Abstract
Maturation in herpesviruses initiates in the nucleus of the infected cell with encapsidation of viral DNA to form nucleocapsids and concludes with envelopment in the cytoplasm to form infectious virions that egress the cell. The entire process of virus maturation is orchestrated by protein-protein interactions and enzymatic activities of viral and host origin. Viral tegument proteins play important roles in maintaining the structural stability of capsids and directing the acquisition of virus envelope. Envelopment occurs at modified host membranes and exploits host vesicular trafficking. In this review, we summarize the current knowledge and concepts in human cytomegalovirus (HCMV) maturation and their parallels in other herpesviruses with an emphasis on viral and host factors regulating this process.
Keywords: Betaherpesviruses, replication, nuclear egress, nucleocapsids, envelope
Overview of cytomegalovirus maturation
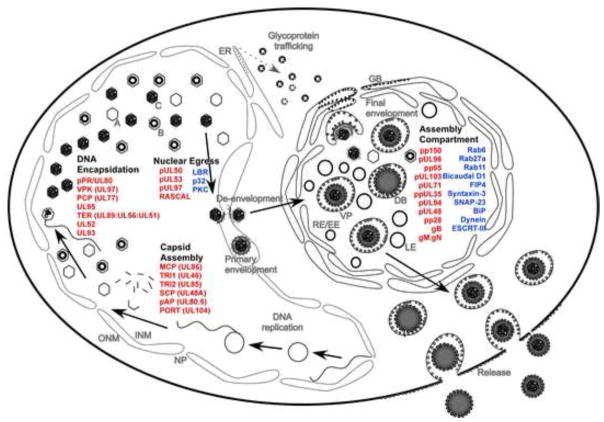
Human cytomegalovirus (HCMV) is a large double stranded DNA virus that belongs to the family herpesviridae [1]. HCMV causes medically significant disease in immunologically compromised individuals such as AIDS patients and organ transplant recipients [2]. HCMV is also the major cause of congenital infections that can lead to developmental abnormalities and fetal death [2]. Similar to other herpesviruses, the replication cycle of HCMV involves nuclear and cytoplasmic stages. Viral capsid assembly, viral DNA synthesis, encapsidation, and initial tegumentation all occur within the host cell nucleus, relying heavily on evolutionarily conserved viral proteins (Figure 1) [2, 3]. Viral DNA encapsidation leads to the formation of nucleocapsids that translocate from the nucleus to the cytoplasm via a nuclear egress complex (NEC), which overcomes the barrier posed by the nuclear lamina and facilitates initial or primary envelopment of nucleocapsids at the inner nuclear membrane followed by de-envelopment at the outer nuclear membrane [3, 4]. In the cytoplasm, a specialized compartment, termed cytoplasmic virus assembly compartment (cVAC or AC) accepts the translocated nucleocapsids, and is the site where remaining tegument as well as a viral envelope are acquired (Figure 1), a process that is generally referred to as secondary (or final) envelopment. The AC is composed of modified and rearranged host organelles that support the final steps in maturation and egress from cells (Figure 1) [5–8]. This review focuses on interplay of major viral (Table 1) and host (Box 1) factors that coordinate the process of HCMV assembly, maturation and egress.
Figure 1.
Summary of HCMV maturation. This model depicts nuclear and cytoplasmic events during HCMV maturation as well as major players contributing to this process. Viral proteins are depicted in red and host proteins are depicted in blue. Abbreviations: DB, dense body; VP, virus particle; EE/RE, early endosome/recycling endosome; LE, late endosome; GB, Golgi body; ER, endosplasmic reticulum; NP, nuclear pore; INM, inner nuclear membrane; ONM, outer nuclear membrane; A, B and C, types of nuclear capsids.
Table 1.
Major viral factors in HCMV maturation
| Common name | HCMV | HSV | Kaposi’fs sarcoma herpesvirus (KSHV) | Conservation in herpesviruses | Key functions |
|---|---|---|---|---|---|
| Major capsid protein (MCP) | UL86 | UL19 | ORF25 | α, β, γ | Capsid assembly [3, 10] |
| Triplex monomer (TR1) | UL46 | UL38 | ORF62 | α, β, γ | Capsid assembly [3, 10] |
| Triplex dimer (TR2) | UL85 | UL18 | ORF26 | α, β, γ | Capsid assembly [3, 10] |
| Small capsid protein (SCP) | UL48A | UL35 | ORF65 | α, β, γ | Capsid assembly [3, 10] |
| Portal protein (PORT) | UL104 | UL6 | ORF43 | α, β, γ | Viral DNA packaging [11] |
| (Putative) capsid vertex specific component-1 (CVSC-1) | UL77 | UL25 | ORF19 | α, β, γ | Stabilize capsid DNA packaging [17] |
| (Putative) capsid vertex specific component-2 (CVSC-2) | UL93 | UL17 | ORF32 | α, β, γ | Stabilize capsid DNA packaging [17] |
| Nuclear egress complex membrane anchoring component 1 (NEC1) | UL50 | UL34 | ORF66 | α, β, γ | Interacts with pUL53 at inner nuclear membrane to control capsid egress [34] |
| Nuclear egress complex component 2 (NEC2) | UL53 | UL31 | ORF69 | α, β, γ | Interacts with UL50 membrane protein and nuclear lamina [34] |
| Nuclear rim-associated cytomegalovirus protein (RASCAL) | c-ORF29 | CMV only | Component of nuclear egress complex [30] | ||
| Viral Protein Kinase (VPK) | UL97 | UL13 | ORF36 | α, β, γ | Regulates phosphorylation of multiple proteins [37] |
| Largest tegument protein (LTP) | UL48 | VP1–2, UL36 | ORF64 | α, β, γ | Release of viral DNA in the nucleus, secondary envelopment [3] |
| LTP binding protein (LTPbp) | UL47 | UL37 | ORF63 | α, β, γ | Secondary envelopment [100] |
| Lower matrix protein (LMP) | pp65, UL83 | CMV only | Major tegument component, dispensible for replication [24] | ||
| Basic phosphoprotein (BPP), Nucleocapsid proximal stabilization protein (NSP) | pp150, UL32 | β | Stabilize nucleocapsids during nuclear to cytoplasmic translocation [7, 8] | ||
| pUL96 | UL96 | β | Stabilize nucleocapsids during nuclear to cytoplasmic translocation [8] | ||
| Cytoplasmic egress tegument protein, Myristoylated tegument protein (CETP, MTP) | pp28, UL99 | UL11 | ORF38 | α, β, γ | Secondary envelopment [56, 101] |
| CETP binding protein (CETPbp) | UL94 | UL16 | ORF33 | α, β, γ | Secondary envelopment [50] |
| Cytoplasmic egress facilitator-1 | UL71 | UL51 | ORF55 | α, β, γ | Cytoplasmic egress [46, 94, 95] |
| Cytoplasmic egress facilitator-2 | UL88 | UL21 | ORF23 | β, γ | Putative cytoplasmic egress |
| Encapsidation and egress protein (EEP) | UL103 | UL7 | ORF42 | α, β, γ | Cytoplasmic egress [45] |
Box 1. Major host factors in HCMV maturation.
Nuclear lamina: is composed of lamins (A, B1, B2 and C) and associated proteins and poses as a major hurdle to the movement of nucleocapsids out of the nucleus.
p32, lamin B receptor (LBR), and protein kinase C (PKC): are part of the nuclear egress complex (NEC) that functions to dissolve nuclear lamina for nuclear egress of capsids.
