk inhibits CML progression, we induced CML and then transduced bone marrow cells, which contain established leukemia cells, with empty vector (MSCV-IRES-hCD4) or Blk (MSCV-Blk-IRES-hCD4). After sorting hCD4+ cells by magnetic-activated cell sorting (MACS), we normalized and transplanted an equal number of GFP+hCD4+ cells into recipient mice (Fig. 1k). We observed that the percentages of GFP+hCD4+ leukemia cells in the two groups were initially similar (data not shown), but Blk-expressing leukemia cells gradually decreased with time (Fig. 1k).
Suppression of LSCs in CML mice by Blk raised the possibility that restoration of Blk expression could synergize with a BCR-ABL kinase inhibitor in CML treatment. We induced CML in mice with BCR-ABL or BCR-ABL-Blk, and treated these mice with a placebo or imatinib. Either overexpression of Blk or imatinib treatment prolonged survival, as expected, but overexpression of Blk in combination of imatinib was much more effective, with about 40% of CML mice surviving longer than 130 days (Fig. 1l). This therapeutic effect correlated with lower white blood cell counts (Fig. 1m) and the disappearance of leukemic cells (Fig. 1n) as confirmed by real time RT-PCR detection of BCR-ABL transcripts in cells from peripheral blood of CML mice (Supplementary Fig. 4).
Blk suppresses LSCs
The down-regulation of Blk by BCR-ABL in LSCs and the ability of Blk to suppress CML development prompted us to test whether Blk suppresses LSCs. Fig. 2a shows that the percentages of total LSCs and long-term (CD34−) or short-term (CD34+) LSCs (LT-LSCs or ST-LSCs, respectively) in bone marrow of recipients of BCR-ABL-transduced Blk−/− donor bone marrow cells were significantly higher than those in bone marrow of recipients of BCR-ABL-transduced WT cells, indicating that Blk suppresses LSCs. By contrast, Blk deficiency did not significantly alter the percentages of the myeloid progenitors CMP (common myeloid progenitor, Lin−Sca-1−Kit+CD34+FcyRII/IIIlo), GMP (granulocyte-macrophage progenitor, Lin−Sca-1−Kit+CD34+FcyRII/IIIhi), and MEP (megakaryocyte-erythroid progenitor, Lin−Sca-1−Kit+CD34−FcyRII/IIIlo) in bone marrow of CML mice (Fig. 2a). To confirm the inhibitory effect of Blk on LSCs, we tested whether Blk overexpression causes a reduction of LSCs in CML mice. We transduced bone marrow cells with BCR-ABL or BCR-ABL-Blk to induce CML, and observed that the percentages and numbers of total LSCs, LT-LSCs and ST-LSCs were significantly lower in recipients of BCR-ABL-Blk-transduced bone marrow cells than in recipients of BCR-ABL-transduced cells (Fig. 2b). To verify the ectopic expression of Blk, we sorted LSCs by fluorescence-activated cell sorting (FACS), isolated total RNA for RT-RCR analysis, and found that Blk expression was largely but not completely restored to the endogenous level in LSCs (Fig. 2c). To further demonstrate the inhibitory effect of Blk on LSCs, we tested their ability to transfer disease to secondary recipient mice. Bone marrow cells were transduced with BCR-ABL or BCR-ABL-Blk to induce primary CML, and then bone marrow cells from these CML mice were transferred into secondary recipient mice. Fig. 2d shows that Blk overexpression caused a significant delay of CML development in the secondary recipients (Fig. 2d).
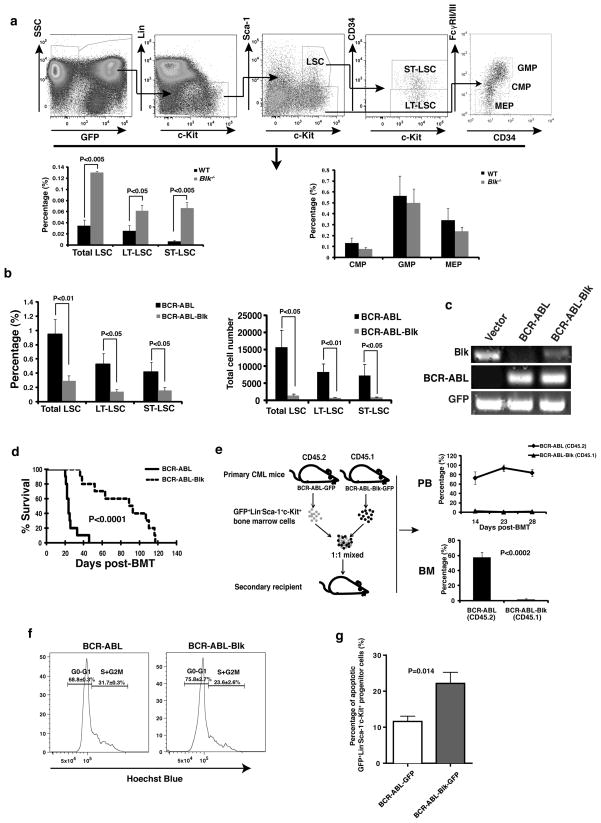
Figure 2. Blk suppresses LSCs.
(a) The percentages of total LSCs, LT-LSCs, ST-LSCs, CMP, GMP and MEP in bone marrow of recipients of BCR-ABL-transduced Blk−/− donor bone marrow cells (n=9) were compared with those in bone marrow of recipients of BCR-ABL-transduced WT donor bone marrow cells (n=5) at day 11 after transplantation. Mean values ± s.e.m. (b) The percentages and numbers of total LSCs, LT-LSCs and ST-LSCs in bone marrow of recipients of BCR-ABL- or BCR-ABL-Blk-transduced bone marrow cells were analyzed at day 15 after transplantation. Mean values ± s.e.m. (n=5). (c) RT-PCR analysis of expression of Blk, BCR-ABL, and GFP in FACS-sorted LSCs at 2 weeks after transplantation. (d) Kaplan-Meier survival curves for secondary CML mice receiving bone marrow cells obtained at day 15 after transplantation from primary CML mice induced by BCR-ABL or BCR-ABL-Blk (n=10 for each group). (e) 103 sorted-LSCs from bone marrow of primary CML mice induced by BCR-ABL-transduced CD45.2 or BCR-ABL-Blk-transduced CD45.1 donor bone marrow cells were mixed at 1:1 ratio, followed by transplantation into recipient mice. The percentages of BCR-ABL- and BCR-ABL-Blk expressing cells in peripheral blood and bone marrow were compared at 2, 3, and 4 weeks after transplantation (n=3 for each time point). (f) Cell cycle analysis of LSCs from bone marrow of CML mice induced by BCR-ABL or BCR-ABL-Blk (n=5 for each group). (P<0.05). (g) The percentage of apoptotic GFP+Lin−Sca-1+c-Kit+ cells in bone marrow from CML mice at day 14 after transplantation. Mean values ± s.e.m.
To more rigorously evaluate the inhibitory effect of Blk on LSC function, we examined whether Blk reduces the ability of LSCs to repopulate. LSCs were sorted by FACS from bone marrow of mice with primary CML induced by transplantation with BCR-ABL-transduced CD45.2 or BCR-ABL-Blk-transduced CD45.1 donor bone marrow cells. The sorted CD45.2 and CD45.1 LSCs were mixed in a 1:1 ratio, and transplanted into recipient mice. At days 14, 23 and 28 after transplantation, fewer than 5% of GFP+Gr-1+ cells in peripheral blood of the mice were CD45.1 leukemia cells that overexpressed Blk, whereas greater than 75%–80% of GFP+Gr-1+ cells were CD45.2 leukemia cells that did not overexpress Blk (Fig. 2e). Consistent with these results, at day 28, the percentage of CD45.1+ leukemia cells that overexpressed Blk in bone marrow was also very low (Fig. 2e). The suppression of LSCs by Blk can be explained, at least in part, by inhibition of cell cycle progression, as there were significantly fewer LSCs that overexpressed Blk in the S+G2M phase of the cell cycle compared to LSCs that did not overexpress Blk (Fig. 2f). In addition, we observed increased apoptosis in LSCs from recipients of BCR-ABL-Blk-transduced bone marrow cells (Fig. 2g).
Blk does not suppress normal hematopoietic stem cells
We ask whether Blk has a similar inhibitory effect on normal HSCs. Using real time RT-PCR, we first assessed Blk expression in different hematopoietic stem/progenitor populations, including LT-HSC (CD34−Flt-3−LSK), ST-HSC (CD34+Flt-3−LSK), MPP (CD34+Flt-3+LSK), CMP, MEP, and GMP. We found that Blk was highly expressed in LT-HSCs but not in ST-HSCs, MPPs and progenitors excluding MEP with a higher level of Blk expression (Supplementary Fig. 5a). Next, we examined the effect of Blk on normal hematopoiesis and HSCs. Fig. 3a–c show that the percentages of total LSK,LT-HSCs, ST-HSCs, CMP and MEP in bone marrow of Blk−/− and WT mice were similar, although the percentage of GMP was higher in Blk−/− (0.19%) than in WT (0.12%) mice. Notably, however, there was no significant difference in more mature myeloid cells (Gr-1+Mac-1+) in bone marrow of Blk−/− and WT mice (Fig. 3d). We also found that Blk deficiency did not affect cell cycle progression (Fig. 3e) or apoptosis (Fig. 3f) of LSK cells.