BiP/GRP78 and dynein: interact with viral proteins to affect nuclear egress as well as the formation of the AC.
Cis-and trans-Golgi networks: are parts of the AC and possibly contribute to virus envelopment.
Endoplasmic reticulum (ER): is excluded from the AC but is essential for viral glycoprotein trafficking.
Endoplasmic reticulum Golgi intermediate compartments (ERGIC): possibly contributes to HCMV envelopment.
Endosomes: contribute to the formation of herpesvirus envelope.
Endosome sorting machinery required for transport (ESCRT): plays important role in the generation of multivesicular bodies (MVB) as well as in virus maturation by providing essential budding functions.
Rab GTPases: are the major determinant of vesicular trafficking in eukaryotic cells and impact viral maturation by affecting the trafficking of viral components to the AC.
Nuclear stage of HCMV maturation
The nuclear events in viral maturation start with the formation of a capsid and encapsidation of viral DNA, steps that are similar across herpesviruses and have significant similarity with HK97 class of DNA bacteriophages [9]. To exit the nucleus, herpesviruses have evolved a specific egress strategy controlled by evolutionarily conserved proteins. Thus, HCMV nuclear steps (Figure 1) can be projected on the backdrop of studies that have been carried out with other herpesviruses, particularly herpes simplex virus (HSV) [9]. Soon after infection, nucleocapsids track to the nucleus and release viral DNA into the nucleus where both RNA transcription and DNA synthesis take place in a tightly regulated manner, starting with the activation of the immediate early genes [2]. Later, following expression of delayed early proteins that include components of viral DNA synthesis machinery, progeny viral DNA accumulates in the nucleus as head-to-tail concatamers and is encapsidated in a directional, head-full (i.e. to the capacity) manner into procapsids through a modified penton that is composed of the portal protein (PORT, pUL104) [10–12]. HCMV DNA encapsidation is dependent on the activities of the terminase complex (pUL89–pUL56–pUL51) [12, 13] as well as pUL52 (Figure 1) [14]. Major events during nuclear stage of virus maturation include capsid assembly, DNA encapsidation, initial tegumentation and nuclear egress.
Capsid assembly and DNA encapsidation
HCMV capsids contain four core components: major capsid protein (MCP/pUL86), triplex monomer (TRI1/pUL46, also called minor capsid binding protein [mcBP]), triplex dimer (TRI2/pUL85, also called minor capsid protein [mCP]) and smallest capsid protein (SCP/pUL48A) [2, 3, 10, 15, 16]. Capsids assemble with the assistance of a pUL80-based scaffold, which translocates MCP into the nucleus and organizes the capsid shell while being cleaved by the viral maturational protease (pUL80a) [10]. Additional viral proteins associate with nuclear capsids as maturation proceeds, including the portal protein PORT and possibly the homologs of HSV capsid vertex specific components (CVSC; pUL25 and pUL17) in HCMV (pUL77 and pUL93, respectively) that are predicted to be required for stable DNA packaging [17]. The encapsidation machinery described above recognizes the cis-acting signals (pac1 and pac2) present at direct terminal repeats of HCMV genome and initiates packaging and direct genome cleavage [18]. The success or failure of DNA packaging determines the fate of capsids and results in three stable virus capsid types normally detected in the nucleus of herpesvirus-infected cells (Figure 2, Table 2) [10]: (i) A-capsids that appear empty, lacking either scaffold of immature capsids or packaged viral DNA of mature nucelocapsids, and are believed to be the result of abortive attempts to package virus DNA. (ii) B-capsids that contain scaffold but no viral DNA, and have been considered dead-end viral replication products in HSV studies [9, 19]. In HCMV, B-capsids may include precursors to nucleocapsids because they accumulate when viral DNA packaging is blocked [11]. (iii) C-capsids contain viral DNA without any scaffold and thus probably represent nucleocapsids in the process of maturation [20]. Spherical procapsid forms that have been visualized in alphaherpesvirus infections [21] have thus far eluded detection in HCMV. C-capsids enjoy a selective preference for nuclear egress during HCMV infection, as in other herpesviruses [22], however, in HCMV infections, B-capsids can also become enveloped to form noninfectious enveloped particles (NIEP) [23]. Soon after assembly, nucleocapsids proceed to the subsequent maturation steps within the nucleus.
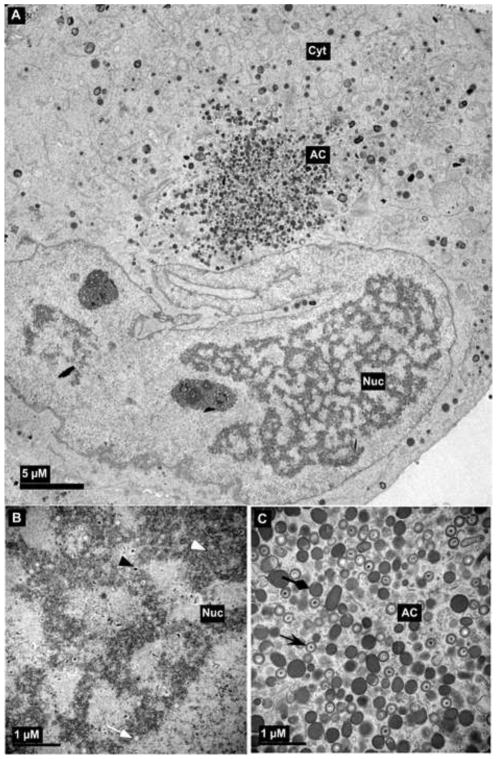
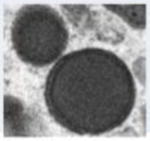
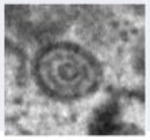
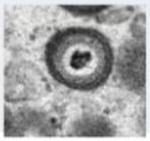
Figure 2.
Transmission electron micrograph of HCMV infected fibroblast at late stage of infection. The whole cell (a), nucleus (b) and cytoplasm (c) show several types of virus particles normally present during infection. Abbreviations: Nuc, nucleus; Cyt, cytoplasm; AC, assembly compartment. Symbols: white arrowheads, B-capsids; black arrowheads, C-capsids; white arrows, A-capsids; black arrows, mature virus particles; black diamondheads, dense bodies. Bars, 5 μm (a) and 1μm (b) and (c).
Table 2.