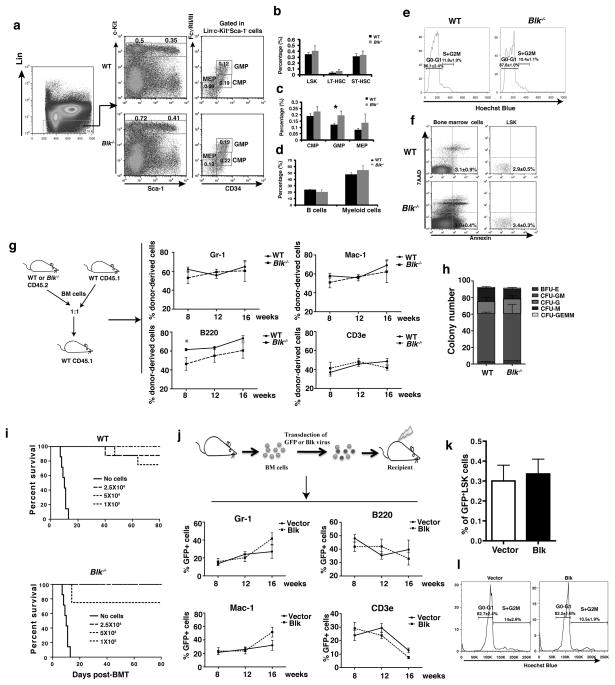
Figure 3. Blk does not suppress normal HSCs.
(a, b and c) FACS analysis of HSCs, CMP, GMP and MEP cells in bone marrow of WT (n=3) or Blk−/− (n=4) mice. (b) The percentages of LSK, LT-HSCs, and ST-HSCs in bone marrow of WT or Blk−/− mice. (c) The percentages of CMP, GMP, and MEP cells in bone marrow of WT (n=3) or Blk−/− (n=4) mice. (d) The percentages of myeloid (Gr-1+Mac-1+) and lymphoid (B220+IgM+) cells in bone marrow of WT or Blk−/− mice. (e) Cell cycle analysis of LSK cells in bone marrow of WT or Blk−/− mice. (f) Apoptosis of bone marrow cells and LSK cells from WT (n=3) or Blk−/− (n=4) mice. (g) FACS analysis of different donor cell lineages in recipient mice at 8, 12 and 16 weeks after transplantation. * P<0.05 (h) Colony forming assay of WT and Blk−/− bone marrow cells. (i) Three doses (1×105, 5×105, 2.5×104) of WT or Blk−/− BM cells were injected into lethally irradiated recipients, and survival of the mice were compared. (j) FACS analysis of cell lineages in peripheral blood of recipients of Blk and vector transduced bone marrow cells at 8, 12, 16 weeks after transplantation. (k) The percentages of GFP+ LSK cells in bone marrow of recipients of vector and Blk transduced bone marrow cells at 16 weeks after transplantation. (l) Cell cycle analysis of LSK cells from bone marrow of recipients of GFP or Blk/GFP transduced bone marrow cells. (P=0.86 for G0-G1; P=0.2 for S+G2M).
To examine whether Blk affects the function of normal HSCs, we performed a competitive repopulation assay. 2×105 bone marrow cells from WT or Blk−/− mice (CD45.2) were transplanted into each lethally irradiated WT recipient (CD45.1) along with an equal number of WT competitor cells (CD45.1). The lineage contribution of WT or Blk−/− cells in recipient mice was evaluated at 8, 12 and 16 weeks after transplantation. We observed similar percentages of donor-derived myeloid (Gr-1+ and Mac-1+) and T lymphoid cells (CD4+ and CD8+) (Fig. 3g), indicating that Blk did not affect the function of normal HSCs. Although Blk deficiency affected the levels of B cells (B220+) (Fig. 3g), this effect is likely due to the known role of Blk in B cell development 17. We also performed a colony-forming assay to examine the effect of Blk on progenitor cell function in vitro using sorted LSK cells from Blk−/− and WT mice bone marrow. Similar numbers and types of colonies were formed in the presence and absence of Blk (Fig. 3h). Further, there was no significant difference in the ability of WT and Blk−/− bone marrow cells to rescue lethally irradiated mice (Fig. 3i).
To provide additional evidence for the role of Blk in regulation of HSC function, we tested whether overexpression of Blk suppresses HSCs. We transduced bone marrow cells from WT mice with Blk-GFP or GFP retrovirus, followed by transplantation into recipient mice (Fig. 3j). Overexpression of Blk in GFP+ LSK cells was confirmed by real time RT-PCR (Supplementary Fig. 5b). The lineage contribution of GFP or Blk-GFP cells in recipient mice was evaluated at 8, 12 and 16 weeks after transplantation. The percentages of mature myeloid cells (GFP+Gr-1+/Mac-1+), B-lymphoid cells (GFP+B220+), and T cells (GFP+CD3e+) in peripheral blood of recipients of Blk-GFP- or GFP-transduced marrow cells were similar (Fig. 3j), and the percentages of GFP+LSK cells in bone marrow of recipients of Blk-GFP- or GFP-transduced marrow cells at 16 weeks were also similar (Fig. 3k). In addition, there was no significant difference in cell cycle progression between Blk-GFP- and GFP-transduced bone marrow cells (Fig. 3l). Next, we conducted an in vivo limiting dilution analysis. At 16 weeks after transplantation, GFP+LSK cells were sorted from recipients of Blk-GFP or GFP transduced marrow cells, and were injected into secondary recipients. After 12 weeks, we analyzed the GFP+ cells. Poisson statistics showed no significant difference in the frequency of long-term repopulation ability among control and Blk-transduced cells (Supplementary Table 1).
Pax5 is an upstream regulator of Blk in LSCs
Pax5 binds to the Blk promoter and stimulates Blk expression 18. We therefore considered the possibility that the down-regulation of Blk expression by BCR-ABL in LSCs is mediated through Pax5. We found that BCR-ABL markedly down-regulated Pax5 expression in LSCs (Fig. 4a). To test whether Pax5 suppresses LSCs and CML development, we generated a retroviral construct that co-expressed BCR-ABL and Pax5 (Fig. 4b). We transduced bone marrow cells with BCR-ABL or BCR-ABL-Pax5 to induce CML. Fourteen days later, bone marrow cells from CML mice were analyzed for the percentages and numbers of LSCs. Fig. 4c shows that Pax5 overexpression caused a marked decrease in total, LT- and ST-LSCs. We next compared survival between the two transplantation groups. All recipients of BCR-ABL-transduced bone marrow cells died of CML within 3 weeks, whereas fewer than 20% of the recipients of BCR-ABL-Pax5-transduced bone marrow cells developed CML and died (Fig. 4d), which correlated with the lower and gradually decreasing percentages of myeloid leukemia cells in peripheral blood during the course of the disease (Fig. 4e) and with less severe splenomegaly and leukemia cell infiltration in the spleen and lung (Fig. 4f).
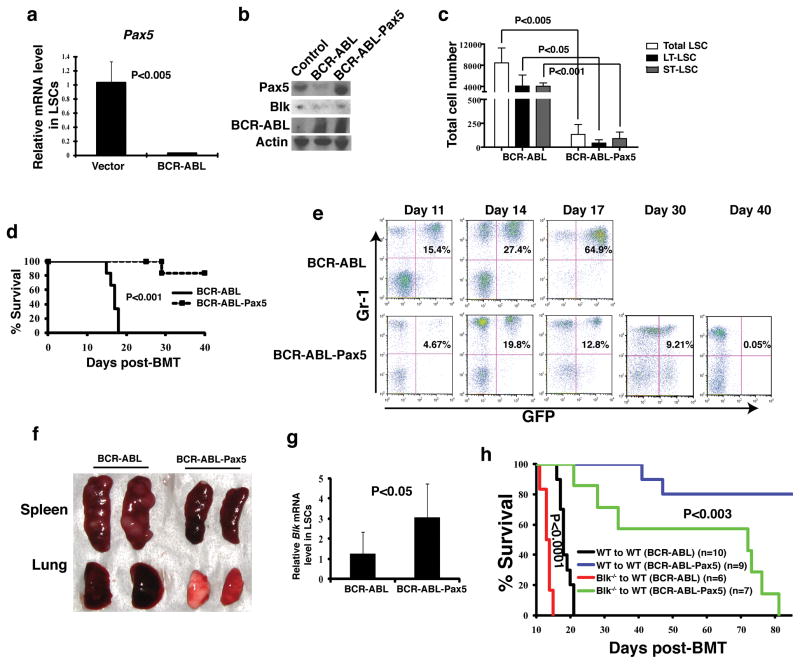
Figure 4. Pax5 is an upstream partner of Blk in LSCs.
(a) Real time RT-PCR analysis showing expression of Pax5 in LSCs as compared to normal HSCs. Results are given as mean ± s.e.m. (b) Western blot analysis showing expression of Pax5, Blk and BCR-ABL in 293T cells transfected with BCR-ABL and BCR-ABL-Pax5. (c) FACS analysis of the numbers of total LSCs, LT-LSCs, and ST-LSCs from recipients of BCR-ABL- or BCR-ABL-Pax5-transduced BM cells. Results are given as mean ± s.e.m. (d) Kaplan-Meier survival curves for recipients of BCR-ABL-(n=7) or BCR-ABL-Pax5- (n=6) transduced bone marrow cells. (e) FACS analysis showing the percentages of GFP+Gr-1+ cells in peripheral blood of recipients of BCR-ABL- or BCR-ABL-Pax5-transduced bone marrow cells at days 11, 14, 17, 30, and 40 after BMT, and gradual disappearance of GFP+Gr-1+ cells in peripheral blood of recipients of BCR-ABL-Pax5-transduced bone marrow cells but not in recipients of BCR-ABL-transduced bone marrow cells. (f) Gross appearance of the lungs and spleens of recipients of BCR-ABL- or BCR-ABL-Pax5-transduced donor bone marrow cells at day 14 after BMT. (g) Real time RT-PCR analysis monitoring Blk expression in LSCs from bone marrow of recipients of BCR-ABL- and BCR-ABL-Pax5-transduced bone marrow cells. Bone marrow cells from mice with CML induced by BCR-ABL or BCR-ABL-Pax5 were cultured under stem cell conditions for 6 days, and LSCs were sorted by FACS for isolation of total RNA for real time PCR analysis. (h) Kaplan-Meier survival curves for recipients of BCR-ABL- or BCR-ABL-Pax5-transduced WT or Blk−/− bone marrow cells.
We also found that ectopically expressed Pax5 caused an increase of Blk expression in LSCs (Fig. 4g), supporting the idea that Pax5 functions upstream of Blk to mediate the down-regulation of Blk by BCR-ABL. To further test this idea, we transduced bone marrow cells from Blk−/− or WT mice with BCR-ABL-Pax5 or BCR-ABL alone to induce CML, and compared to the accelerated CML development in recipients of Blk−/− bone marrow cells transduced by BCR-ABL alone (Fig. 4h), recipients of Blk−/− bone marrow cells transduced with BCR-ABL-Pax5 died of CML much more slowly, although these mice developed CML significantly faster than recipients of BCR-ABL-Pax5-transduced WT bone marrow cells. These results suggest that Blk is one but not only the downstream functional target gene of Pax5 in LSCs.
We tested whether Pax5 suppresses normal HSCs. We first assessed Pax5 expression in different hematopoietic stem/progenitor populations using qRT-PCR, and found that Pax5 was highly expressed in LT-HSCs but not in ST-HSCs, MPPs and progenitors excluding MEP (Supplementary Fig. 6a). Next, we transduced bone marrow cells from normal B6 mice with retrovirus expressing Pax5 and GFP or GFP alone, and the transduced cells were cultured under stem cell conditions for 4 days, followed by FACS analyses of control or Pax5-expressing GFP+LSK. Pax5 overexpression was confirmed by RT-PCR (Supplementary Fig. 6b). Pax5 reduced the number of LSKcells from 45.5% to 29.4% (Supplementary Fig. 6c), suggesting that unlike Blk, Pax5 has some effect on normal HSCs. This result further suggests that besides Blk, Pax5 also regulates other downstream genes. However, when we monitored the distribution of different lineages at 8, 12, and 16 weeks after transplantation, the initial decrease of mature myeloid cells (GFP+Gr-1+/Mac-1+) at 8 and 12 weeks was reversed at 16 weeks (Supplementary Fig. 6d), suggesting that the function of HSCs was not significantly affected. Development of lymphoid cells (GFP+B220+/CD3e+) was affected by Pax5 (Supplementary Fig. 6d), presumably due to the specific role of Pax5 in lymphoid development19.
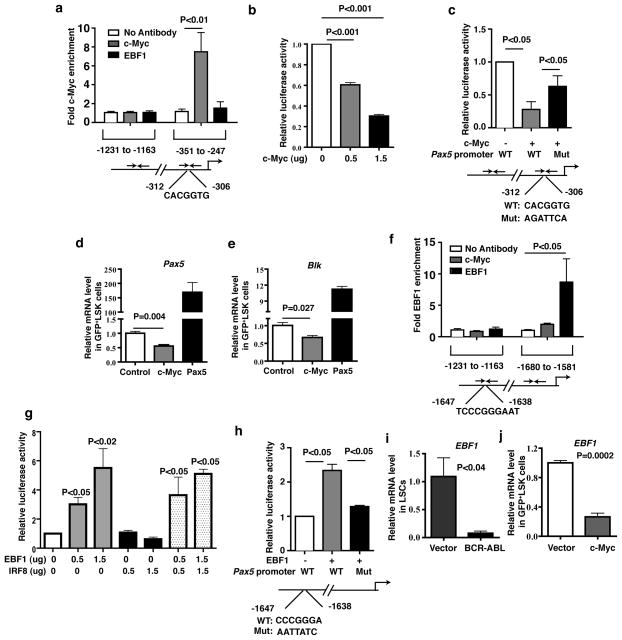
c-Myc and EBF1 mediate down-regulation of Pax5 by BCR-ABL
We studied how BCR-ABL down-regulates Pax5 expression. BCR-ABL induces c-Myc expression 20–22, and analysis of the Pax5 promoter region revealed a consensus c-Myc binding motif at −312 base pair upstream of the transcription start site (Supplementary Fig. 7a). Chromatin immunoprecipitation (ChIP) analysis demonstrated that c-Myc directly binds to this region, but not to a region further upstream (Fig. 5a). Therefore, we investigated whether BCR-ABL down-regulates Pax5 through c-Myc using a luciferase assay. Fig. 5b shows that expression of c-Myc caused a reduction of Pax5 promoter activity in a dose dependent manner in NIH3T3 cells. We mutated the c-Myc binding site in the Pax5 promoter, and found that the suppression of luciferase activity by c-Myc was markedly rescued (Fig. 5c). These results indicate that c-Myc directly binds to the Pax5 promoter to suppress Pax5 expression. To determine whether c-Myc down-regulates Pax5 expression in HSCs, we transduced bone marrow cells with a retrovirus expressing c-Myc (Supplementary Fig. 7b) or, as a control, Pax5. qRT-PCR analysis showed that c-Myc significantly inhibited expression of Pax5 and Blk in LSK cells (Fig. 5d and 5e), whereas Pax5 dramatically enhanced Blk expression in these cells (Fig. 5e).
Figure 5. c-Myc and EBF1 regulate Pax5 expression.
(a) ChIP assay showed c-Myc directly bound to the Pax5 promoter. Results were shown as mean ± s.e.m. (b) A Pax5 promoter luciferase reporter construct was cotransfected with empty vector or c-Myc plasmid into NIH3T3 cells. Cell extracts were analyzed for luciferase activity. Results were shown as mean ± s.e.m.. (c) Luciferase assay showed mutant c-Myc binding site in the Pax5 promoter restored the luciferase activity. Results were shown as mean ± s.e.m. (d and e) Real time RT-PCR analysis monitoring Pax5 and Blk expression in c-Myc-expressing or Pax5-expressing LSK cells. Results were shown as mean ± s.e.m. (f) ChIP assay showed EBF1 directly bind to the Pax5 promoter. Results were shown as mean ± s.e.m. (g) A Pax5 promoter luciferase reporter construct was cotransfected with empty vector, EBF1, or IRF8 plasmids into NIH3T3 cells. Cell extracts were analyzed for luciferase activity. Results were shown as mean ± s.e.m. (h) Luciferase assay showed mutant EBF1 binding site in the Pax5 promoter rescued the luciferase activity. Results were shown as mean ± s.e.m. (i) Real time RT-PCR analysis monitoring EBF1 expression by BCR-ABL in LSCs as compared to normal HSCs. Results are given as mean ± s.e.m. (j) Real time RT-PCR analysis monitoring EBF1 expression in c-Myc-expressing LSK cells. Results are given as mean ± s.e.m.
The transcription factor EBF1 binds to the Pax5 promoter and stimulates Pax5 expression 23,24, and our microarray results indicated that EBF1 was significantly downregulated in LSCs (Supplementary Fig. 8). ChIP analysis showed that EBF1 directly bound to the Pax5 promoter within a region from −1638 to −1647 (Fig. 5f), consistent with previous results 23,24. Also, expression of EBF1 increased Pax5 luciferase activity (Fig. 5g). Although interferon regulatory factor 8 (IRF8) regulates expression of EBF1 and Pax5 25,26, IRF8 had no effect on Pax5 promoter activity (Fig. 5g). We mutated the EBF1 binding site in the Pax5 promoter, and found that the increased luciferase activity by EBF1 was markedly inhibited (Fig. 5h). It remained possible that c-Myc also down-regulates EBF1 expression resulting in decreased Pax5 expression, and qRT-PCR analysis showed that BCR-ABL down-regulated EBF1 expression in LSCs (Fig. 5i), and c-Myc down-regulated EBF1 expression in LSK cells (Fig. 5j).