Types of virus particles produced during wild-type HCMV infection.
| Nuclear | Cytoplasmic | ||||
|---|---|---|---|---|---|
| A-capsids | B-capsids | C-capsids | Dense bodies | NIEP | Virion |
| Lack scaffold as well as viral DNA and are believed to result from abortive viral DNA encapsidation | Contain scaffold but lack viral DNA; likely to result from abortive capsid formation or DNA encapsidation | Contain viral DNA and lack scaffold; likely representing nucleocapsids in the process of maturation | Noninfectious capsidless particles and carry pp65 tegument protein as the main constituent | Noninfectious enveloped particles (NIEP) produced when B-capsids mature | Infectious virus particles produced when C-capsids mature, containing encapsidated viral DNA. |
|

|
|
|
|
|
Tegumentation
Although nuclear localization of several tegument proteins suggest that they may play roles in translocation of nucleocapsids, there is little direct evidence of tegumentation within the nucleus. Some proteins, such as pp65, localize to the nucleus early in infection independent of capsid localization, but are added to the tegument in the cytoplasmic AC later in infection [24]. There is a possibility that proteins acquired in the nucleus may become lost during translocation to the cytoplasm and therefore only transiently associate with nucleocapsids. Interestingly, the HCMV pp150 tegument protein appears to be associated with purified nuclear B-capsids [8]. Incorporation onto C-capsids likely starts in the nucleus and continues in the AC. A cryo-electron microscopy based structural study implicated pp150 binding to nucleocapsids based on very loose criteria [15]. Later, an in vitro binding assay was developed and used to demonstrate that pp150 binds selectively to capsids through its amino terminus, which contains two functionally important conserved regions (CR1 and CR2) [25]. Real-time analysis of movement of HCMV particles labeled by GFP-tagged pp150 confirmed earlier reports of pp150 acquisition in the nucleus [26]. This study demonstrated that fluorescent progeny particles appeared in the nucleus at 44 h after infection. Strict co-localization of pp150 and the MCP within nuclear inclusions indicated that incorporation of pp150 into nascent HCMV particles occurred simultaneously with or immediately after assembly of the capsid [26]. Thus, nuclear steps start the process of tegumentation, but the accumulation of pp150 and other major tegument proteins occurs in the AC and includes acquisition of a thick tegument layer on cytoplasmic capsids. This suggests that most of the final tegument is added to the nucleocapsid in the cytoplasm, immediately prior to final envelopment and egress from the cell.
Translocation from the nucleus to the cytoplasm
Considerable information has been generated on the NEC, which binds and dissolves the nuclear lamina during egress of virus particles from the nucleus to the cytoplasm [2, 4, 27]. In HCMV infections, the transmembrane-domain containing pUL50 anchors the NEC within the inner nuclear membrane and associates with other the other core NEC component, pUL53, as well as pUL97 (viral protein kinase [VPK]) and the recently identified RASCAL (nuclear rim-associated cytomegaloviral protein, the product of cORF-29) [28–31]. All herpesviruses rely on a pUL50–pUL53-like NEC to facilitate the nucleocapsid translocation process by interfacing with viral and cellular protein kinases that phosphorylate nuclear lamins and open the laminar network, thereby allowing egress of a ~130 nM particle to the cytoplasm (Figure 1) [3, 4, 10, 28–36]. Several host proteins including p32, lamin B receptor (LBR), and protein kinase C (PKC) associate with NEC [27, 34]. For HCMV infections, VPK activity, in parallel with host cell-cycle-regulated kinases determine events that follow viral DNA synthesis by regulating the pUL50–pUL53 complex as well as the reorganization of nuclear lamins A and C [29, 37]. It is possible that phosphorylation by VPK or PKC controls the dissociation of pUL53 from pUL50, an event thought to be associated with nuclear egress based on evaluation of herpes simplex virus (HSV) homologs [27]. Other host proteins that may influence nuclear egress and virus maturation in general include BiP/GRP78 and dynein, both of which seem to act via pUL50 [38]. Recent studies suggest that the NEC is also a quality control step where preference is given to DNA-containing C-capsids (nucleocapsids) over A-or B-capsids [22, 39]. An interaction of HSV pUL17–pUL25 complex (homologs of HCMV pUL93 and pUL77, respectively) with pUL31 (a component of the NEC in HSV and homolog of HCMV pUL53) selects for C particles during nuclear egress [39, 40]. The NEC component pUL50 interacts with Bip/GRP78, an endoplasmic reticulum (ER) chaperone protein that accumulates in abundance during HCMV infection [38], although the precise steps that are facilitated by this interaction remain unclear. Bip/GRP78 is also important for the formation and maintenance of the AC [32] and therefore its impact on nuclear egress may be indirect. Thus, the NEC, possibly via pUL50, may play a role in maintaining the continuity of virus maturation between nuclear and cytoplasmic compartments.
Cytoplasmic stage of HCMV maturation
An understanding of virion maturation in the cytoplasm has emerged over the past few years. The data based upon molecular markers and transmission electron microscopy suggests that endosomal membranes are responsible for envelopment [5, 41, 42] with the caveat that cellular organelles and membranes change in character during HCMV infection [6]. Trans-Golgi network (TGN) and ER-Golgi intermediate compartments (ERGIC) also contribute to HCMV envelopment [3, 42]. Similarly, viral proteins are predicted to employ several independent cellular pathways to accumulate into AC during later stages of viral morphogenesis [43, 44] suggesting an intricate interplay of virus and host factors during cytoplasmic maturation. Major events during this stage of HCMV maturation include formation of the AC, tegumentation, envelopment and egress.
Formation of the AC
HCMV is remarkable in the way it modulates host membranes for virus maturation [5, 6]. Importantly, although replication of all herpesviruses include nuclear and cytoplasmic maturation events, the AC is a unique feature of betaherpesvirus-infected cells [5]. Virus particles congregate in the AC during late phases of infection [8] consistent with its important role in controlling final tegumentation, envelopment and egress from cell. The AC is also the site where the appearance of virus particles has facilitated study of replication defective mutants [7, 8, 14, 45, 46]. Both VPK and host protein kinases, as well as host BiP/GRP78 and dynein contribute to the formation of AC [32]. Cellular organelles, including the cis-and trans-Golgi network, ER, and endosomes, become rearranged into concentric cylinders as infection proceeds [5, 6]. Also, many cellular markers of early, recycling-and late-endosomes, as well as the endosomal sorting complex required for transport (ESCRT) and several Rab GTPases localize to the AC (Figure 1) [5, 6, 43, 47]. In addition, viral tegument proteins pp150 (pUL32), pUL96, pp28 (pUL99), pp65 (pUL83), pUL103 and pUL94 as well as envelope constituents, including glycoprotein (g) gM:gN, gB, and gH:gL all accumulate in this same region [7, 8, 45, 48–50]. Host proteins also contribute to these steps, as exemplified by the requirement for Rab6 protein for proper localization of pp150 to the AC [43]. It appears that the movement of pp150-associated nucleocapsids destined for maturation is critically dependent on vesicle transport and targeting regulated via Rab6. Studies of real-time localization of HCMV and cellular proteins during the course of infection will shed light on the dynamics of AC formation and envelopment as suggested [6] once markers for key viral proteins are colocalized with the cellular markers.