p27 functions downstream of Blk to suppress proliferation of LSCs
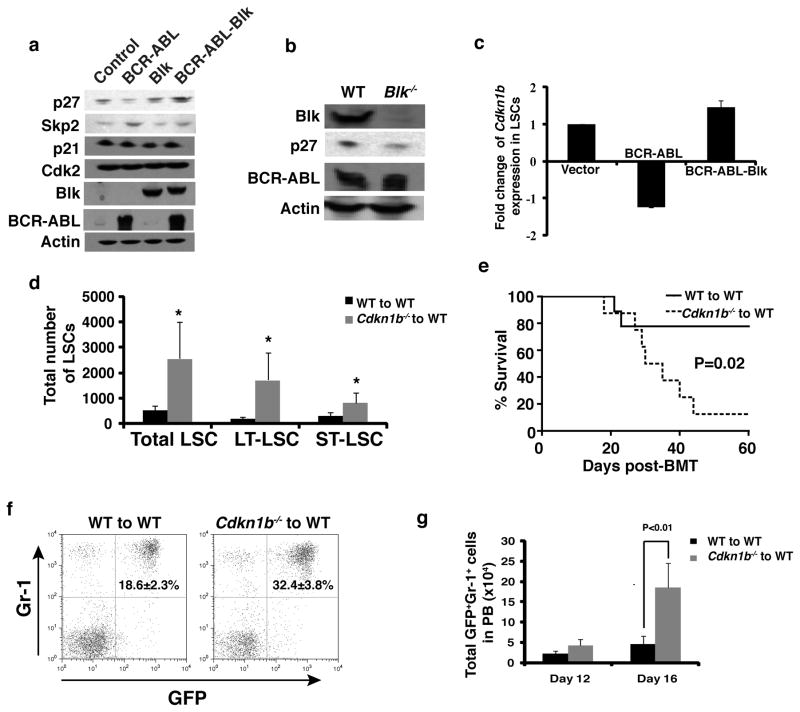
We attempted to identify genes required for Blk to suppress LSC proliferation and CML development. The mammalian cyclin-dependent kinase inhibitor 1B (Cdkn1b) p27 is a negative cell cycle regulator that blocks the G1 to S phase transition 27. BCR-ABL down-regulates Cdkn1b expression through multiple mechanisms 28–32. We compared the levels of p27 in 293T cells transfected with BCR-ABL alone or with both BCR-ABL and Blk, and found that BCR-ABL down-regulated p27 and Blk restored p27 expression through inhibiting S-phase kinase associated protein 2 (Skp2) expression (Fig. 6a). The inhibition of Skp2 by Blk was confirmed by qRT-PCR (Supplementary Fig. 9a). BCR-ABL and Blk did not alter the levels of other cell cycle regulators such as p21 and cyclin-dependent kinase 2 (Cdk2) (Fig. 6a). To confirm that BCR-ABL functions through Blk to reduce p27 expression, we transduced WT or Blk−/− bone marrow cells with BCR-ABL, and found that p27 expression was significantly lower in the absence of Blk (Fig. 6b and Supplementary Fig. 9b). Conversely, we overexpressed Blk in LSCs both to verify that Blk increases p27 expression and to identify other Blk target genes. Bone marrow cells were transduced with GFP, BCR-ABL-GFP or BCR-ABL-Blk-GFP, and fourteen days after transplantation, bone marrow cells were isolated and LSCs were sorted by FACS for isolation of total RNA for DNA microarray analysis. BCR-ABL down-regulated p27 expression, which was reversed by Blk overexpression (Fig. 6c). We also identified other genes that were significantly up- or down-regulated by Blk in LSCs (Supplementary Table 1).
Figure 6. p27 is a downstream partner of Blk in LSCs.
(a) Western blot analysis of the expression of p27, p21, Cdk2 and Skp2 in 293T cells after transfection with BCR-ABL, Blk or BCR-ABL-Blk. (b) Western blot analysis monitoring p27 expression. BCR-ABL-transduced bone marrow cells from WT or Blk−/− mice were grown in Whitlock-Witte culture for 7 days, and protein lysates were isolated for comparing p27 expression regulated by BCR-ABL in the presence and absence of BCR-ABL by Western blotting. (c) Microarray analysis showing Cdkn1b expression in vector-, BCR-ABL- and BCR-ABL-Blk-transduced LSCs. Mean values (± s.e.m) are shown. (d) The total numbers of total LSCs, LT-LSCs, and ST-LSCs in bone marrow of recipients of BCR-ABL-transduced Cdkn1b−/− (n=4) and BCR-ABL-transduced WT (n=3) donor bone marrow cells at day 14 after BMT. Mean values (± s.e.m.) are shown. (*P<0.05) (e) Kaplan-Meier survival curves for recipients of BCR-ABL-transduced bone marrow cells from WT or Cdkn1b−/− donor mice (P=0.02; n=8 for each group). (f) FACS analysis showing the percentages of GFP+Gr-1+ cells in peripheral blood of recipients of BCR-ABL-transduced bone marrow cells from WT or Cdkn1b−/− donor mice at day 12 after BMT. (P<0.02). (g) The total numbers of GFP+Gr-1+ cells in peripheral blood of recipients of BCR-ABL-transduced bone marrow cells from WT or Cdkn1b−/− donor mice at days 12 and 16 after BMT. Mean values (± s.e.m.) are shown.
To test whether p27 suppresses LSC proliferation and CML development, we transduced WT or Cdkn1b−/− bone marrow cells with BCR-ABL, followed by transplantation of a relatively small number of transduced cells (1×105 cells per recipient). After 14 days, bone marrow cells from CML mice were analyzed. p27 deficiency caused a marked increase in total, LT- and ST-LSCs (Fig. 6d). Significantly, only 20% of recipients of BCR-ABL-transduced WT bone marrow cells died by 60 days after transplantation, whereas 90% of recipients of BCR-ABL-transduced Cdkn1b−/− bone marrow cells died by 45 days (Fig. 6e), correlating with a higher percentage and number of myeloid leukemia cells in peripheral blood (Fig. 6f, g). The regulation of p27 by Blk in cell cycle progression was further demonstrated in cultured bone marrow cells from CML mice. Consistent with the results in 293T cells, Blk overexpression increased expression of p27 but not Cdk2 and Cyclin E (Supplementary Fig. 9c). Collectively, these results indicate that p27 functions downstream of Blk to suppress LSC proliferation and CML development. By contrast, although Cdkn1b was induced to a high level by Blk in LSK cells (Supplementary Fig. 9d), the percentage of LSK were still similar in bone marrow of mice receiving control and Blk-transduced marrow cells (Fig. 3k), indicating that p27 did not suppress proliferation of normal HSCs.
Suppression of CML does not require Blk kinase activity
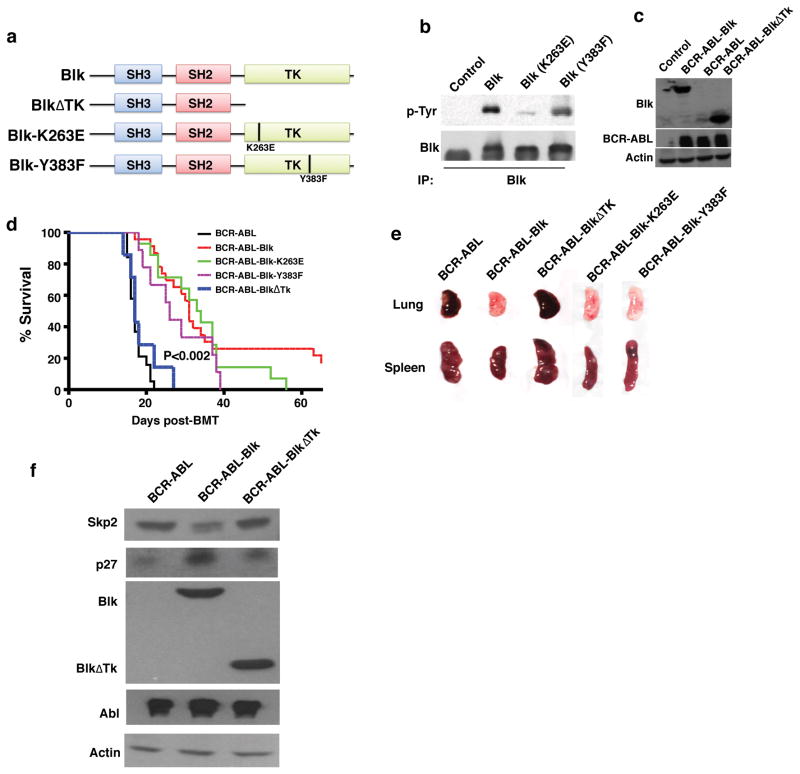
To determine whether Blk kinase activity is required for suppression of CML, we analyzed three Blk mutants: deletion of the entire kinase domain (ΔTk), K263E, and Y383F (Fig. 7a). The K263E mutation causes a loss of Blk kinase activity, and the Y383F mutation reduces Blk autophosphorylation 33. We co-expressed these three Blk mutants with BCR-ABL in 293T cells (Fig. 7b, c), and kinase activity of Blk-K263E was almost completely lost and autophosphorylation of Blk-Y383F was significantly reduced (Fig. 7b). We transduced bone marrow cells with BCR-ABL, BCR-ABL-Blk, BCR-ABL-Blk(ΔTk), BCR-ABL-Blk-K263E, or BCR-ABL-Blk-Y383F, which had similar viral titers (Supplementary Fig. 10a, b). We found that recipients of bone marrow cells transduced with BCR-ABL-Blk (ΔTk) or BCR-ABL alone developed CML similarly, as shown by survival (Fig. 7d) and infiltration of leukemic cells into the lung and spleen (Fig. 7e). Surprisingly, in recipients of BCR-ABL-Blk-K263E- and BCR-ABL-Blk-Y383F-transduced bone marrow cells, CML development was also suppressed (Fig. 7d), correlating with decreased infiltration of leukemic cells in the spleen and lung (Fig. 7e). Thus, suppression of CML development by Blk requires its kinase domain but not its kinase activity, although we cannot rule out the possibility that the very low levels of kinase activity of the Blk mutants are required for CML suppression.