Tegumentation
Tegument proteins are major players in cytoplasmic virus maturation steps that culminate in virus envelopment. The tegument in herpesviruses contributes to viral replication in at least three ways: (i) to package and provide efficient delivery of preformed proteins required for initiation of infection, (ii) to provide structural stability to nucleocapsids and (iii) to mediate the interactions necessary for envelopment and eventual release of virus particles. The HCMV tegument contains at least 32 proteins, most of which are phosphorylated. Ten tegument proteins are conserved across herpesviruses. Although traditionally considered amorphous, the HCMV nucleocapsid proximal inner tegument retains structure that has been dissected using cryo-electron microscopy [15, 16, 51]. A number of tegument proteins exhibit an exclusive cytoplasmic localization, accumulating in the AC during virus maturation (Figure 1) [3], in a pattern that determines function as well as acquisition by maturing virions at this site. Major components of tegument and their roles in virus maturation are described below:
pp28 is a myristoylated and phosphorylated tegument protein encoded by HCMV UL99 from a true late viral transcript [52] and is a counterpart of HSV UL11 [2]. UL99 mutant HCMV fail to replicate efficiently in cell culture and pp28, like the UL11 gene product in HSV [27], facilitates a late step in virus maturation within the AC where it colocalizes with pp150, pp65, gB, gH and gL late in infection [49]. The phosphorylation of pp28 is important for its stability and is affected by the UL26 gene product [53]. The myristoylation at amino acid 2 (glycine) is required for production of infectious virus [3], which along with an acidic cluster (amino acids 44 to 57) govern the trafficking of pp28 to the AC [54, 55]. Somewhat surprisingly, a recombinant virus expressing only the amino terminal one-third of pp28 (as little as 50 aa) is fully replication competent, and exhibits appropriate localization to the AC suggesting that the acidic cluster plays a context dependent role in protein interaction required for the envelopment [55]. Multimerization of pp28 is essential for the generation of enveloped virus and it occurs in AC mediated by the amino terminus of the protein (aa 1 to 43) [54, 56]. pp28 binds to UL94, a HSV UL16 homolog [50, 57]. This binding is reminiscent of UL11–UL16 binding in HSV and suggests that the functions of pp28 are conserved across herpesviruses. pp28 also associate with Bip/GRP78 and is thus expected to play a role in the formation of AC [32]. pp28 may also interact with ubiquitin and the proteins of the ESCRT pathway [44], although functional significance of these interactions is still unclear. Thus, pp28 is an important determinant of virus maturation and appears to help orchestrate virus envelopment, possibly by a process of oligomerization in the AC.
pp150 is the second most abundant constituent of HCMV tegument layer [20, 23, 51, 58] and has been recognized as a major cytomegalovirus structural antigen since the 1980s [59, 60]. UL32 antisense RNA was first used to implicate this protein in the late stage of virus maturation [61], revealing an essential role in virus replication that has been confirmed and extended using viral mutants [7, 62–64]. It was not until the advent of molecular genetic and complementation systems that this protein was subjected to a complete genetic analysis [7, 65]. pp150 localizes to the nucleus early in virus maturation [26, 66] and as maturation proceeds, this protein becomes predominant in the cytoplasm, associating with the AC where, of course, virions as well as capsidless dense bodies accumulate. Dense bodies are non-infectious particles that are released from HCMV infected cells with pp65 tegument protein as a major constituent [10, 23]. Functional studies suggested that pp150 is critical for cytoplasmic virus maturation supported by successful translocation of MCP and viral DNA within the AC of UL32 mutant virus-infected cells [65]. It was also established that both CR1 and CR2 are critical for maturation, whereas most of the carboxyl terminal region including the O-linked glycosylation sites are dispensable [65]. Further analysis of UL32 mutants revealed individual residues in the CR1 and CR2 regions that are critical for pp150 role in viral replication [7]. The location of these residues in the nucleocapsid-binding region of pp150 highlights the functional importance of pp150-capsid interaction in the stabilization of nucleocapsids during maturation. One unexpected consequence of these studies came from ultrastructural analysis, which revealed intact nucleocapsids within the nucleus but abnormal vesicle-like particles in the AC of UL32 mutant virus-infected cells, indicating disintegration of preformed nucleocapsids in the absence of pp150 function [7]. Based upon this data, it was concluded that pp150 provides stability to virus particles during their translocation from the nucleus to the cytoplasm [7]. A recent study confirmed pp150 to be a constituent of the net-like layer of icosahedrally-ordered capsid-bound tegument [51] thus supporting its proposed nucleocapsid-proximal capsid-stabilizing role during maturation. The close association of pp150 with nucleocapsids and the many cytoplasmic steps that are involved in maturation leaves open the possibility that this protein is also involved in later virus maturation steps. Further details about these functions could emerge from studies focused on additional UL32 mutants.
pUL96 is a betaherpesvirus specific tegument component of HCMV [8, 67] and the UL96 mutant HCMV fails to replicate efficiently in cell culture [63, 64]. pUL96 is expressed with early kinetics [8, 68], however functions late in virus maturation where it is implicated in preserving the integrity of pre-formed nucleocapsids during translocation from the nucleus to the cytoplasm [8]. pUL96 can be detected in extracellular virus particles but not in nuclear B-capsids, indicating that it is added to virus particles either immediately after encapsidation in the nucleus or later in the cytoplasm. pUL96 co-localizes with pp150 in the AC during late phases of infection and affects the accumulation of virions and dense bodies in the AC [8]. Based on the common phenotype and the AC properties of UL96, UL32 and dual UL32–UL96 mutant viruses, both pUL96 and pp150 appear to act in a concerted fashion within the maturation pathway [8]. Ultrastructural studies of cells infected with a conditional UL96 mutant provided evidence that pUL96 functions downstream of pp150 and may mediate its effects by influencing pp150 activity [8]. While the disruption of either protein resulted in unstable nucleocapsids during translocation from the nucleus to the cytoplasm, the pUL96 mutant had recognizable cytoplasmic virus particles whereas UL32 mutants had no recognizable virus particles [7, 8].
The herpesvirus-conserved largest tegument protein (LTP; pUL48; also referred to as the high molecular weight virion protein or HMWP) strongly associates with nucleocapsids [51], analogous to the situation in other herpesviruses. The HSV homolog (pUL36) has long been recognized as essential for delivery of viral genome into the infected cell nucleus during entry [69, 70] and has also been implicated in nucleocapsid and virion maturation [71]. LTP binds to LTP binding protein (LTPbp) [20, 72], but this interaction is not required for viral replication, at least in pseudorabies virus (PRV) [73]. A deubiquitinase activity is located at the N-terminal domain of HCMV LTP and is conserved across herpesviruses [74]. This property is not crucial for viral replication in HCMV [75] or PRV [76], however, it affects neuroinvasion of PRV in mice [77], leaving open a role for the HCMV deubiquitinase activity in pathogenesis. The HCMV LTP deubiquitinase activity is biased towards Lys63 linked modifications rather than Lys48 linked modifications and shows no activity towards ubiquitin-like modifications [78]. Therefore, it can be interpreted that this activity does not direct proteins for proteasomal degradation. Abolishing this activity by active site-specific mutations only slightly impedes viral replication and does not alter either protein composition or morphology of virus particles in the nucleus or cytoplasm [75]. The HSV homolog is reported to bind to microtubules [79] suggesting a role in virus trafficking. A temperature sensitive isolate of HSV (tsB7) that has lesions in UL36 [69], with Tyr1453 mutated to Ser responsible for most defects [80], accumulates mutant LTP in cytosolic clusters [81], suggesting a role of LTP also in cytoplasmic virus maturation. Structural analysis of HSV virions stripped of envelope and most of the outer tegument proteins shows that LTP and pUL37 are the components of the tufts and thin strands that extend from capsids and could serve as capsid-proximal organizing features of the tegument, as they have the potential to extend to all parts of the tegument [82]. LTP has also been detected as part of the disulfide linked complexes in HSV that include pUL37 and VP22 [83]. This, along with binding of LTP to HSV pUL25 [17], suggests important roles of LTP in connecting tegument to capsids [84]. Thus LTP is crucial for virus maturation and participates in events ranging from tegumentation to virus particle trafficking. Recent protein interactions studies in HCMV indicate that LTP has the potential of binding with a variety of viral proteins including pUL47, pUL45, pUL88, pUL69, pUL103, pUL50 and pUL132 [85]. Considering that these proteins function at different phases of viral replication, LTP can be predicted to influence several steps in viral replication.
pp65 and pp71 are the other major tegument proteins in HCMV [10, 67]. pp65 is dispensable for virus replication [24] whereas the major impact of pp71 is in viral gene expression where it modulates epigenetic gene regulation by interacting with the host protein DAXX [86]. In summary, HCMV tegumentation confers stability to nucleocapsids and provides a signal for final envelopment through protein-protein interactions that start at the surface of the capsid prior to encapsidation of viral DNA in the nucleus and continue throughout cytoplasmic maturation events.