Figure 7. The inhibitory effect of Blk on CML does not require Blk kinase activity.
(a) Schematic structures of Blk mutants. SH, Src-homology; TK, tyrosine kinase. (b) Western blot analysis monitoring the phosphorylation status of Blk-K263E, and the reduced phosphorylation level of Blk-Y383F, as compared to that of WT Blk. (c) Western blot analysis monitoring expression of Blk, BlkΔTk, and BCR-ABL in 293T cells. (d) Kaplan-Meier survival curves for recipients of BCR-ABL (n=19), BCR-ABL-Blk (n=23), BCR-ABL-BlkΔTk (n=8), BCR-ABL-Blk-K263E (n=14) or BCR-ABL-Blk-Y383F (n=9) transduced bone marrow cells. (e) Gross appearance of the lungs and spleens of recipients of BCR-ABL-, BCR-ABL-Blk- or BCR-ABL-BlkΔTk- BCR-ABL-Blk-K263E- or BCR-ABL-Blk-Y383F-transduced bone marrow cells at 14 days after BMT. (f) Western blot analysis indicated that Blk but not the truncated BlkΔTk regulated Skp2 and p27 expression.
It is possible that the ability of Blk to stimulate p27 expression involves Skp2, because p27 levels are inversely correlated with Skp2 expression 34. Also, BCR-ABL stimulates cell cycle progression by promoting Skp2-mediated degradation of p27 28; and Skp2 is required for BCR-ABL induced myeloproliferative disease 35. Fig. 7f shows that Blk prevented BCR-ABL-induced Skp2 expression, which was dependent on the Blk kinase domain.
Blk functions as a tumor suppressor in human CML cells
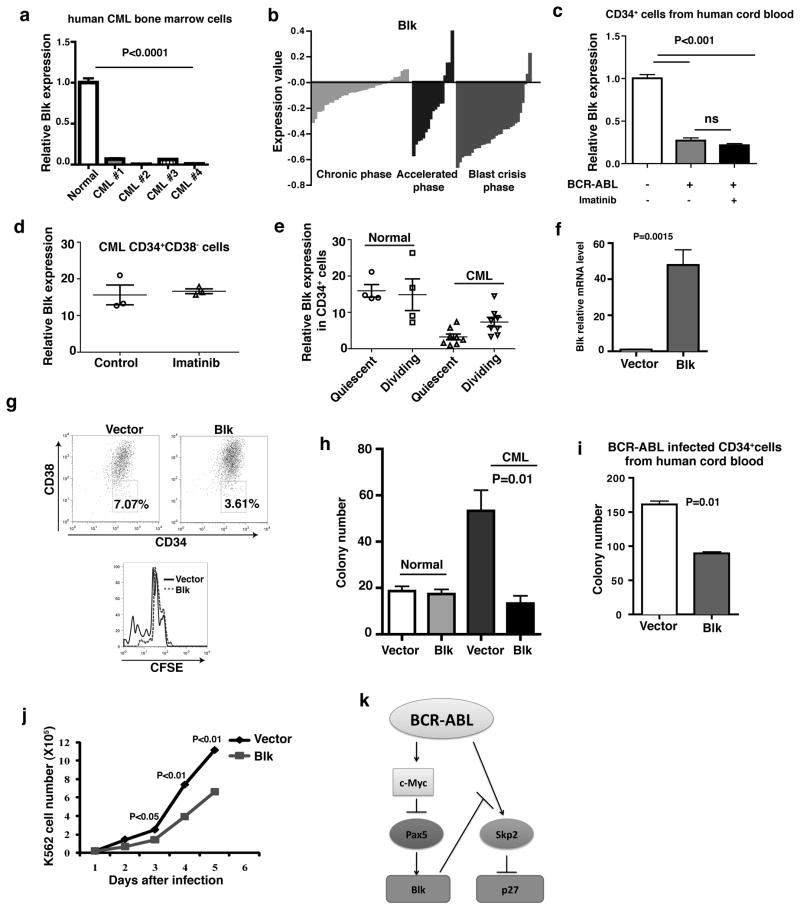
We first asked whether Blk expression was lost in human CML cells. Fig. 8a shows that Blk expression was substantially lower in bone marrow cells from human CML patients compared to normal human bone marrow cells. We also analyzed a publicly available gene expression profiling database derived from analysis of human bulk CD34+ cells in CML patients36, and found that Blk expression was markedly down-regulated in the majority of CML patients in chronic phase, accelerated phase and blast crisis (Fig. 8b). Fig. 8c shows that BCR-ABL significantly lowered Blk expression in human cord blood CD34+ cells transduced by BCR-ABL and this effect was not reversed by imatinib, indicating that downregulation of Blk expression by BCR-ABL in human CML cells does not require BCR-ABL kinase activity. To examine whether the BCR-ABL kinase activity independent regulation of Blk is at the level of CML stem cells, we first analyzed another publicly available DNA microarray study of human CML CD34+CD38− cells37, and found that Blk expression was not altered by imatinib treatment (Fig. 8d). We further analyzed Blk expression in quiescent and dividing CD34+ cells from CML patients based on the results in a public database38. We found that the levels of Blk expression in quiescent and dividing CD34+ human CML stem cells were significantly lower than those in quiescent and dividing normal CD34+ cells (Fig. 8e). In addition, the level of Blk expression in quiescent CML stem cells was significantly lower than that in dividing CML stem cells, but there was no difference in Blk expression between quiescent and dividing normal CD34+ cells (Fig. 8e). Further, we sorted CFSE stained CD34+CD38− human CML stem cells by FACS into quiescent and dividing populations and isolated RNA for qRT-PCR analysis, and confirmed that Blk expression levels were lower in quiescent human CML stem cells than in dividing human CML stem cells (data not shown).
Figure 8. Blk functions as a tumor suppressor in human CML cells.
(a) Real-time RT-PCR analysis of Blk expression in bone marrow cells from CML patients and normal donors. (P<0.0001). (b) Microarray analysis of Blk expression in bone marrow and peripheral blood CD34+ cells from 42 chronic (green), 17 accelerated (blue) and 31 blast crisis phase (red) CML patients. (c) Real-time RT-PCR analysis of Blk expression in BCR-ABL-transduced human cord blood CD34+ cells. BCR-ABL-transduced CD34+ cells were also treated by imatinib (1 μM) for 24 hours. ns, no significance. Mean values (± s.e.m) are shown. (d) Blk expression in human CD34+CD38− CML stem cells was not affected by imatinib. (n=3) (e) The expression of Blk in normal and CML quiescent and dividing CD34+ cells. (f) Real-time RT-PCR analysis of Blk expression in bone marrow cells from chronic phase CML patients transduced with either an empty or Blk lentivirus (pLenti-puro or pLenti-Blk-puro). (P=0.0015). (g) FACS analysis showed inhibition of proliferation of CD34+CD38− CML stem cells. (h) Equal numbers of human CML bone marrow cells transduced with empty or Blk-expressing lentivirus were plated in cytokine-supplemented methylcellulose in the presence of puromycin. (P=0.01). (i) CD34+ cells from human cord blood were co-transduced with BCR-ABL-GFP retrovirus and either an empty lentivirus or lentivirus expressing Blk, and were plated in cytokine-supplemented methylcellulose in the presence of puromycin. (P=0.01) (j) Growth curves of FACS sorted human K562 cells transduced with Blk-GFP or GFP alone. (k) Molecular model of the Blk pathway in LSCs.
Next, we analyzed the functional effect of Blk on human CML stem cells. To infect quiescent cells, we used a lentiviral vector to express Blk in human CML cells. We purified lineage negative cells from human primary CML patients, transduced the cells with Blk (Fig. 8f), and subsequently labeled these transduced cells with CFSE to track quiescent and dividing CML stem cells 39. We found that Blk overexpression inhibited proliferation of CD34+CD38− CML stem cells, as shown by a lesser percentage of CFSElow cells in Blk-infected CML stem cells than in vector-infected CML stem cells (Fig. 8g). Also Blk overexpression induced apoptosis of CD34+CD38− CML stem cells (Supplementary Fig. 11). Further, we performed a colony-forming assay to assess progenitor function40, and found that Blk overexpression inhibited the colony-forming ability of human CML but not normal bone marrow cells (Fig. 8h). Blk expression also inhibited the colony-forming ability of BCR-ABL-transduced human cord blood CD34+ cells (Fig. 8i). Finally, we transduced BCR-ABL+ human K562 cells with a retrovirus co-expressing Blk and GFP or, as a control, GFP alone, and showed that Blk-expressing K562 cells grew significantly slower than cells that did not express Blk (Fig. 8j).
DISCUSSION
We show that Blk functions as a tumor suppressor in CML through a pathway summarized in Fig. 8k and discussed below. Blk is downregulated by BCR-ABL in both mouse and human CML hematopoietic cells. Of particular significance, Blk expression is markedly down-regulated in bulk CD34+ cells from the majority of CML patients in chronic, accelerated and blastic phases. Thus, suppression of Blk expression begins at an early stage of CML and is maintained throughout the course of disease.
Although BCR-ABL kinase inhibitors induce a complete cytogenetic response in the majority of CML patients in chronic phase, they are incapable of eradicating LSCs 6,13,41. We show that Blk suppresses LSCs without affecting normal HSCs or hematopoiesis. Thus, the Blk pathway provides a selective target for eradicating LSCs. CML could be treated through restoring Blk expression or up-regulating other Blk pathway genes such as Pax5 and Cdkn1b. We note, however, that restoration of Blk expression in CML patients would be technically challenging and may require, for example, gene therapy approaches.
The strategy of selectively targeting LSCs contrasts sharply with other therapeutic approaches that inhibit the function of genes essential for both LSCs and normal HSCs 42–46. For example, the Wnt signaling pathway is critical in regulating hematopoietic stem and progenitor cell function 43,44, and deletion of the β-catenin gene causes a profound defect in LSCs and subsequent induction of CML by BCR-ABL 45,47. Inhibition of the hedgehog pathway impairs both LSCs and normal HSCs 42,46.
The finding that Blk functions as a tumor suppressor role is somewhat unexpected because some Src family kinases promote leukemogenesis 48–51. Paradoxically, Blk promotes normal B cell development by cooperating with other Src family members. Pax5, which we show functions upstream of Blk, is also required for normal B cell development 19,52. Deletions and mutations of Pax5 have been identified in human acute lymphoid leukemia 53,54, suggestive of a tumor suppressor function.
We show that Pax5 mediates down-regulation of Blk by BCR-ABL through c-Myc and that p27 mediates the inhibitory effect of Blk on LSCs, although it is likely that there are other downstream Blk target genes. Mechanistically, Blk upregulates p27 through downregulation of Skp2. We have previously shown that LSCs in CML are also positively regulated by the Alox5 gene, which is up-regulated by BCR-ABL 5. Our unpublished data suggest that Blk regulates Alox5 in LSCs, and we should further elucidate the functional relationship between Blk and Alox5 in LSCs.
We found that Blk and p27 do not suppress proliferation of normal HSCs, consistent with a previous finding that p27 has no effect on the number and self-renewal ability of HSCs 55. Blk is a tyrosine kinase but unexpectedly found to have tumor suppressor function not related its kinase activity. Thus, an Src kinase inhibitor such as dasatinib will not inhibit Blk tumor suppressor activity in CML treatment. Down-regulation of Blk by BCR-ABL is not reversed following inhibition of BCR-ABL kinase activity, consistent with the inability of imatinib to kill LSCs. Importantly, Blk inhibits proliferation of human CML stem cells, providing a rationale for targeting LSCs by restoring the Blk pathway.
METHODS
Samples
Cord blood mononuclear cells were isolated by Ficoll-Hypaque density gradient centrifugation. CD34+ cells were enriched using the MACS CD34 progenitor kit (Miltenyi Biotec). Transduction of cord blood CD34+ cells were performed as described previously 56. Six human CML bone marrow samples contained greater than 97% (in average) of leukemia cells based on the karyotyping results for the t(9;22) translocation. This result is supported by the FISH analysis of the chimeric BCR-ABL oncogene in the cells. Human bone marrow CML cells were cultured in Iscoves Modified Dulbecco medium (Sigma) supplemented with a serum substitute (BIT; StemCell), 40μg/mL low-density lipoproteins, 100ng/mL recombinant human Flt3-ligand, 100ng/mL steel factor, 20ng/mL recombinant human interleukin-3 (IL-3), IL-6, and granulocyte-colony-stimulating factor 57. For CML stem cell proliferation assay, sorted human CML lineage negative(Lin−) cells were transduced with vector or Blk-expressing lentivirus, and labeled with 1μM carboxyfluorescein diacetate succinimidyl ester (CFSE), and cultured for 4 days in the presence of puromycin (2.5 μg/ml) for selecting the transduced cells.
Mice
Blk−/− mice were kindly provided by Dr. Alexander Tarakhovsky (Rockefeller University). Cdkn1b−/−, C57BL/6J-CD45.1, C57BL/6J-CD45.2 mice were obtained from The Jackson Laboratory. All mice were in C57BL/6J background
Cell culture
K562 cells were obtained from ATCC, and maintained in RPMI 1640 plus 10% fetal bovine serum (FBS). 293T cells were cultured in DMEM median plus 10% FBS.
Generation of retrovirus and lentivirus Stocks
The retroviral constructs MSCV-IRES-GFP, MSCV-BCR-ABL-IRES-GFP, MSCV-Blk-IRES-GFP, MSCV-BCR-ABL-IRES-Blk-IRES-GFP, and MSCV-BCR-ABL-IRES-Pax5-IRES-GFP were used to generate high-titer, helper-free, replication-defective ecotropic virus stock by transient transfection of 293T cells as previously described 58. In the retroviral constructs MSCV-IRES-hCD4 and MSCV-Blk-IRES-hCD4, human CD4 (hCD4) lacking the cytoplastic domain can be expressed as a cell surface marker. Lentiviral vector (pLenti-Puro) was a kind gift from Dr. Eric Campaus (University of Massachusetts Medical School). Lentiviral particles were produced by cotransfection of 293T cells with pLP1, pLP2, VSV-G and empty vector or pLenti-hBlk-Puro. Lentiviral shRNA vector pLKO.1 were from OpenBiosystems. The targeted BCR-ABL sequences are as follows: sense 5′-CTGACCAACTCGTGTGTGAAA-3′, antisense 5′-TTTCACACACGAGTTGGTCAG-3′.
Bone marrow transduction/transplantation
Eight to twelve week-old C57BL/6 mice were used for bone marrow transduction/transplantation. Retroviral transduction and transplantation of mouse bone marrow cells for inducing CML by BCR-ABL had been described previously 48,58,59.
Flow cytometry analysis
For stem cell analysis, bone marrow cells were suspended in staining medium (Hank’s Balanced Salt Solution (HBSS) with 2% heat-inactivated calf serum), and incubated with biotin-labeled lineage antibody cocktail containing a mixture of antibodies against CD3, CD4, CD8, B220, Gr-1, Mac-1 and Ter119. After washing, the fluorochrome-labeled secondary antibody (APC-Cy7-conjugated Streptavidin) for recognizing biotin and PE-conjugated c-Kit and APC-conjugated Sca-1 antibodies were added to the cells. Long-term and short-term LSCs were distinguished by the CD34 antibody. LSCs were analyzed by FACS. All these antibodies were purchased from eBioscience.
Leukemia stem cell culture
For mouse leukemia stem cell culture, bone marrow cells isolated from CML mice were cultured in vitro in the presence of Stemspan SFEM, SCF, IGF-2, TPO, heparin, and α-FGF as reported previously for culturing hematopoietic stem cells 59.
In Vitro methylcellulose colony formation assay
Human CML cells or BCR-ABL transformed human cord blood CD34+ cells were transduced with vector or Blk lentivirus and cultured in methylcellulose medium containing 2.5 μg/mL puromycin for selection (Methocult GF H4435; Stem Cell Technologies). Colonies were counted under microscope after 7 days.
Chromatin immunoprecipitation
ChIP assays were performed. Briefly, 3×107 ENU or BaF3 cells were incubated with 1% formaldehyde for 10 min at room temperature before crosslinking was quenched by addition of 0.125 M glycine. Cells were collected by centrifugation and lysed in lysis buffer containing 50mM Tris-HCl (pH 8.0), 10 mM EDTA, 0.5% SDS, proteinase inhibitors and phosphatase inhibitors. The cells suspension was sonicated seven times for 10s each with 2-min intervals on ice using a Misonix Sonicator 3000 at output 8. Sonicated chromatin was then incubated at 4°C overnight with 5 μg of the appropriate antibody: α-c-Myc (Santa Cruz), α-EBF1 (Avaon). Immunoprecipitated DNA was amplified by real-time PCR using the primers described in Supplementary Table 3.
Luciferase reporter assays
2×105 NIH3T3 cells were seeded in six-well plates 24h before transfection. 1 μg of the Pax5 promoter luciferase reporter plasmid (pGL3-Pax5), kindly provided by Dr. Kathryn Calame (Columbia University), was cotransfected with various amounts of EBF1, IRF8, c-Myc, or control vectors, into NIH3T3 cells by the calcium phosphate method. Cells were harvested at 48h after transfection and luciferase activity was determined using the Dual-Lucifease Reporter Assay system (Promega).