Envelopment and egress
The envelope is a major determinant of infectivity in herpesviruses and it is derived from the host membranes that have been modified by insertion of viral glycoproteins. The ER, ERGIC and endosomes may all contribute to envelopment. In early 1990s, Tooze et al. [41] reported that HCMV particles utilize endosomal cisternae for the envelopment based on the detection of horseradish peroxidase label in the HCMV envelope. Recently, the role that endosomal compartments play in herpesvirus maturation has become better understood, both structurally and functionally [5, 6, 47]. After initial conflicting reports about the role of ESCRT proteins in herpesvirus maturation [87] it is has become quite clear that herpesviruses employ ESCRT proteins during virus maturation [47, 88, 89] although the precise steps that are facilitated by this machinery remain unresolved. HCMV has also been shown to utilize the SNARE protein syntaxin-3 for morphogenesis [90] supporting the idea that host vesicles contribute to HCMV envelopment.
Major viral factors governing HCMV envelopment are viral glycoproteins. The envelope contains three major glycoprotein complexes (gc): gcI (gB), gcII (gM–gN), and gcIII (gH gL) [3]. Of these, gcI and gcII are disulfide bond linked whereas gcIII is non-covalently associated. Other gc found in minor quantities in the envelope layer include gH–gL–gO and gH–gL–gUL128-gUL130-gUL131A [91]. The acidic motifs on HCMV gB cytoplasmic domain determine its localization to the AC by interacting with host protein PACS-1, a connector protein that is required for the trafficking of proteins containing such motifs from endosomes to the trans-Golgi network. [92]. gM carboxy-terminal cytoplasmic tail (gM-CT) interacts with FIP4, a Rab11-GTPase effector protein, and this interaction determines the trafficking of gM to AC [93]. Thus, trafficking of several viral glycoproteins to the site of virus envelopment is determined by interactions with host proteins.
Other than envelope glycoproteins, HCMV envelopment is also influenced by viral proteins pUL71, pUL103 and ppUL35. pUL71 is a late viral protein that affects viral egress by promoting efficient formation of enveloped virions [46, 94]. It accumulates in the AC during HCMV infection and a UL71 mutant virus is defective in properly organizing the AC [95]. UL71 mutant virus particles accumulate within the cytoplasm of infected cells and are localized to the periphery of large structures with properties of lysosomes [95] or multivesicular bodies [46]. Further investigation is needed to elucidate the mechanism of action of pUL71 in HCMV envelopment. UL103 is a herpesvirus core tegument protein that functions very late in infection and influences the exocytic release of virions and dense bodies [45] opening up the way to understand the role of viral gene products in this late maturation step. Viral DNA and proteins accumulate with normal kinetics in UL103 mutant virus infections, excluding any impact of pUL103 on early stages of virus infection [45]. Thus, pUL103 is the first viral protein identified to specifically affect egress of a herpesvirus [45], suggesting that herpesvirus egress is an active process. UL35 encodes an early 22-kDa phosphoprotein (ppUL35a) as well as a late phosphoprotein of 75 kDa (ppUL35) [96]. The 22-kDa protein localizes to the nucleus, whereas the 75-kDa protein localizes to the AC and is packaged into virus particles [96]. UL35 is essential for virus replication in cell culture at low multiplicity of infection and UL35 mutant virus produces fewer enveloped particles without dense bodies. However, the particles produced by wild-type and mutant viruses do not show significant ultrastructural differences suggesting that ppUL35 specifically affects virus egress [97]. A recent study suggests that ppUL35 (but not ppUL35a) can contribute to viral replication through the manipulation of host responses [98]. The details of these functions are only beginning to emerge.
Concluding remarks
A number of herpesvirus conserved proteins, including components of capsid, tegument and envelope are critical for HCMV maturation and indicate shared aspects of maturation with other herpesviruses. Exploitation of host components for maturation such as ESCRT and Rab proteins by different herpesviruses also stresses an envolutionary conserved pathway governing HCMV maturation. A detailed mechanistic understanding of host-protein interactions during maturation is lacking but is required for better illustration of herpesvirus maturation pathway as well as for interfering with viral replication for therapy purposes. On a different note, HCMV encodes proteins that are essential for efficient maturation but have no homologs in alpha or gamma-herpesviruses. Example of these proteins include pp150 and pUL96. In addition, host metabolism, respiration and protein synthesis are stimulated throughout HCMV infection unlike other herpesviruses, consistent with an intricate host-virus interaction unique to HCMV [3, 53, 99]. Moreover, the arrangement of modified host organelles in AC also appears to be unique for betaherpesviruses. All of these features point to the uniqueness of HCMV maturation pathway. Other herpesviruses must utilize parallel mechanism to mature through similar host cell environment. Modern technologies for fast and efficient manipulation of viral genome combined with advanced microscopy and real-time follow-up of infected cells are likely to yield answers to several important questions in HCMV maturation including (i) how host membranes are manipulated for envelopment? (ii) How virus particles hijack host vesicular trafficking? (iii) How the tegument is acquired and (iv) what components of tegument connect capsids to the envelope?
Acknowledgments
We would like to thank Philip Pellett, Bill Britt, Louise McCormick, Prashant Desai and Jenny Ahlqvist for critical reading of the manuscript. Research in the Mocarski lab is supported by PHS grants (RO1 AI020211 and AI030363).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Pellett PE, Roizman B. The Family Herpesviridae: A Brief Introduction. In: Knipe DM, Howley PM, editors. Fields Virology. 5. Lippincott Williams & Wilkins; Philadelphia: 2006. pp. 2479–2499. [Google Scholar]
- 2.Mocarski ES, Jr, Shenk T, Pass RF. Cytomegaloviruses. In: Knipe DM, Howley PM, editors. Fields Virology. 5. Lippincott Williams & Wilkins; Philadelphia: 2006. pp. 2701–2772. [Google Scholar]
- 3.Britt W. CMV maturation and egress. In: Arvin AM, et al., editors. Human Herpesviruses: Biology, Therapy and Immunoprophylaxis. Cambridge Press; 2007. pp. 311–323. [PubMed] [Google Scholar]
- 4.Muranyi W, et al. Cytomegalovirus recruitment of cellular kinases to dissolve the nuclear lamina. Science. 2002;297:854–857. doi: 10.1126/science.1071506. [DOI] [PubMed] [Google Scholar]
- 5.Das S, et al. Three-dimensional structure of the human cytomegalovirus cytoplasmic virion assembly complex includes a reoriented secretory apparatus. J Virol. 2007;81:11861–11869. doi: 10.1128/JVI.01077-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Das S, Pellett PE. Spatial relationships between markers for secretory and endosomal machinery in human cytomegalovirus-infected cells versus those in uninfected cells. J Virol. 2011;85:5864–5879. doi: 10.1128/JVI.00155-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tandon R, Mocarski ES. Control of cytoplasmic maturation events by cytomegalovirus tegument protein pp150. J Virol. 2008;82:9433–9444. doi: 10.1128/JVI.00533-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tandon R, Mocarski ES. Cytomegalovirus pUL96 Is Critical for the Stability of pp150-Associated Nucleocapsids. J Virol. 2011;85:7129–7141. doi: 10.1128/JVI.02549-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cardone G, et al. Procapsid assembly, maturation, nuclear exit: dynamic steps in the production of infectious herpesvirions. Advances in experimental medicine and biology. 2012;726:423–439. doi: 10.1007/978-1-4614-0980-9_19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gibson W. Structure and formation of the cytomegalovirus virion. Curr Top Microbiol Immunol. 2008;325:187–204. doi: 10.1007/978-3-540-77349-8_11. [DOI] [PubMed] [Google Scholar]
- 11.Dittmer A, et al. Interaction of the putative human cytomegalovirus portal protein pUL104 with the large terminase subunit pUL56 and its inhibition by benzimidazole-D-ribonucleosides. J Virol. 2005;79:14660–14667. doi: 10.1128/JVI.79.23.14660-14667.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mocarski ES. Betaherpesvirus genes and their functions. In: Arvin AM, Mocarski ES, Moore P, Whitley R, Yamanishi K, Campadelli-Fiume G, Roizman B, editors. Human Herpesviruses: Biology, Therapy and Immunoprophylaxis. Cambridge Press; Cambridge: 2007. pp. 202–228. [PubMed] [Google Scholar]
- 13.Goldner T, et al. The novel anticytomegalovirus compound AIC246 (Letermovir) inhibits human cytomegalovirus replication through a specific antiviral mechanism that involves the viral terminase. J Virol. 2011;85:10884–10893. doi: 10.1128/JVI.05265-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Borst EM, et al. The essential human cytomegalovirus gene UL52 is required for cleavage-packaging of the viral genome. J Virol. 2008;82:2065–2078. doi: 10.1128/JVI.01967-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen DH, et al. Three-dimensional visualization of tegument/capsid interactions in the intact human cytomegalovirus. Virology. 1999;260:10–16. doi: 10.1006/viro.1999.9791. [DOI] [PubMed] [Google Scholar]
- 16.Trus BL, et al. Capsid structure of simian cytomegalovirus from cryoelectron microscopy: evidence for tegument attachment sites. J Virol. 1999;73:2181–2192. doi: 10.1128/jvi.73.3.2181-2192.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Toropova K, et al. The herpes simplex virus 1 UL17 protein is the second constituent of the capsid vertex-specific component required for DNA packaging and retention. J Virol. 2011;85:7513–7522. doi: 10.1128/JVI.00837-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang JB, McVoy MA. A 128-base-pair sequence containing the pac1 and a presumed cryptic pac2 sequence includes cis elements sufficient to mediate efficient genome maturation of human cytomegalovirus. J Virol. 2011;85:4432–4439. doi: 10.1128/JVI.02307-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu ST, et al. Synaptic vesicle-like lipidome of human cytomegalovirus virions reveals a role for SNARE machinery in virion egress. Proc Natl Acad Sci U S A. 2011;108:12869–12874. doi: 10.1073/pnas.1109796108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gibson W. Structure and assembly of the virion. Intervirology. 1996;39:389–400. doi: 10.1159/000150509. [DOI] [PubMed] [Google Scholar]
- 21.Newcomb WW, et al. Assembly of the herpes simplex virus procapsid from purified components and identification of small complexes containing the major capsid and scaffolding proteins. J Virol. 1999;73:4239–4250. doi: 10.1128/jvi.73.5.4239-4250.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Klupp BG, et al. Nuclear envelope breakdown can substitute for primary envelopment-mediated nuclear egress of herpesviruses. J Virol. 2011;85:8285–8292. doi: 10.1128/JVI.00741-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Irmiere A, Gibson W. Isolation and characterization of a noninfectious virion-like particle released from cells infected with human strains of cytomegalovirus. Virology. 1983;130:118–133. doi: 10.1016/0042-6822(83)90122-8. [DOI] [PubMed] [Google Scholar]
- 24.Schmolke S, et al. The dominant phosphoprotein pp65 (UL83) of human cytomegalovirus is dispensable for growth in cell culture. J Virol. 1995;69:5959–5968. doi: 10.1128/jvi.69.10.5959-5968.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Baxter MK, Gibson W. Cytomegalovirus basic phosphoprotein (pUL32) binds to capsids in vitro through its amino one-third. J Virol. 2001;75:6865–6873. doi: 10.1128/JVI.75.15.6865-6873.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sampaio KL, et al. Human cytomegalovirus labeled with green fluorescent protein for live analysis of intracellular particle movements. J Virol. 2005;79:2754–2767. doi: 10.1128/JVI.79.5.2754-2767.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Johnson DC, Baines JD. Herpesviruses remodel host membranes for virus egress. Nature reviews Microbiology. 2011;9:382–394. doi: 10.1038/nrmicro2559. [DOI] [PubMed] [Google Scholar]
- 28.Milbradt J, et al. Cytomegaloviral proteins pUL50 and pUL53 are associated with the nuclear lamina and interact with cellular protein kinase C. J Gen Virol. 2007;88:2642–2650. doi: 10.1099/vir.0.82924-0. [DOI] [PubMed] [Google Scholar]
- 29.Milbradt J, et al. Novel mode of phosphorylation-triggered reorganization of the nuclear lamina during nuclear egress of human cytomegalovirus. J Biol Chem. 2010;285:13979–13989. doi: 10.1074/jbc.M109.063628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Miller MS, et al. RASCAL is a new human cytomegalovirus-encoded protein that localizes to the nuclear lamina and in cytoplasmic vesicles at late times postinfection. J Virol. 2010;84:6483–6496. doi: 10.1128/JVI.02462-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sam MD, et al. Biochemical, biophysical, and mutational analyses of subunit interactions of the human cytomegalovirus nuclear egress complex. J Virol. 2009;83:2996–3006. doi: 10.1128/JVI.02441-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Buchkovich NJ, et al. The endoplasmic reticulum chaperone BiP/GRP78 is important in the structure and function of the human cytomegalovirus assembly compartment. J Virol. 2009;83:11421–11428. doi: 10.1128/JVI.00762-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Marschall M, et al. Regulatory roles of protein kinases in cytomegalovirus replication. Advances in virus research. 2011;80:69–101. doi: 10.1016/B978-0-12-385987-7.00004-X. [DOI] [PubMed] [Google Scholar]