DNA microarray and data analysis
Bone marrow cells were isolated from CML mice at 14 days after the induction of the disease. BCR-ABL-expressing or BCR-ABL-Blk-expressing (transduced with the BCR-ABL-GFP or BCR-ABL-Blk-GFP, respectively) GFP+Lin−c-Kit+Sca-1+ cells (representing LSCs) were stored by FACS directly into RNAlater (Ambion) and homogenized in RLT Buffer (RNeasy Micro Kit) (Qiagen). Total RNA was isolated by following the protocol for the RNeasy Micro Kit. RNA was amplified, labeled, and approximately 2.0μg of fragmented and biotin-labeled cDNA was then hybridized onto Mouse Genome 430 2.0 microarray (Affymetrix). Relative fold change (RFC) of a gene between BCR-ABL-expressing and BCR-ABL/Blk-expressing LSCs was calculated using the formula: RFC = (sign(D) + (D == 0)) * 2abs(D). The log2 of the RFC value for the gene was shown. The detailed analysis of the microarray experiment and data were described in the Supplemental Information and the microarray data were deposited into GEO database (GSE36096). The expression of the Blk gene in human CML cells, quiescent and dividing CD34+ CML cells, and imatinib-treated CML stem cells were analyzed independently in publicly available microarray data sets including GSE4170 for human CML bulk CD34+ cells, GSE GSE24739 for quiescent (G0) and dividing (G1) CML stem cells, and GSE20876 for CD34+CD38− human CML stem cells with/without imatinib treatment. The probe-level raw intensity data were normalized and summarized into probes-set level data using Probe Logarithmic Intensity Error (PLIER) method. The Blk gene expression were extracted and further re-normalized at probe-set level by a set of suitable reference genes, the significance of changes between relevant groups was assessed by t-test.
Immunoprecipitation, western blotting and antibodies
Protein lysates were prepared by lysing cells in RIPA buffer containing 25mM TrisHCl, 150mM NaCl, 1%NP-40, 1% sodium deoxycholate, 0.1% SDS. Blk was immunoprecipitated with anti-Blk antibody and blotted with anti-phospho-tyrosine (p-Tyr) antibody. Antibodies against c-Abl, Blk, p-Tyr, Pax5, p27, p21, CDK2, Skp2, c-Myc and β-actin were purchased from Santa Cruz Biotechnology.
Real time-PCR
Total RNA was isolated from GFP+LSK bone marrow cells from mice using the RNeasy Mini kit (Qiagen). cDNA was synthesized using the Ovation-Pico cDNA synthesis method. All real time PCR reactions were done using the Applied Biosystems 7500. 25μL reaction system was composed of 12.5μL SYBR Green, 2.5μL 20uM primer mixture, 10ng cDNA and nuclease-free water. All experiments were performed in triplicate. β-actin was the internal control. For specific primer sequences, see the Supplementary Table 3.
Statistical analysis
Results are given as mean ± s.e.m. Statistical analysis was performed by Students’s t test for all column statistics. For survival curves, p values were obtained using a Log-rank test.
Supplementary Material
Acknowledgments
We thank Dr. Alexander Tarakhovsky at Rockefeller University for providing us with the Blk−/−mouse, and Dr. Kathryn Calame at Columbia University for the pGL3-Pax5 plasmid. We thank Sara Deibler for editorial assistance. This work was supported by the grants from the Leukemia & Lymphoma Society and the National Institute of Health (R01-CA122142, R01-CA114199) to S.L. M.A.B. was supported by National Institutes of Health Grant AI46629. S.L. is a Scholar of the Leukemia & Lymphoma Society.
Footnotes
Accession codes NCBI GEO: DNA microarray results have been deposited under accession code GSE36096.
AUTHOR CONTRIBUTIONS
Contribution: H.Z designed and performed experiments, analyzed data and wrote the paper; C.P., Y.H., H.L., Y.C., C.S., Z. S.; J.C., L.H., A.H., P.M., M.B. helped with experiments; X.Z., D.L. helped to analyze microarray data; M.R.G. helped to design experiments and write the paper; S.L. designed experiments, analyzed the data and wrote the paper.
Authors have no competing financial interests.
References
- 1.Huntly BJ, Gilliland DG. Leukaemia stem cells and the evolution of cancer-stem-cell research. Nat Rev Cancer. 2005;5:311–21. doi: 10.1038/nrc1592. [DOI] [PubMed] [Google Scholar]
- 2.Reya T, Morrison SJ, Clarke MF, Weissman IL. Stem cells, cancer, and cancer stem cells. Nature. 2001;414:105–11. doi: 10.1038/35102167. [DOI] [PubMed] [Google Scholar]
- 3.Visvader JE, Lindeman GJ. Cancer stem cells in solid tumours: accumulating evidence and unresolved questions. Nat Rev Cancer. 2008;8:755–68. doi: 10.1038/nrc2499. [DOI] [PubMed] [Google Scholar]
- 4.Wang JC, Dick JE. Cancer stem cells: lessons from leukemia. Trends Cell Biol. 2005;15:494–501. doi: 10.1016/j.tcb.2005.07.004. [DOI] [PubMed] [Google Scholar]
- 5.Chen Y, Hu Y, Zhang H, Peng C, Li S. Loss of the Alox5 gene impairs leukemia stem cells and prevents chronic myeloid leukemia. Nat Genet. 2009;41:783–92. doi: 10.1038/ng.389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hu Y, et al. Targeting multiple kinase pathways in leukemic progenitors and stem cells is essential for improved treatment of Ph+ leukemia in mice. Proc Natl Acad Sci U S A. 2006;103:16870–5. doi: 10.1073/pnas.0606509103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Melo JV, Barnes DJ. Chronic myeloid leukaemia as a model of disease evolution in human cancer. Nat Rev Cancer. 2007;7:441–53. doi: 10.1038/nrc2147. [DOI] [PubMed] [Google Scholar]
- 8.Ren R. Mechanisms of BCR-ABL in the pathogenesis of chronic myelogenous leukaemia. Nat Rev Cancer. 2005;5:172–83. doi: 10.1038/nrc1567. [DOI] [PubMed] [Google Scholar]
- 9.Wong S, Witte ON. The BCR-ABL story: bench to bedside and back. Annu Rev Immunol. 2004;22:247–306. doi: 10.1146/annurev.immunol.22.012703.104753. [DOI] [PubMed] [Google Scholar]
- 10.Druker BJ, et al. Efficacy and safety of a specific inhibitor of the BCR-ABL tyrosine kinase in chronic myeloid leukemia. N Engl J Med. 2001;344:1031–7. doi: 10.1056/NEJM200104053441401. [DOI] [PubMed] [Google Scholar]
- 11.Kantarjian H, et al. Hematologic and cytogenetic responses to imatinib mesylate in chronic myelogenous leukemia. N Engl J Med. 2002;346:645–52. doi: 10.1056/NEJMoa011573. [DOI] [PubMed] [Google Scholar]
- 12.Talpaz M, et al. Dasatinib in imatinib-resistant Philadelphia chromosome-positive leukemias. N Engl J Med. 2006;354:2531–41. doi: 10.1056/NEJMoa055229. [DOI] [PubMed] [Google Scholar]
- 13.Graham SM, et al. Primitive, quiescent, Philadelphia-positive stem cells from patients with chronic myeloid leukemia are insensitive to STI571 in vitro. Blood. 2002;99:319–25. doi: 10.1182/blood.v99.1.319. [DOI] [PubMed] [Google Scholar]
- 14.Corbin AS, et al. Human chronic myeloid leukemia stem cells are insensitive to imatinib despite inhibition of BCR-ABL activity. J Clin Invest. 2011;121:396–409. doi: 10.1172/JCI35721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lessard J, Sauvageau G. Bmi-1 determines the proliferative capacity of normal and leukaemic stem cells. Nature. 2003;423:255–60. doi: 10.1038/nature01572. [DOI] [PubMed] [Google Scholar]
- 16.Park IK, et al. Bmi-1 is required for maintenance of adult self-renewing haematopoietic stem cells. Nature. 2003;423:302–5. doi: 10.1038/nature01587. [DOI] [PubMed] [Google Scholar]
- 17.Saijo K, et al. Essential role of Src-family protein tyrosine kinases in NF-kappaB activation during B cell development. Nat Immunol. 2003;4:274–9. doi: 10.1038/ni893. [DOI] [PubMed] [Google Scholar]
- 18.Zwollo P, Desiderio S. Specific recognition of the blk promoter by the B-lymphoid transcription factor B-cell-specific activator protein. J Biol Chem. 1994;269:15310–7. [PubMed] [Google Scholar]
- 19.Nutt SL, Heavey B, Rolink AG, Busslinger M. Commitment to the B-lymphoid lineage depends on the transcription factor Pax5. Nature. 1999;401:556–62. doi: 10.1038/44076. [DOI] [PubMed] [Google Scholar]
- 20.Xie S, Lin H, Sun T, Arlinghaus RB. Jak2 is involved in c-Myc induction by Bcr-Abl. Oncogene. 2002;21:7137–46. doi: 10.1038/sj.onc.1205942. [DOI] [PubMed] [Google Scholar]
- 21.Sawyers CL, Callahan W, Witte ON. Dominant negative MYC blocks transformation by ABL oncogenes. Cell. 1992;70:901–10. doi: 10.1016/0092-8674(92)90241-4. [DOI] [PubMed] [Google Scholar]
- 22.Notari M, et al. A MAPK/HNRPK pathway controls BCR/ABL oncogenic potential by regulating MYC mRNA translation. Blood. 2006;107:2507–16. doi: 10.1182/blood-2005-09-3732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.O’Riordan M, Grosschedl R. Coordinate regulation of B cell differentiation by the transcription factors EBF and E2A. Immunity. 1999;11:21–31. doi: 10.1016/s1074-7613(00)80078-3. [DOI] [PubMed] [Google Scholar]