- 34.Milbradt J, et al. Cytomegaloviral proteins that associate with the nuclear lamina: components of a postulated nuclear egress complex. J Gen Virol. 2009;90:579–590. doi: 10.1099/vir.0.005231-0. [DOI] [PubMed] [Google Scholar]
- 35.Rupp B, et al. Random screening for dominant-negative mutants of the cytomegalovirus nuclear egress protein M50. J Virol. 2007;81:5508–5517. doi: 10.1128/JVI.02796-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sanchez V, Spector DH. Virology. CMV makes a timely exit. Science. 2002;297:778–779. doi: 10.1126/science.1075102. [DOI] [PubMed] [Google Scholar]
- 37.Prichard MN. Function of human cytomegalovirus UL97 kinase in viral infection and its inhibition by maribavir. Rev Med Virol. 2009;19:215–229. doi: 10.1002/rmv.615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Buchkovich NJ, et al. Role of the endoplasmic reticulum chaperone BiP, SUN domain proteins, and dynein in altering nuclear morphology during human cytomegalovirus infection. J Virol. 2010;84:7005–7017. doi: 10.1128/JVI.00719-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Leelawong M, et al. A Physical Link between the Pseudorabies Virus Capsid and the Nuclear Egress Complex. J Virol. 2011;85:11675–11684. doi: 10.1128/JVI.05614-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yang K, Baines JD. Selection of HSV capsids for envelopment involves interaction between capsid surface components pUL31, pUL17, and pUL25. Proc Natl Acad Sci U S A. 2011;108:14276–14281. doi: 10.1073/pnas.1108564108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tooze J, et al. Progeny vaccinia and human cytomegalovirus particles utilize early endosomal cisternae for their envelopes. European journal of cell biology. 1993;60:163–178. [PubMed] [Google Scholar]
- 42.Cepeda V, et al. Human cytomegalovirus final envelopment on membranes containing both trans-Golgi network and endosomal markers. Cell Microbiol. 2010;12:386–404. doi: 10.1111/j.1462-5822.2009.01405.x. [DOI] [PubMed] [Google Scholar]
- 43.Indran SV, Britt WJ. A role for the small GTPase Rab6 in assembly of human cytomegalovirus. J Virol. 2011;85:5213–5219. doi: 10.1128/JVI.02605-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Moorman NJ, et al. A targeted spatial-temporal proteomics approach implicates multiple cellular trafficking pathways in human cytomegalovirus virion maturation. Mol Cell Proteomics. 2010;9:851–860. doi: 10.1074/mcp.M900485-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ahlqvist J, Mocarski E. Cytomegalovirus UL103 controls virion and dense body egress. J Virol. 2011;85:5125–5135. doi: 10.1128/JVI.01682-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schauflinger M, et al. The tegument protein UL71 of human cytomegalovirus is involved in late envelopment and affects multivesicular bodies. J Virol. 2011;85:3821–3832. doi: 10.1128/JVI.01540-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tandon R, et al. Human cytomegalovirus exploits ESCRT machinery in the process of virion maturation. J Virol. 2009;83:10797–10807. doi: 10.1128/JVI.01093-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mach M, et al. Complex formation by glycoproteins M and N of human cytomegalovirus: structural and functional aspects. J Virol. 2005;79:2160–2170. doi: 10.1128/JVI.79.4.2160-2170.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sanchez V, et al. Accumulation of virion tegument and envelope proteins in a stable cytoplasmic compartment during human cytomegalovirus replication: characterization of a potential site of virus assembly. J Virol. 2000;74:975–986. doi: 10.1128/jvi.74.2.975-986.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Phillips SL, Bresnahan WA. The human cytomegalovirus tegument protein UL94 is essential for secondary envelopment of HCMV virions. J Virol. 2012;86:2523–2532. doi: 10.1128/JVI.06548-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yu X, et al. Biochemical and structural characterization of the capsid-bound tegument proteins of human cytomegalovirus. J Struct Biol. 2011;174:451–460. doi: 10.1016/j.jsb.2011.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Depto AS, Stenberg RM. Functional analysis of the true late human cytomegalovirus pp28 upstream promoter: cis-acting elements and viral trans-acting proteins necessary for promoter activation. J Virol. 1992;66:3241–3246. doi: 10.1128/jvi.66.5.3241-3246.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Munger J, et al. UL26-deficient human cytomegalovirus produces virions with hypophosphorylated pp28 tegument protein that is unstable within newly infected cells. J Virol. 2006;80:3541–3548. doi: 10.1128/JVI.80.7.3541-3548.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jones TR, Lee SW. An acidic cluster of human cytomegalovirus UL99 tegument protein is required for trafficking and function. J Virol. 2004;78:1488–1502. doi: 10.1128/JVI.78.3.1488-1502.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Seo JY, Britt WJ. Sequence requirements for localization of human cytomegalovirus tegument protein pp28 to the virus assembly compartment and for assembly of infectious virus. J Virol. 2006;80:5611–5626. doi: 10.1128/JVI.02630-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Seo JY, Britt WJ. Multimerization of tegument protein pp28 within the assembly compartment is required for cytoplasmic envelopment of human cytomegalovirus. J Virol. 2008;82:6272–6287. doi: 10.1128/JVI.02345-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Liu Y, et al. The tegument protein UL94 of human cytomegalovirus as a binding partner for tegument protein pp28 identified by intracellular imaging. Virology. 2009;388:68–77. doi: 10.1016/j.virol.2009.03.007. [DOI] [PubMed] [Google Scholar]
- 58.Gibson W. Structural and nonstructural proteins of strain Colburn cytomegalovirus. Virology. 1981;111:516–537. doi: 10.1016/0042-6822(81)90354-8. [DOI] [PubMed] [Google Scholar]
- 59.Jahn G, et al. The two major structural phosphoproteins (pp65 and pp150) of human cytomegalovirus and their antigenic properties. J Gen Virol. 1987;68 (Pt 5):1327–1337. doi: 10.1099/0022-1317-68-5-1327. [DOI] [PubMed] [Google Scholar]
- 60.Gibson W, Irmiere A. Selection of particles and proteins for use as human cytomegalovirus subunit vaccines. Birth defects original article series. 1984;20:305–324. [PubMed] [Google Scholar]
- 61.Meyer HH, et al. Human cytomegalovirus late-phase maturation is blocked by stably expressed UL32 antisense mRNA in astrocytoma cells. J Gen Virol. 1997;78 (Pt 10):2621–2631. doi: 10.1099/0022-1317-78-10-2621. [DOI] [PubMed] [Google Scholar]
- 62.Zipeto D, et al. Identification of a human cytomegalovirus mutant in the pp150 matrix phosphoprotein gene with a growth-defective phenotype. J Gen Virol. 1993;74 (Pt 8):1645–1648. doi: 10.1099/0022-1317-74-8-1645. [DOI] [PubMed] [Google Scholar]
- 63.Dunn W, et al. Functional profiling of a human cytomegalovirus genome. Proc Natl Acad Sci U S A. 2003;100:14223–14228. doi: 10.1073/pnas.2334032100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yu D, et al. Functional map of human cytomegalovirus AD169 defined by global mutational analysis. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:12396–12401. doi: 10.1073/pnas.1635160100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.AuCoin DP, et al. Betaherpesvirus-conserved cytomegalovirus tegument protein ppUL32 (pp150) controls cytoplasmic events during virion maturation. J Virol. 2006;80:8199–8210. doi: 10.1128/JVI.00457-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hensel G, et al. Nuclear localization of the human cytomegalovirus tegument protein pp150 (ppUL32) J Gen Virol. 1995;76 (Pt 7):1591–1601. doi: 10.1099/0022-1317-76-7-1591. [DOI] [PubMed] [Google Scholar]
- 67.Varnum SM, et al. Identification of proteins in human cytomegalovirus (HCMV) particles: the HCMV proteome. J Virol. 2004;78:10960–10966. doi: 10.1128/JVI.78.20.10960-10966.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wing BA, Huang ES. Analysis and mapping of a family of 3′-coterminal transcripts containing coding sequences for human cytomegalovirus open reading frames UL93 through UL99. J Virol. 1995;69:1521–1531. doi: 10.1128/jvi.69.3.1521-1531.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Batterson W, et al. Molecular genetics of herpes simplex virus. VIII. further characterization of a temperature-sensitive mutant defective in release of viral DNA and in other stages of the viral reproductive cycle. J Virol. 1983;45:397–407. doi: 10.1128/jvi.45.1.397-407.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Jovasevic V, et al. Proteolytic cleavage of VP1–2 is required for release of herpes simplex virus 1 DNA into the nucleus. J Virol. 2008;82:3311–3319. doi: 10.1128/JVI.01919-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Desai PJ. A null mutation in the UL36 gene of herpes simplex virus type 1 results in accumulation of unenveloped DNA-filled capsids in the cytoplasm of infected cells. J Virol. 2000;74:11608–11618. doi: 10.1128/jvi.74.24.11608-11618.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Mettenleiter TC. Herpesvirus assembly and egress. J Virol. 2002;76:1537–1547. doi: 10.1128/JVI.76.4.1537-1547.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Fuchs W, et al. Essential function of the pseudorabies virus UL36 gene product is independent of its interaction with the UL37 protein. J Virol. 2004;78:11879–11889. doi: 10.1128/JVI.78.21.11879-11889.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Schlieker C, et al. A deubiquitinating activity is conserved in the large tegument protein of the herpesviridae. J Virol. 2005;79:15582–15585. doi: 10.1128/JVI.79.24.15582-15585.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wang J, et al. High-molecular-weight protein (pUL48) of human cytomegalovirus is a competent deubiquitinating protease: mutant viruses altered in its active-site cysteine or histidine are viable. J Virol. 2006;80:6003–6012. doi: 10.1128/JVI.00401-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Bottcher S, et al. Identification of functional domains within the essential large tegument protein pUL36 of pseudorabies virus. J Virol. 2007;81:13403–13411. doi: 10.1128/JVI.01643-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Bottcher S, et al. Mutagenesis of the active-site cysteine in the ubiquitin-specific protease contained in large tegument protein pUL36 of pseudorabies virus impairs viral replication in vitro and neuroinvasion in vivo. J Virol. 2008;82:6009–6016. doi: 10.1128/JVI.00280-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kim ET, et al. Cleavage specificity of the UL48 deubiquitinating protease activity of human cytomegalovirus and the growth of an active-site mutant virus in cultured cells. J Virol. 2009;83:12046–12056. doi: 10.1128/JVI.00411-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Shanda SK, Wilson DW. UL36p is required for efficient transport of membrane-associated herpes simplex virus type 1 along microtubules. J Virol. 2008;82:7388–7394. doi: 10.1128/JVI.00225-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Abaitua F, et al. A single mutation responsible for temperature-sensitive entry and assembly defects in the VP1–2 protein of herpes simplex virus. J Virol. 2011;85:2024–2036. doi: 10.1128/JVI.01895-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Abaitua F, et al. Characterization of the herpes simplex virus (HSV)-1 tegument protein VP1–2 during infection with the HSV temperature-sensitive mutant tsB7. J Gen Virol. 2009;90:2353–2363. doi: 10.1099/vir.0.012492-0. [DOI] [PubMed] [Google Scholar]
- 82.Newcomb WW, Brown JC. Structure and capsid association of the herpesvirus large tegument protein UL36. J Virol. 2010;84:9408–9414. doi: 10.1128/JVI.00361-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Szczepaniak R, et al. Disulfide bond formation contributes to herpes simplex virus capsid stability and retention of pentons. J Virol. 2011;85:8625–8634. doi: 10.1128/JVI.00214-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Coller KE, et al. The capsid and tegument of the alphaherpesviruses are linked by an interaction between the UL25 and VP1/2 proteins. J Virol. 2007;81:11790–11797. doi: 10.1128/JVI.01113-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.To A, et al. Yeast two hybrid analyses reveal novel binary interactions between human cytomegalovirus-encoded virion proteins. PLoS One. 2011;6:e17796. doi: 10.1371/journal.pone.0017796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Nicholson IP, et al. Properties of virion transactivator proteins encoded by primate cytomegaloviruses. Virology journal. 2009;6:65. doi: 10.1186/1743-422X-6-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Fraile-Ramos A, et al. The ESCRT machinery is not required for human cytomegalovirus envelopment. Cell Microbiol. 2007;9:2955–2967. doi: 10.1111/j.1462-5822.2007.01024.x. [DOI] [PubMed] [Google Scholar]
- 88.Calistri A, et al. Intracellular trafficking and maturation of herpes simplex virus type 1 gB and virus egress require functional biogenesis of multivesicular bodies. J Virol. 2007;81:11468–11478. doi: 10.1128/JVI.01364-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Crump CM, et al. Herpes simplex virus type 1 cytoplasmic envelopment requires functional Vps4. J Virol. 2007;81:7380–7387. doi: 10.1128/JVI.00222-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Cepeda V, Fraile-Ramos A. A role for the SNARE protein syntaxin 3 in human cytomegalovirus morphogenesis. Cell Microbiol. 2011;13:846–858. doi: 10.1111/j.1462-5822.2011.01583.x. [DOI] [PubMed] [Google Scholar]
- 91.Revello MG, Gerna G. Human cytomegalovirus tropism for endothelial/epithelial cells: scientific background and clinical implications. Rev Med Virol. 2010;20:136–155. doi: 10.1002/rmv.645. [DOI] [PubMed] [Google Scholar]
- 92.Crump CM, et al. Role of PACS-1 in trafficking of human cytomegalovirus glycoprotein B and virus production. J Virol. 2003;77:11105–11113. doi: 10.1128/JVI.77.20.11105-11113.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Krzyzaniak MA, et al. HCMV-encoded glycoprotein M (UL100) interacts with Rab11 effector protein FIP4. Traffic. 2009;10:1439–1457. doi: 10.1111/j.1600-0854.2009.00967.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Meissner CS, et al. A leucine zipper motif of a tegument protein triggers final envelopment of human cytomegalovirus. J Virol. 2012;86:3370–3382. doi: 10.1128/JVI.06556-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Womack A, Shenk T. Human cytomegalovirus tegument protein pUL71 is required for efficient virion egress. MBio. 2010;1:e00282–10. doi: 10.1128/mBio.00282-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Liu Y, Biegalke BJ. The human cytomegalovirus UL35 gene encodes two proteins with different functions. J Virol. 2002;76:2460–2468. doi: 10.1128/jvi.76.5.2460-2468.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Schierling K, et al. Human cytomegalovirus tegument protein ppUL35 is important for viral replication and particle formation. J Virol. 2005;79:3084–3096. doi: 10.1128/JVI.79.5.3084-3096.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Salsman J, et al. Proteomic Profiling of the Human Cytomegalovirus UL35 Gene Products Reveals a Role for UL35 in the DNA Repair Response. J Virol. 2012;86:806–820. doi: 10.1128/JVI.05442-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Stanton RJ, et al. Cytomegalovirus destruction of focal adhesions revealed in a high-throughput Western blot analysis of cellular protein expression. J Virol. 2007;81:7860–7872. doi: 10.1128/JVI.02247-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Bechtel JT, Shenk T. Human cytomegalovirus UL47 tegument protein functions after entry and before immediate-early gene expression. J Virol. 2002;76:1043–1050. doi: 10.1128/JVI.76.3.1043-1050.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Seo JY, Britt WJ. Cytoplasmic envelopment of human cytomegalovirus requires the postlocalization function of tegument protein pp28 within the assembly compartment. J Virol. 2007;81:6536–6547. doi: 10.1128/JVI.02852-06. [DOI] [PMC free article] [PubMed] [Google Scholar]