- 24.Lin H, Grosschedl R. Failure of B-cell differentiation in mice lacking the transcription factor EBF. Nature. 1995;376:263–7. doi: 10.1038/376263a0. [DOI] [PubMed] [Google Scholar]
- 25.Wang H, et al. IRF8 regulates B-cell lineage specification, commitment, and differentiation. Blood. 2008;112:4028–38. doi: 10.1182/blood-2008-01-129049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang H, Morse HC., 3rd IRF8 regulates myeloid and B lymphoid lineage diversification. Immunol Res. 2009;43:109–17. doi: 10.1007/s12026-008-8055-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xia ZB, et al. The MLL fusion gene, MLL-AF4, regulates cyclin-dependent kinase inhibitor CDKN1B (p27kip1) expression. Proc Natl Acad Sci U S A. 2005;102:14028–33. doi: 10.1073/pnas.0506464102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Andreu EJ, et al. BCR-ABL induces the expression of Skp2 through the PI3K pathway to promote p27Kip1 degradation and proliferation of chronic myelogenous leukemia cells. Cancer Res. 2005;65:3264–72. doi: 10.1158/0008-5472.CAN-04-1357. [DOI] [PubMed] [Google Scholar]
- 29.Chu I, et al. p27 phosphorylation by Src regulates inhibition of cyclin E-Cdk2. Cell. 2007;128:281–94. doi: 10.1016/j.cell.2006.11.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Grimmler M, et al. Cdk-inhibitory activity and stability of p27Kip1 are directly regulated by oncogenic tyrosine kinases. Cell. 2007;128:269–80. doi: 10.1016/j.cell.2006.11.047. [DOI] [PubMed] [Google Scholar]
- 31.Jonuleit T, et al. Bcr-Abl kinase down-regulates cyclin-dependent kinase inhibitor p27 in human and murine cell lines. Blood. 2000;96:1933–9. [PubMed] [Google Scholar]
- 32.Rangatia J, Bonnet D. Transient or long-term silencing of BCR-ABL alone induces cell cycle and proliferation arrest, apoptosis and differentiation. Leukemia. 2006;20:68–76. doi: 10.1038/sj.leu.2403999. [DOI] [PubMed] [Google Scholar]
- 33.Oda H, Kumar S, Howley PM. Regulation of the Src family tyrosine kinase Blk through E6AP-mediated ubiquitination. Proc Natl Acad Sci U S A. 1999;96:9557–62. doi: 10.1073/pnas.96.17.9557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Frescas D, Pagano M. Deregulated proteolysis by the F-box proteins SKP2 and beta-TrCP: tipping the scales of cancer. Nat Rev Cancer. 2008;8:438–49. doi: 10.1038/nrc2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Agarwal A, et al. Absence of SKP2 expression attenuates BCR-ABL-induced myeloproliferative disease. Blood. 2008;112:1960–70. doi: 10.1182/blood-2007-09-113860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Radich JP, et al. Gene expression changes associated with progression and response in chronic myeloid leukemia. Proc Natl Acad Sci U S A. 2006;103:2794–9. doi: 10.1073/pnas.0510423103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang B, et al. Effective targeting of quiescent chronic myelogenous leukemia stem cells by histone deacetylase inhibitors in combination with imatinib mesylate. Cancer Cell. 2010;17:427–42. doi: 10.1016/j.ccr.2010.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Affer M, et al. Gene Expression Differences between Enriched Normal and Chronic Myelogenous Leukemia Quiescent Stem/Progenitor Cells and Correlations with Biological Abnormalities. J Oncol. 2011;2011:798592. doi: 10.1155/2011/798592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nordon RE, Ginsberg SS, Eaves CJ. High-resolution cell division tracking demonstrates the FLt3-ligand-dependence of human marrow CD34+CD38- cell production in vitro. Br J Haematol. 1997;98:528–39. doi: 10.1046/j.1365-2141.1997.2823097.x. [DOI] [PubMed] [Google Scholar]
- 40.Heaney NB, et al. Bortezomib induces apoptosis in primitive chronic myeloid leukemia cells including LTC-IC and NOD/SCID repopulating cells. Blood. 2010;115:2241–50. doi: 10.1182/blood-2008-06-164582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bhatia R, et al. Persistence of malignant hematopoietic progenitors in chronic myelogenous leukemia patients in complete cytogenetic remission following imatinib mesylate treatment. Blood. 2003;101:4701–7. doi: 10.1182/blood-2002-09-2780. [DOI] [PubMed] [Google Scholar]
- 42.Dierks C, et al. Expansion of Bcr-Abl-positive leukemic stem cells is dependent on Hedgehog pathway activation. Cancer Cell. 2008;14:238–49. doi: 10.1016/j.ccr.2008.08.003. [DOI] [PubMed] [Google Scholar]
- 43.Kirstetter P, Anderson K, Porse BT, Jacobsen SE, Nerlov C. Activation of the canonical Wnt pathway leads to loss of hematopoietic stem cell repopulation and multilineage differentiation block. Nat Immunol. 2006;7:1048–56. doi: 10.1038/ni1381. [DOI] [PubMed] [Google Scholar]
- 44.Scheller M, et al. Hematopoietic stem cell and multilineage defects generated by constitutive beta-catenin activation. Nat Immunol. 2006;7:1037–47. doi: 10.1038/ni1387. [DOI] [PubMed] [Google Scholar]
- 45.Zhao C, et al. Loss of beta-catenin impairs the renewal of normal and CML stem cells in vivo. Cancer Cell. 2007;12:528–41. doi: 10.1016/j.ccr.2007.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhao C, et al. Hedgehog signalling is essential for maintenance of cancer stem cells in myeloid leukaemia. Nature. 2009;458:776–9. doi: 10.1038/nature07737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hu Y, Chen Y, Douglas L, Li S. beta-Catenin is essential for survival of leukemic stem cells insensitive to kinase inhibition in mice with BCR-ABL-induced chronic myeloid leukemia. Leukemia. 2009;23:109–16. doi: 10.1038/leu.2008.262. [DOI] [PubMed] [Google Scholar]
- 48.Hu Y, et al. Requirement of Src kinases Lyn, Hck and Fgr for BCR-ABL1-induced B-lymphoblastic leukemia but not chronic myeloid leukemia. Nat Genet. 2004;36:453–61. doi: 10.1038/ng1343. [DOI] [PubMed] [Google Scholar]
- 49.Warmuth M, et al. The Src family kinase Hck interacts with Bcr-Abl by a kinase-independent mechanism and phosphorylates the Grb2-binding site of Bcr. J Biol Chem. 1997;272:33260–70. doi: 10.1074/jbc.272.52.33260. [DOI] [PubMed] [Google Scholar]
- 50.Wu J, et al. Lyn regulates BCR-ABL and Gab2 tyrosine phosphorylation and c-Cbl protein stability in imatinib-resistant chronic myelogenous leukemia cells. Blood. 2008;111:3821–9. doi: 10.1182/blood-2007-08-109330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Xiao W, Hong H, Kawakami Y, Lowell CA, Kawakami T. Regulation of myeloproliferation and M2 macrophage programming in mice by Lyn/Hck, SHIP, and Stat5. J Clin Invest. 2008;118:924–34. doi: 10.1172/JCI34013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cobaleda C, Schebesta A, Delogu A, Busslinger M. Pax5: the guardian of B cell identity and function. Nat Immunol. 2007;8:463–70. doi: 10.1038/ni1454. [DOI] [PubMed] [Google Scholar]
- 53.Mullighan CG, et al. Genome-wide analysis of genetic alterations in acute lymphoblastic leukaemia. Nature. 2007;446:758–64. doi: 10.1038/nature05690. [DOI] [PubMed] [Google Scholar]
- 54.Mullighan CG, et al. Deletion of IKZF1 and prognosis in acute lymphoblastic leukemia. N Engl J Med. 2009;360:470–80. doi: 10.1056/NEJMoa0808253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cheng T, Rodrigues N, Dombkowski D, Stier S, Scadden DT. Stem cell repopulation efficiency but not pool size is governed by p27(kip1) Nat Med. 2000;6:1235–40. doi: 10.1038/81335. [DOI] [PubMed] [Google Scholar]
- 56.Ramaraj P, et al. Effect of mutational inactivation of tyrosine kinase activity on BCR/ABL-induced abnormalities in cell growth and adhesion in human hematopoietic progenitors. Cancer Res. 2004;64:5322–31. doi: 10.1158/0008-5472.CAN-03-3656. [DOI] [PubMed] [Google Scholar]
- 57.Copland M, et al. BMS-214662 potently induces apoptosis of chronic myeloid leukemia stem and progenitor cells and synergizes with tyrosine kinase inhibitors. Blood. 2008;111:2843–53. doi: 10.1182/blood-2007-09-112573. [DOI] [PubMed] [Google Scholar]
- 58.Li S, Ilaria RL, Jr, Million RP, Daley GQ, Van Etten RA. The P190, P210, and P230 forms of the BCR/ABL oncogene induce a similar chronic myeloid leukemia-like syndrome in mice but have different lymphoid leukemogenic activity. J Exp Med. 1999;189:1399–412. doi: 10.1084/jem.189.9.1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Peng C, et al. Inhibition of heat shock protein 90 prolongs survival of mice with BCR-ABL-T315I-induced leukemia and suppresses leukemic stem cells. Blood. 2007;110:678–85. doi: 10.1182/blood-2006-10-054098. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.